Abstract
Consensus covariation-based secondary structural models for the 5′ 140 nucleotides of the 5′ untranslated regions (5′UTRs) from mouse hepatitis virus (MHV) and severe acute respiratory syndrome coronavirus (SCoV) were developed and predicted three major helical stem-loop structures, designated stem-loop 1 (SL1), SL2, and SL4. The SCoV 5′UTR was predicted to contain a fourth stem-loop, named SL3, in which the leader transcriptional regulatory sequence (TRS) is folded into a hairpin loop. cDNAs corresponding to MHV/SCoV chimeric genomes were constructed by replacing the complete MHV 5′UTR with the corresponding SCoV sequence and by separately replacing MHV 5′UTR putative SL1, putative SL2, TRS, and putative SL4 with the corresponding SCoV sequences. Chimeric genomes were transcribed in vitro, and viruses were recovered after electroporation into permissive cells. Genomes in which the MHV 5′UTR SL1, SL2, and SL4 were individually replaced by their SCoV counterparts were viable. Chimeras containing the complete SCoV 5′UTR or the predicted SCoV SL3 were not viable. A chimera containing the SCoV 5′UTR in which the SCoV TRS was replaced with the MHV TRS was also not viable. The chimera containing the entire SCoV 5′UTR failed to direct the synthesis of any virus-specific RNA. Replacing the SCoV TRS with the MHV TRS in the MHV/5′UTR SCoV chimera permitted the synthesis of minus-sense genome-sized RNA but did not support the production of positive- or minus-sense subgenomic RNA7. A similar phenotype was obtained with the MHV/SCoV SL3 chimera. These results suggest a role for the TRS in the replication of minus-sense genomic RNA in addition to its known function in subgenomic RNA synthesis.
The coronavirus (CoV) family is a group of enveloped RNA viruses that infect diverse species of mammals, causing respiratory, gastrointestinal, and neurologic disease (18). Prior to 2002, human coronaviruses (HCoV) were primarily thought of as upper respiratory pathogens, accounting for approximately 30% of common colds. In late 2002, a novel CoV emerged in southern China to cause severe acute respiratory syndrome, a disease with 10% mortality. This outbreak rapidly spread to all continents and, in addition to its threat to public health, produced large economic dislocations in affected regions (31). Although no human cases of severe acute respiratory syndrome CoV (SCoV) infection have been reported since 2004, the virus remains a threat to reemerge by transmission from its likely natural host, wild bats (19). Even more recently, two new human coronaviruses, HCoV-NL63 and HCoV-HKU1, were identified as common causes of upper and lower respiratory disease (34, 36).
Coronavirus-infected cells contain 6 to 11 distinct RNA species (18). The intracellular RNAs form a nested set with common 3′ ends, with the longest RNA being the intracellular counterpart of the virion RNA (1). For mouse hepatitis virus (MHV), the 21-kb-long gene 1 contains two open reading frames (ORF1a and ORF1b); a −1 ribosomal frameshift during translation (2) results in the synthesis of a putative 7,178-amino-acid ORF1ab polyprotein. The resulting 740-kDa polypeptide contains a conserved array of functional domains: papain-like cysteine protease(s), ADP-ribose-1"-monophosphate-processing enzyme (X domain), a two-β-barrel-fold (picornavirus 3C-like) main protease, RNA-dependent RNA polymerase, zinc-binding domain-containing helicase, putative exoribonuclease, endoribonuclease, and ribose-2′-O-methyltransferase domains (23).
The coronaviruses have been classified into three subgroups, termed group 1, group 2, and group 3, based on serologic relatedness, genetic organization, and sequence similarity (30). SCoV was originally placed in its own unique group by primary genomic analysis (27). Although somewhat controversial, more recent phylogenetic studies using a variety of software tools and comparing a more extensive set of amino acid sequences than those used in the original analysis (ORF1a, ORF1b, spike [S], envelope [E], membrane [M], and nucleocapsid [N]) revealed that SCoV is best placed as an early branch of the group 2 coronaviruses (9, 30, 38). This group contains MHV (one of the best-characterized coronaviruses), human respiratory coronavirus (HCoV-OC43), and bovine coronavirus (BCoV), among others.
Coronavirus untranslated regions (UTRs) contain cis-acting sequences and structures known to play essential roles in RNA synthesis, gene expression, and virion assembly (6, 10, 16). Although coronaviruses of the same species recombine with high frequency (22), only a few studies have examined the ability of different coronaviruses to recombine (10, 13). Based on the conservation of secondary structures (a bulged stem-loop and an adjacent pseudoknot) in the 3′ untranslated regions (3′UTRs) of all group 2 coronaviruses, functional studies have shown that the MHV 3′UTR can be functionally replaced with the 3′UTRs from BCoV (13) and SCoV (10). Previously, others have partially characterized four stem-loop structures (I to IV) in the BCoV 5′UTR (5, 24, 25). Many of the features of this secondary structural model of the BCoV 5′UTR are supported by enzymatic structure probing and defective interfering (DI) RNA replication experiments. Somewhat surprisingly, stem-loops I and II do not have precise higher-order counterparts in the group 1 and group 3 coronaviruses. In this report, we present an alternative consensus secondary structural model of the coronavirus 5′UTRs (Fig. 1A) (17, 21). The model was developed by covariation analysis of a multiple sequence alignment of the 5′ 140 nucleotides (nt) of the 5′UTRs from nine coronaviruses from all three subgroups and is supported by a variety of RNA secondary structure prediction algorithms (7, 12, 26, 40) as well as nuclear magnetic resonance (NMR) analyses and genetic studies (P. Liu et al., unpublished data; L. Li and D. Giedroc, unpublished data). This model predicts three stem-loops, SL1, SL2, and SL4, that are structurally conserved among the group 1 and group 2 coronaviruses, including SCoV. Putative SL2 contains a (C/U)UUG(U/C) pentaloop sequence that is the most highly conserved contiguous run of nucleotides in the entire 5′UTR outside of the core transcriptional regulatory sequence (TRS), and covariation analysis reveals that this loop is always stacked on a 5-bp helix (Fig. 1B). SCoV is predicted to have an additional putative stem-loop structure (SL3) that contains both the SCoV TRS (ACGAAC) (15, 32) and MHV TRS (UCUAAAC) core sequences (CSs) (3, 28). Although 5′UTRs of the group 2 coronaviruses have a high degree of sequence similarity and are predicted to have similar secondary structures, to date, no study has investigated the ability of various 5′UTRs or portions of the UTRs from different species of coronaviruses to function in the context of a heterologous coronavirus genome. Studies on the ability of the SCoV 5′UTR to function in the background of the MHV genome are likely to be informative regarding the suggested phylogenetic relationship between these two viruses (30) and may indicate the potential for recombination between a significant human pathogen, SCoV, and related animal coronaviruses.
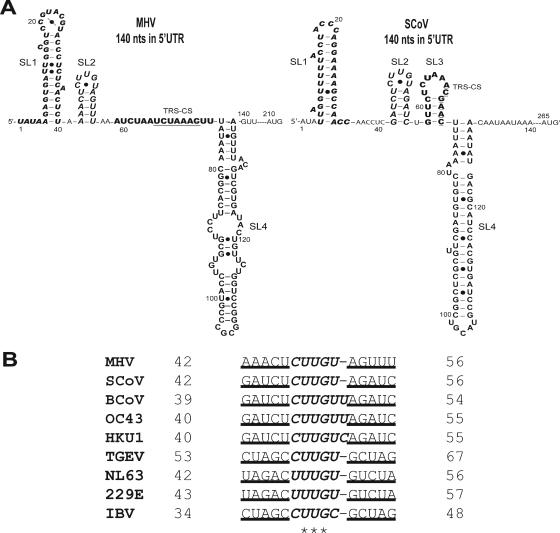
FIG. 1.
Predicted secondary structures within the 140-nt 5′UTR of the MHV and SCoV genomes (see the text for details). Previously described predicted stem-loops 1 (SL1) (boldface and italic type), 2 (SL2) (large font and italics), and 4 (SL4) (large font) in the 5′UTRs are noted (17, 21). MHV leader TRS CS (boldface, underlined type) is contained within the linear sequence between SL2 and SL4. However, the SCoV leader TRS CS (boldface, underlined type) is contained within stem-loop 3 (SL3) (boldface type). The AUGs shown represent the start codons of nsp1 in the MHV (nt 210 to 212) and SCoV (nt 265 to 267) genomes. • indicates noncanonical base pairings. (B) SL2 sequence alignment of group 2 coronaviruses. The stem portion of SL2 is underlined; the U-turn motif is italicized; * indicates absolutely conserved nucleotides. SL2 sequences of MHV (GenBank accession no. NC_001846), SCoV (accession no. AY278741), BCoV (accession no. NC_003045), HCoV-OC43 (accession no. NC_005147), HCoV-HKU1 (accession no. NC_006577), HCoV-NL63 (accession no. NC_005831), HCoV-229E (accession no. NC_002645), TGEV (accession no. NC_002306), and IBV (accession no. NC_001451) are relative to the corresponding viral genomes in GenBank.
In this work, we examine the ability of SCoV/MHV chimeric genomes to support viral replication. Chimeric genomes were created by individually inserting SCoV-encoded putative stem-loops SL1, SL2, and SL4 and the TRS CS in place of their MHV counterparts in the context of an otherwise wild-type (WT) MHV-A59 genome. Using a recently developed reverse genetic system (37), full-length cDNAs of the MHV-A59 genome were assembled to construct SCoV/MHV chimeric genomes. The chimeric genomes were generated in vitro, and their viability, growth kinetics, and ability to direct the synthesis of virus-specific RNAs were determined.
MATERIALS AND METHODS
Virus and cells.
DBT cells were maintained at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% calf serum (HyClone, Logan, UT). L2 cells were maintained at 37°C and 3% CO2 in DMEM supplemented with 10% calf serum. Baby hamster kidney 21 cells expressing the MHV receptor (BHK-R) were grown in DMEM supplemented with 10% calf serum and G418 (800 μg/ml) to select for cells expressing the MHV receptor (37). MHV-A59 1000 (37) was used as a wild-type control virus for comparison with chimeric viruses.
Assembly of a full-length MHV-A59 infectious construct.
The reverse genetic system for MHV-A59 used in this study was initially described by Yount et al. (37). cDNAs representing the entire MHV-A59 genome with either the wild-type sequence or the MHV/SCoV chimeric sequences were constructed by ligation of the A (or G) fragments to fragments (A)B to G(F) as described previously (37). Chimeric and wild-type MHV genomes were transcribed and electroporated into cells as previously described (37). Cultures were observed for up to 72 h for the development of a cytopathic effect (CPE) (cell fusion) and harvested by freezing at −70°C. Cultures that did not develop a CPE were blind passaged three times through DBT cells in a further attempt to recover infectious virus. At least three independent experiments, including at least one experiment in which electroporated cells were incubated at 34°C and 40°C, were performed before a mutant genome was considered nonviable.
Plasmid constructions.
The primers used in this study are listed in Table 1. The A plasmid of the MHV reverse genetic system described previously by Yount et al. (37) was utilized as a basis for constructing a fusion of the SCoV 5′UTR to the MHV gene 1 coding sequence. The strategy employed to construct this fusion exploited “no see'm” technology to eliminate a BsmBI restriction site engineered into the end of DNA fragments by PCR (37). Briefly, the entire SCoV 5′UTR was amplified from purified SCoV RNA (Urbani strain) (27), obtained from the Center for Disease Control and Prevention (CDC), by reverse transcription (RT)-PCR using primers that contained additional BsmBI sites (S 5′UTR swap oligonucleotides 3 and 7). The resultant RT-PCR product was TA cloned into the pGEM-T vector (Promega) to produce plasmid 0A-1, maintaining the BmsBI sites. Plasmid 0A-2 was produced by subcloning a BamHI fragment from the A plasmid of the MHV reverse genetic system into the PCR-XL-TOPO vector (Invitrogen). This BamHI fragment contained a small portion of the A plasmid backbone, a T7 promoter, and the first 252 nt of the MHV genome, extending into the MHV gene 1 coding region. The MHV 5′UTR was removed from plasmid 0A-2 by inverse PCR with Pfx DNA polymerase (Invitrogen) using primers that contained BsmBI sites (S 5′UTR swap oligonucleotides 5 and 6), followed by self-ligation to produce plasmid 0A-3. The SCoV 5′UTR was excised from plasmid 0A-1 by BsmBI digestion and ligated with BsmBI-digested plasmid 0A-3 to produce plasmid 7074. This ligation eliminated the BsmBI sites and precisely fused the SCoV 5′UTR to the MHV gene 1 coding sequence. After sequencing to verify the construction, a restriction fragment exchange was performed to replace the 550-bp BamHI fragment in MHV plasmid A with the 620-bp BamHI fragment containing a T7 promoter plus the SCoV 5′UTR fused to 252 nt of the MHV gene 1 coding sequence. This chimeric plasmid was called pHK0A and could be used in the MHV reverse genetic system in place of plasmid A. A similar strategy was used to replace the MHV 3′UTR with the SCoV 3′UTR (27) in the MHV reverse genetic system G plasmid. The SCoV 3′UTR and 15 nt of the poly(A) tail were amplified from purified SCoV RNA (27) by RT-PCR using primers that contained BsmBI sites (oligonucleotides 1 and 2), which could subsequently be eliminated during ligation, and cloned (plasmid 0G-1). In plasmid 0G-1, the MHV-A59 sequences between an ApaI site at position 30072 (relative to the MHV-A59 genome) (GenBank accession no. NC_001846) (39) and a PacI site inserted just downstream of the poly(A) tail were derived from plasmid B36 (14). Inverse PCR (oligonucleotides 3 and 4) was utilized to produce plasmid 0G-2, replacing the MHV 3′UTR with two BsmBI sites separated by a small spacer sequence. After digestion by the BsmBI restriction enzyme, the plasmid 0G-1 fragment containing the SCoV 3′UTR was ligated into BsmBI-digested plasmid 0G-2 to eliminate the BsmBI sites, producing plasmid 0G-3. This plasmid contained a fusion of the last 969 nt of the N coding sequence to the SCoV 3′UTR. This was then transferred to the MHV genetic system G plasmid by restriction fragment exchange to produce a plasmid called pMF0G.
TABLE 1.
Primers used in this study
| Primer | Sequence (5′-3′)a | Location(s) |
|---|---|---|
| Oligonucleotide A59(+) 14639-14658 | GTGGATACACATCGTTATCG | MHV 14639-14658 |
| Oligonucleotide A59(+) 16038-16059 | ATGAAGTCTACCTTCCATACCC | MHV 16038-16059 |
| Oligonucleotide A59(−) 16596-16577 | TACTGTGGTTTATGGTCCTC | MHV 16596-16577 |
| MHV 5′ 1-20 (+) primer | TATAAGAGTGATTGGCGTCC | MHV 1-20 |
| SCoV 5′ 1-20 (+) primer | TTATTAGGTTTTTACCTACCCAG | SCoV 1-20 |
| 7065 (−) primer | CATTGCAGGAATAGTACCC | MHV 31288-31270 |
| N (−) primer | GGCACTCCTTGTCCTTCT | MHV 29937-29920 |
| 7059 (−) oligonucleotide 8 | CTGGTGTTACACAGGCAGCGCGTCGGCATGTAATACGACTCACTATAGA | MHV 611-628 |
| P1A-Forc,e,f | TAAGAGTGATTGGCGTCCGTACGTACCC | MHV 1-30 |
| P2A-Forc | pTCTCAACTCTAAAACTCTTGTAGTTTAAATCTA | MHV 31-63 |
| P3A-Ford,f | pATCTAAACTTTATAAACGGCACTTCCTGCGTGTCCATGCCCGC | MHV 64-106 |
| P1A-Reve,f | pTCACTCTTATCTATAGTGAGTCGTATTACATGCCGA | T7 promoter in plasmid A |
| P2A-Rev | pCGTTTATAAAGTTTAGATTAGATTTAAACTACAAGAGTTTTAGAGTTGAGAGGGTACGTACGGACGCCAA | MHV 81-12 |
| P3A-Revd,e,f | GGGCATGGACACGCAGGAAG | MHV 104-85 |
| P2A-For (swap TRS)e | pTCTCAACTCTAAAACTCTTGTAGTTTAAGTTCT | MHV 31-58, SCoV 58-62 |
| P3A-For (swap TRS)e | pCTAAACGAACTATAAACGGCACTTCCTGCGTGTCCATGCCCGC | MHV 74-106, SCoV 63-73 |
| P2A-Rev (swap TRS)e | pTGCCGTTTATAGTTCGTTTAGAGAACTTAAACTACAAGAGTTTTAGAGTTGAGAGGGTACGTACGGACGCCAA | MHV 74-84 and 12-58, SCoV 58-72 |
| P2A-SL2-Forf | pTCTCAACTCTAGATCTCTTGTAGATCAAATCTA | MHV 31-41 and 57-63, SCoV 42-56 |
| P2A-SL2-Revf | pTGCCGTTTATAAAGTTTAGATTAGATTTGATCTACAAGAGATCTAGAGTTGAGAGGGTACGTACGGACGCCAA | MHV 84-57 and 41-12, SCoV 56-42 |
| MluI-T7-F | AAAACGCGTCGGCATGTA | T7 promoter |
| SL4-only-BsmBI-Fg | TTTTCGTCTCTGTTCCTTGACTTTCGTTCTCT | MHV 140-160 |
| SL4-only-BsmBI-Rg | TTTCGTCTCGGAACTTTATACTGCGTAGGTGC | MHV 140-143, SCoV 130-113 |
| b/wSL4-AUG-BsmBI-Fh | CTTCGTCTCACAATAATAAATTTTACTG | SCoV 131-148 |
| b/wSL4-AUG-BsmBI-Rh | AAACGTCTCAATTGTACAAATGTCAGCACTAT | MHV 122-139, SCoV 134-131 |
| 7131-5-U1 (SL1)d | CGCGTCGGCATGTAATACGACTCACTATAGTTATTAGGTTTTTACCTACC | SCoV 1-20 |
| 7131-5-U2 (SL1)d | CAGGAAAAGCCAACCAAAACTCTTGTAGTTTAAATCTA | MHV 41-63, SCoV 21-35 |
| 7131-5-D1 (SL1)d | TTTTGGTTGGCTTTTCCTGGGTAGGTAAAAACCTAATAACTATAGTGAGTCGTATTACATGCCGA | MHV 44-41, SCoV 35-2 |
| 7131-5-D2 (SL1)d | TGCCGTTTATAAAGTTTAGATTAGATTTAAACTACAAGAG | MHV 84-45 |
| 17TG | TTTTTTTTTTTTTTTTTG | MHV 31351-31334 |
| S 5′UTR RT primer | CGTTCACGACTCAGTATCTC | SCoV 1637-1618 |
| 7131-2-U1b | CGCGTCGGCATGTAATACGACTCACTATAGATATTAGGTTTTTAC | SCoV 1-15 |
| 7131-2-U2b | pCTACCCAGGAAAAGCCAACCAACCTCGATCTCTTGTAGATCTGTTCTCTAATCTAAAC | MHV 65-71, SCoV 16-66 |
| 7131-2-U3b | pTTTAAAATCTGTGTAGCTGTCGCTCGGCTGCATGCCTAGTGCACC | SCoV 73-117 |
| 7131-2-U4b | pTACGCAGTATAAACAATAATAAATTTTACTGTCGTTGACAAGAAACGAGTAACTCGT | SCoV 118-174 |
| 7131-2-U5b | pCCCTCTTCTGCAGACTGCTTACGGTTTCGTCCGTGTTGCAGTCGATCATCAGCATAC | SCoV 175-231 |
| 7131-2-D1b | TGGCTTTTCCTGGGTAGGTAAAAACCTAATAACTATAGTGAGTCGTATTACATGCCGA | SCoV 32-1 |
| 7131-2-D2b | pCTACACAGATTTTAAAGTTTAGATTAGAGAACAGATCTACAAGAGATCGAGGTTGGT | MHV 71-65, SCoV 88-73 and 66-33 |
| 7131-2-D3b | pTTGTTTATACTGCGTAGGTGCACTAGGCATGCAGCCGAGCGACAG | SCoV 133-89 |
| 7131-2-D4b | pAGTCTGCAGAAGAGGGACGAGTTACTCGTTTCTTGTCAACGACAGTAAAATTTATTA | SCoV 190-134 |
| 7131-2-D5b | CTAGGTATGCTGATGATCGACTGCAACACGGACGAAACCGTAAGC | SCoV 235-191 |
| 7131-4-U1c | pATCTAAACTTTTTAAAATCTGTGTAGCTGTCGCTCGGCTGCATGCCTAGTGCACC | MHV 64-73, SCoV 73-117 |
| 7131-4-D1c | pCTACACAGATTTTAAAAAGTTTAGATTAGATTTAAACTACAAGAGTTTTAGAGTTGAGAGGGTACGTACGGA | MHV 18-73, SCoV 73-88 |
| Oligonucleotide 1 | CCCCCCCGTCTCTTTACACATTAGAGTCATCTTCTA | MHV 31011-31034 |
| Oligonucleotide 2 | CCCCCCCGTCTCATGACAAAAAAAAAAAAAAAAAA | SCoV 29727-29723 |
| Oligonucleotide 3 | CCCCCCCGTCTCTGTAAACACTCATGATGACCACA | pGEM-T vector region |
| Oligonucleotide 4 | CCCCCCCGTCTCAGTCATTCTCCTAAGAAGCTATTA | pGEM-T vector region |
| S 5′UTR swap oligonucleotide 3 | TTTCCCCGTCTCTTATTAGGTTTTTACCTACCCAG | SCoV 1-23 |
| S 5′UTR swap oligonucleotide 7 | TTTCCCCGTCTCACATCTTACCTTTCGGTCACACCC | SCoV 245-267 |
| S 5′UTR swap oligonucleotide 5 | (C)7GTCTCAAATAACTATAGTGAGTCGTATTACATGC | PCR-XL-TOPO region |
| S 5′UTR swap oligonucleotide 6 | (T)6CGTCTCAGATGGCAAAGATGGGCAAATACGGT | PCR-XL-TOPO region |
p indicates the 5′-phosphorylated nucleotide in each primer. SCoV sequence regions are set in italics.
The HK1A mutated region (MluI-AvrII fragment) was produced by the annealing of primers 7131-2-U1 through 7131-2-U5 and 7131-2-D1 through 7131-2-D5.
HK4A; P1A-For, P2A-For, 7131-4-U1, 7131-2-U4, 7131-2-U5, P1A-Rev, 7131-4-D1, 7131-2-D3, 7131-2-D4, and 7131-2-D5.
HK5A; 7131-5-U1, 7131-5-U2, P3A-For, 7131-5-D1, 7131-5-D2, and P3A-Rev.
HK12A; P1A-For, P2A-For (swap TRS), P3A-For (swap TRS), P1A-Rev, P2A-Rev (swap TRS), and P3A-Rev.
HK18A; P1A-For, P2A-SL2-For, P3A-For, P1A-Rev, P2A-SL2-Rev, and P3A-Rev.
No see'm technology was conducted to generate the HK8A mutated region using these primers.
No see'm technology was conducted to generate the HK11A mutated region using these primers.
Site-directed mutagenesis.
Site-directed mutagenesis was performed to introduce mutations into pHK0A or plasmid A using an oligonucleotide assembly strategy (8). Overlapping oligonucleotides were designed to span the sequence regions between MluI and SacII (131 nt, which includes the MHV-A59 sequence at positions 1 to 106) in plasmid A or between MluI and AvrII (256 nt, which includes the SCoV sequence at positions 1 to 231) in pHK0A and, after annealing, to have MluI and SacII or MluI and AvrII overhangs at their 5′ and 3′ ends. The annealed oligonucleotides were ligated, and the resulting DNA fragment was purified and then ligated into MluI- and SacII-cut plasmid A or MluI- and AvrII-cut pHK0A to create chimeric plasmids. Schematic drawings of the various chimeric plasmids are shown in Fig. 2. pHK5A, pHK18A, and pHK12A were derived from plasmid A by replacing MHV sequences containing the predicted SL1, SL2, and SL3 structures with their predicted SCoV counterparts, respectively (Table 2). pHK4A was generated from pHK0A by replacing SCoV SL1, SL2, and SL3 with their MHV counterparts (Table 2). pHK8A was generated using “no see'm” technology that replaced MHV SL4 with its SCoV counterpart (Table 2) (37). To replace MHV SL4 with SCoV SL4, a 5′ DNA fragment spanning MHV SL1, SL2, and TRS and SCoV SL4 was amplified from pHK4A using primers containing MluI (sense) and BsmBI (antisense) restriction sites (MluI-T7-F and SL4-only-BsmBI-R). A 3′ DNA fragment spanning the sequence between the MHV SL4 and a BamHI site in ORF1a was amplified from plasmid A using primers containing BsmBI (sense) and BamHI (antisense) sites (7059 antisense oligonucleotide 8 and SL4-only-BsmBI-R). These 5′ and 3′ fragments were cloned into the pGEM-T easy vector, subsequently retrieved by double digestion of MluI and BsmBI or BamHI and BsmBI, and ligated into MluI and BamHI sites in plasmid A to eventually construct pHK8A. pHK11A was also generated using “no see'm” technology that replaced the MHV sequence between SL4 and the ORF1a start codon with its SCoV counterpart (Table 2). A 5′ DNA fragment spanning MHV SL1, SL2, TRS, and SL4 was amplified from plasmid A using primers containing MluI (sense primer MluI-T7-F) and BsmBI (antisense primer b/w SL4-AUG-BsmBI-R) sites. A 3′ DNA fragment spanning both (i) SCoV sequences between SL4 and the MHV ORF1a start codon and (ii) MHV sequences between the MHV ORF1a start codon and a BamHI site in MHV ORF1a was amplified from pHK4A using primers containing BsmBI (sense primer b/w SL4-AUG-BsmBI-F) and BamHI (7059 antisense oligonucleotide 8) sites. These 5′ and 3′ fragments were cloned into the pGEM-T easy vector, subsequently retrieved by double digestion of MluI and BsmBI or BamHI and BsmBI, and ligated into MluI and BamHI sites in plasmid A to eventually construct pHK11A. A frameshift mutation in ORF1b was created by digesting plasmid F (nt 398) with MunI, filling in the 4-base overhang with Klenow fragment, and religating the plasmid. This produced a plasmid called pLP0F, containing a frameshift mutation that should knock out the expression of functional NSP12 (RdRp) to NSP16 (Table 2).
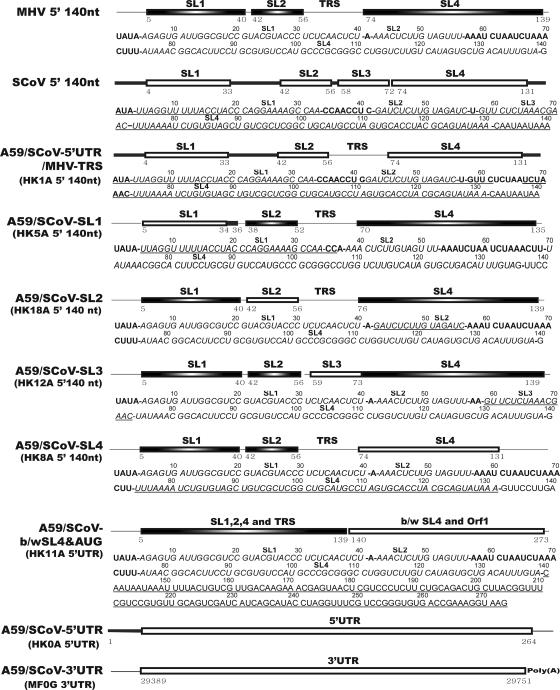
FIG. 2.
Schematic diagram depicting the major constructs used throughout this study. Both the names of the recombinant genomes and plasmids (in parentheses) used to construct these chimeric genomes are shown. The predicted stem-loop structures in the 5′ 140 nt of MHV and SCoV are indicated in the schematic. The positions of the first and last nucleotides are indicated below the boxes depicting the predicted stem-loops. The complete 5′UTR and 3′UTR of SCoV are represented by open rectangles. Single-stranded regions derived from SCoV and MHV are indicated by the thick and thin lines, respectively. The sequences of the 5′UTRs of MHV, SCoV, and each of the chimeric genomes used in this study are shown. SCoV-derived nucleotides are underlined. Boldface type indicates a predicted single-stranded region between the first nucleotide and the predicted 3′ end of SL4. Italicized sequenced are predicted to fold into the stem-loop structures as labeled above the italicized sequences. Bold dashes indicate transitions from predicted stem-loops to single-stranded regions. Sequences 3′ to SL4 are shown in lightface type and are not encompassed by our structural model.
TABLE 2.
Plasmids used in this study
| Plasmid or 3′UTR chimera | Characteristics | Sequence of 5′ regiona |
|---|---|---|
| pHK1A | pHK0A replaced SCoV TRS core sequence (SCoV 67-72)b with MHV TRS core sequence (MHV 65-71)c | SCoV 1-66/MHV 65-71/SCoV 73-264 |
| pHK4A | pHK0A replaced SCoV SL1, SL2, and SL3 (SCoV 1-72) with their MHV counterpart (MHV 1-68) | MHV 1-68/SCoV 73-264 |
| pHK5A | Plasmid A replaced MHV 5′UTR SL1 (MHV 1-40) with SCoV 5′UTR SL1 (SCoV 1-35) | SCoV 1-35/MHV 41-209 |
| pHK8A | Plasmid A replaced MHV 5′UTR SL4 (MHV 67-139) with SCoV 5′UTR SL4 (SCoV 73-130) | MHV 1-66/SCoV 73-130/MHV 140-209 |
| pHK11A | Plasmid A replaced MHV sequence (MHV 140-209) between SL4 and start codon of ORF1 with its SCoV counterpart (SCoV 136-264) | MHV 1-139/SCoV 136-264 |
| pHK12A | Plasmid A replaced MHV TRS core and flanking sequence (MHV 59-73) with SCoV 5′UTR SL3 (SCoV 58-72) | MHV 1-58/SCoV 58-72/MHV 74-209 |
| pHK18A | Plasmid A replaced MHV 5′UTR SL2 (MHV 42-56) with SCoV 5′UTR SL2 (SCoV 42-57) | MHV 1-41/SCoV 42-57/MHV 57-209 |
| pLP0F | Plasmid F containing a frameshift mutation at nt 398 of nsp12 (RdRp) | |
| p0A-1 | pGEM-T vector carrying SCoV 5′UTR with BsmBI site at 5′ and 3′ terminals | |
| p0A-2 | PCR-XL-TOPO carrying BamHI fragment of plasmid A | |
| p0A-3 | p0A-2 deleted MHV 5′UTR by inverse PCR with primers containing BsmBI sites | |
| p7077 | p0A-3 ligated with BsmBI fragment from p0A-1 | |
| pHK0A | Plasmid A replaced with the BamHI fragment of p7077 by restriction fragment exchange, which ultimately replaced MHV 5′UTR (MHV 1-209) with SCoV 5′UTR (SCoV 1-264) | SCoV 1-264 |
| pB36 | T7 transcription vector encoding an MHV A59 DI RNA consisting of 467 nt of the 5′ end of the genome connected in frame, via a 48-nt linker, to the entire N gene and 3′UTR, followed by a poly(A) tail of approximately 115 residuesd | |
| p0G-1 | pGEM-T vector carrying SCoV 3′UTR and 15 nt of poly(A) tail | |
| p0G-2 | pB36 deleted MHV 3′UTR by inverse PCR with primers containing BsmBI sites | |
| p0G-3 | p0G-2 ligated with BsmBI fragment from p0G-1 | |
| pMF1G | G plasmid replaced with the NheI and PacI fragments of p0G-3 by restriction fragment exchange, which ultimately replaced MHV 3′UTR (MHV 31034-31357) with SCoV 3′UTR (SCoV 29389-29751) | SCoV 29389-29751 |
The SCoV or MHV origin of sequences in the chimeric portion of the plasmids are given.
Indicates SCoV sequence relative to the SCoV genome reported under GenBank accession no. AY278741 (27).
Indicates MHV sequence relative to the MHV genome reported under GenBank accession no. NC_001846 (39).
See reference 14.
Plaque purification, RT-PCR, and sequencing.
Mutant viruses were subjected to one round of plaque purification and were expanded once in DBT cells. The sequences of recovered viruses were confirmed by RT-PCR of the 5′UTR and 3′UTR, followed by direct sequencing of the amplified products.
Growth curves.
DBT cells were grown in 96-well plates, and replicate wells were infected at a multiplicity of infection (MOI) of 3 with mutant or wild-type MHV-A59 1000 virus. After washing away the inocula, cultures were incubated until 0, 4, 8, 12, 16, and 24 h postinfection, when they were frozen at −70°C. Triplicate samples were obtained at all time points. Virus production was quantitated by plaque assay on L2 cell monolayers.
Metabolic labeling.
DBT cells (2.25 × 105 cells/well) were seeded into 24-well plates and incubated at 37°C for 15 h to reach approximately 2.5 × 105 cells. The DBT cells were infected at an MOI of 1 (2.5 × 105 PFU) or mock infected, further incubated for 6 h, washed two times with phosphate-free DMEM, fed with DMEM supplemented with 2% dialyzed calf serum and 10 μg/ml of actinomycin D, and incubated at 37°C for 15 min. At the end of 15 min, the medium was replaced with phosphate-free medium containing 10 μg/ml actinomycin D, 2% dialyzed serum, and 200 μCi/ml 32PO4 and incubated at 37°C for 5.5 h, by which time 90% of the cells infected with wild-type MHV-A59 1000 had formed syncytial giant cells. The labeled cultures were washed twice with cold phosphate-buffered saline, and RNA was extracted using an RNeasy Mini kit (QIAGEN). The amount of RNA in each sample was measured using the RediPlate 96 RiboGreen RNA quantitation kit (Invitrogen). Equal amounts of radiolabeled viral RNA (10 μg) were denatured in formaldehyde gel loading buffer containing ethidium bromide (20 μg/ml) at 65°C for 15 min and then electrophoresed in a 1% formaldehyde-agarose gel at 100 V for 5 h. Following electrophoresis, the gel was illuminated with UV light, the image was captured with a FluorChem 8900 (AlphaInotech) imaging system, and the relative amount of 18S rRNA bands was determined by densitometry. The gel was soaked in 70% methanol for 30 min, dried over a vacuum, and exposed to X-ray film. The amount of the individual subgenomic RNAs (sgRNAs) relative to genome-sized RNA (gRNA) and the relative amount of radiolabeled RNA in each sample were determined by exposing the dried gel to a Molecular Dynamics PhosphorImager equipped with Storm 8.2 software. The amount of 18S rRNA in each sample was used to normalize the PhosphorImager signals to account for small differences in the total amounts of RNA loaded per sample.
Detection of gRNA and sgmRNA.
A series of nested RT-PCR assays was performed to analyze RNAs produced by nonviable chimeric genomes. Chimeric genomes or wild-type MHV-A59 1000 RNA was electroporated into BHK-R cells, and total RNAs were extracted at 8 and 24 h postelectroporation (p.e.). In order to determine if the input RNAs, plus any replicated genome RNA, were present in the electroporated cells, the extracted RNAs were primed for reverse transcription by 7059 antisense oligonucleotide 8, followed by PCR using SCoV or MHV 5′ 1-20 (+) primers and 7059 (−) oligonucleotide 8. The RNA species present in cells electroporated with these chimeric genomes were characterized using nested RT-PCR methods described previously (16, 17, 21). For analyzing the synthesis of minus-sense gRNA by the chimeric viruses, the extracted RNAs were primed for reverse transcription by oligonucleotide A59(+) 14639-14658 and followed by the first PCR using oligonucleotide A59(+) 14639-14658 and oligonucleotide A59(−) 16596-16577. The resultant PCR products were further amplified by nested PCR using oligonucleotide A59(+) 16038-16059 and oligonucleotide A59(−) 16596-16577. Parallel reactions (without RT) in which reverse transcriptase was omitted from the cDNA step were always performed to ensure that the PCRs did not detect residual DNA transcription templates that entered the cells during electroporation. To detect plus- or minus-sense subgenomic mRNA7 (sgmRNA7), the extracted RNAs were primed for reverse transcription by antisense primer 7065 to detect minus-sense sgmRNA7 and by SCoV (GenBank accession no. AY278741) or MHV 5′ 1-20 sense primers to detect minus-sense sgmRNA7, respectively. The resultant cDNAs were used as templates for the first PCR using SCoV or MHV 5′ 1-20 sense primers and the 7065 antisense primer. The first PCR products were further amplified by nested PCR using SCoV or MHV 5′ 1-20 sense primers and the N antisense primer, and the nested PCR products were displayed by gel electrophoresis.
RESULTS
Comparison of secondary structures for the 5′-most 140 nt of the 5′UTRs of MHV and SCoV.
Consensus secondary structural models for the 5′-most 140 nt of the 5′UTR from MHV and SCoV were developed using the secondary structure prediction algorithms ViennaRNA 1.5 (12) and mfold 3.1 (40) that were informed by a multiple sequence alignment of nine CoV 5′UTRs (MHV [GenBank accession no. NC_001846], SCoV [accession no. NC_004718], BCoV [accession no. NC_003045], HCoV-OC43 [accession no. NC_005147], HCoV-HKU1 [accession no. NC_006577], HCoV-NL63 [accession no. NC_005831], HCoV-229E _accession no. NC_002645], transmissible gastroenteritis virus (TGEV) [accession no. NC_002306], and infectious bronchitis virus (IBV) [accession no. NC_001451]) and examined for potential sequence covariations that might support the secondary structure prediction. The predicted secondary structural models from nine CoV genomes are strikingly similar and are characterized by three major putative helical stem-loops, named SL1, SL2, and SL4 (Fig. 1). The consensus model is anchored by SL2, which contains a highly conserved and previously unrecognized (C/U)UUG(U/C) U-turn containing a pentaloop (4) sequence stacked on a 5-bp stem of variable composition. While the predicted SL2 stem differs in sequence from virus to virus (Fig. 1B), SL2 is the most conserved structural element in the MHV and SCoV 5′UTR sequences (53.4% overall sequence identity in the first 140 nt) (21). Provided that the stem of SL2 is forced to be base paired, the consensus model is independent of the secondary structure prediction algorithm used (both mfold 3.1 and PKNOTS were tested) (26, 40). The existence of this base pairing is supported by covariation in the stem sequences (Fig. 1B; see Fig. S1 in the supplemental material). Extending these predictions to encompass the entire MHV and SCoV 5′UTRs also predicted the structures SL1 to SL4 (data not shown).
In contrast to SL2, the predicted structures of SL1 and SL4 differ in detail between MHV and SCoV. MHV SL1 has a longer hairpin loop and has two consecutive pyrimidine-pyrimidine mismatches in the stem that are not predicted to be present in SCoV SL1. The predicted MHV SL4 stem contains two internal loops not present in the SCoV SL4 stem. The SCoV 140-nt sequence is predicted to contain a fourth putative stem-loop, named SL3, spanning the SCoV TRS 5′-flanking sequence (5′FS) and CSs. Consistent with our data, van den Born et al. previously predicted that the SCoV TRS CS was contained in the loop portion of a stem-loop structure (33). The counterpart MHV TRS sequence is located between SL2 and SL4 but is predicted to be single stranded or weakly folded at 25°C. The predicted single-stranded region between SCoV SL1 and SL2 was longer (8 nt) than the corresponding region in MHV (1 nt). The SCoV sequence region between the putative SL4 and the start codon of nsp1 (nt 131 to 264) is 65 nt longer than its MHV counterpart (nt 141 to 209), and no significant similarity was found between the two RNA sequences in this region. ViennaRNA 1.5 predicted that this SCoV sequence region forms four stem-loop structures but that its MHV counterpart forms just two stem-loop structures.
The SCoV 5′UTR cannot functionally replace the MHV 5′UTR.
An examination of the sequences contained in the SCoV 5′UTR SL3 revealed that in addition to the SCoV TRS CS (ACGAAC [nt 67 to 72]), this structure also contained an overlapping MHV TRS CS (UCUAAAC [nt 62 to 67]) (Fig. 1). This led us to investigate whether the SCoV 5′UTR could functionally replace its MHV counterpart. In the reverse genetic system for MHV-A59 described previously by Yount et al. (37), plasmid A contains the 5′UTR plus an additional 4,672 nt of the ORF1a coding sequence. As described in Materials and Methods, we generated a modified plasmid A in which the entire MHV 5′UTR was precisely replaced by the SCoV 5′UTR (pHK0A) (Table 2 and Fig. 2). The chimeric cDNA contained in this plasmid was excised and ligated to cloned cDNAs B to G (37) and transcribed in vitro to generate chimeric MHV-A59 genome RNAs in which the SCoV 5′UTR had replaced the corresponding MHV sequences (MHV/SCoV-5′UTR). Three attempts to recover infectious virus after electroporation into MHV-permissive BHK-R cells were unsuccessful, including one attempt in which replicate electroporated cultures were incubated at 34°C and 40°C. To confirm that these chimeric genomes were nonviable, the electroporated cell cultures were frozen at −70°C to release cell-associated virus and blind passaged three times on DBT cells without developing cytopathic effects. Lysates from the third blind passage were subjected to plaque assay using L2 cells. No plaques were observed in all three independent experiments. Thus, we concluded that the MHV/SCoV-5′ UTR chimeric genome was nonviable (Fig. 3).
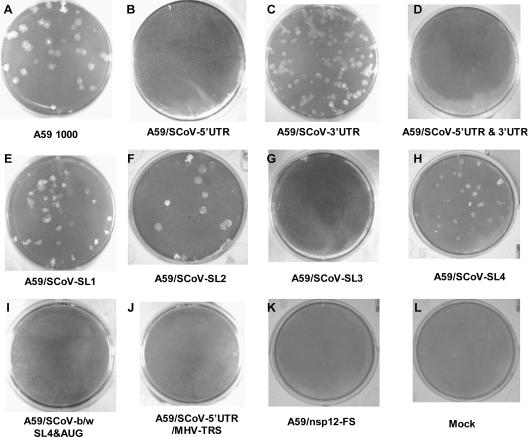
FIG. 3.
Morphologies of plaques formed by MHV and SCoV chimeric genomes. Cultures of BHK-R cells were electroporated with MHV/SCoV chimeric or MHV-A59 full-length transcripts and seeded onto DBT cells in 75-cm2 flasks. If a genome produced viable virus progeny, the progeny was plaque purified and amplified once in DBT cells. The plaques shown are wild-type MHV-A59 1000 virus (A), MHV/SCoV-SL1 (E), MHV/SCoV-SL2 (F), MHV/SCoV-SL4 (H), and MHV/SCoV-3′UTR (C) chimeric viruses. Nonviable chimeric genomes did not form any visible plaque (B, D, G, I, J, and K).
We next determined if a modified MHV/SCoV-5′UTR chimera in which the SCoV TRS CS was replaced by the MHV TRS CS (MHV/SCoV-5′UTR/MHV-TRS) (Fig. 2) was viable. Chimeric genomes were transcribed from ligated cDNAs and electroporated into BHK-R cells, and virus isolation from these cultures was attempted as described above. After three unsuccessful attempts to recover recombinant virus, we concluded that this chimeric genome was also not viable (Fig. 3).
SCoV 5′UTR SL1, SL2, and SL4 were functionally exchangeable with their MHV counterparts.
We next determined whether individual predicted stem-loop structures in the SCoV 5′UTR could functionally replace their MHV counterparts. The following MHV/SCoV chimeric genomes were generated as described in Materials and Methods (Table 2 and Fig. 2): (i) A59/SCoV-SL1 (MHV SL1 was replaced with SCoV SL1 using pHK5A), (ii) A59/SCoV-SL2 (MHV SL2 was replaced with SCoV SL2 using pHK18A), (iii) A59/SCoV-SL3 (MHV TRS CS and 8 nt of the 5′FS were replaced with SCoV SL3 using pHK12A), (iv) A59/SCoV-SL4 (MHV SL4 was replaced with SCoV SL4 using pHK8A), (v) A59/SCoV-b/w SL4&AUG (the MHV sequence region between SL4 and the start codon [AUG] of nsp1 was replaced with its SCoV counterpart using pHK11A), and (vi) MHV/SCoV-3′UTR (MHV 3′UTR was replaced with the SCoV 3′UTR using pMF1G). As a negative control, A59/nsp12-FS was generated using pLP1F, a plasmid that contained a cDNA construct harboring a frameshift mutation in the RdRp domain (nsp12) that should abrogate the translation of downstream nsp13 to nsp16. Cultures electroporated with A59/SCoV-SL1, A59/SCoV-SL2, A59/SCoV-SL4, and A59/SCoV-3′UTR chimeric genomes developed a CPE after 24 to 48 h. Viable virus was recovered from the medium, plaque purified, and expanded in DBT cells (Fig. 3). Sequencing analyses confirmed that there were no additional mutations in the 3′UTRs and 5′UTRs of the recovered viruses. The cultures electroporated with A59/SCoV-SL3 and A59/SCoV-b/w SL4&AUG chimeric genomes and the A59/nsp12-FS mutant genome failed to develop CPE. To confirm that these viral genomes were nonviable, the electroporated cell cultures were frozen at −70°C to release cell-associated virus and blind passaged three times on DBT cells. Lysates from the third blind passage were subjected to plaque assay using L2 cells. No plaques were observed in three independent experiments for each of these viruses. Thus, the A59/SCoV-SL3 and A59/SCoV-b/w SL4&AUG chimeric genomes and the A59/nsp12-FS mutant genome were judged to be nonviable (Fig. 3).
Phenotypic properties of the MHV/SCoV chimeric viruses.
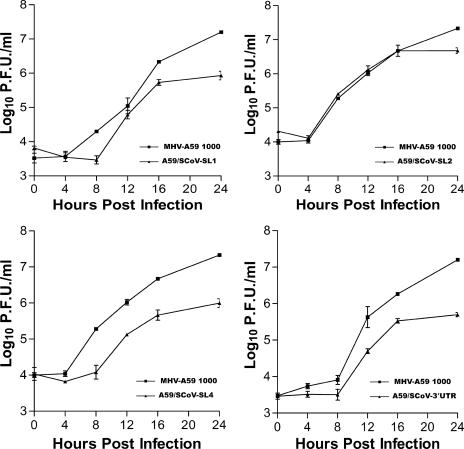
Plaque size and growth kinetics of the recovered chimeric viruses were compared to those of parental MHV-A59 1000. As shown in Fig. 3, the MHV/SCoV chimeric viruses made smaller plaques than the parental MHV-A59 1000 virus. Average plaque sizes of A59/SCoV-SL1, A59/SCoV-SL2, A59/SCoV-SL4, and A59/SCoV-3′UTR chimeric viruses were 1.2 (±0.07), 1.8 (±0.09), 1.1 (±0.04), and 1.4 (±0.07) mm in diameter, respectively (Fig. 4). These sizes corresponded to 39% (P < 0.05) to 64% (P < 0.05) of the average plaque size of MHV-A59 1000, which is 2.8 (±0.07) mm in diameter. A59/SCoV-SL1, A59/SCoV-SL4, and A59/SCoV-3′UTR chimeric viruses grew to lower titers, had significantly delayed growth kinetics relative to those of MHV-A59 1000, and achieved maximal titers 15- to 40-fold less than those achieved by the parental MHV-A59 1000 virus (Fig. 5). However, A59/SCoV-SL2 grew almost as well as the parental virus, achieving a titer only fourfold lower than that of MHV-A59 1000 (Fig. 5).
FIG. 4.
Average plaque sizes of wild-type MHV-A59 1000 and MHV/SCoV-SL1, MHV/SCoV-SL2, MHV/SCoV-SL4, and MHV/SCoV-3′UTR chimeric viruses. The middle bar in each column indicates the mean plaque size of the corresponding virus.
FIG. 5.
One-step growth kinetics for wild-type MHV-A59 1000 and MHV/SCoV-SL1 (A), MHV/SCoV-SL2 (B), MHV/SCoV-SL4 (C), and MHV/SCoV-3′UTR (D) chimeric viruses. Triplicate DBT cell cultures in a 96-well plate were infected at an MOI of 3 and harvested at the designated hours postinfection, and viral titers were determined by plaque assay.
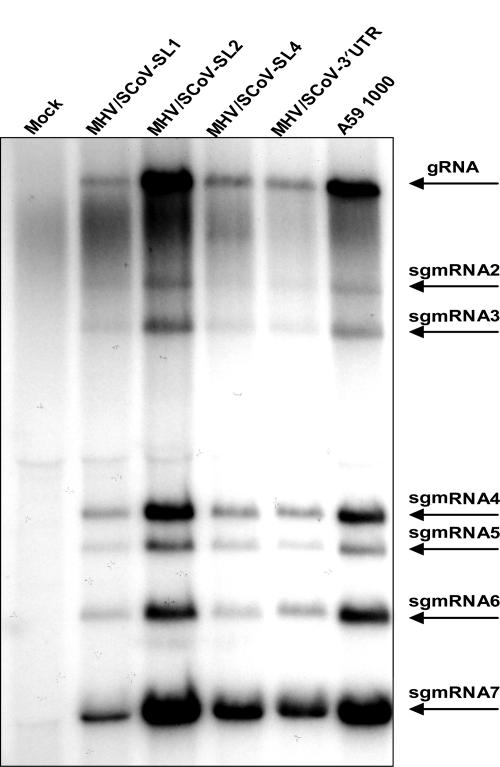
To determine whether the altered growth phenotypes of the chimeric 5′UTR viruses are a result of deficits in genome replication or subgenomic RNA synthesis, virus-specific RNAs were metabolically radiolabeled and analyzed by gel electrophoresis (Fig. 6). DBT cells were either mock infected, infected with MHV-A59 1000 (WT), or infected with MHV/SCoV chimeric viruses. Starting at 5 h postinfection, MHV-specific RNAs were labeled for 5.5 h with 32PO4 in the presence of actinomycin D. After labeling, total viral and cellular RNAs were extracted from cell lysates and quantitated, and equal amounts of RNA were electrophoresed in formaldehyde-agarose gels. Total RNAs were visualized by ethidium bromide staining, and labeled MHV-specific RNAs were visualized by autoradiography. The amount of each MHV-specific RNA as well as the total amount of labeled virus-specific RNA in each sample were quantitated with a PhosphorImager (Fig. 6). All seven species of MHV-specific RNA were detected in cells infected with the MHV/SCoV chimeric viruses and MHV-A59 1000. The total amount of radiolabeled MHV-specific RNA (gRNA [genome] through sgmRNA7) in MHV/SCoV-SL2-infected cells was 92% of that detected in MHV-A59 1000-infected cells, a result that correlates with the relatively modest impairment of viral replication that we observed for this chimera. However, the amount of virus-specific RNA synthesis in cells infected with MHV/SCoV-SL1 (32% relative to WT), MHV/SCoV-SL4 (46%), and MHV/SCoV-3′UTR (42%) was significantly lower than that observed in cells infected with MHV-A59 1000 (Fig. 6). This decrease in RNA synthesis correlates with the decrease in replication efficiency that we observed in one-step growth curves (Fig. 5). The relative molar ratios of gRNA to sgmRNAs present in cells infected with MHV/SCoV chimeric viruses were generally similar to those found in cells infected with the wild type, with some exceptions for individual sgmRNAs (Table 3). The largest difference observed was an almost threefold increase in the relative molar ratio of sgmRNA2 to genome RNA in cells infected with A59/SCoV-SL1.
FIG. 6.
RNA synthesis in cells infected with MHV/SCoV chimeric viruses. Cells were either mock infected or infected with MHV/SCoV-SL1, MHV/SCoV-SL2, MHV/SCoV-SL4, MHV/SCoV-3′UTR, or MHV-A59 1000, and viral RNAs were metabolically labeled as described in Materials and Methods. The labeled viral RNAs were resolved by formaldehyde agarose gel electrophoresis and visualized by autoradiography. Brightness and contrast have been adjusted to enhance the visibility of the bands of the MHV/SCoV chimeric viruses using Adobe Photoshop 6.0. Arrows indicate the positions of bands of virus-specific RNA1 to RNA7.
TABLE 3.
Relative molar amounts of virus-specific RNAs synthesized in cells infected with MHV/SCoV chimeric viruses
| RNA species | Relative molar amt
|
||||
|---|---|---|---|---|---|
| MHV-A59 1000 | A59/SCoV- SL1 | A59/SCoV- SL2 | A59/SCoV- SL4 | A59/SCoV- 3′UTR | |
| RNA1 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| sgmRNA2 | 1.12 | 3.01 | 1.32 | 1.69 | 1.47 |
| sgmRNA3 | 1.57 | 2.82 | 1.68 | 1.75 | 1.63 |
| sgmRNA4 | 6.83 | 7.33 | 7.93 | 7.38 | 7.52 |
| sgmRNA5 | 3.41 | 4.42 | 3.69 | 4.41 | 3.87 |
| sgmRNA6 | 10.49 | 7.40 | 7.21 | 6.02 | 9.52 |
| sgmRNA7 | 38.95 | 31.97 | 33.59 | 51.47 | 45.36 |
Nonviable MHV/SCoV chimeric genomes have defects in RNA synthesis.
We examined the stage at which RNA replication or transcription of the nonviable chimeric genomes was blocked using a series of nested RT-PCR assays (summarized in Table 4). Replicate cultures of BHK-R cells were separately electroporated with in vitro-synthesized genomic RNAs corresponding to A59/SCoV-5′UTR, A59/SCoV-5′UTR/MHV-TRS, and A59/SCoV-SL3 chimeric genomes; the A59/nsp12-FS mutant genome (a frameshift mutant expected to abrogate the expression of proteins required for RNA replication); and parental MHV-A59 1000. RNAs were extracted from the electroporated cultures at 8 and 24 h p.e. As expected, nested RT-PCR analyses of RNA extracted from cells electroporated with the A59/nsp12-FS (Fig. 7B, lanes 1 and 2) mutant genome failed to detect minus-sense gRNA at either time point, whereas this RNA species was readily detected at both time points in cells electroporated with the MHV-A59 1000 genome (Fig. 7B, lanes 9 and 10). Minus-sense gRNA was undetectable in cells electroporated with the A59/SCoV-5′UTR chimera (Fig. 7B, lanes 3 and 4) but was detectable in cells electroporated with the A59/SCoV-5′UTR/MHV-TRS and A59/SCoV-SL3 chimeras (Fig. 7B, lanes 5 to 8). Assays to detect plus- or minus-sense sgmRNA7s were similarly negative with RNAs obtained from cells electroporated with the A59/SCoV-5′UTR chimera (Fig. 7D, lanes 5 to 8) or the A59/nsp12-FS mutant (Fig. 7D, lanes 1 to 4), indicating that these genomes had global defects in RNA synthesis. The nested RT-PCR assay showed that at 24 h p.e., the A59/SCoV-5′UTR/MHV-TRS (Fig. 7D, lanes 9 to 12) and A59/SCoV-SL3 (Fig. 7D, lanes 13 to 16) chimeric genomes directed the synthesis of minus-sense gRNA at levels that appeared to be roughly similar to those observed in cells electroporated with wild-type MHV-A59 1000. Semiquantitative nested RT-PCR assays with serially diluted RNA templates confirmed that the cultures electroporated with the A59/SCoV-5′UTR/MHV-TRS and A59/SCoV-SL3 genomes contained amounts of minus-sense gRNA that were roughly equivalent to those of cells electroporated with MHV-A59 (data not shown). However, neither minus- nor plus-sense sgmRNA7 could be detected in cells electroporated with these two chimeric genomes (Fig. 7D, lanes 9 to 16), whereas these RNAs were easily detectable in cells electroporated with the parental MHV-A59 1000 genome (Fig. 7D, lanes 17 to 20).
TABLE 4.
Characterization of RNA species produced by nonviable MHV/SCoV chimeric genomes
| Genome | Detection of:
|
||
|---|---|---|---|
| Minus-strand gRNAa | Minus-strand sgmRNA7a | Positive-strand sgmRNA7a | |
| MHV-A59 1000 | Yes | Yes | Yes |
| A59/nsp12-FS | No | No | No |
| MHV/SCoV-5′UTR | No | No | No |
| MHV/SCoV-5′UTR/ MHV-TRS | Yes | No | No |
| MHV/SCoV-SL3 | Yes | No | No |
| A59/SCoV-b/w SL4&AUG | Yes | No | No |
Indicates whether the RNA species was detected in cells electroporated with wild-type or chimeric genomes.
FIG. 7.
Nested RT-PCR assays for minus-sense gRNA and sgmRNA7. (A) Schematic drawing of the assay used to detect minus-sense (−) gRNA and the relative locations of the primers within the genome. (B) RT-PCR of RNAs extracted from MHV-A59 1000 (WT) and nonviable MHV/SCoV chimeric genome-electroporated cells at 8 and 24 h p.e. The arrow indicates the specifically amplified fragment from minus-sense gRNA. W/RT and W/O RT indicate products of nested RT-PCR (left) and the corresponding no-RT control reactions (right), respectively. (C) Schematic representation of the RT-nested PCR strategy and primers used to detect minus- and plus-sense sgmRNA7s. (D) Representative nested RT-PCR for minus- and plus-sense sgmRNA7s. The arrow indicates the specifically amplified sgmRNA7 fragment. The relative sizes of the leader sequences in sgmRNA7 and the body of sgmRNA7 are not to scale. The sizes of the marker DNAs in base pairs are shown to the right of electropherograms. The amplified products corresponding to minus-sense sgmRNA7 are shown in lanes 1, 3, 5, 7, 9, 11, 13, 15, 17, and 19. Products corresponding to plus-sense sgmRNA7 are shown in lanes 2, 4, 6, 8, 10, 12, 14, 16, 18, and 20. The position of each primer in the MHV-A59 genome (GenBank accession no. NC_001846) is given in parentheses as part of the primer name. Primer sequences are listed in Table 1. A 1-kbp ladder was used as a molecular size marker.
DISCUSSION
The reverse genetic studies described above investigated the possibility that the SCoV 5′UTR, or selected regions of the 5′UTR, can functionally replace MHV counterparts. These studies were guided by a phylogenetic analysis that places both SCoV and MHV in the group 2 coronaviruses (30) and on a novel secondary structure model that predicts similar secondary structures for all of the group 2 coronaviruses in the first 140 nt of their 5′UTRs (17, 21). This model is supported by NMR, thermal denaturation, and reverse genetic studies that provide support for the presence and functional importance of the highly conserved, but previously overlooked, predicted SL2 stem-loop containing a U-turn motif (Liu et al., unpublished) (21). NMR studies and reverse genetic studies introducing mutations into putative SL1 indicate that this structure is also functionally important for MHV replication (H. Kang et al., unpublished results). Although this model differs from a prior model of the BCoV 5′UTR, another group 2 coronavirus (5), in several key respects, both models predict a stem-loop structure that is part of SL4 in our model (nt 97 to 117) and that was previously designated stem-loop III in the BCoV model (24). The integrity of the stem portion of stem-loop III is required for BCoV DI RNA replication (24). Taken together, these results support the designation of SL1, SL2, and SL4 structures as putative cis-acting elements for MHV genome replication.
Our model differs from prior models of coronavirus 5′UTRs in one important respect. The BCoV model referenced above predicts that the leader TRS CS will be in the loop portion of stem-loop II (5). Similarly, van den Born et al. (33) predicted that in addition to BCoV, other coronaviruses, including MHV, will fold such that the TRS is in the loop portion of a stem-loop structure. In our model, the MHV TRS is unstructured or is contained in a weakly stable stem-loop, SL3 (Fig. 1). mfold 3.1 (http://www.bioinfo.rpi.edu/applications/mfold/old/rna/form1.cgi) generates four secondary structures for the MHV 5′ 140 nt. These four structures have free energies of between −33.6 kcal/mol and −32.3 kcal/mol. Two of the four structures (with free energies of −33.4 and −32.8 kcal/mol) position the TRS sequence in the loop portion of a stem-loop. Our secondary structure model corresponds to an mfold structure with a free energy of −32.3 kcal/mol, only slightly less stable than the two structures putting the TRS into a hairpin loop. However, if folding is constrained to take into account the covariation that strongly predicts the existence of SL2 (Fig. 1B), our model is the most stable structure generated by mfold. None of the data rule out the possibility that the secondary structures that we propose for the 5′UTR are in equilibrium with additional structures similar to those proposed previously by van den Born et al. (33) and Chang et al. (5).
We show here that the SCoV 5′UTR cannot functionally replace the MHV 5′UTR, but we confirm data from a previous study where functional replacement of the MHV 3′UTR by the SCoV 3′UTR was observed in a recombinant virus generated by targeted RNA recombination (10). Although there is little sequence identity between the MHV and SCoV 3′UTRs, both 3′UTRs can be folded into similar secondary structures that contain stem-loop and pseudoknot elements characteristic of group 2 coronaviruses (10). However, the full MHV and SCoV 5′UTRs are significantly different in terms of sequences and predicted secondary structures. The SCoV 5′UTR sequence is 55 nt longer than the MHV 5′UTR. Relative to the MHV counterpart, the region downstream of the putative SL4 in the SCoV 5′UTR was expanded to 65 nt in our secondary structure model. This region of the SCoV 5′UTR from 131 to 264 nt is predicted to fold quite differently than the corresponding region of the MHV 5′UTR (nt 141 to 209) and contains two additional stem-loops relative to its MHV counterpart (data not shown). Considering the 5′-most 140-nt sequences of the MHV and SCoV 5′UTRs, which are predicted to fold quite similarly, the SCoV sequence region is predicted to contain an additional stem-loop, called SL3, that is absent from its MHV counterpart. Any or all of these differences in the sequences and secondary structures might contribute to the nonexchangeability of the SCoV 5′UTR for its MHV counterpart.
In the reverse genetic studies reported here, we demonstrate that sequences containing the predicted SCoV SL1, SL2, and SL4 structures can functionally replace their MHV counterparts. The predicted MHV SL1 structure contains two consecutive noncanonical pyrimidine-pyrimidine base pairs in the stem that are not apparent in the predicted SCoV SL1 structure. The SCoV SL1 substitution produced spacing with 2 nt between the predicted MHV SL1 and SL2. The predicted MHV and SCoV SL2 structures both contain identical U-turn motifs in the loop (CUUGU). This U-turn motif is associated with a sharp turn in the phosphate backbone between U0 and N+1 (11). Moreover, both stems of the MHV and SCoV SL2s were identical except for one base pair at the base of the stem. As the MHV and SCoV SL4s contained potential homologs of the BCoV stem-loop III (24), both ViennaRNA 1.5 and Zuker mfold algorithms predict approximately the same secondary structure in the 5′-most 80- to 140-nt sequence regions of MHV, SCoV, and BCoV, with SL4 and stem-loop III structures conserved in all of these viruses and, in fact, in all coronaviruses (24). Overall, these slight differences in the predicted secondary structural configurations of MHV and SCoV stem-loops did not affect viral viability, but they may have contributed to the reduced viral replication efficiency.
In contrast, the MHV sequence region spanning the TRS CS, 6 nt of 5′FS, and 2 nt of 3′FS could not be functionally replaced by its SCoV counterpart, SL3 (chimeric genome A59/SCoV-SL3). Although the SCoV sequence region contains both the MHV and SCoV TRS CSs, the ViennaRNA 1.5 algorithm predicts that the SCoV sequence TRS region forms a stem-loop structure (SL3), while its MHV counterpart adopts an unpaired structure or one that is predicted to be weakly folded at 25°C. NMR and thermal denaturation experiments using an SCoV RNA encompassing SL2 and SL3 (nt 42 to 72) support the presence of a double-hairpin structure in SCoV (Li and Giedroc, unpublished). This structural difference might contribute to the functional nonexchangeability of TRS sequence regions. Similarly, the SCoV sequence region between SL4 and the start codon of nsp1 is also different in both sequence and secondary structure from its MHV counterpart, which might consequently contribute to the nonviability of the A59/SCoV-b/w SL4&AUG chimeric genome.
An attempt to generate a viable A59/SCoV-5′UTR chimeric genome led to the generation of the A59/SCoV-5′UTR/MHV-TRS chimeric genome, a construct that contains the correct MHV leader TRS. However, the resultant chimeric genome was also nonviable and failed to produce minus- or plus-sense sgmRNA7s. Based on these results, the inability of the SCoV 5′UTR to replace the MHV 5′UTR was not a single consequence of a mismatch of the leader TRS with downstream intergenic TRS sequences. Thus, other differences in sequences or secondary structures, especially the difference in sequences between SL4 and the start codon of nsp1, likely contribute to the observed defects in genome replication and subgenomic RNA synthesis.
Minus-sense gRNA was synthesized in the cells electroporated with A59/SCoV-5′UTR/MHV-TRS and A59/SCoV-SL3 chimeric genomes but not in cells electroporated with the A59/SCoV-5′UTR chimera. It was impossible to determine if electroporated cultures containing minus-sense gRNA but not sgmRNA also synthesized a plus-sense genome. Input genome RNA persisted at easily detectable levels in cells electroporated with a genome unable to direct the synthesis of RNA replicase genes (A59/nsp12-FS, a frameshift mutant expected to abrogate the expression of proteins required for RNA replication) until 24 h p.e., a time when cell fusion was apparent in cultures electroporated with MHV-A59 1000 (data not shown). Real-time RT-PCR assays could not detect a quantitative difference in the amounts of plus-sense gRNAs present in cells electroporated with MHV-A59 1000, A59/nsp12-FS, and the nonviable chimeric genomes at 8 h p.e. At 24 h p.e., the amount of plus-sense gRNA present in cells electroporated with MHV-A59 1000 was significantly greater than that present in cells electroporated with A59/nsp12-FS, A59/SCoV-5′UTR/MHV-TRS, or A59/SCoV-SL3 genomes (data not shown). We concluded that the genome RNAs detected in cells electroporated with nonviable chimeras represented electroporated RNA genomes rather than replicated molecules, since they were found in amounts similar to those for the A59/nsp12-FS genome at 8 and 24 h p.e. Thus, in the absence of subgenomic RNA synthesis and cell fusion, it was impossible to determine if plus-sense gRNA synthesis was directed by the nonviable chimeric genomes, because the persistence of the electroporated RNAs would obscure low levels of genome replication.
The nonviable chimeric genomes (A59/SCoV-5′UTR, A59/SCoV-5′UTR/MHV-TRS, and A59/SCoV-SL3) produced neither minus- nor plus-sense sgmRNA7. Thus, the replacement of the entire MHV 5′UTR with the complete SCoV 5′UTR resulted in a genome that failed to synthesize any viral RNA species. Replacement of the SCoV TRS CS with the MHV TRS CS plus 5 nt of 5′FS and 4 nt of 3′FS in the SCoV 5′UTR background allowed the synthesis of minus-sense gRNA but not sgmRNA7s of either sense. In addition, the insertion of the SCoV region spanning the TRS CS and 5′FS 9 nt in place of the corresponding MHV sequences in the MHV 5′UTR background did not disrupt the synthesis of minus-sense gRNA. A possible explanation for the synthesis of this RNA species in cells electroporated with these nonviable chimeric genomes can be found in the three-step working model for coronavirus mRNA transcription described previously (41), consisting of 5′-to-3′ complex formation, base-pairing scanning, and template switching. The A59/SCoV-5′UTR chimeric genome might be blocked in the first step, 5′-to-3′ complex formation. Both the MHV N protein and the polypyrimidine tract binding (PTB) protein bind to the MHV leader TRS region (20, 35). These proteins might not efficiently bind to the leader MHV TRS CS in the SCoV 5′UTR background, or poor complementarity of sequences flanking the MHV TRS CS may preclude the template jumping step needed for sgmRNA synthesis (29). It has been hypothesized that an interaction between PTB and hnRNP A1, which binds to a site in the 3′UTR, plays an important role in RNA replication (20, 29, 35). If this hypothesis is correct, the poor binding activity of PTB with the TRS region would result in a failure to mediate the formation of an RNP complex involving the 5′- and 3′-end fragments of the A59/SCoV-5′UTR chimeric genome (29). If so, the chimeric genome would not be able to synthesize minus-sense gRNA. Alternatively, the N protein and PTB might efficiently bind to the 5′ ends of A59/SCoV-5′UTR/MHV-TRS and A59/SCoV-SL3 chimeric genomes due to a good fit with the TRS, including flanking sequences. This binding activity would enable 5′-to-3′ complex formation and consequently allow the synthesis of minus-sense gRNA. However, they would be blocked at the leader-body joining step of subgenomic RNA synthesis, where a base-pairing scanning step is thought to regulate this event (41). A lack of sufficient complementary sequence domains would then be predicted to result in a failure to produce sgmRNA7s. Alternatively, differences in the structures of MHV and SCoV 5′UTRs and the various chimeras could have affected the translational efficiencies of the electroporated genomes and hence the levels of replicase proteins. In this instance, differences in the amounts of replicase proteins produced in cells electroporated with different chimeras would determine the particular RNA phenotype observed. We currently have no data on the effects (if any) that the different chimeric 5′UTRs might have on the translation of ORF1a and ORF1b, and thus, we cannot rule out this possibility. Further studies are needed to determine if either of these two hypotheses are correct.
In spite of considerable differences in sequence, the putative cis-acting structural elements are highly conserved and in some cases have been demonstrated to be functionally exchangeable among the group 2 coronaviruses. The inability of the complete SCoV 5′UTR to functionally replace the MHV 5′UTR suggests that recombination events between these two viruses that yield viable recombinants will be uncommon and typically will require two crossover events. However, the compatibility of putative cis-acting elements within the 5′UTRs and 3′UTRs from related coronaviruses suggests that with some additional changes (mutations), viral interspecies recombination could be facilitated. The functional studies reported here provide data that are critical to our understanding of both the phylogenetic relationship of SCoV to other group 2 coronaviruses and the natural evolution of coronaviruses.
Supplementary Material
Acknowledgments
We gratefully acknowledge support from the NIH (AI040187 to D.P.G. and AI51493 to J.L.L.).
We thank Lichun Li in the Department of Biochemistry and Biophysics for help in generating output from secondary structure prediction algorithms. We also thank Lindsay Patty for constructing the plasmid used to generate A59/nsp12-FS mutant genomes and the other members of the Leibowitz laboratory for their help and encouragement.
Footnotes
Published ahead of print on 18 August 2006.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Bonilla, P. J., A. E. Gorbalenya, and S. R. Weiss. 1994. Mouse hepatitis virus strain A59 RNA polymerase gene ORF 1a: heterogeneity among MHV strains. Virology 198:736-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bredenbeek, P. J., C. J. Pachuk, A. F. Noten, J. Charite, W. Luytjes, S. R. Weiss, and W. J. Spaan. 1990. The primary structure and expression of the second open reading frame of the polymerase gene of the coronavirus MHV-A59; a highly conserved polymerase is expressed by an efficient ribosomal frameshifting mechanism. Nucleic Acids Res. 18:1825-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brian, D. A., and R. S. Baric. 2005. Coronavirus genome structure and replication. Curr. Top. Microbiol. Immunol. 287:1-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell, D. O., and P. Legault. 2005. Nuclear magnetic resonance structure of the Varkud satellite ribozyme stem-loop V RNA and magnesium-ion binding from chemical-shift mapping. Biochemistry 44:4157-4170. [DOI] [PubMed] [Google Scholar]
- 5.Chang, R. Y., R. Krishnan, and D. A. Brian. 1996. The UCUAAAC promoter motif is not required for high-frequency leader recombination in bovine coronavirus defective interfering RNA. J. Virol. 70:2720-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dalton, K., R. Casais, K. Shaw, K. Stirrups, S. Evans, P. Britton, T. D. Brown, and D. Cavanagh. 2001. cis-Acting sequences required for coronavirus infectious bronchitis virus defective-RNA replication and packaging. J. Virol. 75:125-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding, Y. 2006. Statistical and Bayesian approaches to RNA secondary structure prediction. RNA 12:323-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flavell, R. A., D. L. Sabo, E. F. Bandle, and C. Weissmann. 1974. Site-directed mutagenesis: generation of an extracistronic mutation in bacteriophage Q. beta RNA. J. Mol. Biol. 89:255-272. [DOI] [PubMed] [Google Scholar]
- 9.Gibbs, A. J., M. J. Gibbs, and J. S. Armstrong. 2004. The phylogeny of SARS coronavirus. Arch. Virol. 149:621-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goebel, S. J., J. Taylor, and P. S. Masters. 2004. The 3′ cis-acting genomic replication element of the severe acute respiratory syndrome coronavirus can function in the murine coronavirus genome. J. Virol. 78:7846-7851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gutell, R. R., J. J. Cannone, D. Konings, and D. Gautheret. 2000. Predicting U-turns in ribosomal RNA with comparative sequence analysis. J. Mol. Biol. 300:791-803. [DOI] [PubMed] [Google Scholar]
- 12.Hofacker, I. L. 2003. Vienna RNA secondary structure server. Nucleic Acids Res. 31:3429-3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsue, B., T. Hartshorne, and P. S. Masters. 2000. Characterization of an essential RNA secondary structure in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 74:6911-6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsue, B., and P. S. Masters. 1997. A bulged stem-loop structure in the 3′ untranslated region of the genome of the coronavirus mouse hepatitis virus is essential for replication. J. Virol. 71:7567-7578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hussain, S., J. Pan, Y. Chen, Y. Yang, J. Xu, Y. Peng, Y. Wu, Z. Li, Y. Zhu, P. Tien, and D. Guo. 2005. Identification of novel subgenomic RNAs and noncanonical transcription initiation signals of severe acute respiratory syndrome coronavirus. J. Virol. 79:5288-5295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, R. F., M. Feng, P. Liu, J. J. Millership, B. Yount, R. S. Baric, and J. L. Leibowitz. 2005. Effect of mutations in the mouse hepatitis virus 3′(+)42 protein binding element on RNA replication. J. Virol. 79:14570-14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang, H., M. Feng, M. E. Schroeder, D. P. Giedroc, and J. L. Leibowitz. 2006. Stem-loop I in the 5′ UTR of the SARS coronavirus can substitute for its counterpart in mouse hepatitis virus. Adv. Exp. Med. Biol. 581:105-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kathryn, V. H., and C. L. Michael. 1996. Coronaviridae: the viruses and their replication, p. 1075-1094. In B. N. Fields, D. N. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 19.Lau, S. K., P. C. Woo, K. S. Li, Y. Huang, H. W. Tsoi, B. H. Wong, S. S. Wong, S. Y. Leung, K. H. Chan, and K. Y. Yuen. 2005. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA 102:14040-14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, H. P., P. Huang, S. Park, and M. M. Lai. 1999. Polypyrimidine tract-binding protein binds to the leader RNA of mouse hepatitis virus and serves as a regulator of viral transcription. J. Virol. 73:772-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, P., J. Millership, L. Li, D. P. Giedroc, and J. L. Leibowitz. 2006. A previously unrecognized UNR stem-loop structure in the coronavirus 5′ untranslated region plays a functional role in replication. Adv. Exp. Med. Biol. 581:25-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makino, S., J. G. Keck, S. A. Stohlman, and M. M. Lai. 1986. High-frequency RNA recombination of murine coronaviruses. J. Virol. 57:729-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Putics, Á., W. Filipowicz, J. Hall, A. E. Gorbalenya, and J. Ziebuhr. 2005. ADP-ribose-1"-monophosphatase: a conserved coronavirus enzyme that is dispensable for viral replication in tissue culture. J. Virol. 79:12721-12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raman, S., P. Bouma, G. D. Williams, and D. A. Brian. 2003. Stem-loop III in the 5′ untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA replication. J. Virol. 77:6720-6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raman, S., and D. A. Brian. 2005. Stem-loop IV in the 5′ untranslated region is a cis-acting element in bovine coronavirus defective interfering RNA replication. J. Virol. 79:12434-12446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rivas, E., and S. R. Eddy. 1999. A dynamic programming algorithm for RNA structure prediction including pseudoknots. J. Mol. Biol. 285:2053-2068. [DOI] [PubMed] [Google Scholar]
- 27.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 28.Sawicki, S. G., and D. L. Sawicki. 2005. Coronavirus transcription: a perspective. Curr. Top. Microbiol. Immunol. 287:31-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi, S. T., and M. M. Lai. 2005. Viral and cellular proteins involved in coronavirus replication. Curr. Top. Microbiol. Immunol. 287:95-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. Poon, Y. Guan, M. Rozanov, W. J. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stadler, K., V. Masignani, M. Eickmann, S. Becker, S. Abrignani, H. D. Klenk, and R. Rappuoli. 2003. SARS—beginning to understand a new virus. Nat. Rev. Microbiol. 1:209-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thiel, V., K. A. Ivanov, A. Putics, T. Hertzig, B. Schelle, S. Bayer, B. Weissbrich, E. J. Snijder, H. Rabenau, H. W. Doerr, A. E. Gorbalenya, and J. Ziebuhr. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 84:2305-2315. [DOI] [PubMed] [Google Scholar]
- 33.van den Born, E., A. P. Gultyaev, and E. J. Snijder. 2004. Secondary structure and function of the 5′-proximal region of the equine arteritis virus RNA genome. RNA 10:424-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Hoek, L., K. Pyrc, M. F. Jebbink, W. Vermeulen-Oost, R. J. Berkhout, K. C. Wolthers, P. M. Wertheim-van Dillen, J. Kaandorp, J. Spaargaren, and B. Berkhout. 2004. Identification of a new human coronavirus. Nat. Med. 10:368-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, Y., and X. Zhang. 1999. The nucleocapsid protein of coronavirus mouse hepatitis virus interacts with the cellular heterogeneous nuclear ribonucleoprotein A1 in vitro and in vivo. Virology 265:96-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Woo, P. C., S. K. Lau, C. M. Chu, K. H. Chan, H. W. Tsoi, Y. Huang, B. H. Wong, R. W. Poon, J. J. Cai, W. K. Luk, L. L. Poon, S. S. Wong, Y. Guan, J. S. Peiris, and K. Y. Yuen. 2005. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 79:884-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yount, B., M. R. Denison, S. R. Weiss, and R. S. Baric. 2002. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J. Virol. 76:11065-11078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu, G., and H. W. Chen. 2004. Monophyletic relationship between severe acute respiratory syndrome coronavirus and group 2 coronaviruses. J. Infect. Dis. 189:1676-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ziebuhr, J., E. J. Snijder, and A. E. Gorbalenya. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81:853-879. [DOI] [PubMed] [Google Scholar]
- 40.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zuniga, S., I. Sola, S. Alonso, and L. Enjuanes. 2004. Sequence motifs involved in the regulation of discontinuous coronavirus subgenomic RNA synthesis. J. Virol. 78:980-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.