Abstract
A standard panel of subtype C human immunodeficiency virus type 1 (HIV-1) Env-pseudotyped viruses was created by cloning, sequencing, and characterizing functional gp160 genes from 18 acute and early heterosexually acquired infections in South Africa and Zambia. In general, the gp120 region of these clones was shorter (most evident in V1 and V4) and less glycosylated compared to newly transmitted subtype B viruses, and it was underglycosylated but no different in length compared to chronic subtype C viruses. The gp120s also exhibited low amino acid sequence variability (12%) in V3 and high variability (39%) immediately downstream of V3, a feature shared with newly transmitted subtype B viruses and chronic viruses of both subtypes. When tested as Env-pseudotyped viruses in a luciferase reporter gene assay, all clones possessed an R5 phenotype and resembled primary isolates in their sensitivity to neutralization by HIV-1-positive plasmas. Results obtained with a multisubtype plasma panel suggested partial subtype preference in the neutralizing antibody response to infection. The clones were typical of subtype C in that all were resistant to 2G12 (associated with loss of N-glycosylation at position 295) and most were resistant to 2F5, but all were sensitive to 4E10 and many were sensitive to immunoglobulin G1b12. Finally, conserved neutralization epitopes in the CD4-induced coreceptor binding domain of gp120 were poorly accessible and were difficult to induce and stabilize with soluble CD4 on Env-pseudotyped viruses. These results illustrate key genetic and antigenic properties of subtype C HIV-1 that might impact the design and testing of candidate vaccines. A subset of these gp160 clones are suitable for use as reference reagents to facilitate standardized assessments of vaccine-elicited neutralizing antibody responses.
Neutralizing antibody (NAb) responses are associated with the clinical success of many approved vaccines (65) and are a high priority for human immunodeficiency virus type 1 (HIV-1) vaccine development (25, 43, 46). To be effective against HIV-1, NAbs will need to overcome the extensive genetic diversity and complex escape mechanisms that typify the surface gp120 and transmembrane gp41 envelope glycoproteins (Env) of the virus (40, 84, 89). At least nine different genetic subtypes and a growing number of circulating recombinant forms (CRFs) of group M HIV-1 account for the majority of infections worldwide (42). Unfortunately, little progress has been made in designing a vaccine immunogen that elicits NAbs against multiple variants within a single genetic subtype, let alone cross-subtype NAbs (4, 5, 10, 49). A variety of new approaches aim to solve this difficult problem by acquiring new knowledge about Env structure, function, and immunobiology and using this knowledge to design better immunogens (12, 34). As new immunogens undergo preclinical and clinical testing, it will be important to compare them to prototypic immunogens and to each other with respect to the magnitude and breadth of the NAb response each generates.
To facilitate these comparative studies, it has been recommended that separate panels of HIV-1 reference strains be devised for each major genetic subtype and CRF; these panels are needed to acquire standardized data sets that may be used to rank vaccine potency and to identify promising candidates for further development (47). The degree of accuracy in predicting vaccine potency with standard reference strains could depend on the particular genetic, antigenic, and biologic properties of these viruses (54). Deciding which viral properties are most suitable is a complicated task that would best be guided by information on an HIV-1 vaccine that is at least partially effective. Because no such vaccine is currently available, the process of selecting appropriate reference strains has instead relied on an alternate set of scientific judgments (47, 54). These judgments place a heavy emphasis on the use of early “transmitted” strains from sexually acquired infections with the rationale that sexual contact is the major route of HIV-1 transmission in the world (31, 79) and that a bottleneck occurs at sexual transmission that selects a subset of viral variants (18, 23, 29, 82, 86, 93, 94). In principle, these sexually transmissible variants are the major targets for vaccination and also represent suitable reference strains for immune monitoring assays.
Recently, a panel of 12 subtype B gp160 reference clones from acute/early sexually acquired infections was described that may be used as Env-pseudotyped viruses for standardized assessments of NAb responses (45). There is an urgent need to develop a separate panel for subtype C, as this is the most abundant subtype in countries that carry the heaviest burden of infections (50). Previous studies of HIV-1 neutralization have focused on the less-prevalent subtype B. Moreover, what little is known about subtype C is derived mostly from studies of chronic infection. In general, subtype C viruses have been shown to be sensitive to neutralization by subtype C and non-subtype C serum samples where only occasional subtype-specific neutralization has been observed (11, 39, 55). Subtype C viruses also are unusually resistant to the gp120 glycan-specific monoclonal Ab 2G12 and to the gp41-specific monoclonal Ab 2F5 (7, 11, 33). Another general feature of subtype C is a low level of amino acid diversity in the V3 loop of gp120 and a high level of diversity in the region immediately downstream of V3 (24, 59).
A recent study of heterosexual subtype C HIV-1 transmission in Zambia showed that gp120 from newly transmitted viruses were shorter, less glycosylated, and more sensitive to neutralization by plasma from the chronically infected partner compared to the transmitted virus (23). Similar features have been associated with the transmission of subtype A viruses in Kenya (18) but were not seen in subtype B transmission (18, 29). Another recent study found a more robust autologous neutralizing antibody response during early subtype C virus infection in Zambia compared to early subtype B virus infection in the United States, where the robust response to subtype C infection was associated with viruses that contained shorter and less-glycosylated gp120s (44). Additional work is needed to confirm and extend these early observations, as they have important implications for vaccine design and testing. Here we describe the cloning, sequencing, biologic phenotype, and neutralization profile of functional gp160 genes from acute/early, heterosexually acquired subtype C HIV-1 infections in South Africa and Zambia.
MATERIALS AND METHODS
Plasma samples, soluble CD4 (sCD4), and monoclonal Abs.
Plasma samples from chronically HIV-1-infected blood donors in South Africa who were presumed to be treatment naive were obtained from the South African National Blood Services. These individual plasma samples were designated BB8, BB12, BB28, BB55, BB70, and BB106 and were selected from a larger set of plasma samples based on their greater neutralizing activities in an initial screen against three subtype C Env-pseudotyped viruses (Du151.2, Du156.12, and Du172.17). All donors were infected with HIV-1 subtype C viruses as determined by gag and env sequences. Plasma pools for subtypes A, B, C, and D were described previously (45). These latter plasma pools were each comprised of plasma from 6 to 10 subjects who had pure subtype infections as verified by full HIV-1 genome sequencing of DNA from cryopreserved peripheral blood mononuclear cells (PBMC). The plasma samples and plasma pools were derived from chronically infected individuals and did not include the HIV-1-positive subjects who served as a source of env clones. A normal plasma pool was prepared from Leukopaks from four HIV-1-negative subjects (BRT Laboratories, Inc., Baltimore, MD). All plasma samples were heat inactivated at 56°C for 1 h prior to use. Informed consent was obtained from all study participants as approved by local institutional review boards and biosafety committees; the blood bank samples were obtained from anonymous donors, with all sample codes delinked from subject identifiers.
Recombinant sCD4 comprising the full-length extracellular domain of human CD4 and produced in Chinese hamster ovary cells was obtained from Progenics Pharmaceuticals, Inc. (Tarrytown, NY). Human monoclonal Ab immunoglobulin G1b12 (IgG1b12) was kindly provided by Dennis Burton (The Scripps Research Institute, La Jolla, CA). Human monoclonal Abs 2G12, 2F5, and 4E10 were obtained from the NIH AIDS Research and Reference Reagent Program (NIH ARRRP; Rockville, MD) as contributed by Hermann Katinger. Monoclonal Abs 17b, 23e, 31H, 21c, E51, 48d, 112d, 412d, and ED10 against CD4-induced (CD4i) epitopes on gp120 were isolated from Epstein-Barr virus-transformed B-cell lines established from subtype B HIV-1-infected individuals as described previously (66, 90). TriMab is a mixture of three monoclonal Abs (IgG1b12, 2G12, and 2F5) prepared as a 1-mg/ml stock solution containing 333 μg of each monoclonal Ab/ml in phosphate-buffered saline, pH 7.4.
Cells.
TZM-bl (JC53-bl) cells were obtained from the NIH ARRRP as contributed by John Kappes and Xiaoyun Wu. This is a genetically engineered HeLa cell clone that expresses CD4, CXCR4, and CCR5 and contains Tat-responsive reporter genes for firefly luciferase (Luc) and Escherichia coli β-galactosidase under regulatory control of an HIV-1 long terminal repeat (60, 83). 293T/17 cells were obtained from the American Type Culture Collection (catalog no. 11268). Both cell lines were maintained in Dulbecco's modified Eagle's medium (Gibco BRL Life Technologies) containing 10% heat-inactivated fetal bovine serum and 50 μg gentamicin/ml in vented T-75 culture flasks (Corning-Costar). Cultures were incubated at 37°C in a humidified 5% CO2-95% air environment. Cell monolayers were split 1:10 at confluence by treatment with 0.25% trypsin, 1 mM EDTA (Invitrogen).
Amplification and cloning of env/rev DNA cassettes.
Specimens for env/rev gene cloning were obtained from 18 HIV-1-infected individuals living in either South Africa or Zambia and who were judged to be in acute/early stages of infection as determined by either their last known seronegative clinic visit, time of onset of acute retroviral syndrome, or a combination of these two clinical parameters. All infections occurred through heterosexual contact. env-rev cassettes were cloned from either plasma viral RNA, uncultured PBMC DNA, or cultured PBMC DNA. In the latter case, fresh phytohemagglutinin-stimulated PBMC from healthy HIV-1-negative donors were inoculated with low-passage (<5 passages) primary isolates, and the infected PBMC were used as the source of DNA for PCR amplification of HIV-1 env-rev cassettes as described elsewhere (45). Virus-containing culture supernatants were clarified from cells by 0.45-μm filtration and stored at −80°C in 1-ml aliquots as archives of the immediate uncloned parent of the corresponding molecularly cloned Env-pseudotyped viruses. In some cases, uncultured PBMC direct from study subjects were used for DNA extraction and PCR amplification. In other cases, virion-associated plasma RNA was purified and subjected to cDNA synthesis prior to PCR amplification as described elsewhere (23, 44, 45). The inner antisense primer EnvNF (5′-GGCACCTGAGGTCTGACTGGAAAGCC-3′) replaced antisense primer EnvN in the second-round PCR and was used in conjunction with inner sense primer EnvA (23) to amplify full-length env from ZM197M. PCR products from cultured PBMC DNA were inserted directly into pcDNA 3.1D/V5-His-TOPO (Invitrogen Corp., Carlsbad, CA). PCR products from uncultured PBMC DNA and from plasma viral RNA were inserted into either pcDNA 3.1/V5-His-TOPO or pCR3.1 (Invitrogen Corp., Carlsbad, CA) by TA cloning.
Plasmid minipreps from multiple colonies of transformed JM109 cells were screened by restriction enzyme digestion for full-length inserts. Clones with inserts in the correct orientation were screened by cotransfection with an env-deficient HIV-1 (SG3Δenv) backbone in 293T cells to produce Env-pseudotyped viruses that were subsequently titrated for infectivity in TZM-bl cells as described previously (45). Env clones conferring the highest infectivity were selected for further characterization. Twelve of these molecularly cloned gp160 genes were chosen to comprise a new panel of standard reference reagents and have been donated to the NIH ARRRP (item no. 11326).
Other viruses.
A functional gp160 clone for the R5 subtype B virus SF162.LS (17, 73) was obtained from Leonidas Stamatatos. T-cell line-adapted HIV-1MN (30) was obtained from Robert Gallo and was propagated in H9 cells as described elsewhere (53). Molecularly cloned gp160 genes from a standard panel of subtype B reference strains was described previously (45). The CD4-independent strain NL-ADArs (38) was obtained from Joseph Sodroski and was propagated in PBMC.
DNA sequence and phylogenetic analysis.
Sequence analysis was performed by cycle sequencing and BigDye terminator chemistry with automated DNA Sequenators (models 3100 and 3730; Applied Biosystems, Inc.) as recommended by the manufacturer. Individual sequence fragments for each env clone were assembled and edited using the Sequencher program 4.2 (Gene Codes Corp., Ann Arbor, MI). Nucleotide and deduced Env amino acid sequences were initially aligned using CLUSTAL W (35, 76) and manually adjusted for an optimal alignment using MASE (27). Pair-wise evolutionary distances were estimated using Kimura's two-parameter method (37) to correct for superimposed substitutions; sequence gaps and ambiguous areas within the alignment were excluded from all comparisons. Phylogenetic trees were constructed by the neighbor-joining method (67), and the reliability of branching orders was assessed by bootstrap analysis using 1,000 replicates (28). The sequences of all new gp160 genes are available from GenBank and the Los Alamos HIV Sequence Database (see Table 1 for accession numbers).
TABLE 1.
Demographic and biologic properties of molecularly cloned, Env-pseudotyped strains of subtype C HIV-1
| Env clonea | Panel designation | Subtype | CoR | Gender | Mode of transmission | Mo/yr isolated | Wks after est. infection date | Location | Plasma VL | CD4 count | Sourceb | Accession no. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Du123.6 | SVPC1 | C | R5 | F | M-F | 11/1998 | 12 | Durban, South Africa | 19,331 | 841 | ccPBMC | DQ411850 |
| Du151.2 | SVPC2 | C | R5 | F | M-F | 11/1998 | 6 | Durban, South Africa | >500,000 | 367 | ccPBMC | DQ411851 |
| Du156.12* | SVPC3* | C | R5 | F | M-F | 02/1999 | <4 | Durban, South Africa | 22,122 | 404 | ccPBMC | DQ411852 |
| Du172.17* | SVPC4* | C | R5 | F | M-F | 11/1998 | 12 | Durban, South Africa | 1,916 | 793 | ccPBMC | DQ411853 |
| Du422.1* | SVPC5* | C | R5 | F | M-F | 11/1998 | 8 | Durban, South Africa | 17,118 | 409 | ccPBMC | DQ411854 |
| ZM197M.PB7* | SVPC6* | C | R5 | M | F-M | 10/2002 | <15 | Lusaka, Zambia | NAc | NA | ucPBMC | DQ388515 |
| ZM214M.PL15* | SVPC7* | C | R5 | M | F-M | 07/2003 | <13 | Lusaka, Zambia | 198,800 | NA | Plasma | DQ388516 |
| ZM215F.PB8 | SVPC8 | C | R5 | F | M-F | 10/2002 | <15 | Lusaka, Zambia | 608,560 | NA | ucPBMC | DQ422948 |
| ZM233M.PB6* | SVPC9* | C | R5 | M | F-M | 12/2002 | <15 | Lusaka, Zambia | NA | NA | ucPBMC | DQ388517 |
| ZM249M.PL1* | SVPC10* | C | R5 | M | F-M | 08/2003 | <1 | Lusaka, Zambia | 1,143,760 | NA | Plasma | DQ388514 |
| ZM53M.PB12* | SVPC11* | C | R5 | M | F-M | 02/2000 | <14 | Lusaka, Zambia | 26,643 | NA | ucPBMC | AY423984 |
| ZM55F.PB28a | SVPC12 | C | R5 | F | M-F | 08/1998 | <13 | Lusaka, Zambia | 88,544 | NA | ucPBMC | AY423971 |
| ZM109F.PB4* | SVPC13* | C | R5 | F | M-F | 03/2000 | <14 | Lusaka, Zambia | 887,586 | NA | ucPBMC | AY424138 |
| ZM106F.PB9 | SVPC14 | C | R5 | F | M-F | 06/1998 | <18 | Lusaka, Zambia | 48,442 | NA | ucPBMC | AY424163 |
| ZM135M.PL10a* | SVPC15* | C | R5 | M | F-M | 06/1998 | <15 | Lusaka, Zambia | 202,999 | NA | Plasma | AY424079 |
| CAP45.2.00.G3* | SVPC16* | C | R5 | F | M-F | 05/2005 | 5 | Durban, South Africa | 236,000 | 974 | Plasma | DQ435682 |
| CAP210.2.00.E8* | SVPC17* | C | R5 | F | M-F | 05/2005 | 5 | Durban, South Africa | 127,000 | 461 | Plasma | DQ435683 |
| CAP244.2.00.D3 | SVPC18 | C | R5 | F | M-F | 05/2005 | 8 | Durban, South Africa | 19,200 | 557 | Plasma | DQ435684 |
Clones selected as standard reference strains are marked with an asterisk.
ccPBMC, cocultured PBMC; ucPBMC, uncultured PBMC.
NA, not available.
Analysis of coreceptor usage.
Coreceptor usage of 15 Env-pseudotyped viruses was determined in TZM-bl cells by measuring reductions in infectivity in the presence of the CXCR4 antagonist AMD 3100 and the CCR5 antagonist TAK-779 as described elsewhere (45). For the three CAP clones, coreceptor usage was determined by green fluorescence protein (GFP) reporter gene activation in GHOST cells that expressed either CCR5 or CXCR4 (19).
Neutralization assay.
Neutralization was measured as a reduction in Luc reporter gene expression after a single round of virus infection in TZM-bl cells as described previously (52). This assay is a modified version of the assay used by Wei et al. (83, 84). The 50% inhibitory dose (ID50) was defined as either the plasma dilution or sample concentration (in the case of sCD4 and monoclonal Abs) at which relative luminescence units (RLU) were reduced 50% compared to virus control wells after subtraction of background RLU in cell control wells.
Statistical analyses.
Differences in the neutralizing potencies of subtype C plasma samples (six samples with a BB prefix) were determined by comparing their geometric mean titer (GMT) against each virus. Differences in neutralization sensitivity between subtype B and C viruses were compared using a two-sided Wilcoxon matched pairs test with a 95% confidence interval. Differences were considered significant if P was <0.05. Differences in envelope lengths and number of potential N-linked glycans (PNLG) between subtype B and C viruses were compared using the Wilcoxon two-sided rank test. The two-sided Fisher's exact test was used to determine the relative loss of specific PNLG in one HIV-1 subtype compared to another. Both of these latter tests were performed using the R Project for Statistical Computing (www.r-project.org).
Nucleotide sequence accession numbers.
GenBank accession numbers for the sequences identified in this study are DQ411850 to DQ411854, DQ388514 to DQ388517, DQ422948, DQ435682 to DQ435684, AY423971, AY423984, AY424079, AY424138, and AY424163.
RESULTS
Demographics and biologic properties of the molecularly cloned gp160 genes.
Candidate gp160 reference clones were obtained from studies of acute and early sexually acquired subtype C infections in South Africa and Zambia (Table 1). Five clones with a Du prefix were obtained in 1998 from commercial sex workers recruited from truck stops between Durban and Johannesburg (11); these women were participating in a multicenter clinical trial of a potential vaginal microbicide (81). Clones with a ZM prefix were obtained between June 1998 and July 2003 from a study of HIV-1-discordant couples in Lusaka, Zambia (1, 23). Clones with a CAP prefix were obtained in 2005 from a study of acute/early heterosexual HIV-1 transmission organized by the Center for the AIDS Program of Research in South Africa (CAPRISA). Twelve strains arose from male-female transmission and six strains arose from female-male transmission. All molecularly cloned Env-pseudotyped viruses used CCR5 but not CXCR4 for cell entry.
Sequence analysis.
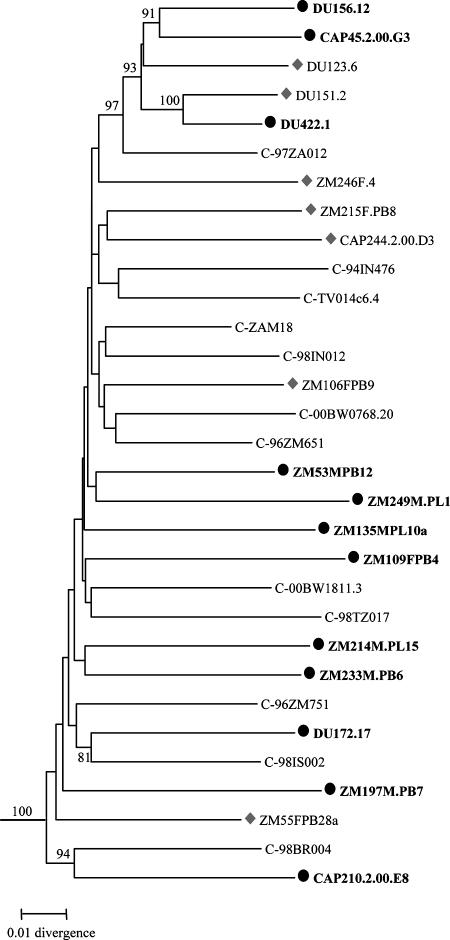
Phylogenetic analysis of full-length gp160 nucleotide sequences confirmed that all 18 functional clones grouped within subtype C (Fig. 1). The clones comprised a wide spectrum of genetic diversity with only two clones (Du151.2 and Du422.1) that clustered with a high bootstrap value.
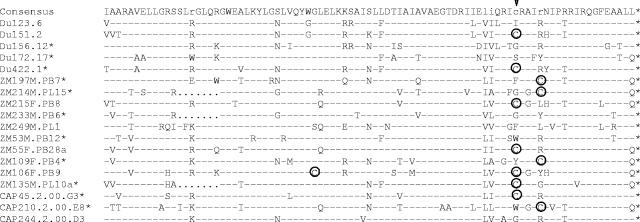
FIG. 1.
Phylogenetic relationships of subtype C reference env clones. The newly characterized sequences are indicated by solid circles and shaded diamonds, and the 12 env clones that are recommended as standard reference reagents are represented by the solid circles and are bolded. Horizontal branch lengths are drawn to scale (the scale bar represents 0.01 nucleotide substitutions per site), but vertical separation is for clarity only. Values at nodes indicate the percentage of bootstraps in which the cluster to the right was found; only values of 80% or greater are shown. The phylogenetic tree was rooted with subtype D env sequences (NDK, Z2Z3, and 94UG114).
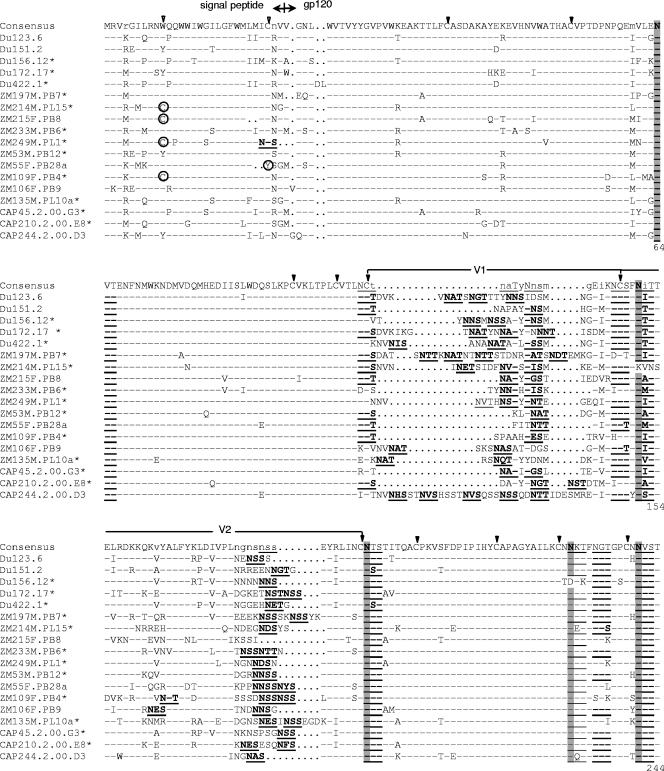
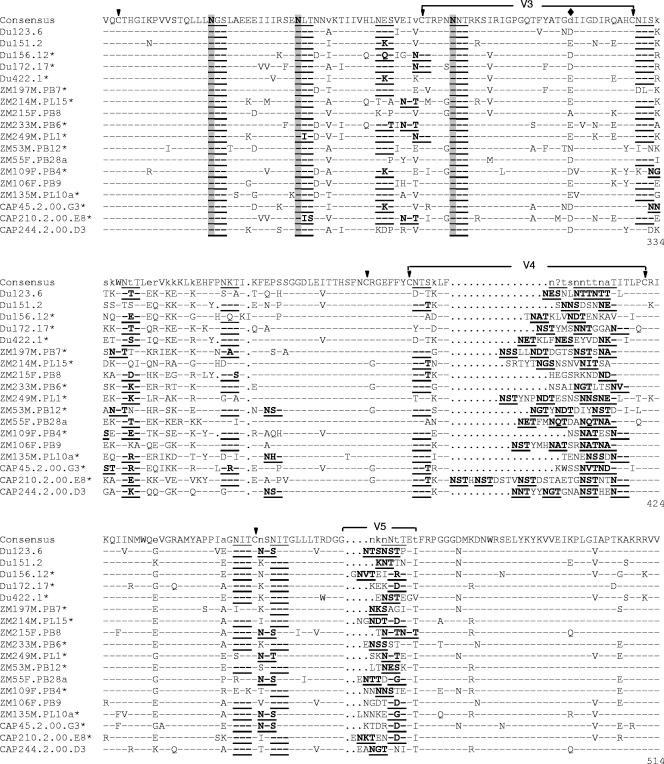
Deduced amino acid sequences showed that all 18 clones had an uninterrupted env open reading frame and 100% conservation of the cysteine residues that form the V1, V2, V3, and V4 loops of gp120 (Fig. 2). Considerable amino acid sequence variability was seen in V1, V2, V4, V5, and a region of approximately 34 amino acids immediately downstream of V3. V1, V2, V4, and V5 also exhibited substantial length variation (Table 2). V3 did not vary in length (35 amino acids in all clones) and exhibited only minor sequence variability that included position 320 (numbered as in Fig. 2) flanking the tip of the loop; this same position also shows greatest variability in the V3 loop of subtype B gp160 reference clones (45). Compared to a subtype C consensus sequence, amino acids in the V3 loop of these clones exhibited an average of 12% variability, whereas the 34 amino acids immediately downstream varied by an average of 39%.
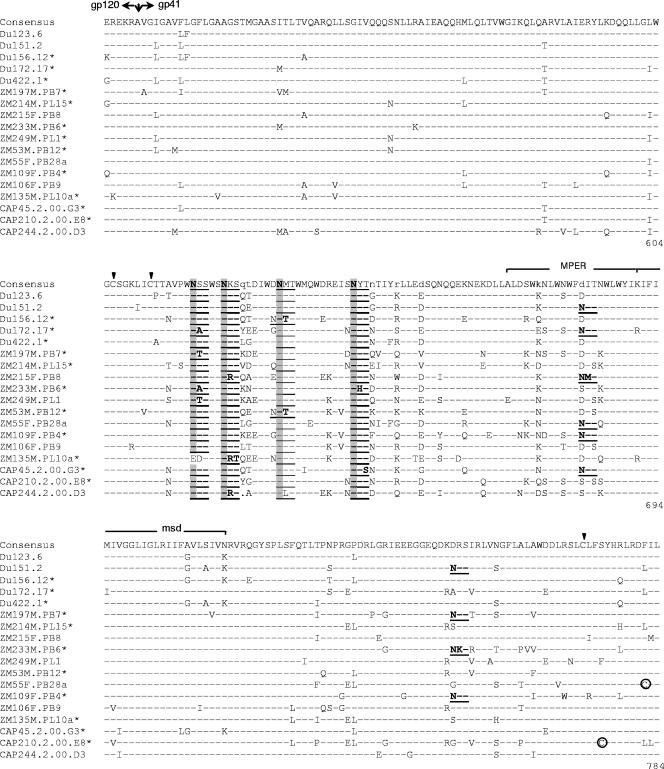
FIG.2.
Alignment of deduced amino acid sequences from acute/early subtype C HIV-1 env genes. Nucleotide sequences of newly derived env genes were translated, aligned, and compared with a consensus sequence generated by MASE. Numbering of amino acid residues begins with the first residue of gp120 and does not include the signal peptide. Dashes denote sequence identity, while dots represent gaps introduced to optimize alignments. Small letters in the consensus sequence indicate sites at which fewer than 50% of the viruses share the same amino acid residue. Triangles above the consensus sequence denote cysteine residues (solid triangles indicate sequence identity, while open triangles indicate sequence variation). V1, V2, V3, V4, and V5 regions designate hypervariable HIV-1 gp120 domains as previously described. The signal peptide and Env precursor cleavage sites are indicated; msd denotes the membrane-spanning domain in gp41; asterisks mark in-frame stop codons. Open circles highlight altered cysteine residues. Potential N-linked glycosylation sites (NXYX motif, where X is any amino acid other than proline and Y is either serine or threonine) are bolded and underlined. Highly conserved sites of potential N-linked glycosylation are shaded. The solid diamond denotes a highly variable amino acid position in the V3 loop.
TABLE 2.
Lengths of gp120 variable regions
| Env clonea | No. of amino acid residues
|
||||
|---|---|---|---|---|---|
| V1 | V2 | V3 | V4 | V5 | |
| Du123.6 | 30 | 40 | 35 | 24 | 10 |
| Du151.2 | 18 | 45 | 35 | 22 | 8 |
| Du156.12* | 25 | 42 | 35 | 27 | 12 |
| Du172.17* | 30 | 46 | 35 | 25 | 9 |
| Du422.1* | 27 | 44 | 35 | 28 | 8 |
| ZM197M.PB7* | 39 | 49 | 35 | 31 | 9 |
| ZM214M.PL15* | 28 | 43 | 35 | 30 | 11 |
| ZM215F.PB8 | 17 | 38 | 35 | 23 | 8 |
| ZM233M.PB6* | 17 | 42 | 35 | 23 | 10 |
| ZM249M.PL1* | 23 | 42 | 35 | 31 | 9 |
| ZM53M.PB12* | 15 | 42 | 35 | 26 | 9 |
| ZM55F.PB28a | 13 | 45 | 35 | 28 | 11 |
| ZM109F.PB4* | 17 | 45 | 35 | 20 | 10 |
| ZM106F.PB9 | 25 | 42 | 35 | 29 | 10 |
| ZM135M.PL10a* | 23 | 49 | 35 | 21 | 11 |
| CAP45.2.00.G3* | 17 | 45 | 35 | 22 | 10 |
| CAP210.2.00.E8* | 19 | 45 | 35 | 39 | 12 |
| CAP244.2.00.D3 | 42 | 40 | 35 | 29 | 11 |
| Mean | 23.6 | 43.6 | 35 | 26.6 | 9.9 |
Clones selected as standard reference strains are marked with an asterisk.
Moderate variation was seen in the number and position of PNLG, where V1, V2, and V4 of gp120 were the most heavily glycosylated regions (Fig. 2; Table 3). All clones contained at least one PNLG in the relatively short stretch of amino acids comprising V5. Six PNLG in gp120 and three in gp41 were 100% conserved in all 18 clones (Fig. 2). Another three PNLG were conserved in all but one clone (Fig. 2). One highly conserved PNLG in the N terminus of V3 has been shown to mask neutralization epitopes in the V3 loop of subtype B viruses (2, 71).
TABLE 3.
Potential N-linked glycosylation sites on gp120 and gp41
| Env clonea | No. of N-linked glycosylation sites
|
||
|---|---|---|---|
| gp120 | gp41 | gp41 ectodomain | |
| Du123.6 | 25 | 4 | 4 |
| Du151.2 | 22 | 6 | 5 |
| Du156.12* | 24 | 5 | 5 |
| Du172.17* | 27 | 5 | 5 |
| Du422.1* | 24 | 4 | 4 |
| ZM197M.PB7* | 27 | 5 | 4 |
| ZM214M.PL15* | 23 | 4 | 4 |
| ZM215F.PB8 | 21 | 5 | 5 |
| ZM233M.PB6* | 24 | 5 | 4 |
| ZM249M.PL1* | 24 | 4 | 4 |
| ZM53M.PB12* | 25 | 5 | 5 |
| ZM55F.PB28a | 23 | 5 | 5 |
| ZM109F.PB4* | 23 | 6 | 5 |
| ZM106F.PB9 | 25 | 4 | 4 |
| ZM135M.PL10a* | 23 | 3 | 3 |
| CAP45.2.00.G3* | 24 | 5 | 5 |
| CAP210.2.00.E8* | 29 | 4 | 4 |
| CAP244.2.00.D3 | 26 | 4 | 4 |
| Mean | 24.4 | 4.6 | 4.4 |
Clones selected as standard reference strains are marked with an asterisk.
Neutralization phenotype.
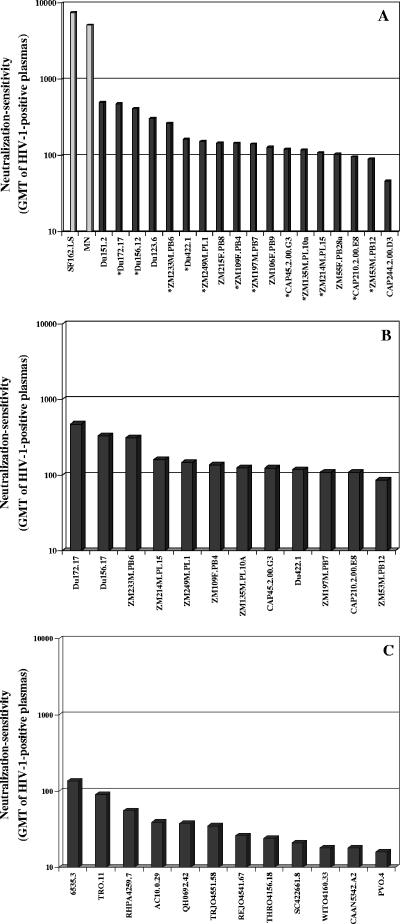
Neutralization phenotypes were characterized with six subtype C plasma samples, four subtype-specific plasma pools, sCD4, and the broadly neutralizing monoclonal Abs IgG1b12, 2G12, 2F5, and 4E10 (Table 4). They were also characterized with a series of monoclonal Abs against CD4i epitopes on gp120 (Table 5). Each subtype C Env-pseudotyped virus was broadly sensitive to neutralization by individual subtype C plasma samples and by subtype-specific plasma pools. Some subtype C viruses were clearly more sensitive than others, but the level of sensitivity in all cases was at least 10-fold lower than that of MN and SF162.LS (Fig. 3A). This diminished sensitivity relative to MN and SF162.LS distinguishes the subtype C viruses from easily neutralized “tier 1” viruses (47). Another interesting property of the subtype C Env-pseudotyped viruses was their greater sensitivity to neutralization by a subtype C plasma pool compared to plasma pools of subtypes A, B, and D (P = 0.0002). The overall potencies of these plasma pools were ranked in the following order: subtype C > subtype A > subtype D > subtype B pool. Significant differences also were seen for the subtype A pool compared to the subtype B (P = 0.0078) and subtype D (P = 0.0443) pools but not when the subtype B pool was compared to the subtype D pool (P = 0.107).
TABLE 4.
Neutralization phenotypes as determined with serum samples from HIV-1-infected individuals, sCD4, and monoclonal Abs
| Virusa | ID50 in TZM-bl cells based onb:
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Reciprocal serum dilution
|
Concn (μg/ml)
|
||||||||||||||||
| BB8 | BB12 | BB28 | BB55 | BB70 | BB106 | Pool A | Pool B | Pool C | Pool D | Normal Pool | sCD4 | IgG1b12 | 2G12 | 2F5 | 4E10 | TriMab | |
| MN | 5,107 | 4,573 | 537 | 2,964 | 43,740 | 203 | 9,013 | 43,740 | 5,550 | 16,952 | <20 | ||||||
| SF162.LS | 15,548 | 5,480 | 2,629 | 20,302 | 43,740 | 552 | 5,597 | 43,264 | 8,066 | 2,438 | <20 | ||||||
| Du123.6 | 207 | 352 | 165 | 84 | 147 | 198 | 306 | 182 | 18,845 | 220 | 90 | 0.3 | 0.2 | >50 | >50 | 0.1 | 1.6 |
| Du151.2 | 196 | 1,555 | 2,529 | 818 | 241 | 487 | 347 | 518 | 527 | 123 | 20 | 3.0 | 1.4 | >50 | >50 | 0.8 | 13.5 |
| Du156.12* | 369 | 426 | 238 | 336 | 406 | 258 | 810 | 231 | 2,368 | 228 | 52 | 13.4 | 0.8 | >50 | >50 | 0.2 | 1.2 |
| Du172.17* | 429 | 884 | 499 | 196 | 549 | 550 | 462 | 315 | 629 | 562 | 91 | 1.7 | 1.0 | >50 | >50 | 0.3 | 2.6 |
| Du422.1* | 134 | 354 | 193 | 91 | 51 | 63 | 335 | 114 | 2,138 | 65 | 90 | 9.1 | 0.2 | >50 | >50 | 0.7 | 0.8 |
| ZM197M.PB7* | 79 | 103 | 76 | 117 | 348 | 68 | 163 | 185 | 332 | 176 | <20 | 3.9 | 19.9 | >50 | 12.3 | 0.5 | 24.8 |
| ZM214M.PL15* | 259 | 366 | 104 | 90 | 206 | 96 | 127 | <20 | 162 | 64 | <20 | 8.0 | 3.0 | >50 | >50 | 4.0 | 9.7 |
| ZM215F.PB8 | 231 | 694 | 122 | 384 | 90 | <20 | 165 | 51 | 706 | 108 | 32 | 14.0 | >50 | >50 | >50 | 0.4 | >25 |
| ZM233M.PB6* | 295 | 153 | 214 | 413 | 2,899 | 80 | 127 | 108 | 640 | 218 | <20 | 2.9 | >50 | >50 | >50 | 1.2 | >25 |
| ZM249M.PL1* | 160 | 105 | 287 | 158 | 112 | 117 | 193 | 63 | 383 | 156 | 33 | 9.7 | 3.2 | >50 | >50 | 2.1 | 7.9 |
| ZM53M.PB12* | 76 | 73 | 411 | 86 | <20 | 98 | 57 | 91 | 566 | 65 | <20 | 8.3 | 25.9 | >50 | >50 | 7.0 | >25 |
| ZM55F.PB28a | 96 | 206 | 106 | 169 | 47 | 110 | 99 | 56 | 250 | 59 | 27 | 24.0 | >50 | >50 | 33.6 | 8.0 | >25 |
| ZM109F.PB4* | 160 | 188 | 89 | 160 | 238 | 61 | 185 | 82 | 365 | 119 | <20 | 0.2 | >50 | >50 | >50 | 0.6 | >25 |
| ZM106F.PB9 | 249 | 378 | 287 | 262 | 68 | 71 | 101 | 76 | 433 | <20 | <20 | 23.0 | >50 | >50 | >50 | 7.2 | >25 |
| ZM135M.PL10a* | 138 | 179 | 93 | 190 | 50 | 177 | 108 | 57 | 202 | 109 | 29 | 6.1 | >50 | >50 | >50 | 0.6 | >25 |
| CAP45.2.00.G3* | 61 | 437 | 351 | 44 | 170 | 52 | 142 | 47 | 453 | 60 | <20 | 26.0 | 0.7 | >50 | >50 | 2.6 | 1.5 |
| CAP210.2.00.E8* | 111 | 75 | 82 | 113 | 272 | 75 | 101 | 29 | 293 | 51 | <20 | 3.4 | 20.4 | >50 | >50 | 1.2 | 20.8 |
| CAP244.2.00.D3 | 53 | 47 | 32 | 51 | 91 | 30 | 38 | 27 | 94 | 40 | <20 | 8.9 | >50 | >50 | >50 | 1.9 | >25 |
| GMTc | 151 | 227 | 177 | 151 | 142 | 97 | 158 | 79 | 514 | 98 | 5.3 | 1.1 | |||||
Clones selected as standard reference strains are marked with an asterisk.
Values are the dilution or concentration at which RLU were reduced 50% compared to virus control wells. BB8, BB12, BB28, BB70, and BB106 are plasma samples from individuals infected with subtype C HIV-1.
Geometric mean titer against the 18 pseudoviruses containing cloned primary isolate Env (excludes MN and SF162.LS). Serum/plasma titers of <20 were assigned a value of 10 for calculations. Because multiple ID50 values were >50, the GMT was not determined for IgG1b12, 2G12, 2F5, or TriMab.
TABLE 5.
Neutralization with CD4i monoclonal Abs in the presence and absence of sCD4
| Virusa | ID50 in TZM-bl cellsb
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 17b | 23e | 31H | 21c | E51 | 48d | 112d | 412d | ED10 | |
| NL-ADArs | 0.002 | 0.002 | 0.002 | 0.002 | <0.0001 | 0.2 | 6.1 | 0.002 | 2.9 |
| MN | 3.5 | 4.4 | — | 14.4 | 0.3 | 0.4 | — | 16.5 | — |
| SF162.LS | 0.9/2.3 | 1.3/3.7 | 1.0/2.9 | 1.1/3.5 | 0.1/0.6 | 4.6/12.6 | —/— | 0.2/1.4 | 3.0/5.9 |
| Du156.12* | —/— | —/— | —/— | —/— | —/— | —/— | —/— | —/— | —/— |
| Du172.17* | —/— | —/— | —/— | —/— | —/4.7 | —/— | —/— | —/— | —/— |
| Du422.1* | —/— | —/— | —/— | —/— | —/— | —/— | —/— | —/— | —/— |
| ZM197M.PB7* | —/— | —/— | —/— | —/— | 19.5/21.6 | —/— | —/— | 16.7/— | —/— |
| ZM214M.PL15* | —/— | —/— | —/— | —/— | —/— | —/— | —/— | —/— | —/— |
| ZM233M.PB6* | —/— | —/— | —/— | —/— | 6.5/10.2 | —/— | —/— | —/— | —/— |
| ZM249M.PL1* | —/— | —/— | —/— | —/— | —/— | —/— | —/— | —/— | —/— |
| ZM53M.PB12* | —/— | —/— | —/— | —/— | —/— | —/— | —/— | —/— | —/— |
| ZM109F.PB4* | —/— | —/— | —/— | —/— | 9.9/16.8 | —/— | —/— | 19.3/19.1 | —/— |
| ZM135M.PL10a* | 17.0/— | 5.9/— | —/— | —/— | 17.6/— | —/— | —/— | —/20.7 | —/— |
| CAP45.2.00.G3* | —/— | —/— | —/— | —/— | —/— | —/— | —/— | —/— | —/— |
| CAP210.2.00.E8* | —/— | —/— | —/— | —/— | —/— | —/— | —/— | —/— | —/— |
Clones selected as standard reference strains are marked with an asterisk. NL-ADArs and MN were uncloned viruses.
Values are the monoclonal Ab concentration at which RLU were reduced 50% compared to virus control wells. Neutralization was measured in the absence/presence of sCD4. sCD4 was present in all wells (including cell control and virus control wells) at a concentration equal to the ID50 for each virus. Dashes signify no neutralization at the highest concentration of monoclonal Ab tested (25 μg/ml). NL-ADArs and MN were assayed in the absence of sCD4 only.
FIG. 3.
Neutralization sensitivity of acute/early subtype C and subtype B HIV-1 Env-pseudotyped viruses as determined with plasma samples from HIV-1-infected individuals. Bar height represents the GMT of neutralizing Abs against the indicated Env-pseudotyped viruses. A. GMT for all HIV-1-positive plasma samples shown in Table 4 (BB8, BB12, BB28, BB55, BB70, and BB106 and pool A, pool B, pool C, and pool D). Clones selected as standard subtype C reference strains are marked with an asterisk. B. GMT of neutralizing Abs for the individual subtype C plasma samples BB8, BB12, BB28, BB55, BB70, and BB106 assayed against the 12 subtype C reference strains. C. GMT of neutralizing Abs in the individual subtype C plasma samples BB8, BB12, BB28, BB55, BB70, and BB106 assayed against 12 subtype B reference strains.
All subtype C Env-pseudotyped viruses were sensitive to inhibition by sCD4. ID50 doses ranged from 0.2 μg/ml to 26 μg/ml, suggesting a broad spectrum of epitope exposure in and around the CD4 binding site of gp120. IgG1b12, which targets a complex epitope on gp120 that affects CD4 binding (13, 58), neutralized 10 of the 17 subtype C Env clones. No clear association was seen between the neutralizing activity of this monoclonal Ab and sensitivity to inhibition by sCD4.
All 18 gp160 clones were highly resistant to neutralization by the glycan-specific monoclonal Ab 2G12. Similar broad resistance to 2G12 has been reported for other subtype C viruses (7, 11, 33) and is associated with an absence of PNLG at critical positions that affect the 2G12 epitope (7, 14, 33, 69, 70). Each of our clones lacked a PNLG at one or more of these critical positions; most often this was position 295 at the N-terminal base of V3. Loss of a PNLG at position 295 has been associated with the general 2G12-resistant phenotype of subtype C viruses from chronically infected individuals (7) and pediatric infections (33). Our results indicate that the same is true for newly transmitted, heterosexually acquired subtype C viruses.
Monoclonal Abs 2F5 and 4E10, which recognize adjacent epitopes in the membrane-proximal external region of gp41 (3, 56, 57, 61, 74, 95), were quite different from one another in their ability to neutralize these subtype C viruses: 2F5 neutralized only 2 viruses, whereas 4E10 neutralized all 18 viruses. Neutralization by these two monoclonal Abs was highly predicted by amino acid sequence. Thus, both 2F5-sensitive viruses contained a DKW motif that has been reported to be a minimum requirement for 2F5 recognition (7, 45). Two additional clones (ZM214M.PL15 and CAP210.2.00.E8) contained this motif but had changes elsewhere in the 2F5 epitope. All clones except ZM215F.PB8 contained the WFXI motif that has been reported to be important for 4E10 recognition (7, 45). Clone ZM215F.PB8 contained a different motif (WFNM) yet it was highly susceptible to neutralization by 4E10, suggesting additional flexibility in the 4E10 core epitope. Six clones contained a potential PNLG in this core epitope that might be expected to mask the virus from 4E10, but biochemical evidence suggests this site is not glycosylated (41).
Consistent with the general resistance of these viruses to 2G12 and 2F5, their ability to be neutralized by TriMab (mixture of IgG1b12, 2G12, and 2F5) tracked with their sensitivity to IgG1b12. We saw no clear evidence of synergism with this triple combination of monoclonal Abs.
Neutralization phenotypes were further assessed by using CD4i monoclonal Abs to probe epitopes in the coreceptor binding domain of gp120. These are some of the most highly conserved epitopes on the gp120 molecule (22), but they are often concealed from Abs unless sCD4 is present to induce their exposure prior to virus-cell engagement (22, 68, 75). The CD4-independent NL-ADArs virus on which CD4i epitopes are spontaneously exposed (38) was highly sensitive to all of these monoclonal Abs, confirming that they possessed potential neutralizing activity (Table 5). Most of these CD4i monoclonal Abs also neutralized SF162.LS and, to a lesser degree, MN, although much higher concentrations were usually needed in these cases compared to NL-ADArs. In general, the subtype C Env-pseudotyped viruses were resistant to these CD4i monoclonal Abs regardless of whether subinhibitory doses of sCD4 were present (Table 5). An exception was E51, which neutralized 5 of 12 subtype C viruses but only at high concentrations. These latter results agree with a previous study in which E51 was superior to other CD4i antibodies (91). We observed two cases where sCD4 augmented the neutralizing activity of a CD4i monoclonal Ab: one case produced at least a fivefold increase in sensitivity (neutralization of Du172.17 by E51), whereas the other case was much less dramatic (neutralization of ZM135M.PL10a by 412d). Notably, this latter virus was the only new strain that was neutralized by four CD4i monoclonal Abs, including two that did not neutralize the other subtype C viruses. Although the potency of neutralization against ZM135M.PL10a was relatively weak, there appears to be greater exposure of CD4i epitopes on this virus compared to other subtype C viruses. Overall, the results suggest that epitopes in the coreceptor binding domain on newly transmitted subtype C viruses are mostly concealed from Abs.
Noting that our gp160 genes were cloned from either cocultured PBMC virus, uncultured PBMC, or directly from plasma, we performed an analysis to determine whether the source of gp160 influenced the general neutralization sensitivity of the corresponding Env-pseudotyped viruses. For this analysis, combined neutralization data from the subtype C plasma samples (BB samples) and subtype-specific plasma pools in Table 4 were compared between the three categories of clones. The results showed a trend in neutralization sensitivity such that cocultured PBMC clones (GMT, 344) > uncultured PBMC clones (GMT, 129) > plasma clones (GMT, 101), but this trend was not significant (P > 0.05). Also, greater resistance to IgG1b12 was seen in the uncultured PBMC group of clones, especially when compared to the cocultured PBMC clones. We caution that a larger number of clones will be needed to confirm this latter observation.
Selection of standard reference strains.
The demographic, biologic, genetic, and neutralization properties of many of the Env-pseudotyped viruses described in this report appear suitable for standardized assessments of vaccine-elicited NAb responses. For adequate statistical power, it has been recommended that each standard panel be comprised of 12 reference strains (47). Among the 18 candidate reference strains described here, 12 were selected to comprise a panel that represents the greatest genetic and antigenic diversity while avoiding strains that are unusually sensitive or resistant to neutralization. None of the 18 strains would be considered unusually sensitive to neutralization; however, 1 strain was omitted for being relatively insensitive to neutralization by HIV-1-positive plasma samples (CAP244.2.00.D3). Another strain (ZM215F.PB8) was omitted because careful analysis showed that all bulk-derived env genes from this patient (who had a heterogenous early infection) were in vitro recombinants. Also, the Du123.6 strain was excluded because it was easily neutralized by pool C plasma, with titers higher than those seen for MN and SF162.LS viruses. On the other hand, two strains (Du422.1 and Du151.2) were phylogenetically closer to each other than any other pair of strains, clustering together with a high bootstrap value (Fig. 1); to maximize the genetic diversity among members in the panel, only Du422.1 was selected while Du151.2 was excluded. Of the 12 selected strains, 7 originated in Zambia and 5 in South Africa (Table 1). Also, six arose by male-female transmission, while the remaining six arose by female-male transmission (Table 1).
Genetic comparisons between subtype B and C reference strains.
The variable loops and number and position of PNLG in gp120 have been shown to influence the neutralization phenotype of HIV-1 (8, 26, 48, 56, 62, 84). Recent results also suggest that acute/early subtype B and C strains differ from one another in their neutralization properties (23, 29). For these reasons, amino acid sequences of the 12 subtype C gp160 reference genes were compared to the sequences of 12 subtype B gp160 reference genes described previously (45). Comparisons also were made between subtype B and C sequences in the LANL database. These comparisons focused on the number of amino acids that comprise gp120 and its variable regions. They also focused on the number and position of PNLG in gp120.
LANL database sequence comparisons showed that subtype C gp120 was generally shorter and contained fewer PNLG compared to subtype B (Table 6). Differences in gp120 length were evident by shorter V1, V3, and V4 regions despite a slightly longer V2 region in subtype C. Fewer PNLG were present in V1, V3, V4, and V5 of subtype C gp120, whereas the number of PNLG in V2 remained constant in both subtypes. Many PNLG sites were common in both subtypes, but four were prevalent in only one subtype. Subtype-specific PNLG included positions 230 (between V2 and V3) and 442 (between V4 and V5) that were diminished in subtype B (HXB2 numbering). They also included positions 295 (proximal to N terminus of V3) and 362 (between V3 and V4) that were diminished in subtype C (HXB2 numbering).
TABLE 6.
Comparison of sequence lengths and PNLG between HIV-1 subtypes B and C gp120
| Genomic region | Analysis groupa | Sequence length interquartile | Glycan no. interquartile | Comparison |
P value
|
|
|---|---|---|---|---|---|---|
| Sequence length comparisonb | Glycan no. comparisonb | |||||
| gp120 | B-ref | 509-519 | 26-28 | B-ref/C-ref | 0.01 | 0.01 |
| C-ref | 492-509 | 24-26 | B-db/C-db | 8.44 × 10−15 | 0.004 | |
| B-db | 507-515 | 24-27 | B-ref/B-db | 0.44 | 0.02 | |
| C-db | 499-510 | 23-26 | C-ref/C-db | 0.45 | 0.86 | |
| V1 | B-ref | 25-31 | 4-5 | B-ref/C-ref | 0.03 | 0.06 |
| C-ref | 17-27 | 3-4 | B-db/C-db | 3.37 × 10−13 | 0.0008 | |
| B-db | 26-32 | 4-5 | B-ref/B-db | 0.75 | 0.43 | |
| C-db | 22-28 | 3-5 | C-ref/C-db | 0.31 | 0.45 | |
| V2 | B-ref | 40-46 | 4-5 | B-ref/C-ref | 0.52 | 0.85 |
| C-ref | 42-45 | 4-5 | B-db/C-db | 3.33 × 10−5 | 0.50 | |
| B-db | 40-44 | 4-5 | B-ref/B-db | 0.23 | 0.52 | |
| C-db | 41-45 | 4-5 | C-ref/C-db | 0.16 | 0.88 | |
| V3 | B-ref | 35-35 | 3-3 | B-ref/C-ref | 0.36 | 0.0036 |
| C-ref | 35-35 | 1-2 | B-db/C-db | 0.02 | <2.2 × 10−16 | |
| B-db | 35-35 | 2-3 | B-ref/B-db | 0.61 | 0.4259 | |
| C-db | 35-35 | 2-2 | C-ref/C-db | 0.60 | 0.7318 | |
| V4 | B-ref | 31-33 | 4-5 | B-ref/C-ref | 0.005 | 0.02 |
| C-ref | 23-30 | 3-4 | B-db/C-db | 1.74 × 10−12 | 0.02 | |
| B-db | 30-33 | 4-5 | B-ref/B-db | 0.50 | 0.75 | |
| C-db | 27-31 | 4-5 | C-ref/C-db | 0.06 | 0.01 | |
| V5 | B-ref | 9-10 | 2-2 | B-ref/C-ref | 0.57 | 0.09 |
| C-ref | 9-11 | 1-2 | B-db/C-db | 0.13 | 0.004 | |
| B-db | 9-11 | 1-2 | B-ref/B-db | 0.26 | 0.12 | |
| C-db | 9-11 | 1-2 | C-ref/C-db | 0.83 | 0.71 | |
B-ref, 12 selected sequences from the subtype B panel of reference strains; B-db, 176 well-characterized subtype B database sequences; C-ref, 12 selected sequences from the C panel of reference strains; C-db, 122 well-characterized subtype C database sequences.
Bolded P values (P < 0.002) are significant differences after Bonferroni correction. Underlined P values (0.002 < P < 0.05) are significantly different trends after Bonferroni correction.
No significant differences were seen in the lengths of V2 and V3 and the number of PNLG in V1 and V5 when sequences of the subtype B and C reference clones were compared. Thus, overall the subtype C panel of reference clones contained shorter gp120 regions (V1 and V4) and fewer PNLG in gp120 (V1, V3, and V4) than the subtype B panel of reference clones. Differences observed only in database (mostly chronic virus) sequences might be an indication that gp120 of certain subtypes evolved over time in infected individuals to give rise to viral variants that are rarely transmitted. In this regard, our intrasubtype comparisons suggest that transmission of subtype C favors variants with fewer PNLG in V4 compared to chronic subtype C variants (P = 0.01). The comparisons also suggest that transmission of subtype B favors variants with a greater number of PNLG in gp120 compared to chronic subtype B variants (P = 0.02).
Neutralization phenotype comparisons between subtype B and C reference strains.
Results obtained with the subtype C plasma samples from South Africa, subtype-specific plasma pools, sCD4, and monoclonal Abs were used to compare the neutralization phenotypes of subtype B and C reference strains. Overall, the subtype C reference strains were more sensitive to neutralization by subtype C plasma samples (P = 0.0005) (Fig. 3). Additional comparisons were made with published data on a panel of 12 subtype B reference strains (45). In general, the subtype C panel was less sensitive to neutralization by a subtype B plasma pool (P = 0.009), but no significant difference was found between the two panels of reference strains when compared in terms of their overall sensitivity to CD4, IgG1b12, 4E10, and the subtype A, C, and D plasma pools. The most dramatic difference between the two panels of reference strains was the uniform resistance of subtype C to neutralization by 2G12 and their infrequent neutralization by 2F5.
DISCUSSION
We describe here the first extensive characterization of the neutralization properties of newly transmitted subtype C HIV-1 variants from heterosexually acquired infections. The viruses were characterized as molecularly cloned Env-pseudotyped viruses by using a wide range of antibody specificities and in the context of genetic features that are known to influence epitope exposure on the complex gp120 molecule. Comparisons were made to subtype B, the most extensively studied HIV-1 subtype.
The R5 biologic phenotype (6) of all 18 subtype C gp160 clones in this report is typical of newly transmitted subtype B viruses (21, 93, 94) and appears to be common for subtype C regardless of the stage of infection (16, 59, 85). A general distinguishing feature was the shorter and less glycosylated gp120 on these subtype C viruses compared to newly transmitted subtype B viruses. A similar observation was made previously for both subtypes C and A where, unlike subtype B (29), the gp120 of subtypes C and A might expand and add PNLG over the course of infection (18, 23, 44). Li et al. recently reported that early autologous NAb responses in subtype C-infected individuals might be more potent than the early responses in subtype B-infected individuals (44). These authors suggested that increased exposure of epitopes on the shorter, less-glycosylated gp120 of newly transmitted subtype C viruses could have resulted in enhanced immunogenicity, greater antigenicity, or both. This interpretation is based on evidence that the variable loops and number and position of PNLG on gp120 are used by the virus to mask epitopes as a very effective means to evade NAbs (15, 26, 62, 84, 88). It has been suggested that the shorter, less-glycosylated gp120 of newly transmitted subtype C viruses might enhance the accessibility of certain epitopes to vaccine-elicited NAbs (23). The subtype C gp160 clones described here should be useful in future studies that address this possibility in greater detail.
It has been reported that the gp120 region of envelopes from newly transmitted subtype C viruses is shorter and less glycosylated compared to subtype C variants from chronically infected individuals (23) and that these differences are not seen in subtype B (18, 29). We performed a similar analysis by comparing the newly derived subtype B and C reference env sequences to subtype-matched gp120 controls in the LANL database, which is known to mostly contain sequences from chronic viruses. Results of this comparison showed a significant trend toward fewer PNLG in the V4 of newly transmitted subtype C viruses, but we found no evidence that these viruses contained shorter gp120 regions than chronic subtype C viruses. We also found a significant trend toward a greater number of PNLG on the gp120 of newly transmitted subtype B viruses compared to chronic subtype B viruses. We caution that our comparisons did not have strong statistical support because of the small sample size. Thus, a larger number of gp120 sequences from acutely and chronically infected individuals will be needed to resolve whether or not newly transmitted viruses have unique genetic features.
A general feature of the newly transmitted subtype C viruses was their relatively conserved V3 loop that was followed immediately downstream by a region of approximately 34 amino acids that was highly variable. A similar observation has been made for subtype C viruses from chronically infected individuals (24, 59) and for the mostly chronic subtype C viruses in the Los Alamos sequence database (32). Moreover, Milich et al. reported that the V3 loop of R5 but not X4 subtype B viruses also exhibits low sequence variability (51). In this regard, the V3 loop of our subtype C viruses was no more variable than the V3 loop of newly transmitted R5 subtype B reference clones (45). Notably, the region immediately downstream of V3 in the subtype B reference clones was highly variable (45). Thus, low amino acid sequence variability in V3 and high sequence variability immediately downstream of V3 appear to be features that are shared between newly transmitted and chronic R5 viruses of both subtypes.
The V3 loop is of considerable interest because it plays a central role in receptor and coreceptor interactions that determine viral tropism and entry, making this region an attractive target for NAb-based vaccines. Unfortunately, the cryptic nature of V3 (8) makes it extremely difficult for Abs to gain access to this region. Thus, HIV-1 has evolved to escape V3-specific NAbs by masking epitopes through strategic placement of PNLG (2, 71) and by the conformation of its V2 loop (15). We observed fewer PNLG on the V3 loop of newly transmitted subtype C viruses compared to subtype B, making it possible that enhanced epitope exposure on subtype C viruses could result in a stronger V3-directed NAb response compared to subtype B. An exposed V3 loop can in fact be a major target for NAbs (36, 80). Although we did not examine V3-specific neutralization here, we found no evidence in a previous study that newly transmitted subtype C viruses, including some of the same viruses used here, were unusually sensitive to V3-specific Abs (11). All of our subtype C viruses retain a highly conserved PNLG at a key site (position 301, HXB2 numbering) that is known to mask V3 epitopes on subtype B viruses (2, 71). They also possessed a V2 loop that on average was the same size as newly transmitted subtype B reference clones. These structural features might contribute to the effective masking of V3 epitopes on newly transmitted subtype C viruses.
Another interesting and potentially important neutralization target on the gp120 molecule is the coreceptor binding domain that lies in a region within and around the bridging sheet and might also involve a portion of the V3 loop (20, 63, 64, 72, 77, 78, 87). This region contains some of the most highly conserved neutralization epitopes on the virus; however, exposure of these epitopes in most cases requires conformational changes in gp120 that occur upon CD4 ligation (22, 68, 75). Viruses that possess a spontaneously exposed coreceptor binding domain have been observed in vivo (22, 92), making it possible that conditions exist where this property affords a fitness advantage. One example would be to provide a selective advantage for transmission and early virus replication before CD4i antibodies are made. We tested this hypothesis by using a series of monoclonal Abs to probe CD4i epitopes on our newly transmitted subtype C viruses and found very few cases of positive neutralization regardless of whether subinhibitory doses of sCD4 were present. This outcome is discordant with a previous study in which CD4i epitopes were shown to be potent targets for neutralization on a subset of HIV-2 viruses (22). Based on our results, CD4i epitopes on newly transmitted subtype C viruses appear to be effectively masked and difficult to induce and stabilize with sCD4. We acknowledge that, because our viruses were obtained several weeks after infection, it remains possible that initial transmission involves viruses on which CD4i epitopes are spontaneously exposed.
We found it interesting that SF162.LS and MN, two strains of HIV-1 that are highly sensitive to neutralization by HIV-1-positive sera, were both sensitive to neutralization by CD4i monoclonal Abs. T-cell-line-adapted strains, including MN, are thought to be highly sensitive to HIV-1-positive sera because the V3 loop on these viruses is exposed (36, 80). Our results suggest that exposure of CD4i epitopes is another reason why certain strains of HIV-1 are highly sensitive to HIV-1-positive sera. Indeed, serum samples from most HIV-1-infected individuals have been shown to contain high titers of CD4i antibodies (22).
A major goal of this study was to create a well-characterized panel of subtype C gp160 reference clones to facilitate standardized assessments of vaccine-elicited NAb responses. A similar panel of gp160 reference clones for subtype B was described recently (47). We considered it important that the envelope glycoproteins of reference strains exhibit general antigenic features that will not overestimate the potential value of Abs against cryptic epitopes, such as those in the V3 loop and coreceptor binding domain, that tend to be poor targets on primary isolates (45). The fact that our subtype C Env-pseudotyped viruses were a great deal less sensitive to neutralization by HIV-1-positive plasma samples than MN and SF162.LS suggests they possess appropriate antigenic properties. These antigenic properties were characterized in greater detail by using a set of broadly neutralizing monoclonal Abs whose epitopes are of major interest for vaccine development. As expected, the 2G12 epitope was completely absent and the 2F5 epitope was rarely detected on these subtype C Env-pseudotyped viruses. Similar results have been reported previously for newly transmitted subtype C viruses (11) and for subtype C viruses from chronically infected individuals (7) and pediatric infections (33). Also in agreement with these previous reports, all of our subtype C Env-pseudotyped viruses contained the 4E10 epitope and many contained the IgG1b12 epitope.
The overall neutralization phenotype and inferred structural properties of these HIV-1 subtype C Env clones appear to be suitably representative of subtype C to support their use as reference reagents. For uniformity, we have designated a subset of 12 of these gp160 clones to recommend as standards for assessing the NAb responses generated by the current pipeline of candidate vaccines. This panel complements an existing panel of subtype B gp160 reference clones (45) and a multisubtype panel of viruses from chronically infected individuals (9), all of which may need to be modified in the future as new information emerges on how to improve their correlative value (47). Additional panels of newly transmitted gp160 reference clones are needed for other major subtypes of HIV-1, including A, D, CRF01_AE, and CRF02_AG. The subtype preferences of HIV-1-positive plasma samples seen here, combined with genetic differences between subtypes that might affect neutralization, support the need for separate panels. We also encourage the development of multiple panels of gp160 clones within each major subtype to more completely represent intrasubtype diversity at the geographic level (11). Considering the prevalence and global distribution of subtype C, it will be especially important to develop additional panels of reference gp160 clones from other parts of the world where this subtype dominates the epidemic, including China, India, and other African countries. These additional panels should facilitate vaccine discovery by enhancing the standardized assessment of vaccine-elicited NAb responses on a global scale.
Acknowledgments
We thank Gita Ramjee and the CAPRISA clinical and laboratory staff for sample collection. We also thank Florette Treunicht, Isaac Choge, and Penny Moore for generating functional env clones from the CAP isolates and Natasha Taylor-Meyer, Eleanor Cave, and Isaac Choge for characterizing the BB plasma samples. Finally, we thank Opendra Sharma and his staff for their assistance in making the reference clones available through the NIH ARRRP.
This work was supported by National Institutes of Health grants AI30034 and AI46705 (D.C.M.), AI055386 (F.G.), AI51231 and AI64060 (E.H.), and AI54497, AI85338, AI41530, and AI27767 (B.H.H.). CAPRISA is supported by NIH grant AI51794.
Footnotes
Published ahead of print on 13 September 2006.
REFERENCES
- 1.Allen, S., J. Meinzen-Derr, M. Kautzman, I. Zulu, S. Trask, U. Fideli, R. Musonda, F. Kasolo, F. Gao, and A. Haworth. 2003. Sexual behavior of HIV discordant couples after HIV counseling and testing. AIDS 17:733-740. [DOI] [PubMed] [Google Scholar]
- 2.Back, N. K. T., L. Smit, J.-J. de Jong, W. Keulen, M. Schutten, J. Goudsmit, and M. Tersmette. 1994. An N-glycan within the human immunodeficiency virus type 1 gp120 V3 loop affects virus neutralization. Virology 199:431-438. [DOI] [PubMed] [Google Scholar]
- 3.Barbato, G., E. Bianchi, P. Ingallinella, W. H. Hurn, M. D. Miller, G. Ciliberto, R. Cortese, R. Bazzo, J. W. Shiver, and A. Pessi. 2003. Structural analysis of the epitope of the anti-HIV antibody 2F5 sheds light into its mechanism of neutralization and HIV fusion. J. Mol. Biol. 330:1101-1115. [DOI] [PubMed] [Google Scholar]
- 4.Beddows, S., S. Lister, R. Cheingsong, C. Bruck, and J. Weber. 1999. Comparison of the antibody repertoire generated in healthy volunteers following immunization with a monomeric recombinant gp120 construct derived from a CCR5/CXCR4-using human immunodeficiency virus type 1 isolate with sera from naturally infected individuals. J. Virol. 73:1740-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belshe, R. B., G. J. Gorse, M. J. Mulligan, T. G. Evans, M. C. Keefer, J.-L. Excler, A.-M. Duliege, J. Tartaglia, W. I. Cox, J. McNamara, K.-L. Hwang, A. Bradney, D. Montefiori, and K. J. Weinhold. 1998. Induction of immune responses to HIV-1 canarypox virus (ALVAC) HIV-1 and gp120 SF-2 recombinant vaccines in uninfected volunteers. AIDS 12:2407-2415. [DOI] [PubMed] [Google Scholar]
- 6.Berger, E. A., R. W. Doms, E.-M. Fenyö, B. T. M. Korber, D. R. Littman, J. P. Moore, Q. J. Sattentau, H. Schuitemaker, J. Sodrsoki, and R. A. Weiss. 1998. HIV-1 phenotypes classified by co-receptor usage. Nature 391:240. [DOI] [PubMed] [Google Scholar]
- 7.Binley, J., T. Wrin, B. Korber, M. Zwick, M. Wang, C. Chappey, G. Stiegler, R. Kunert, S. Zolla-Pazner, H. Katinger, C. Petropoulos, and D. Burton. 2004. Comprehensive cross-subtype neutralization analysis of a panel of anti-human immunodeficiency virus type 1 monoclonal antibodies. J. Virol. 78:13232-13252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bou-Habib, D. C., G. Roderiquez, T. Oravecz, P. W. Berman, P. Lusso, and M. A. Norcross. 1994. Cryptic nature of envelope V3 region epitopes protects primary monocytotropic human immunodeficiency virus type 1 from antibody neutralization. J. Virol. 68:6006-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, B. K., J. M. Darden, S. Tovanabutra, T. Oblander, J. Frost, E. Sanders-Buell, M. S. DeSouza, D. L. Birx, F. E. McCutchan, and V. R. Polonis. 2005. Biologic and genetic characterization of a panel of 60 human immunodeficiency virus type 1 (HIV-1) isolates, representing clades A, B, C, D, CRF01_AE, and CRF02_AG, for the development and assessment of candidate vaccines. J. Virol. 79:6089-6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bures, R., A. Gaitan, T. Zhu, C. Graziosi, K. M. McGrath, J. Tartaglia, P. Caudrelier, R. El Habib, M. Klein, A. Lazzarin, D. M. Stablein, M. Deers, L. Corey, M. L. Greenberg, D. H. Schwartz, and D. C. Montefiori. 2000. Immunization with recombinant canarypox vectors expressing membrane-anchored gp120 followed by gp160 protein boosting fails to generate antibodies that neutralize R5 primary isolates of human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 16:2019-2035. [DOI] [PubMed] [Google Scholar]
- 11.Bures, R., L. Morris, C. Williamson, G. Ramjee, M. Deers, S. A. Fiscus, S. A. Karim, and D. C. Montefiori. 2002. Regional clustering of shared neutralization determinants on primary isolates of subtype C human immunodeficiency virus type 1 from South Africa. J. Virol. 76:2233-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burton, D., R. Desrosiers, R. Doms, W. Koff, P. Kwong, J. Moore, G. Nabel, J. Sodroski, I. Wilson, and R. Wyatt. 2004. HIV vaccine design and the neutralizing antibody problem. Nat. Immunol. 5:233-236. [DOI] [PubMed] [Google Scholar]
- 13.Burton, D. R., J. Pyati, R. Koduri, S. J. Sharp, G. B. Thornton, P. W. Parren, L. S. Sawyer, R. M. Hendry, N. Dunlop, P. L. Nara, et al. 1994. Efficient neutralization of primary isolates of HIV-1 by a recombinant human monoclonal antibody. Science 266:1024-1027. [DOI] [PubMed] [Google Scholar]
- 14.Calarese, D. A., C. N. Scanlan, M. B. Zwick, S. Deechongkit, Y. Mimura, R. Kunert, P. Zhu, M. R. Wormald, R. L. Stanfield, K. H. Roux, J. W. Kelly, P. M. Rudd, R. A. Dwek, H. Katinger, D. R. Burton, and I. A. Wilson. 2003. Antibody domain exchange is an immunologic solution to carbohydrate cluster recognition. Science 300:2065-2071. [DOI] [PubMed] [Google Scholar]
- 15.Cao, J., N. Sullivan, E. Desjardin, C. Parolin, J. Robinson, R. Wyatt, and J. Sodroski. 1997. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J. Virol. 71:9808-9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cecilia, D., S. S. Kulkarni, S. P. Tripathy, R. R. Gangakhedkar, R. S. Paranjape, and D. A. Gadkari. 2000. Absence of coreceptor switch with disease progression in human immunodeficiency virus infections in India. Virology 271:253-258. [DOI] [PubMed] [Google Scholar]
- 17.Cheng-Mayer, C., R. Liu, N. R. Landau, and L. Stamatatos. 1997. Macrophage tropism of human immunodeficiency virus type 1 and utilization of the CC-CKR5 coreceptor. J. Virol. 71:1657-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chohan, B., D. Lang, M. Sagar, B. Korber, L. Lavreys, B. Richardson, and J. Overbaugh. 2005. Selection for human immunodeficiency virus type 1 envelope glycosylation variants with shorter V1-V2 loop sequences occurs during transmission of certain genetic subtypes and may impact viral RNA levels. J. Virol. 79:6528-6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cilliers, T., J. Nhlapo, M. Coetzer, D. Orlovic, T. Ketas, W. C. Olson, J. P. Moore, A. Trkola, and L. Morris. 2003. The CCR5 and CXCR4 coreceptors are both used by human immunodeficiency virus type 1 primary isolates from subtype C. J. Virol. 77:4449-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cocchi, F., A. L. DeVico, A. Garzino-Demo, A. Cara, R. C. Gallo, and P. Lusso. 1996. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat. Med. 2:1244-1247. [DOI] [PubMed] [Google Scholar]
- 21.Conner, R. I., K. E. Sheridan, D. Ceradini, S. Choe, and N. R. Landau. 1997. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J. Exp. Med. 185:621-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Decker, J., F. Bibollet-Ruche, X. Wei, S. Wang, D. N. Levy, W. Wang, E. Delaporte, M. Peeters, C. A. Derdeyn, S. Allen, E. Hunter, M. S. Saag, J. A. Hoxie, B. H. Hahn, P. D. Kwong, J. E. Robinson, and G. M. Shaw. 2005. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J. Exp. Med. 201:1407-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Derdeyn, C. A., J. M. Decker, F. Bibollet-Ruche, J. L. Mokili, M. Muldoon, S. A. Denham, M. L. Heil, F. Kasolo, R. Musonda, B. H. Hahn, G. M. Shaw, B. T. Korber, S. Allen, and E. Hunter. 2004. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science 303:2019-2022. [DOI] [PubMed] [Google Scholar]
- 24.Engelbrecht, S., T. de Villiers, C. C. Sampson, J. Z. Megede, S. W. Barnett, and E. J. Van Rensburg. 2001. Genetic analysis of the complete gag and env genes of HIV type 1 subtype C primary isolates from South Africa. AIDS Res. Hum. Retrovir. 17:1533-1547. [DOI] [PubMed] [Google Scholar]
- 25.Esparza, J., R. Klausner, and the Coordinating Committee of the Global HIV/AIDS Vaccine Enterprise. 2005. The Global HIV/AIDS Vaccine Enterprise: scientific strategic plan. Policy Forum, vol. 2. [Online.] doi: 10.1371/journal.pmed.0020025. [DOI]
- 26.Etemad-Moghadam, B., G. B. Karlsson, M. Halloran, Y. Sun, D. Schenten, M. Fernandes, N. L. Letvin, and J. Sodroski. 1998. Characterization of simian-human immunodeficiency virus envelope glycoprotein epitopes recognized by neutralizing antibodies from infected macaques. J. Virol. 72:8437-8445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Faulkner, D. M., and J. Jurka. 1988. Multiple aligned sequence editor (MASE). Trends Biochem. Sci. 13:321-322. [DOI] [PubMed] [Google Scholar]
- 28.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the boostrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 29.Frost, S. D. W., Y. Liu, S. L. Kosakovsky Pond, C. Chappey, T. Wrin, C. J. Petropoulos, S. J. Little, and D. D. Richman. 2005. Characterization of human immunodeficiency virus type 1 (HIV-1) envelope variation and neutralizing antibody responses during transmission of HIV-1 subtype B. J. Virol. 79:6523-6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gallo, R. C., S. Z. Salahuddin, M. Popovic, G. M. Shearer, M. Kaplan, B. F. Haynes, T. J. Palker, R. Redfield, J. Oleske, B. Safai, G. White, P. Foster, and P. D. Markham. 1984. Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science 224:500-503. [DOI] [PubMed] [Google Scholar]
- 31.Galvin, S. R., and M. S. Cohen. 2006. Genital tract reservoirs. Curr. Opin. HIV AIDS 1:162-166. [DOI] [PubMed] [Google Scholar]
- 32.Gaschen, B., J. Taylor, K. Yusim, B. Foley, F. Gao, D. Lang, V. Novitsky, B. Haynes, B. H. Hahn, T. Bhattacharya, and B. Korber. 2003. Diversity of considerations in HIV-1 vaccine selection. Science 296:2354-2360. [DOI] [PubMed] [Google Scholar]
- 33.Gray, E. S., T. Meyers, G. Gray, D. C. Montefiori, and L. Morris. 2006. Insensitivity of pediatric HIV-1 subtype C viruses to broadly neutralizing monoclonal antibodies raised against subtype B. PloS Med. 3:1023-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haynes, B. F., and D. C. Montefiori. 2006. Aiming to induce broadly reactive neutralizing antibody responses with HIV-1 vaccine candidates. Exp. Rev. Vaccines 5:347-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Higgins, D. G., and P. M. Sharp. 1989. Fast and sensitive multiple sequence alignments on a microcomputer. Comput. Appl. Biosci. 5:151-153. [DOI] [PubMed] [Google Scholar]
- 36.Javaherian, K., A. J. Langlois, C. McDanal, K. L. Ross, L. I. Echkler, C. L. Jellis, A. T. Profy, J. R. Rusche, D. P. Bolognesi, S. D. Putney, and T. J. Matthews. 1989. Principal neutralizing domain of the human immunodeficiency virus type 1 envelope protein. Proc. Natl. Acad. Sci. USA 86:6768-6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 38.Kolchinsky, P., E. Kiprilov, and J. Sodsroski. 2001. Increased neutralization sensitivity of CD4-independent human immunodeficiency virus variants. J. Virol. 75:2041-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kostrikis, L. G., Y. Cao, H. Ngai, J. P. Moore, and D. D. Ho. 1996. Quantitative analysis of serum neutralization of human immunodeficiency virus type 1 from subtypes A, B, C, D, E, F, and I: lack of direct correlation between neutralization serotypes and genetic subtypes and evidence for prevalent serum-dependent infectivity enhancement. J. Virol. 70:445-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kwong, P. D., M. L. Doyle, D. J. Casper, C. Cicala, S. A. Leavitt, S. Majeed, T. D. Steenbeke, M. Venturi, I. Chaiken, M. Fung, H. Katinger, P. W. H. I. Parren, J. Robinson, D. Van Ryk, L. Wang, D. R. Burton, E. Freire, R. Wyatt, J. Sodroski, W. A. Hendrickson, and J. Arthos. 2002. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature 420:678-682. [DOI] [PubMed] [Google Scholar]
- 41.Lee, W. R., X. F. Yu, W. J. Syu, M. Essex, and T. H. Lee. 1992. Mutational analysis of conserved N-linked glycosylation sites of human immunodeficiency virus type 1 gp41. J. Virol. 66:1799-1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leitner, T., B. Korber, M. Daniels, C. Calef, and B. Foley. 2005. HIV-1 subtype and circulating recombinant form (CRF) reference sequences, 2005, p. 41-48. In T. Leitner, B. Foley, B. Hahn, P. Marx, F. McCutchan, J. W. Mellors, S. Wolinski, and B. Korber (ed.), HIV Sequence Compendium 2005. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 43.Letvin, N. L., D. H. Barouch, and D. C. Montefiori. 2002. Prospects for vaccine protection against HIV-1 infection and AIDS. Annu. Rev. Immunol. 20:73-99. [DOI] [PubMed] [Google Scholar]
- 44.Li, B., J. M. Decker, R. W. Johnson, F. Bibollet-Ruche, X. Wei, J. Mulenga, S. Allen, E. Hunter, B. H. Hahn, G. M. Shaw, J. L. Blackwell, and C. A. Derdeyn. 2006. Evidence for potent autologous neutralizing antibody titers and compact envelopes in early infection with subtype C human immunodeficiency virus type 1. J. Virol. 80:5211-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li, M., F. Gao, J. R. Mascola, L. Stamatatos, V. R. Polonis, M. Koutsoukos, G. Voss, P. Goepfert, P. Gilbert, K. M. Greene, M. Bilska, D. L. Kothe, J. F. Salazar-Gonzalez, X. Wei, J. M. Decker, B. H. Hahn, and D. C. Montefiori. 2005. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J. Virol. 79:10108-10125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mascola, J. R. 2003. Defining the protective antibody response for HIV-1. Curr. Mol. Med. 3:211-218. [DOI] [PubMed] [Google Scholar]
- 47.Mascola, J. R., P. D'Souza, P. Gilbert, B. Hahn, N. L. Haigwood, L. Morris, C. J. Petropoulos, V. R. Polonis, M. Sarzotti-Kelsoe, and D. C. Montefiori. 2005. Recommendations for the design and use of standard virus panels to assess the neutralizing antibody response elicited by candidate human immunodeficiency virus type 1 vaccines. J. Virol. 79:10103-10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mascola, J. R., and D. C. Montefiori. 2003. HIV: nature's master of disguise. Nat. Med. 9:393-394. [DOI] [PubMed] [Google Scholar]
- 49.Mascola, J. R., S. W. Snyder, O. S. Weislow, S. M. Belay, R. B. Belshe, D. H. Schwartz, M. L. Clements, R. Dolin, B. S. Graham, G. J. Gorse, M. C. Keefer, M. J. McElrath, M. C. Walker, K. F. Wagner, J. G. McNeil, F. E. McCutchan, and D. S. Burke. 1996. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratory-adapted but not primary isolates of human immunodeficiency virus type 1. J. Infect. Dis. 173:340-348. [DOI] [PubMed] [Google Scholar]
- 50.McCutchan, F. E. 2000. Understanding the genetic diversity of HIV-1. AIDS 14(Suppl. 3):S31-S44. [PubMed] [Google Scholar]
- 51.Milich, L., B. H. Margolin, and R. Swanstrom. 1997. Patterns of amino acid variability in NSI-like and SI-like V3 sequences and a linked change in the CD4-binding domain of the HIV-1 Env protein. Virology 239:108-118. [DOI] [PubMed] [Google Scholar]
- 52.Montefiori, D. C. 2004. Evaluating neutralizing antibodies against HIV, SIV and SHIV in luciferase reporter gene assays, p. 12.11.1-12.11.15. In J. E. Coligan, A. M. Kruisbeek, D. H. Margulies, E. M. Shevach, W. Strober, and R. Coico (ed.), Current protocols in immunology. John Wiley & Sons, New York, N.Y. [DOI] [PubMed]
- 53.Montefiori, D. C., W. E. Robinson, Jr., S. S. Schuffman, and W. M. Mitchell. 1988. Evaluation of antiviral drugs and neutralizing antibodies to human immunodeficiency virus by a rapid and sensitive microtiter infection assay. J. Clin. Microbiol. 26:231-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moore, J. P., and D. R. Burton. 2004. Urgently needed: a filter for the HIV-1 vaccine pipeline. Nat. Med. 10:769-771. [DOI] [PubMed] [Google Scholar]
- 55.Moore, J. P., Y. Cao, J. Leu, L. Qin, B. Korber, and D. D. Ho. 1996. Inter- and intrasubtype neutralization of human immunodeficiency virus type 1: genetic subtypes do not correspond to neutralization serotypes but partially correspond to gp120 antigenic serotypes. J. Virol. 70:427-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moore, J. P., and J. Sodroski. 1996. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J. Virol. 70:1863-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Muster, T., F. Steindl, M. Purtscher, A. Trkola, A. Klima, G. Himmler, F. Ruker, and H. Katinger. 1993. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. J. Virol. 67:6642-6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pantophlet, R., E. O. Saphire, P. Poignard, P. W. H. I. Parren, I. A. Wilson, and D. R. Burton. 2003. Fine mapping of the interaction of neutralizing and nonneutralizing monoclonal antibodies with the CD4 binding site of human immunodeficiency virus type 1 gp120. J. Virol. 77:642-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ping, L.-H., J. A. E. Nelson, I. F. Hoffman, J. Schock, S. L. Lamers, M. Goodman, P. Vernazza, P. Kazembe, M. Maida, D. Zimba, M. M. Goodenow, J. J. Eron, Jr., S. A. Fiscus, M. S. Cohen, and R. Swanstrom. 1999. Characterization of V3 sequence heterogeneity in subtype C human immunodeficiency virus type 1 isolates from Malawi: underrepresentation of X4 variants. J. Virol. 73:6271-6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Platt, E. J., K. Wehrly, S. E. Kuhmann, B. Chesebro, and D. Kabat. 1998. Effects of CCR5 and CD4 cell surface concentrations on infection by macrophage tropic isolates of human immunodeficiency virus type 1. J. Virol. 72:2855-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Purtscher, M., A. Trkola, G. Gruber, A. Buchacher, R. Predl, F. Steindl, C. Tauer, R. Berger, N. Barrett, A. Jungbauer, and H. Katinger. 1994. A broadly neutralizing human monoclonal antibody against gp41 of human immunodeficiency virus. AIDS Res. Hum. Retrovir. 10:1651-1658. [DOI] [PubMed] [Google Scholar]
- 62.Reitter, J. N., R. E. Means, and R. C. Desrosiers. 1998. A role for carbohydrates in immune evasion in AIDS. Nat. Med. 4:679-684. [DOI] [PubMed] [Google Scholar]
- 63.Rizzuto, C., and J. Sodroski. 2000. Fine definition of a conserved CCR5-binding region on the human immunodeficiency virus type 1 glycoprotein gp120. AIDS Res. Hum. Retrovir. 16:741-749. [DOI] [PubMed] [Google Scholar]
- 64.Rizzuto, C. D., R. Wyatt, N. Hernández-Ramos, Y. Sun, P. D. Kwong, W. A. Hendrickson, and J. Sodroski. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280:1949-1953. [DOI] [PubMed] [Google Scholar]
- 65.Robbins, J. B., R. Schneerson, and S. C. Szu. 1995. Perspective: serum IgG antibody is sufficient to confer protection against infectious diseases by inactivating the inoculum. J. Infect. Dis. 171:1387-1398. [DOI] [PubMed] [Google Scholar]
- 66.Robinson, J. E., D. Holton, J. Liu, H. McMurdo, A. Murciano, and R. Gohd. 1990. A novel enzyme-linked immunosorbent assay (ELISA) for the detection of antibodies to HIV-1 envelope glycoproteins based on immobilization of viral glycoproteins in microtiter wells coated with concanavalin A. J. Immunol. Methods 132:63-71. [DOI] [PubMed] [Google Scholar]
- 67.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 68.Salzwedel, K., E. D. Smith, B. Dey, and E. A. Berger. 2000. Sequential CD4-coreceptor interactions in human immunodeficiency virus type 1 Env function: soluble CD4 activates Env for coreceptor-dependent fusion and reveals blocking activities of antibodies against cryptic conserved epitopes on gp120. J. Virol. 74:326-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sanders, R. W., M. Venturi, L. Schiffer, R. Kalyanaraman, H. Katinger, K. O. Lloyd, P. D. Kwong, and J. P. Moore. 2002. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J. Virol. 76:7293-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Scanlan, C. N., R. Pantophlet, M. R. Wormald, E. O. Saphire, R. Stanfield, I. A. Wilson, H. Katinger, R. A. Dwek, P. M. Rudd, and D. R. Burton. 2002. The broadly neutralizing anti-HIV-1 antibody 2G12 recognizes a cluster of α1-2 mannose residues on the outer face of gp120. J. Virol. 76:7306-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schønning, K., B. Jansson, S. Olofsson, J. O. Neilsen, and J.-E. S. Hansen. 1996. Resistance to V3-directed neutralization caused by an N-linked oligosaccharide depends on the quaternary structure of the HIV-1 envelope oligomer. Virology 218:134-140. [DOI] [PubMed] [Google Scholar]
- 72.Speck, R. F., K. Wehrly, E. J. Platt, R. E. Atchison, I. F. Charo, D. Kabat, B. Chesebro, and M. A. Goldsmith. 1997. Selective employment of chemonike receptors as human immunodeficiency virus type 1 coreceptors determined by individual amino acids within the envelope V3 loop. J. Virol. 71:7136-7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stamatatos, L., M. Wiskerchen, and C. Cheng-Mayer. 1998. Effect of major deletions in the V1 and V2 loops of a macrophage-tropic HIV-1 isolate on viral envelope structure, cell-entry and replication. AIDS Res. and Hum. Retrovir. 14:1129-1139. [DOI] [PubMed] [Google Scholar]
- 74.Stiegler, G., R. Kunert, M. Purtscher, S. Wolbank, R. Voglauer, F. Steindl, and H. Katinger. 2001. A potent cross-subtype neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS Res. Hum. Retrovir. 17:1757-1765. [DOI] [PubMed] [Google Scholar]
- 75.Sullivan, N., Y. Sun, Q. Sattentau, M. Thali, D. Wu, G. Denisova, J. Gershoni, J. Robinson, J. Moore, and J. Sodroski. 1998. CD4-induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J. Virol. 72:4694-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Trkola, A., T. Dragic, J. Arthos, J. M. Binley, W. C. Olson, G. P. Allaway, C. Cheng-Mayer, J. Robinson, P. J. Maddon, and J. P. Moore. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184-187. [DOI] [PubMed] [Google Scholar]
- 78.Ugolini, S., I. Mondor, P. W. H. I. Parren, D. R. Burton, S. A. Tilley, P. J. Klasse, and Q. J. Sattentau. 1997. Inhibition of virus attachment to CD4+ target cells is a major mechanism of T cell line-adapted HIV-1 neutralization. J. Exp. Med. 186:1287-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.UNAIDS. 2005. UNAIDS/WHO AIDS epidemic update: December 2005. [Online.] http://www.unaids.org/epi/2005/.
- 80.Vancott, T. C., V. R. Polonis, L. D. Loomis, N. L. Michael, P. L. Nara, and D. L. Birx. 1995. Differential role of V3-specific antibodies in neutralization assays involving primary and laboratory-adapted isolates of HIV type 1. AIDS Res. Hum. Retrovir. 11:1379-1390. [DOI] [PubMed] [Google Scholar]
- 81.van Damme, L., C. Chandeying, G. Ramjee, H. Rees, P. Sirivongrangson, M. Laga, and J. Perriens. 2000. Safety of multiple daily applications of COL-1492, a nonoxynol-9 vaginal gel, among female sex workers. AIDS 14:85-88. [DOI] [PubMed] [Google Scholar]
- 82.van't Wout, A. B., N. A. Kootstra, G. A. Mulder-Kampinga, N. Albrecht-van Lent, H. J. Scherpbier, J. Veenstra, K. Boer, R. A. Coutinho, F. Miedema, and H. Schuitemaker. 1994. Macrophage-tropic variants initiate human immunodeficiency virus type 1 infection after sexual, parenteral, and vertical transmission. J. Clin. Investig 94:2060-20607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 85.Williamson, C., L. Morris, M. F. Maughan, L. H. Ping, S. A. Dryga, R. Thomas, E. A. Reap, T. Cilliers, J. van Harmelen, A. Pascual, G. Ramjee, G. Gray, R. Johnston, S. A. Karim, and R. Swanstrom. 2003. Characterization and selection of HIV-1 subtype C isolates for use in vaccine development. AIDS Res. Hum. Retrovir. 19:133-144. [DOI] [PubMed] [Google Scholar]
- 86.Wolfs, T. F., G. Zwart, M. Bakker, and J. Goudsmit. 1992. HIV-1 genomic RNA diversification following sexual and parenteral virus transmission. Virology 189:103-110. [DOI] [PubMed] [Google Scholar]
- 87.Wu, L., N. P. Gerard, R. Wyatt, H. Choe, C. Parolin, N. Ruffing, A. Borsetti, A. A. Cardoso, E. Desjardin, W. Newman, C. Gerard, and J. Sodroski. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179-183. [DOI] [PubMed] [Google Scholar]
- 88.Wyatt, R., J. Moore, M. Accola, E. Desjardin, J. Robinson, and J. Sodroski. 1995. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J. Virol. 69:5723-5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]
- 90.Xiang, S.-H., N. Doka, R. K. Choudhary, J. Sodroski, and J. E. Robinson. 2002. Characterization of CD4-induced epitopes on the HIV type 1 gp120 envelope glycoprotein recognized by neutralizing human monoclonal antibodies. AIDS Res. Hum. Retrovir. 18:1207-1217. [DOI] [PubMed] [Google Scholar]
- 91.Xiang, S.-H., L. Wang, M. Abreu, C.-C. Huang, P. D. Kwong, E. Rosenberg, J. E. Robinson, and J. Sodroski. 2003. Epitope mapping and characterization of a novel CD4-induced human monoclonal antibody capable of neutralizing primary HIV-1 strains. Virology 315:124-134. [DOI] [PubMed] [Google Scholar]
- 92.Zhang, P. F., P. Bouma, E. J. Park, J. B. Margolick, J. E. Robinson, S. Zolla-Pazner, M. N. Flora, and G. V. Quinnan, Jr. 2002. A variable region 3 (V3) mutation determines a global neutralization phenotype and CD4-independent infectivity of a human immunodeficiency virus type 1 envelope associated with a broadly cross-reactive, primary virus-neutralizing antibody response. J. Virol. 76:644-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang, L. Q., P. MacKenzie, A. Cleland, E. C. Holmes, A. J. Brown, and P. Simmonds. 1993. Selection for specific sequences in the external envelope protein of human immunodeficiency virus type 1 upon primary infection. J. Virol. 67:3345-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhu, T., H. Mo, N. Wang, D. S. Nam, Y. Cao, R. A. Koup, and D. D. Ho. 1993. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science 261:1179-1181. [DOI] [PubMed] [Google Scholar]
- 95.Zwick, M. B., A. F. Labrijn, M. Wang, C. Spenlehauer, E. O. Saphire, J. M. Binley, J. P. Moore, G. Stiegler, H. Katinger, D. R. Burton, and P. W. H. I. Parren. 2001. Broadly neutralizing antibodies targeted to the membrane-proximal external region of human immunodeficiency virus type 1 glycoprotein gp41. J. Virol. 75:10892-10905. [DOI] [PMC free article] [PubMed] [Google Scholar]