Abstract
In the hematopoietic system the bZip transcription factor MafB is selectively expressed at high levels in monocytes and macrophages and promotes macrophage differentiation in myeloid progenitors, whereas a dominant-negative allele can inhibit this process. To analyze the requirement of MafB for macrophage development, we generated MafB-deficient mice and, due to their neonatal lethal phenotype, analyzed macrophage differentiation in vitro, in the embryo, and in reconstituted mice. Surprisingly we observed in vitro differentiation of macrophages from E14.5 fetal liver (FL) cells and E18.5 splenocytes. Furthermore we found normal numbers of F4/80+/Mac-1+ macrophages and monocytes in fetal liver, spleen, and blood as well as in bone marrow, spleen, and peritoneum of adult MafB−/− FL reconstituted mice. MafB−/− macrophages showed intact basic macrophage functions such as phagocytosis of latex beads or Listeria monocytogenes and nitric oxide production in response to lipopolysaccharide. By contrast, MafB−/− macrophages expressed increased levels of multiple genes involved in actin organization. Consistent with this, phalloidin staining revealed an altered morphology involving increased numbers of branched protrusions of MafB−/− macrophages in response to macrophage colony-stimulating factor. Together these data point to an unexpected redundancy of MafB function in macrophage differentiation and a previously unknown role in actin-dependent macrophage morphology.
The bZip transcription factor MafB belongs to a larger family of Maf-type transcription factors, characterized by a conserved extended bZip DNA binding domain that recognizes Maf recognition element (MARE)-type DNA consensus sequences, comprising characteristic 3-bp extensions of a an AP-1 core (6, 26). Gene inactivation studies or mutant analysis of MafB and its Drosophila melanogaster homologue Traffic Jam have demonstrated important roles in development and differentiation processes of the brain, gonads, and kidney (4, 8, 23, 32).
In the hematopoietic system MafB is selectively expressed at high levels in monocytes and macrophages but not in other myeloid or lymphoid lineages. This expression profile is conserved across species and was observed in avian and human cell lines as well as myeloid progenitor or monocyte-derived primary macrophages (3, 10, 12, 13, 20, 34). By contrast, MafB expression in closely related myeloid lineages such as granulocytes or dendritic cells is very low or absent (3, 20, 34). In vivo, MafB expression was observed in resident macrophages of several tissues (10) and transgenic mouse reporter lines indicated MafB expression in peritoneal and bone marrow F4/80+/Mac-1+ macrophages but not in other hematopoietic lineages (13).
Both in avian and in human differentiation systems MafB expression is strongly upregulated after inducing macrophage differentiation of myeloid progenitor cells (3, 10, 12, 20). Consistent with a functional importance of this expression profile, ectopic retroviral expression of MafB in these cells promotes phenotypic and functional monocyte/macrophage differentiation (3, 12, 20). In addition, MafB inhibits differentiation along other lineages such as the erythroid (34) or dendritic (3) cell lineage. Furthermore, a MafB deletion mutant lacking N-terminal effector domains and consisting only of the DNA binding domain acts as a dominant-negative mutant for MafB transactivation and inhibits monocyte/macrophage differentiation in avian myeloid progenitors (20). Together these results strongly suggested a critical function of MafB in macrophage differentiation. To further analyze the requirement for MafB in this process, we generated MafB-deficient mice. Since these mice die at birth from defects of respiratory neurons in the hindbrain and subsequent central breathing failure (4, 5), we analyzed macrophage development by in vitro differentiation and in the embryo and in adult mice reconstituted with MafB-deficient fetal liver cells. Surprisingly, we observed normal numbers of Mac-1+/F4/80+ monocytes and macrophages in all analyzed tissues. Macrophages could also be derived by vitro differentiation from MafB−/− fetal liver or spleen. On the other hand MafB−/− macrophages showed an increased expression of genes involved in modulation of actin organization and dramatically amplified morphological changes in response to monocyte colony-stimulating factor (M-CSF), involving the rapid formation of multiple, branched protrusions. Together our results indicated an unexpected redundancy of MafB function in macrophage differentiation and suggest a previously unknown role of MafB in influencing actin organization in macrophages.
MATERIALS AND METHODS
Mice.
We described previously the generation of mafB-deficient mice on a 129Sv-C57BL/6 background and their genotyping by PCR with primers for mafB and gfp, replacing mafB in the knockout allele (4). Congenic C57BL/6SJL (Ly5.1 allele) mice were purchased from the Charles River Laboratory (L'Arbresle, France). All experiments were performed in accordance with institutional guidelines using mice maintained under specific-pathogen-free conditions.
Cell preparations and FACS analysis.
Bone marrow cells were flushed from femurs and tibias in Iscove modified Dulbecco medium (IMDM)-20% fetal calf serum (FCS), and splenocytes and fetal liver cells were obtained by mechanical dissociation of organs. Peripheral blood was obtained by heparinized microcapillary aspiration from decapitated embryos. Cell preparations containing red cells were treated with lysis buffer (0.15 M NH4Cl, 17 mM Tris, pH 7.2) prior to fluorescence-activated cell sorting (FACS) analysis. For antibody staining cells were resuspended in FACS medium (0.2% bovine serum albumin [BSA] and 0.1% NaN3 in phosphate-buffered saline [PBS]) at a concentration of 1 × 106 to 1 × 107 cells/ml followed by incubation at 4°C for 20 min with properly diluted fluorochrome- or biotin-labeled monoclonal antibodies, followed by secondary incubation with fluorochrome-coupled streptavidin where required. Flow cytometry was performed on a FACScalibur cytometer (Becton Dickinson, San Jose, CA) with CellQuest (Becton Dickinson) software.
Bone marrow-reconstituted mice.
Fetal liver cells (1 × 106) from E14.5 control and mafB−/− Ly5.2 embryos were injected into the tail vein of lethally irradiated (900- to 1,000-rad) age- and sex-matched Ly5.1 recipient mice. Irradiation was done at least 4 h before cell transfer, and mice were kept on antibiotics in the drinking water for 4 weeks posttransplantation.
Colony and plasma clot assays.
Fetal liver cells (1.5 × 104) were seeded in 1 ml of IMDM containing 1% methylcellulose supplemented with 10% FCS, 450 μM monothioglycerol (Sigma), 10 μg/ml insulin (Sigma), 50 U/ml penicillin, 50 mg/ml streptomycin, and 2 mM glutamine (all from Invitrogen); 2.5% interleukin 3 (IL-3)-containing conditioned medium; 5% granulocyte-macrophage colony-stimulating factor (GM-CSF)-containing conditioned medium; and 5% M-CSF-containing conditioned medium (CA medium). At day 11, differentiated cells were washed out of methylcellulose and stained as described below for cytospins.
In plasma clot assays the morphology of the colony can be preserved during fixation and staining. Cells (104 to 105) from mouse fetal spleen were seeded directly in plasma clot media: CA medium and 10% bovine citrated plasma (Sigma) clotted by the addition of 10 U/ml of thrombin (Sigma). Assays were analyzed at day 6, and clotted colonies were compressed onto slides using Whatman paper and stained as described below for cytospins.
Cytospins, blood smears, and morphological staining and immunostaining.
Cytocentrifugation of cells was carried out using 10 to 100,000 cells per well in a Shandon cytocentrifuge. Samples were air dried and fixed in 100% methanol for 4 min, followed by 2 min of incubation in neutral benzidine (1% O-dianisidine in methanol) to stain erythrocytes. Slides were then rinsed in hydrogen peroxide (0.5% in 50% ethanol) for 1.5 min followed by a 30-s rinse in water. Leukocytes were stained with Diff-Quick (Baxter), 3 min in eosin G in phosphate buffer, pH 6.6, and 2 min in thiazine dye in phosphate buffer, pH 6.6.
Peripheral blood smears were fixed in 100% methanol for 5 min and stained with Giemsa stain (Sigma Diagnostics; GS-5000) for 30 min at room temperature. Differential leukocyte counting was done on a Leitz DMRBE microscope and images acquired with Act-1 software. Nucleated red cells were counted per 100 leukocytes and used for correcting total leukocyte count. Whole blood was diluted (1:40) in Turk solution (Merck, catalog no. 9277), and total nucleated cells were counted in a Neubauer chamber.
For immunostaining, 18.5-day-postcoitum spleens were fixed in 3% paraformaldehyde for 120 min followed by O.C.T. (Tissue-Tek, The Netherlands) embedding. Cells were stained with biotin-conjugated anti-mouse antibody against F4/80 (eBioscience) and counterstained with the nuclear dye DAPI (4′,6′-diamidino-2-phenylindole; 5 μg/ml for 45 min; Sigma-Aldrich). Photomicrographs were taken with a multifluorescence Zeiss Axioplan 2 microscope and acquired with SmartCapture 2 software.
In vitro-derived primary macrophages.
Control or mafB−/− fetal liver cells were seeded at 2 × 105 cells/ml on two 15-cm bacterial dishes in complete medium (IMDM-10% FCS; 450 μM monothioglycerol σ; 10 μg/ml insulin σ; 50 U/ml penicillin, 50 mg/ml streptomycin, and 2 mM glutamine [all from Invitrogen]; 2.5% IL-3-containing conditioned medium; 5% GM-CSF-containing conditioned medium; 5% M-CSF-containing conditioned medium). From day 6, medium was changed to M-CSF medium (MM: complete medium without IL-3 and with GM-CSF and M-CSF only). At day 10, adherent cells were scraped off and harvested for RNA or protein extraction or split into new dishes for functional analysis.
Microarray and quantitative real-time reverse transcription-PCR (RT-PCR) analysis.
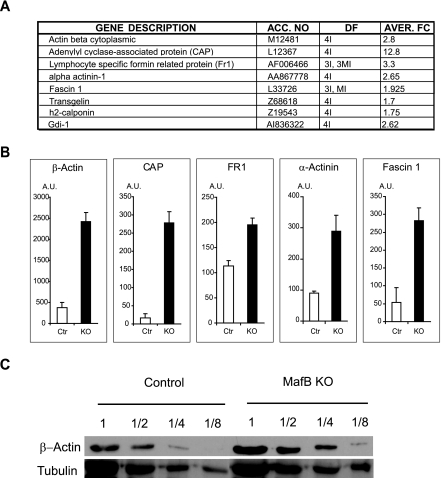
Total RNA was extracted using Trizol (Invitrogen) reagent from equal numbers of in vitro M-CSF-derived macrophages and digested with DNase I (QIAGEN). RNA was pooled from two MafB+/+ control and two MafB−/− macrophage samples derived from independent individual fetal livers and processed by a commercial service to generate probes for Affymetrix HG_U95Av2 chip hybridization with Microarray Suite software. Results from two independent arrays each containing duplicate data points were analyzed with Gene Chip EASI expression analysis software. For single gene expression analysis 1- to 2-μg RNA samples of macrophages derived from individual mafB+/+ or mafB−/− fetal livers were reverse transcribed with SuperScript II (Invitrogen) and subjected to quantitative real-time PCR using the SYBR green PCR master mix (PE Applied Biosystems, Foster City, CA), following the manufacturer's instructions. Reactions were performed in a 7500 Fast quantitative real-time PCR detection system (PE-Applied Biosystems, Foster City, CA). Tested expression levels were normalized to hypoxanthine phosphoribosyltransferase (HPRT) expression. Primers used were as follows: Fr1, plus strand, CCGACCGGAAACAGACGTT, and minus strand, AACCCGTGAGCTGTGGGTACT; adenylate cyclase-associated protein (CAP), plus strand, GAAAAGTGCCAACCATTTCCA, and minus strand, CACAGTCCAGGGAGTTCTTGCT; cytoplasmic β-actin, plus strand, GACGGCCAAGTCATCACTATTG, and minus strand, CAAGAAGGAAGGCTGGAAAAGA; fascin 1, plus strand, TCAGTCCTCCTGTTATCCTTACTCATC, and minus strand, CCGTTTTCTCTTGGGTTTCCA; α-actinin-1, plus strand, AGCCAGGAACAGATGAACGAA, and minus strand, CCAACGTGCCGGAGTGAT; c-Maf, plus strand, GGATGGCTTCAGAACTGGCA, and minus strand, AACATATTCCATGGCCAGGG; HPRT, plus strand, AGCCCTCTGTGTGCTAAGG, and minus strand, CTGATAAAATCTACAGTCCATAGGAATGGA.
Western blot analysis.
Proteins were extracted from in vitro-differentiated macrophages. Western analysis was performed as described previously (20). Primary antibodies were rabbit anti-mouse c-Maf (Santa Cruz Biotechnology; sc-7866; 1:500), mouse anti-β-actin (Sigma; A5316; 1:20,000), and mouse anti-β-tubulin 1 (Sigma; T7816; 1:20,000). Secondary antibodies were horseradish peroxidase-coupled goat anti-rabbit immunoglobulin G (Santa Cruz Biotechnology; sc-2054; 1/1,000) and anti-mouse immunoglobulin G (Santa Cruz Biotechnology; sc-2055; 1/1,000).
Macrophage stimulation and assays for NO.
Day 12 in vitro-differentiated macrophages at 106 cells/ml were incubated for 24 h with 100 ng/ml lipopolysaccharide (LPS) (Sigma) and 50 U/ml of gamma interferon (IFN-γ) (mouse recombinant; Sigma). Nitric oxide (NO) production was determined by measurement of nitrite, which forms in a reaction of NO with the culture medium. Cell supernatant was cleared of cell debris by centrifugation, and 200 μl was combined with 600 μl of Griess-Ilovays reagent (Merck) and incubated at room temperature for 10 min as recommended elsewhere (1). Absorbance was measured at 546 nm and compared to a standard curve prepared with sodium nitrite.
Phagocytosis assay.
Fluorescent beads (Molecular Probes; 1 μM; F-8851) were washed once in sterile PBS, resuspended in Dulbecco modified Eagle medium-10% FCS, and sonicated for 10 intervals at 10 seconds each. Twenty-five microliters of bead solution was incubated with day 12 differentiated macrophages in 24-well plates for 2 h. Cells were then extensively washed with PBS, fixed with 1% paraformaldehyde, and analyzed by flow cytometry on a FACScalibur cytometer (Becton Dickinson).
M-CSF stimulation and actin filament staining.
Day 10 in vitro-derived macrophages were plated in MM at 4 × 105 cells/ml on alcian blue-treated coverslips. After 24 h cells were cultured in M-CSF-free MM for 12 h and then restimulated with 5% M-CSF-conditioned medium for 5 min. Cells were fixed in 4% paraformaldehyde-PBS and stained with tetramethyl rhodamine isocyanate-conjugated phalloidin (Sigma; 25 ng/ml) in PBS-0.1% saponin-2% BSA. Peritoneal exudate cells were harvested from adult reconstituted mice. Cells (2 × 106) were plated for 2 h in IMDM-10% FCS, 50 U/ml penicillin, and 50 mg/ml streptomycin on alcian blue-treated coverslips. Coverslips were then washed once in PBS and stimulated with 5% M-CSF-conditioned medium for 5 min. Cells were fixed and stained with tetramethyl rhodamine isocyanate-conjugated phalloidin as above and labeled with biotin anti-F4/80 (eBiosciences; 1/500) in PBS-0.1% saponin-2% BSA and fluorescein isothiocyanate-coupled streptavidin (eBiosciences; 1/500). Photomicrographs were taken with a multifluorescence Zeiss Axioplan 2 microscope and acquired with SmartCapture 2 software.
Listeria infection.
Day 12 in vitro-differentiated macrophages were prestimulated by treatment with 100 ng/ml LPS overnight (16 h). Listeria monocytogenes, strain 10403S cells were cultured overnight in brain heart infusion medium (DIFCO) at room temperature until they were in the exponential growth phase. A serial dilution of bacteria covering a range of 1,000 bacteria per cell to 10 bacteria per cell was added to macrophages in Dulbecco modified Eagle medium-10% FCS (without antibiotics) for 35 min. The wells were rinsed three times with medium, and remaining bacteria were killed by the addition of gentamicin at 5 μg/ml. Actin filaments were stained with phalloidin as described above, and bacterial and cellular nuclei were stained with 10 μg/ml DAPI (Roche) for 10 min. Photomicrographs were taken with an Axiophot fluorescence microscope.
RESULTS
Normal numbers of macrophages in MafB−/− fetal liver.
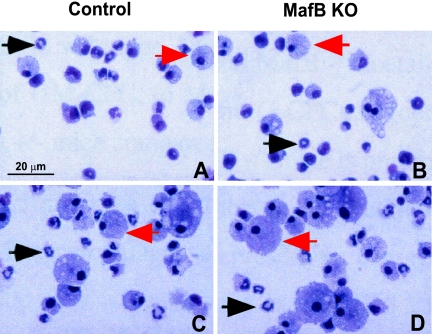
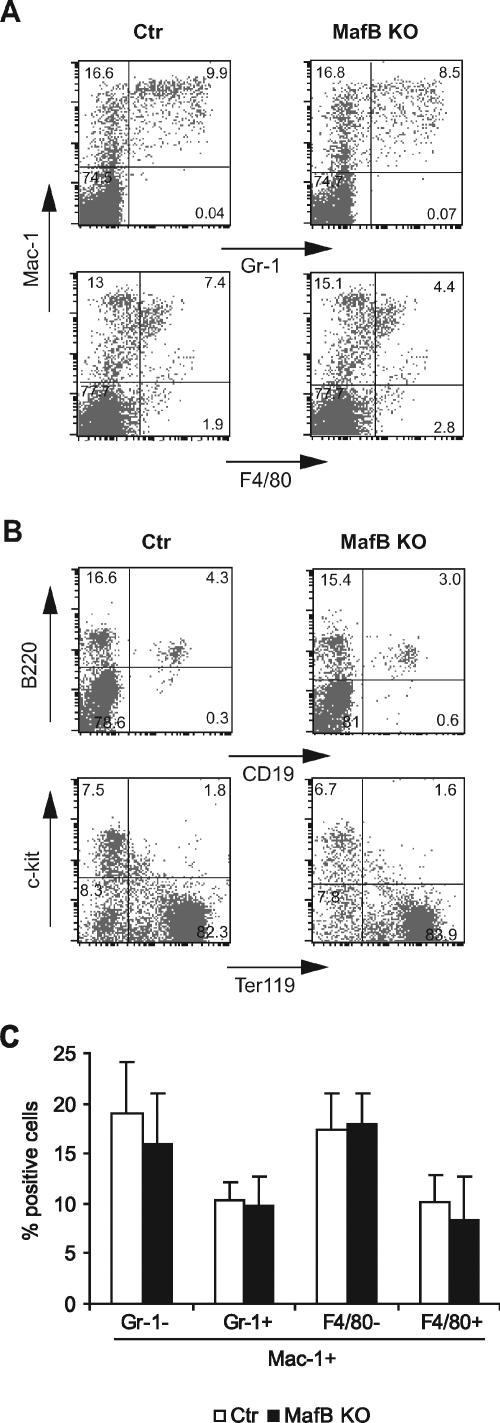
We have described previously the generation of MafB-deficient mice by homologous recombination and verified the absence of MafB mRNA and protein expression in these mice (4). Because gene expression data and gain-of-function and dominant-negative experiments (3, 10, 12, 13, 20, 34) indicated a critical role of MafB in macrophage differentiation, we investigated the effect of MafB deletion on macrophage development. Due to the neonatal death of MafB−/− mice from central respiratory defects, we initially analyzed the fetal liver of E14.5 MafB−/− embryos as the major site of hematopoiesis at this stage of development. Surprisingly we could detect macrophages in cytocentrifuged preparations of MafB−/− fetal liver cell suspensions (Fig. 1A and B). Furthermore, we also observed robust development of mature macrophages of typical morphology (Fig. 1C and D) by in vitro differentiation of MafB−/− fetal liver cells in IL-3, GM-CSF, and M-CSF, cytokines that support myeloid progenitor proliferation and macrophage differentiation. To investigate whether MafB deficiency had quantitative effects on macrophage development or on the relative proportions of different lineages in the hematopoietic system, we analyzed E14.5 fetal liver of MafB−/− mice by FACS. As shown in Fig. 2A and B, we did not observe any differences in the relative abundance of Mac-1+/F4/80+ macrophages or other myeloid and lymphoid cell populations between MafB−/− and wild-type (wt) control embryos of the same litter. The analysis of a larger number of MafB−/− and wt control embryos from several litters also did not reveal any significant difference in the number of Mac-1+/F4/80+ macrophages or other myeloid cell populations between MafB−/− (n = 7) and wt control (n = 7) E14.5 fetal livers (Fig. 2C). Together these observations indicated that normal numbers of fetal macrophages could develop in MafB-deficient embryos.
FIG. 1.
Analysis of myeloid cells from control and MafB−/− E14.5 fetal liver. A and B. Cell suspensions of control MafB+/− (A) and MafB−/− (B) E14.5 fetal liver were cytocentrifuged and stained with Diff-Quick histological stain. C and D. Cells from control MafB+/− (C) and MafB−/− (D) E14.5 fetal livers were cultured in M-CSF-, GM-CSF-, and IL-3-containing semisolid medium for 11 days, washed out from methylcellulose, and stained with Diff-Quick histological stain. Black and red arrows indicate granulocytes and macrophages, respectively.
FIG. 2.
Flow cytometric analysis of hematopoietic cells from control and MafB−/− E14.5 fetal liver. A and B. FACS profiles of MafB+/+ control and MafB−/− E14.5 fetal liver cells identifying myelomonocytic and granulocytic (Mac-1/Gr-1 and Mac-1/F4/80) populations (A) or B-lymphoid (B-220/CD19) and erythroid (Terr-119/c-kit) populations (B). C. Quantification of different populations of Mac-1-positive myeloid cells from MafB+/+ control (n = 7) and MafB−/− (n = 7) E14.5 fetal liver cells, shown as percentages of Terr-119-negative cells. Statistical analysis by two-tailed Mann-Whitney test revealed no statistical significance of the observed minor differences (P values from left to right: 0.16, 0.33, 0.37, and 0.20). Error bars indicate standard errors of the means.
Normal numbers of macrophages in MafB−/− spleen and blood at birth.
During late gestation hematopoiesis shifts sequentially from the fetal liver to the spleen and eventually to the bone marrow, which coincides with the generation of adult-type macrophages (7). To test whether macrophage defects might become apparent in MafB−/− mice only at this stage of development, we analyzed the presence of macrophages in the spleen at E18.5 just prior to birth. As shown in Fig. 3A and C histological staining of cytospin preparations from cell suspensions of E18.5 spleen did not reveal any apparent differences between MafB−/− and wt control samples and the presence of monocytic and granulocytic cells for both genotypes. Since mature macrophages can survive for a long time in tissues, it could not be formally excluded that the observed monocytic cells were at least in part of embryonic origin. We therefore performed plasma clot assays with E18.5 spleen cells to analyze the presence of myeloid progenitors that could give rise to adult-type macrophages. Both macrophages (Fig. 3B and D) and granulocytes (not shown) developed from MafB-deficient spleen cells after 6 days in GM-CSF, which supports the growth of both cell types. Furthermore immunofluorescence staining with anti-F4/80 antibody on frozen sections of E18.5 spleen revealed normal numbers and tissue distribution of macrophages in MafB-deficient samples (Fig. 3E and F). To further analyze whether quantitative differences between macrophage populations might exist, we analyzed MafB−/− and wt control E18.5 fetal spleen by FACS. As shown in Fig. 3G, Mac-1/F4/80 profiles did not reveal any differences between MafB−/− and wt control samples and the quantification of both Mac-1+/F4/80+ and Mac-1+/F4/80− populations did not show any statistically significant differences between MafB−/− (n = 5) and wt control (n = 11) samples (Fig. 3I).
FIG. 3.
Analysis of myeloid cells from control and MafB−/− E18.5 fetal spleen and newborn blood. A and C. Cell suspensions of control MafB+/− (A) and MafB−/− (C) E18.5 fetal spleen were cytocentri fuged and stained with Diff-Quick histological stain. Black and white arrows indicate granulocytes and monocytes, respectively. B and D. Dried and Diff-Quick-stained colonies from day 6 plasma clot assays of control MafB+/− (B) and MafB−/− (D) E18.5 fetal spleen in the presence of GM-CSF, showing a macrophage colony. White arrows indicate examples of individual macrophages. E and F. Immunofluorescence staining for spleen macrophages with anti-F4/80 antibody on frozen sections of control MafB+/+ (E) and MafB−/− (F) E18.5 fetal spleen. G. Flow cytometric analysis of myelomonocytic cell populations from MafB+/+ control (left) and MafB−/− (right) E18.5 fetal spleen. H. Flow cytometric analysis of myelomonocytic cell populations in MafB+/+ (left) control and MafB−/− (right) newborn blood. I. Quantification of different Mac-1-positive myelomonocytic cell populations from MafB+/+ control (n = 11) and MafB−/− (n = 5) E18.5 fetal spleen cells, shown as percentage of total cells. Statistical analysis by two-tailed Mann-Whitney test revealed low or no statistical significance of the observed minor differences (P values from left to right: 0.02 and 0.08). J. Quantification of Mac-1-positive myeloid cell populations from MafB+/+ control (n = 9) and MafB−/− (n = 5) newborn blood after lysis of erythrocytes. Statistical analysis by two-tailed Mann-Whitney test revealed no statistical significance of the observed small differences (P values from left to right: 0.13 and 0.08). Error bars in panels I and J indicate standard errors of the means. K. Histological staining of monocytes, lymphocytes, and granulocytes from blood smears of MafB+/+ control and MafB−/− newborn blood.
We also analyzed the blood from newborn mice and found normal monocytes, granulocytes, and lymphocytes in blood smears from MafB-deficient samples (Fig. 3K) as well as similar total leukocyte counts in wt control (5,925 ± 990/μl) and MafB−/− samples (6,380 ± 930/μl). Furthermore, FACS staining also did not reveal any statistically significant alterations in the relative frequency of both Mac-1+/F480+ and Mac-1+/F4/80− populations between MafB−/− (n = 5) and wt control (n = 9) samples (Fig. 3H and J).
Together these results indicated that normal numbers of adult-type monocytes and macrophages could develop in MafB-deficient embryos.
Normal numbers of MafB−/− macrophages in reconstituted adult mice.
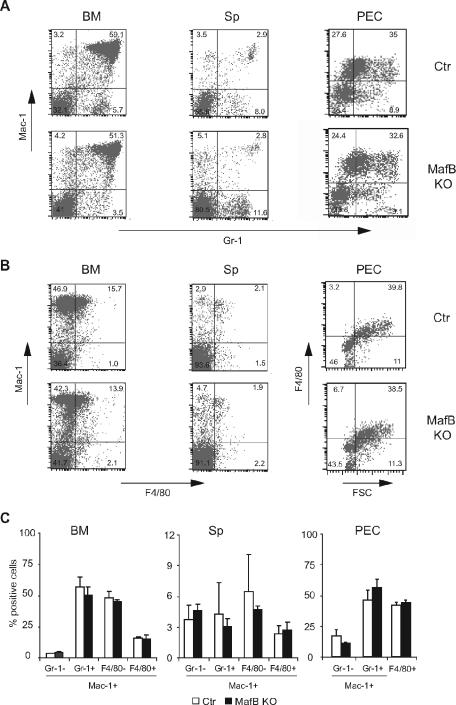
Despite the apparently normal development of adult-type macrophages in MafB-deficient late gestational embryos, it could not be excluded that defects may become evident only later during adult life or in specific macrophage populations that could not be analyzed in the embryo. To test this possibility, we reconstituted lethally irradiated mice with MafB−/− and wt control fetal liver as a source of hematopoietic cells and analyzed macrophage populations in these mice after 8 to 12 weeks, when a complete hematopoietic system had been regenerated. Only mice with quantitative donor reconstitution and less than 2% remaining host contribution to the hematopoietic system were used for analysis. As shown in Fig. 4, we observed normal Mac-1/Gr-1 and Mac-1/F4/80 profiles of myelomonocytic populations in the bone marrow and spleen of MafB−/− reconstituted mice as well as elicited macrophages in the peritoneum 96 h after thioglycolate injection. No statistically significant differences could be detected between the relative proportions of MafB−/− (n = 4) and wt control (n = 4) myelomonocytic and macrophage populations in the analyzed organs (Fig. 4C). Together this indicated that also in adult mice MafB deficiency did not result in compromised macrophage differentiation.
FIG. 4.
Flow cytometric analysis of myeloid cells from adult lethally irradiated mice reconstituted with MafB+/+ control and MafB−/− E14.5 fetal liver cells. A and B. FACS profiles of Mac-1/Gr-1 myeloid (A) and Mac-1/F4/80 myelomonocytic (B) populations from bone marrow (BM), spleen (Sp), and peritoneal exudate cells (PEC), 4 days after thioglycolate stimulation, from irradiated mice reconstituted with a MafB+/+ control or MafB−/− hematopoietic system. C. Quantification of Mac-1-positive myeloid cell populations from MafB+/+ control (n = 4) and MafB−/− (n = 4) bone marrow, spleen, and peritoneal exudate cells. Statistical analysis by two-tailed Mann-Whitney test revealed low or no statistical significance of the observed minor differences (P values from left to right: bone marrow, 0.014, 0.24, 0.17, and 0.17; spleen, 0.17, 0.56, 0.56, and 0.44; peritoneal exudate cells, 0.1, 0.2, and 0.5). Error bars indicate standard errors of the means.
Preserved macrophage functions in MafB−/− macrophages.
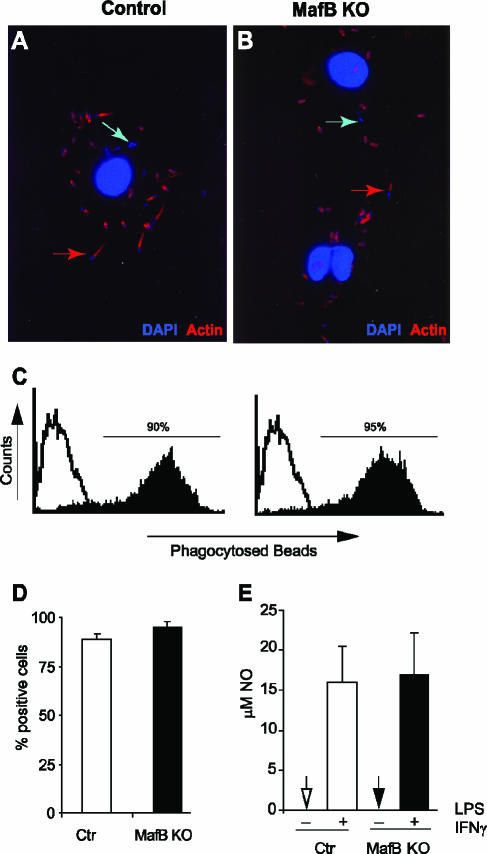
To further analyze whether MafB-deficient macrophages might have defects in characteristic macrophage functions, we derived macrophages by in vitro differentiation from fetal liver and subjected them to functional assays. A defining characteristic of macrophages is the ability to phagocytose pathogens and cellular debris. To investigate whether this function might be affected by MafB deficiency, we infected MafB−/− and control macrophages with L. monocytogenes and stained the cells with phalloidin for actin and with DAPI for cellular and bacterial DNA. As shown in Fig. 5A and B both MafB−/− and control macrophages had phagocytosed bacteria that had no actin tail (blue arrows), indicative of killed bacteria in the lysosomes, as well as actin-tailed bacteria (red arrows), indicative of bacteria that escaped the phagolysosome and used the cellular actin machinery to move through the host cell, a typical characteristic of L. monocytogenes infection (9). To further quantitatively analyze phagocytic ability, we incubated MafB−/− and wt control macrophages with fluorescent latex beads and subjected them to FACS analysis. As shown in Fig. 5C and D, no differences were found in the percentage of phagocytic cells or the average of phagocytosed beads per cell. Together this indicated that MafB is not required for the general phagocytic machinery of macrophages. Upon activation by IFN-γ and bacterial cell wall components such as LPS, macrophages secrete large quantities of NO, which is thought to contribute to bacterial killing. As shown in Fig. 5E, we observed the same level of NO production after LPS-IFN-γ stimulation for MafB−/− and wt control macrophages, indicating that MafB is not required for the signaling and effector mechanisms of this pathway.
FIG. 5.
Functional analysis of macrophages differentiated in vitro from control and MafB−/− E14.5 fetal liver cells. A and B. Confocal images of MafB+/− control (A) or MafB−/− (B) macrophages infected with Listeria monocytogenes showing DAPI-stained bacterial and cellular DNA (in blue) and phalloidin-stained actin (in red). Both killed bacteria without actin tail (blue arrows indicate examples) and infectious actin-tailed bacteria that have escaped the phagolysosome (red arrows indicate examples) are visible in both genotypes. C. FACS analysis of MafB+/+ control (left) and MafB−/− (right) macrophage cultures incubated for 2 h with fluorescent latex beads to quantify phagocytic capacity. D. Quantification of percent phagocytic cells from panel C (n = 2). E. NO production measured as μM nitrite accumulated in the medium of MafB+/+ control and MafB−/− macrophage cultures (n = 3) with 1 × 106 cells, 24 h after stimulation with 100 ng/ml LPS and 50 U/ml IFN-γ. Error bars indicate standard errors of the means.
Increased expression of c-Maf in MafB−/− macrophages.
Given the surprising absence of a macrophage differentiation phenotype in MafB-deficient hematopoietic cells, we pursued the hypothesis that MafB function in this process might be compensated for by other, closely related members of the Maf family such as c-Maf, which similarly to MafB can induce monocytic differentiation in human myeloid cell lines (14). As shown in Fig. 6, we indeed observed increased c-Maf expression on both the message (Fig. 6A) and the protein (Fig. 6B) level in MafB-deficient macrophages, suggesting that upregulation of c-Maf might rescue MafB function in macrophage differentiation of MafB−/− cells.
FIG. 6.
Expression of c-Maf in control and MafB−/− macrophages. A. Quantitative real-time RT-PCR for c-Maf on mRNA isolated from pooled MafB+/+ control (n = 3) and MafB−/− (n = 2) in vitro-differentiated macrophage cultures shown as arbitrary units of expression levels. Samples were normalized to HPRT expression, and expression data were confirmed in two independent experiments. Error bars indicate standard errors of the means. B. Western blot analysis for c-Maf expression in pooled MafB+/+ control (n = 3, lane 1) and MafB−/− (n = 2, lane 2) in vitro-differentiated macrophages. Lanes 3 and 4 show extracts from untransfected or c-Maf expression plasmid-transfected HEK 293 cells, respectively.
Increased expression of actin organizing factors in MafB−/− macrophages.
To reveal potential unsuspected functions of MafB in macrophages, we decided to monitor global changes in gene expression resulting from MafB deficiency that might group directly and indirectly controlled genes into functionally related classes. Therefore, we isolated mRNAs from in vitro-differentiated fetal liver-derived MafB−/− and wt control macrophages and subjected them to hybridization with Affymetrix microarray DNA chips. Detailed analysis of the expression data revealed the coregulation and increased expression of a group of genes involved in actin organization (Fig. 7A). To verify the altered expression of these genes in MafB-deficient macrophages, we used quantitative real-time RT-PCR with specific primers for β-actin, CAP, formin-related protein 1 (Fr1), fascin, and α-actinin. As shown in Fig. 7B, a significantly increased expression, ranging from 2- to over 12-fold, was observed in MafB-deficient macrophages for all of the analyzed genes, thus confirming the gene array data. Increased expression of β-actin in MafB−/− macrophages was further confirmed on the protein level by Western blotting (Fig. 7C).
FIG. 7.
Differential gene expression in macrophages differentiated in vitro from control and MafB−/− E14.5 fetal liver cells. A. Actin skeleton-related genes differentially expressed in MafB+/+ control and MafB−/− macrophages by Affymetrix gene array analysis. Gene accession number, number of quadruplicate samples that differed (I, increased; MI, moderately increased), and average change (n-fold) are indicated. B. Quantitative real-time RT-PCR for selected genes on mRNA isolated from pooled MafB+/+ control (n = 3) and MafB−/− (n = 2) in vitro-differentiated macrophage cultures shown as arbitrary units of expression levels. All samples were normalized to HPRT expression, and all expression data were confirmed in at least two independent experiments. Error bars indicate standard errors of the means. C. Western blot analysis of β-actin in serial diluted extracts from MafB+/+ control (n = 3) and MafB−/− (n = 2) in vitro-differentiated macrophage cultures. Tubulin expression is shown as a normalization control.
Altered morphology and actin organization in MafB−/− macrophages.
To identify potential phenotypic consequences from the altered expression of actin-modulating genes, we investigated processes in macrophages known to involve actin metabolism.
When Listeria monocytogenes cells are phagocytosed by macrophages, some bacteria can escape the phagolysosome and rearrange cellular actin to propel themselves through the cell and infect neighboring cells (9). Our studies with L. monocytogenes shown in Fig. 5A and B demonstrated that both MafB−/− and control macrophages contained actin-tailed bacteria that used the cellular actin machinery to move through the host cell. The altered expression of actin-modulating genes in MafB−/− macrophages thus did not appear to prevent the typical actin rearrangements induced by L. monocytogenes infection.
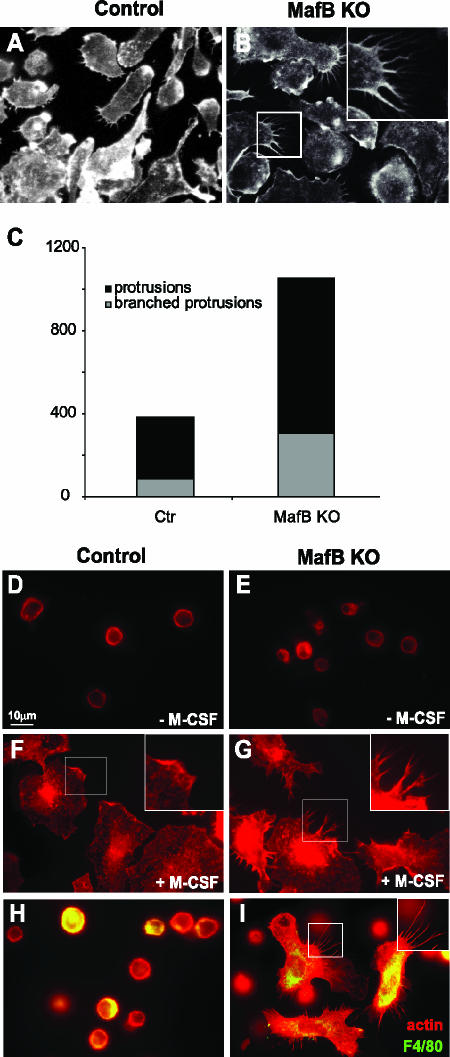
Macrophages are also highly motile cells and extend filopodia from the cell body during their movements, a process that equally involves extensive actin rearrangement. At least three of the genes that we found to be upregulated in MafB−/− macrophages, the formin, fascin, and α-actinin genes, have been shown to participate in the formation of filopodia (30, 31, 33, 35, 36). We therefore carefully analyzed cellular protrusions of phalloidin-stained control and MafB−/− macrophages at high magnification under the microscope. In contrast to MafB+/+ control macrophages (Fig. 8A), we observed multiple and frequently branched protrusions on a high proportion of MafB-deficient cells (Fig. 8B). Although control macrophages also had protrusions, they were less prominent, less frequent, and rarely branched. This difference was confirmed by counting straight and branched protrusions per 100 cells on control and MafB−/− macrophages (Fig. 8C).
FIG. 8.
Analysis of actin organization in control and MafB−/− macrophages. A and B. Immunofluorescence images of macrophage cultures differentiated in vitro from E14.5 fetal liver cells of MafB+/+ control (A, n = 2) or MafB−/− (B, n = 2) embryos. Phalloidin staining revealed prominent, often branched actin-containing protrusions in MafB-deficient macrophages (B). C. Quantification of actin-containing protrusions observed in panels A and B, expressed as total numbers of branched and straight protrusions per 100 cells. D to G. MafB+/+ control (D and F) or MafB−/− (E and G) in vitro-differentiated macrophages from three age-matched embryos each were cultured overnight without M-CSF (D and E), restimulated for 5 min with M-CSF (F and G), and analyzed by phalloidin staining for actin organization, showing rapid spreading in both cases but mainly lamellopodial extensions in MafB+/+ control (inset in panel F) and mainly filopodial, often branched, protrusions in MafB−/− macrophages (inset in panel G). H and I. Peritoneal exudate cells from two MafB+/+ control (H) or two MafB−/− (I) reconstituted mice were pooled and cultured for 5 min in M-CSF-containing medium, fixed, and stained with phalloidin to reveal actin organization and anti-F4/80 antibody to identify macrophages. Both F4/80− lymphocytes and F4/80+ macrophages were detected in both samples. MafB−/− macrophages revealed prominent filopodial, often branched, protrusions (I, inset).
Macrophages respond to M-CSF by massive morphological changes involving actin reorganization (29). To test whether this response might be involved in the observed morphological differences of control and MafB−/− macrophages, we M-CSF starved macrophage cultures overnight (Fig. 8D and E) and analyzed their morphology and actin organization by phalloidin staining after restimulation for 5 min with M-CSF. As shown in Fig. 8F and G, stimulation of both control and MafB−/− macrophages resulted in dramatic morphological reorganization and spreading on the culture dish, but whereas MafB+/+ control macrophage were spreading mainly by lamellopodial extensions (Fig. 8F), MafB−/− macrophages showed multiple filopodial and frequently branched protrusions as observed before (Fig. 8G). Even though MafB+/+ control cells eventually also developed protrusions at later time points, they were less prominent, less frequent, and rarely branched.
To analyze whether this phenotype was also observed in macrophage populations directly isolated from tissues in vivo, we prepared peritoneal exudate cells from wt control or MafB−/− reconstituted mice, cultured them for 5 min in M-CSF, and stained them with antibody for F4/80 to detect macrophages and phalloidin to reveal the actin skeleton. As shown in Fig. 8H and I, actin+ F4/80− lymphocytes with a typical round morphology were detected in both control and MafB−/− samples. By contrast, macrophages still had a round morphology in the control samples (Fig. 8H) but had undergone dramatic shape changes in the MafB−/− samples (Fig. 8I). The macrophages had spread out and formed multiple extensive and often branched protrusions, similar to those observed before with in vitro-differentiated macrophages.
Together these results suggested that the increased expression of actin-modulating genes in MafB−/− macrophages caused changes in actin organization that altered the rapidity and extent of the formation of cellular, possibly filopodial, protrusions in response to M-CSF.
DISCUSSION
Based on the selective hematopoietic expression of MafB in monocytes and macrophages as well as its ability to specifically control macrophage differentiation upon ectopic expression in myeloid progenitors (3, 12, 20), we tested whether it was also required for macrophage differentiation by analyzing MafB-deficient mice. Despite the previously known inhibitory role of a dominant-negative MafB for macrophage differentiation (20), unexpectedly we did not observe any loss or reduction of fetal or adult monocyte and macrophage populations. In vitro-differentiated MafB−/− macrophages were also uncompromised for typical macrophage functions such as NO production and phagocytosis. By contrast, gene expression analysis of MafB−/− macrophages revealed increased expression of multiple genes involved in the modulation of actin organization. Consistent with this, phalloidin staining revealed a high frequency of characteristic branched actin-containing protrusions on MafB-deficient macrophages. Together our results revealed an unexpected redundancy of MafB function in macrophage differentiation and a previously unknown role in actin organization and macrophage morphology.
Early fetal and adult-type macrophage populations develop along distinct pathways in the embryo (7, 24, 27) and differ in their expression profile for Mac-1 and F4/80 (24). MafB deficiency also did not appear to have selective effects on either of these populations, as we did not detect any differences of these populations in E12.5 interdigital macrophages (unpublished results), E14.5 fetal liver, prenatal spleen, or blood and adult reconstituted mice.
Although the original gain-of-function experiments were performed in an avian system (3, 20), the unexpected absence of a macrophage differentiation phenotype in MafB-deficient mice did not appear to be due to species-dependent differences of MafB function. The original observations in chicken cells have been confirmed in human cells now (3, 12), and retroviral overexpression of MafB in murine lineage negative bone marrow progenitors also promoted macrophage differentiation (S. Sarrazin and M. Sieweke, unpublished observations). Our data thus appear to indicate that MafB function in macrophage differentiation may be compensated for by other, closely related members of the Maf family such as c-Maf, which similarly to MafB can induce monocytic differentiation in human myeloid cell lines (14). Consistent with such a role, we observed that c-Maf expression is increased in MafB-deficient macrophages. The effect of a dominant-negative MafB construct on macrophage differentiation (20) might then be explained by its leucine zipper specificity, which permits dimerization with both MafB and c-Maf (18, 22) and thus inhibition of both molecules. These observations highlight the importance of the relative expression levels and functional interaction of different Maf family members in hematopoietic differentiation processes (25, 28). It may therefore be interesting in the future to combine MafB-deficient mice with other knockout alleles and overexpression strategies both for c-Maf and for other family members.
Despite the surprising absence of a macrophage differentiation phenotype, we found several genes to be differentially expressed between MafB+/+ control and MafB−/− macrophages that are related to the control of actin organization. At the moment it is unclear whether the identified genes are direct MafB targets. A preliminary inspection of the presumed promoter sequences of confirmed upregulated genes with MatInspector software (Genomatix), focusing on sequences between −2,000 bp and +500 bp from the transcriptional start site, did not identify high-homology sites with fewer than three mismatches to the TGCTGACTCAGCA MARE consensus binding site (19, 21). However, this does not necessarily exclude direct regulation of these genes by MafB. Firstly, Maf binding sites have been identified that deviate significantly from the MARE consensus but have high biological significance (16, 37). Furthermore, it is possible that more distant enhancer elements influence the expression of these genes. Finally, besides directly activating promoters via MARE binding sites, Maf factors including MafB can also repress gene expression by inhibitory interactions with other transcription factors (15, 34). It is thus possible that protein-protein interactions rather than direct DNA binding via MARE sequences mediate the repression of negative target genes, such as the potential candidates identified here. Irrespectively of whether these genes are direct MafB targets or whether their increased expression represents a more indirect consequence of an altered cellular state induced by MafB deficiency, their coregulation and functional relation suggest that they belong to a MafB-dependent transcriptional program of biological significance.
Essential monocyte/macrophage functions in patrolling and infiltrating tissues under physiological and pathological conditions require dynamic morphological changes (29). On the cellular level this involves the formation of lamellopodial and filopodial protrusions, processes that are highly dependent on the remodeling of the actin cytoskeleton (30). Of the genes upregulated in MafB-deficient macrophages a majority belonged to the family of actin binding proteins, which are known to regulate remodeling of the actin skeleton and dynamic changes in cell morphology (30). At least three of these have been shown to be directly involved in filopodium formation. The actin-bundling protein fascin has been shown to be required for filopodium formation in activated dendritic cells, neurons, and metastatic cancer cells (31, 35), cell types whose morphology is characterized by prominent cellular processes. Formins are a family of proteins that are also found in filopodia, including in macrophages (36), and some members have been shown to be required for filopodium formation (33). Interestingly, overexpression of FRL, a formin protein, has been shown to alter actin organization, morphology, and motility in macrophages (36). Finally α-actinins also contribute to the remodeling of the actin skeleton (30) and are found at the base of filopodial structures (35).
Besides actin binding proteins, we also found upregulation of CAP, a highly conserved monomeric actin binding protein that regulates the cellular G-actin/F-actin ratio and the dynamic equilibrium of actin polymerization and depolymerization in response to extracellular signals. Experiments in several model systems indicate that the perturbation of CAP levels influences actin dynamics and cellular morphology in developmental processes that involve migration, invasion, and polarity (17). Interestingly, increasing cellular CAP levels by microinjection promoted the formation of actin filaments (11).
Together with the observed upregulation of β-actin itself, the increased expression of these genes suggests that they may contribute to the formation of the characteristic branched actin-containing filopodial protrusion in MafB-deficient macrophages by stimulating actin reorganization. Macrophages undergo rapid morphological changes in response to M-CSF that involve both lamellopodial and filopodial extensions (29), which are under the control of communicating but distinct signaling pathways (2). It is possible that the increased expression of actin remodeling genes in MafB−/− macrophages alters these signaling pathways in ways that lead to aberrant interpretation of M-CSF signaling. Alternatively our results are also consistent with the hypothesis that MafB deficiency may change the intracellular sensitivity to M-CSF and thus lead to a grossly exaggerated but in principle normal response to M-CSF.
In summary, whereas MafB function in macrophage differentiation can unexpectedly be completely compensated for by other family members, it appears to be specifically required for the control of M-CSF-induced changes in macrophage morphology and actin organization.
Acknowledgments
We thank I. Lafon, B. Blanchi, and T. Fukao for technical help, advice, and preliminary results.
A. Aziz was supported by the Kind Philipp Stiftung für Leukämieforschung; P. Mohideen, C. Otto, and L. Vanhille by the French Ministère de l'éducation nationale, de l'enseignement supérieur et de la recherche; C. Otto by La Ligue Nationale contre le Cancer; Y. Bakri by the Centre National de la Recherche Scientifique (CNRS) and the Fondation pour la Recherche Médicale (FRM); L. M. Kelly by the Association pour la Recherche sur le Cancer (ARC); and S. Sarrazin by ARC, the Societé Francaise d'Hématologie (SFH), and the Fondation de France (FdF). The work was supported by the ATIPE program of the CNRS; an installation grant from FRM; grant 2004004150 from FdF; and grants 5387, 8453, and 3422 from ARC.
REFERENCES
- 1.Aggarwal, B. B., and K. Mehta. 1996. Determination and regulation of nitric oxide production from macrophages by lipopolysaccharides, cytokines, and retinoids. Methods Enzymol. 269:166-171. [DOI] [PubMed] [Google Scholar]
- 2.Allen, W. E., G. E. Jones, J. W. Pollard, and A. J. Ridley. 1997. Rho, Rac and Cdc42 regulate actin organization and cell adhesion in macrophages. J. Cell Sci. 110:707-720. [DOI] [PubMed] [Google Scholar]
- 3.Bakri, Y., S. Sarrazin, U. P. Mayer, S. Tillmanns, C. Nerlov, A. Boned, and M. H. Sieweke. 2005. Balance of MafB and PU.1 specifies alternative macrophage or dendritic cell fate. Blood 105:2707-2716. [DOI] [PubMed] [Google Scholar]
- 4.Blanchi, B., L. M. Kelly, J. C. Viemari, I. Lafon, H. Burnet, M. Bevengut, S. Tillmanns, L. Daniel, T. Graf, G. Hilaire, and M. H. Sieweke. 2003. MafB deficiency causes defective respiratory rhythmogenesis and fatal central apnea at birth. Nat. Neurosci. 6:1091-1100. [DOI] [PubMed] [Google Scholar]
- 5.Blanchi, B., and M. H. Sieweke. 2005. Mutations of brainstem transcription factors and central respiratory disorders. Trends Mol. Med. 11:23-30. [DOI] [PubMed] [Google Scholar]
- 6.Blank, V., and N. C. Andrews. 1997. The Maf transcription factors: regulators of differentiation. Trends Biochem. Sci. 22:437-441. [DOI] [PubMed] [Google Scholar]
- 7.Bonifer, C., N. Faust, H. Geiger, and A. M. Muller. 1998. Developmental changes in the differentiation capacity of haematopoietic stem cells. Immunol. Today 19:236-241. [DOI] [PubMed] [Google Scholar]
- 8.Cordes, S. P., and G. S. Barsh. 1994. The mouse segmentation gene kr encodes a novel basic domain-leucine zipper transcription factor. Cell 79:1025-1034. [DOI] [PubMed] [Google Scholar]
- 9.de Chastellier, C., and P. Berche. 1994. Fate of Listeria monocytogenes in murine macrophages: evidence for simultaneous killing and survival of intracellular bacteria. Infect. Immun. 62:543-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eichmann, A., A. Grapin-Botton, L. Kelly, T. Graf, N. M. Le Douarin, and M. Sieweke. 1997. The expression pattern of the mafB/kr gene in birds and mice reveals that the kreisler phenotype does not represent a null mutant. Mech. Dev. 65:111-122. [DOI] [PubMed] [Google Scholar]
- 11.Freeman, N. L., and J. Field. 2000. Mammalian homolog of the yeast cyclase associated protein, CAP/Srv2p, regulates actin filament assembly. Cell Motil. Cytoskelet. 45:106-120. [DOI] [PubMed] [Google Scholar]
- 12.Gemelli, C., M. Montanari, E. Tenedini, T. Zanocco Marani, T. Vignudelli, M. Siena, R. Zini, S. Salati, E. Tagliafico, R. Manfredini, A. Grande, and S. Ferrari. 3 February 2006. Virally mediated MafB transduction induces the monocyte commitment of human CD34+ hematopoietic stem/progenitor cells. Cell Death Differ. [Online.] doi: 10.1038/sj.cdd.4401860. [DOI] [PubMed]
- 13.Hamada, M., T. Moriguchi, T. Yokomizo, N. Morito, C. Zhang, and S. Takahashi. 2003. The mouse mafB 5′-upstream fragment directs gene expression in myelomonocytic cells, differentiated macrophages and the ventral spinal cord in transgenic mice. J. Biochem. (Tokyo) 134:203-210. [DOI] [PubMed] [Google Scholar]
- 14.Hedge, S., J. Zhao, R. Ashmun, and L. Shapiro. 1999. c-Maf induces monocytic differentiation and apoptosis in bipotent myeloid progenitors. Blood 94:1578-1589. [PubMed] [Google Scholar]
- 15.Hedge, S. P., A. Kumar, C. Kurschner, and L. H. Shapiro. 1998. c-Maf interacts with c-Myb to regulate transcription of an early myeloid gene during differentiation. Mol. Cell. Biol. 18:2729-2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ho, I. C., M. R. Hodge, J. W. Rooney, and L. H. Glimcher. 1996. The proto-oncogene c-maf is responsible for tissue-specific expression of interleukin-4. Cell 85:973-983. [DOI] [PubMed] [Google Scholar]
- 17.Hubberstey, A. V., and E. P. Mottillo. 2002. Cyclase-associated proteins: CAPacity for linking signal transduction and actin polymerization. FASEB J. 16:487-499. [DOI] [PubMed] [Google Scholar]
- 18.Kataoka, K., K. T. Fujiwara, M. Noda, and M. Nishizawa. 1994. MafB, a new Maf family transcription activator that can associate with Maf and Fos but not with Jun. Mol. Cell. Biol. 14:7581-7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kataoka, K., M. Noda, and M. Nishizawa. 1994. Maf nuclear oncoprotein recognizes sequences related to an AP-1 site and forms heterodimers with both Fos and Jun. Mol. Cell. Biol. 14:700-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelly, L. M., U. Englmeier, I. Lafon, M. H. Sieweke, and T. Graf. 2000. MafB is an inducer of monocytic differentiation. EMBO J. 19:1987-1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kerppola, T. K., and T. Curran. 1994. A conserved region adjacent to the basic domain is required for recognition of an extended DNA binding site by Maf/Nrl family proteins. Oncogene 9:3149-3158. [PubMed] [Google Scholar]
- 22.Kerppola, T. K., and T. Curran. 1994. Maf and Nrl can bind to AP-1 sites and form heterodimers with Fos and Jun. Oncogene 9:675-684. [PubMed] [Google Scholar]
- 23.Li, M. A., J. D. Alls, R. M. Avancini, K. Koo, and D. Godt. 2003. The large Maf factor Traffic Jam controls gonad morphogenesis in Drosophila. Nat. Cell Biol. 5:994-1000. [DOI] [PubMed] [Google Scholar]
- 24.Lichanska, A. M., C. M. Browne, G. W. Henkel, K. M. Murphy, M. C. Ostrowski, S. R. McKercher, R. A. Maki, and D. A. Hume. 1999. Differentiation of the mononuclear phagocyte system during mouse embryogenesis: the role of transcription factor PU.1. Blood 94:127-138. [PubMed] [Google Scholar]
- 25.Motohashi, H., F. Katsuoka, J. A. Shavit, J. D. Engel, and M. Yamamoto. 2000. Positive or negative MARE-dependent transcriptional regulation is determined by the abundance of small Maf proteins. Cell 103:865-875. [DOI] [PubMed] [Google Scholar]
- 26.Motohashi, H., T. O'Connor, F. Katsuoka, J. D. Engel, and M. Yamamoto. 2002. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene 294:1-12. [DOI] [PubMed] [Google Scholar]
- 27.Naito, M., S. Umeda, T. Yamamoto, H. Moriyama, H. Umezu, G. Hasegawa, H. Usuda, L. D. Shultz, and K. Takahashi. 1996. Development, differentiation, and phenotypic heterogeneity of murine tissue macrophages. J. Leukoc. Biol. 59:133-138. [DOI] [PubMed] [Google Scholar]
- 28.Onodera, K., J. A. Shavit, H. Motohashi, M. Yamamoto, and J. D. Engel. 2000. Perinatal synthetic lethality and hematopoietic defects in compound mafG::mafK mutant mice. EMBO J. 19:1335-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pixley, F. J., and E. R. Stanley. 2004. CSF-1 regulation of the wandering macrophage: complexity in action. Trends Cell Biol. 14:628-638. [DOI] [PubMed] [Google Scholar]
- 30.Revenu, C., R. Athman, S. Robine, and D. Louvard. 2004. The co-workers of actin filaments: from cell structures to signals. Nat. Rev. Mol. Cell Biol. 5:635-646. [DOI] [PubMed] [Google Scholar]
- 31.Ross, R., H. Jonuleit, M. Bros, X. L. Ross, S. Yamashiro, F. Matsumura, A. H. Enk, J. Knop, and A. B. Reske-Kunz. 2000. Expression of the actin-bundling protein fascin in cultured human dendritic cells correlates with dendritic morphology and cell differentiation. J. Investig. Dermatol. 115:658-663. [DOI] [PubMed] [Google Scholar]
- 32.Sadl, V., F. Jin, J. Yu, S. Cui, D. Holmyard, S. Quaggin, G. Barsh, and S. Cordes. 2002. The mouse Kreisler (Krml1/MafB) segmentation gene is required for differentiation of glomerular visceral epithelial cells. Dev. Biol. 249:16-29. [DOI] [PubMed] [Google Scholar]
- 33.Schirenbeck, A., T. Bretschneider, R. Arasada, M. Schleicher, and J. Faix. 2005. The Diaphanous-related formin dDia2 is required for the formation and maintenance of filopodia. Nat. Cell Biol. 7:619-625. [DOI] [PubMed] [Google Scholar]
- 34.Sieweke, M. H., H. Tekotte, J. Frampton, and T. Graf. 1996. MafB is an interaction partner and repressor of Ets-1 that inhibits erythroid differentiation. Cell 85:49-60. [DOI] [PubMed] [Google Scholar]
- 35.Svitkina, T. M., E. A. Bulanova, O. Y. Chaga, D. M. Vignjevic, S. Kojima, J. M. Vasiliev, and G. G. Borisy. 2003. Mechanism of filopodia initiation by reorganization of a dendritic network. J. Cell Biol. 160:409-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yayoshi-Yamamoto, S., I. Taniuchi, and T. Watanabe. 2000. FRL, a novel formin-related protein, binds to Rac and regulates cell motility and survival of macrophages. Mol. Cell. Biol. 20:6872-6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yoshida, T., T. Ohkumo, S. Ishibashi, and K. Yasuda. 2005. The 5′-AT-rich half-site of Maf recognition element: a functional target for bZIP transcription factor Maf. Nucleic Acids Res. 33:3465-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]