Abstract
The P19 protein of Tomato bushy stunt virus is a potent suppressor of RNA silencing and, depending on the host species, is required for short- and long-distance virus movement and symptom production. P19 interacts with plant ALY proteins and relocalizes a subset of these proteins from the nucleus to the cytoplasm. Here we showed that coexpression by agroinfiltration in Nicotiana benthamiana of P19 and the subset of ALY proteins that are not relocalized from the nucleus interfered with the ability of P19 to suppress RNA silencing. We demonstrated that this interference correlates with the relocation of P19 from the cytoplasm into the nucleus, and by constructing and analyzing chimeric ALY genes, we showed that the C-terminal part of the central, RNA recognition motif of ALY is responsible for interaction with P19, relocalization or nonrelocalization of ALY, and inhibition of silencing suppression by P19. We studied the interaction of ALY and P19 by using the technique of bimolecular fluorescence complementation to show that these proteins associate physically in the nucleus but not detectably in the cytoplasm, and we present a model to explain the dynamics of this interaction.
The Tomato bushy stunt virus (TBSV) P19 is one of a class of plant and animal virus proteins that are known to interfere with a host defense mechanism, referred to as RNA silencing, that targets foreign (virus and retrotransposon) RNAs in a sequence-specific manner for degradation. RNA silencing is triggered by double-stranded RNA (dsRNA) that may be formed during virus replication or by secondary structure in viral single-stranded RNA (11). In plants, the dsRNA is cleaved by (one or more) Dicer-like (DCL) proteins that possess an RNase III domain to produce short (21- to 25-nucleotide) dsRNA molecules known as small interfering RNAs (siRNAs) (26). The siRNA duplex is unwound, and one strand becomes incorporated into the RNA-induced silencing complex (RISC) where it guides the complex by base pairing with the complementary (viral) target RNA (1). Subsequently, an RNase activity of RISC cleaves the target RNA, leading to its further degradation. The silencing suppression activity of P19 has been studied in some detail and involves the direct binding of ds siRNA to a P19 dimer, preventing incorporation of siRNA into RISC (5, 10, 24, 28). It is not known how P19 comes into contact with siRNAs and whether P19 is recycled during this process. However, for this mechanism to be effective, P19 must be expressed early in virus infection and to high levels so as to inhibit the RNA silencing process before the pool of siRNA, which is itself amplified by host-encoded RNA-dependent RNA polymerase, becomes too large (14, 16, 19). RNA silencing spreads through the vascular tissue from the place of initiation to distant parts of the plant, and sequestration of siRNAs by P19 may block the spread of a systemic silencing signal (10).
In the context of virus infection, P19, like some other plant virus-encoded silencing suppressors, apparently is responsible for several different phenotypes. TBSV causes lethal systemic necrosis in Nicotiana benthamiana at about 2 weeks after infection. Virus in which the P19 gene was mutated to prevent its expression moved systemically in these plants but did not induce necrosis (17). Other experiments showed that whereas viral RNA of a P19 mutant initially accumulated to wild-type levels in systemic N. benthamiana leaves, after about 7 days the mutant RNA levels began to fall and were only about 1/200 of the level of wild-type TBSV by 14 days postinoculation (15). In situ immunohistochemical analysis of N. benthamiana infected with wild-type and P19 mutant Cymbidium ringspot virus, which is related to TBSV, revealed that the mutant virus moved into systemic leaves but was confined to the vascular tissue and only a few cells of the surrounding mesophyll tissue (8). In spinach, the P19 mutant replicated and spread in inoculated leaves but did not move into systemic leaves (17), whereas in pepper and cowpea, the P19 mutant was unable to move even a short distance in the inoculated leaf (22). In N. tabacum, TBSV induced a hypersensitive response and the formation of necrotic lesions on the inoculated leaf, which were replaced by large, spreading chlorotic lesions in the presence of the TBSV P19 mutant (18). Whether these behaviors all resulted from the failure of the P19 mutant to inhibit RNA silencing is not known.
Although the current model for P19 silencing suppression activity proposes that it functions in isolation by direct interaction with siRNAs, we and others previously found that P19 also interacts with members of the ALY family of plant RNA-binding proteins (13, 23). Animal (mouse, Drosophila, and human) and yeast ALY proteins are known to be involved in the export of mRNAs from the nucleus prior to translation and are incorporated into the exon junction complex of proteins that mark mRNAs during splicing (7, 29). In addition, animal ALY is also known to be a transcriptional coactivator that possibly functions as a chaperone to promote the interaction of DNA-binding proteins (25). Animals encode only one or two ALY proteins, whereas Arabidopsis thaliana encodes four ALYs and there are four or more ALYs in many monocots (23). The function of the plant ALYs is not known, although in one study, two of the Arabidopsis ALY proteins were found to interact in yeast cells with the DNA-binding domain of poly(ADP-ribose) polymerase (21). Evidence in plants from cell fractionation and green fluorescent protein (GFP) tagging suggested that P19 is a cytoplasmic protein (17), whereas the GFP-tagged ALY proteins are nuclear (21, 23). However, animal ALY proteins are known to shuttle between the nucleus and cytoplasm, and we hypothesized that P19 interacts with ALY in the cytoplasm (23). Furthermore, we showed that as a result of this interaction, two of the four Arabidopsis ALYs and one of two N. benthamiana ALYs were retained in the cytoplasm in the presence of P19.
We have extended our study of the P19-ALY interaction to demonstrate a previously undetected influence of ALY expression on P19 silencing suppression activity. This effect correlates with changes to the subcellular distribution of P19, which is itself directed by interaction with a specific domain of ALY.
MATERIALS AND METHODS
Cloning of additional N. benthamiana ALY genes.
Previously two ALY genes (ALY615 and ALY617) were identified and cloned from N. benthamiana (23) and a partial ALY gene, referred to as Hin19, was cloned from Nicotiana tabacum (13). BLAST searches of The Institute for Genomic Research (Rockville, MD) gene indices (http://www.tigr.org/tigr-scripts/tgi/T_reports.cgi?species=N.benthamiana) identified two additional N. benthamiana ALY homologues (TC1693 and TC916). PCR amplification of cDNA synthesized from total RNA isolated from leaves of N. benthamiana was carried out using primers 871 (5′-AAACATGTCAAATCTCGATGTATCCCTCG-3′) and 828 (5′-TTTCAGTTAGTCTCCATTGCTTCTGAATG-3′) to clone NbALY1693 and primers 833 (5′-AACCATGGCTTCCTTGGATATGACCCTCG-3′) and 834 (5′-TTTTAGGTCTGCATAGCATCTGCAGT-3′) to clone NbALY916.
Construction and expression of ALY and P19 proteins in plants.
The Arabidopsis and N. benthamiana ALY genes and TBSV P19 were cloned by restriction fragment ligation into the binary vector pROK2 to allow expression in plants. These genes were also cloned in pROK2, in sequential or three-fragment ligations with the green fluorescent protein (GFP) gene and, for P19, also with the monomeric red fluorescent protein (mRFP) gene (3), to allow expression in plants of the ALY and P19 proteins each with a C-terminal GFP or mRFP extension. The ALY and P19 genes were also cloned into the gateway entry vectors pDONR201 and pDONR207 (Invitrogen) to allow their subsequent mobilization by recombination into vectors pLX222, pBAT-TL-K-GFP, pBAT-TL-K-sRFP-N, and pBAT-TL-K-sRFP-C for expression in plants as either unfused proteins or proteins fused at the C terminus with GFP or fused at the C terminus with N- or C-truncated mRFP for bimolecular fluorescence complementation, respectively (2). Plasmids were transformed into Agrobacterium tumefaciens strains LBA4404 and C58C1 for delivery into N. benthamiana plants by agroinfiltration (27). 4′,6′-Diamidino-2-phenylindole (DAPI) staining of nuclei was achieved by infiltration of a 1-μg ml−1 aqueous solution as described previously (6).
Imaging of fluorescent proteins.
All imaging was conducted using either a Leica TCS-SP1 confocal laser-scanning microscope (CLSM) or a Leica TCS-SP2 acoustical optical beam splitter (AOBS) (Leica Microsystems Heidelberg GmbH, Germany). All images were obtained by sequential scanning, either frame by frame (SP1) or line by line (SP2) using the following excitation (EX) and emission (EM) settings: for GFP, EX of 488 nm and EM of 500 to 530 nm; for DsRed and mRFP, EX of 568 nm and EM of 580 to 600 nm (SP1) or EX of 561 nm and EM of 590 to 630 nm (SP2); for chlorophyll-associated autofluorescence, EX of 488 nm and EM of 660 to 700 nm; and for DAPI, EX of 405 nm and EM of 440 to 475 nm.
Silencing assays.
N. benthamiana plants were infiltrated with a culture of Agrobacterium containing the binary plasmid pGPTV(+35,+NosT) expressing β-glucuronidase (GUS) (4) either alone or mixed with other cultures expressing the ALY proteins, either with or without a GFP tag, or P19 either untagged or with GFP or mRFP tags. Where possible, expression of these proteins was confirmed by confocal fluorescence microscopy. Infiltrated tissue was collected after 6 days, and RNA samples were extracted, processed for mRNA and siRNA extraction, and analyzed by Northern blotting using digoxigenin-labeled GUS-specific probes as described before (4).
Arabidopsis ALY chimeras.
For the synthesis of chimeras between AtALY2 and AtALY3, the ALY protein was divided into three domains: an N-terminal domain (amino acids [aa] 1 to 100); a central, RNA recognition motif (RRM) domain (aa 101 to 195 of AtALY2; aa 101 to 191 of AtALY3); and a C-terminal domain (aa 196 to 292 of AtALY2; aa 192 to 293 of AtALY3). Each gene was modified by PCR to introduce a SpeI restriction site between the N-terminal and central domains and a NgoMIV restriction site between the central and C-terminal domains. Thus, for AtALY2, aa 101 and 102, Gly (GGC) and Ser (TCG) became Thr (ACT) and Ser (AGT), and aa 196 and 197, Ala (GCT) and Thr (ACA) became Ala (GCC) and Gly (GGC). For AtALY3, aa 100 and 101, Val (GTG) and Ser (TCA) became Thr (ACT) and Ser (AGT), and aa 192 and 193, Ala (GCA) and Ala (GCT) became Ala (GCC) and Gly (GGC). Using these new restriction sites and existing, unique sites (HpaI and XbaI) located upstream or downstream, respectively, of the ALY genes, AtALY2 and AtALY3 chimeras were assembled carrying all eight possible combinations of the three domains of the ALY protein. The chimeras were named according to their construction; thus, chimera 2-2-2 carries all three domains derived from AtALY2, whereas, for example, chimera 2-3-2 carries the N- and C-terminal domains from AtALY2 together with the central domain from AtALY3. The chimeras were cloned into either Tobacco rattle virus or pROK2 as described before (23) for expression in plants and analysis by confocal microscopy.
To create chimeras of the central domain, an EagI site (CGGCCG) was introduced at nucleotides 432 to 437 (spanning aa 144 to 146) in the AtALY2 gene and nucleotides 429 to 434 in the AtALY3 gene (spanning aa 143 to 145). These nucleotide changes did not alter the amino acid sequences of the ALY proteins but separated the N-terminal part of the RRM domain, including the highly conserved RNP2 motif, from the C-terminal part containing the RNP1 motif. The SpeI-EagI and EagI-NgoMIV fragments of the central domain were assembled with the N- and C-terminal domains of AtALY3 to construct two chimeras. Chimera 3-23-3 has the N-terminal part of the central domain from AtALY2 and the C-terminal part of the central domain from AtALY3, whereas chimera 3-32-3 has the N-terminal part of the central domain from AtALY3 and the C-terminal part of the central domain from AtALY2. The chimeras were cloned into pROK2 for analysis in plants.
Nucleotide sequence accession numbers.
The sequences of two N. benthamiana ALY genes were deposited with the EMBL/GenBank data libraries (NbALY1693, accession number AM167905; NbALY916, accession number AM167906).
RESULTS
Effect of ALY expression on P19 suppressor function.
In our earlier work we did not observe any link between the interaction of P19 protein with ALY protein and the known function of P19 as a silencing suppressor protein (23). As we had cloned only two ALY genes (ALY617 and ALY615) from N. benthamiana, the plant species that was used for the virus-induced gene silencing and agroinfiltration experiments, we suggested that examination of additional genes would be useful. Consequently, for this work we have cloned two more N. benthamiana ALY genes (ALY1693 and ALY916). ALY916 is the orthologue of N. tabacum Hin19 (13) and is 275 aa long, whereas ALY1693 is 246 aa long. Sequence comparison of the four N. benthamiana ALY proteins shows that ALY615 and ALY1693 are 93% identical, ALY615 and ALY617 are 57% identical, ALY615 and ALY916 are 38% identical, ALY617 and ALY916 are 41% identical, ALY617 and ALY1693 are 39% identical, and ALY916 and ALY1693 are 25% identical.
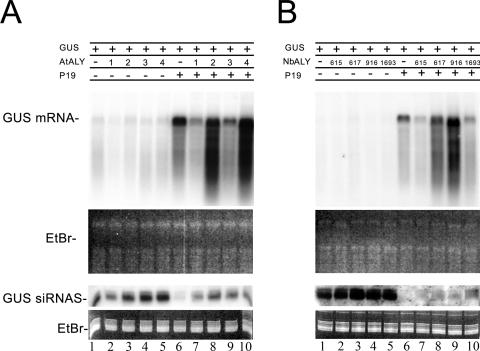
To test whether any of the ALY proteins could affect silencing suppression by P19, we used agroinfiltration of leaves to transiently express the P19 protein together with or without each of the four Arabidopsis or four N. benthamiana ALY proteins, and examined whether P19 was able to suppress silencing of a coexpressed marker gene (β-glucuronidase). As expected, accumulation of the GUS mRNA increased when coexpressed with P19 due to the silencing suppressor activity of P19 (Fig. 1A and B, lane 6 versus lane 1). Without P19, transient expression of each of the ALY genes had no effect on the level of GUS mRNA accumulation (Fig. 1A and B, lanes 2 to 5 versus lane 1). However, when both ALY and P19 were expressed simultaneously, four of the ALYs (AtALY1, AtALY3, NbALY615, and NbALY1693) partially inhibited the suppressor activity of P19, resulting in reduced levels of GUS mRNA accumulation compared to that found with P19 alone (Fig. 1A, lanes 7 and 9 versus lane 6; Fig. 1B, lanes 7 and 10 versus lane 6), whereas the other four ALYs (AtALY2, AtALY4, NbALY617, and NbALY916) did not reduce the level of accumulated GUS mRNA (Fig. 1A, lane 8 and 10 versus lane 6; Fig. 1B, lanes 8 and 9 versus lane 6). This result was obtained consistently when coexpressing native ALY and P19 (six independent experiments), ALY-GFP and P19-mRFP (two experiments), and native ALY with P19-mRFP (one experiment).
FIG. 1.
Expression of a subset of ALY proteins inhibits P19 silencing suppressor function. Northern blot analysis of RNA extracted from plants probed for GUS. The blots show GUS mRNA and GUS siRNAs, with ethidium bromide (EtBr) staining as a loading control. All plants infiltrated with a mixture of three Agrobacterium cultures containing binary vectors expressing the GUS gene, together with empty binary vector only (lane 1), empty binary vector plus ALY (lanes 2 to 5), P19 plus empty binary vector (lane 6), and P19 plus ALY (lanes 7 to 10). (A) Arabidopsis ALYs; (B) N. benthamiana ALYs. The presence (+) or absence (−) of GUS, P19, and ALY proteins (AtALY1 to AtALY4 and NbALY615, NbALY617, NbALY916, and NbALY1693) is shown above the blots.
Fusion of GFP to P19 affects its function.
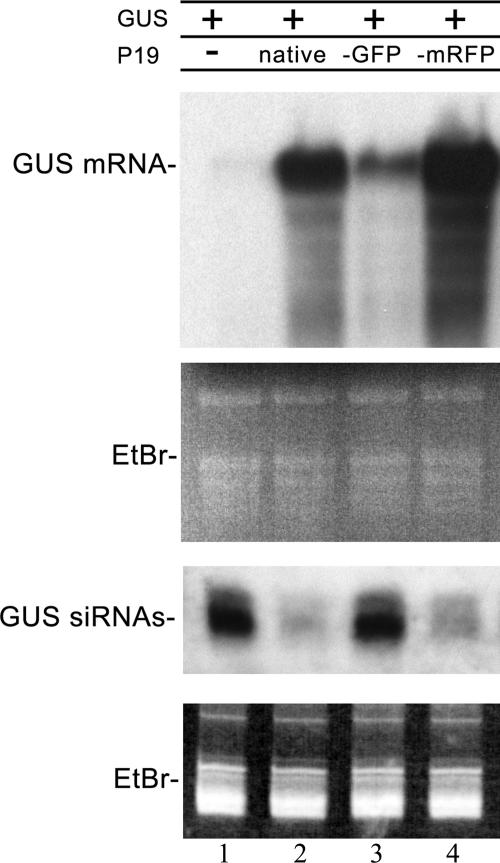
Our previous studies, focusing on the subcellular distribution of ALY, have all used native P19; however, to examine in greater detail the interaction between P19 and the different ALY proteins, we decided to tag P19 at the C terminus by fusion either to GFP or mRFP. To assess whether tagging compromised the activity of P19, we examined whether P19-GFP and P19-mRFP retained the ability to suppress RNA silencing. Plants were agroinfiltrated with a binary plasmid encoding GUS, together with P19, either untagged, GFP tagged or mRFP tagged, or with an empty binary plasmid as a control. Initially, GUS mRNA accumulated, but after 3 or 4 days, it began to decline in amount as RNA silencing was initiated, and GUS-specific siRNAs accumulated instead. In the presence of native P19, full-length GUS mRNA was not degraded and siRNAs accumulated only to a low level (Fig. 2, lane 2). However, Western blot experiments showed that P19-GFP accumulated poorly in infiltrated tissue (data not shown), and there was a significant impairment in the suppression of silencing of GUS, leading to the degradation of GUS mRNA and accumulation of GUS-derived siRNAs (Fig. 2, lane 3). P19-mRFP accumulated to the same level as native P19 in infiltrated tissue (data not shown) and was an effective suppressor of silencing, allowing full-length GUS mRNA to accumulate (Fig. 2, lane 4). Consequently, all other experiments included mRFP-tagged P19, rather than GFP-tagged P19.
FIG. 2.
Tagging of P19 with mRFP does not alter its silencing suppression activity. Northern blots of GUS mRNA and GUS siRNAs extracted from plants infiltrated with the GUS gene together with the empty binary vector (lane 1), native P19 (lane 2), P19-GFP (lane 3), and P19-mRFP (lane 4). RNA loading controls are stained with ethidium bromide (EtBr).
Subcellular relocalization of P19 by ALY.
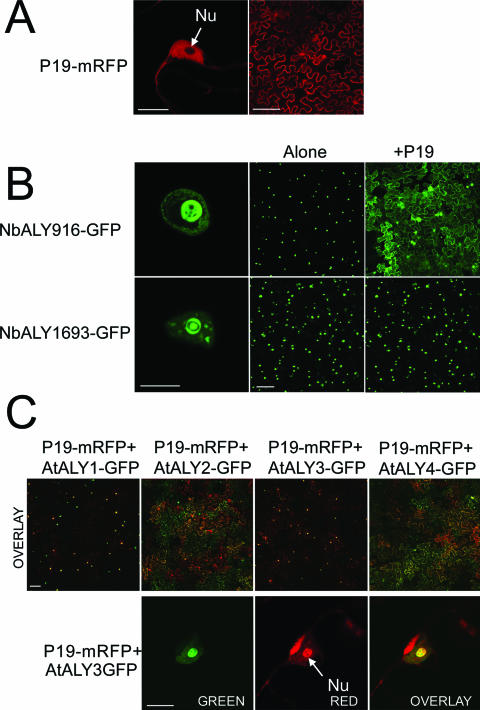
P19 is considered primarily to be a cytoplasmic protein (17), although some P19-GFP was also found to be present in the nucleus (but not the nucleolus) of agroinfiltrated plants (23) in a pattern similar to the distribution of free (unfused) GFP. In this work, fluorescence derived from P19-mRFP had the same distribution (Fig. 3A). We showed previously that when coexpressed with native P19, three of the ALY-GFP proteins (AtALY2, AtALY4, and NbALY617) were localized to the cytoplasm, whereas the remaining three ALY-GFP proteins (AtALY1, AtALY3, and NbALY615) were localized to the nucleus (23). The two newly identified N. benthamiana ALY proteins, NbALY916 and NbALY1693, were both found to be nuclear (Fig. 3B, left panels).
FIG. 3.
(A) Fluorescence derived from the suppressor P19-mRFP is mostly cytoplasmic (the right panel shows a view of a field of N. benthamiana epidermal cells) and nuclear but not nucleolar (the left panel shows a view of a nucleus). (B) Fluorescence from the two newly identified N. benthamiana ALY proteins tagged with GFP (NbALY916-GFP and NbALY1693-GFP) accumulated in the nucleus (the left and central panels show views of individual nuclei and of fields of epidermal cells, respectively). However, in the presence of the suppressor P19, NbALY916-GFP relocated to the cytoplasm, whereas NbALY1693-GFP remained nuclear (right panels). (C) Fluorescence from the suppressor P19-mRFP accumulated in the nucleus/nucleolus when coexpressed with ALY-GFPs that remain nuclear in its presence (top row, panels with AtALY1 and AtALY3). P19-mRFP remained mainly cytoplasmic when coexpressed with ALY-GFPs that delocalize to the cytoplasm in the presence of the suppressor (top row, AtALY2 and AtALY4). The images show mRFP- and GFP-derived fluorescence as red and green, respectively. In places where both fluorescences are present, the resulting color is yellow-orange. The panels in the bottom row show a nucleus from an epidermal cell in which both AtALY3-GFP and P19-mRFP are being expressed. The left and central panels show GFP- and mRFP-derived fluorescence, respectively. The rightmost panel is an overlay of both images. Fluorescence from P19-mRFP accumulates strongly in the nucleolus in the presence of AtALY3-GFP. The bars are 10 μm for nucleus panels and 100 μm for field panels. Nu, nucleolus.
However, NbALY916-GFP was relocalized to the cytoplasm by P19, whereas NbALY1693-GFP was retained in the nucleus (Fig. 3B, right panels). The distribution patterns of the ALY-GFP proteins remained consistent when P19-mRFP was substituted for P19 (data not shown).
An unexpected observation in these experiments was that an alteration in the subcellular distribution pattern of P19-mRFP also occurred. When coexpressed with those ALY-GFP proteins that were delocalized to the cytoplasm, P19-mRFP was also localized mostly to the cytoplasm (Fig. 3C and data not shown). In contrast, a significant proportion of P19-mRFP accumulated inside the nucleus, and specifically in the nucleolus, when coexpressed with those ALY-GFP proteins that themselves remained nuclear/nucleolar (Fig. 3C and data not shown). The same pattern of P19 distribution occurred when untagged ALY rather than ALY-GFP was included in the infiltrations (data not shown). A summary of the localization patterns of each of the ALY proteins and P19 is shown in Table 1.
TABLE 1.
Subcellular locations of ALY-GFP and P19-mRFP in infiltrated leaves
| Protein | Subcellular location of ALY-GFPa
|
Subcellular location of P19-mRFP in the presence of ALYa | |
|---|---|---|---|
| In the absence of P19 | In the presence of P19 | ||
| AtALY1 | N, Nu | N, Nu | N, Nu |
| AtALY2 | N | Cyt | Cyt |
| AtALY3 | N, Nu | N, Nu | N, Nu |
| AtALY4 | N, Nu | Cyt | Cyt |
| NbALY615 | N, Nu | N, Nu | N, Nu |
| NbALY617 | N, Nu | Cyt | Cyt |
| NbALY916 | N, Nu | Cyt | Cyt |
| NbALY1693 | N, Nu | N, Nu | N, Nu |
The predominant localization is shown. N, nuclear; Nu, nucleolar; Cyt, cytoplasmic.
Mapping of ALY domains involved in nucleolar targeting and delocalization induced by P19.
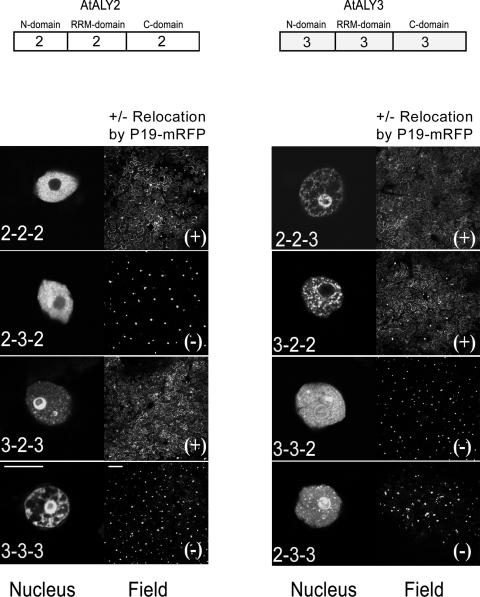
In order to map the ALY domains involved in nucleolar targeting and the nucleus-to-cytoplasm delocalization induced by P19, chimeras were made between two GFP-tagged A. thaliana ALY proteins, AtALY2, which does not accumulate in the nucleolus and is relocated to the cytoplasm in the presence of P19 (or P19-mRFP), and AtALY3, which accumulates in the nucleolus and remains there in the presence of P19 (or P19-mRFP). For synthesis of the chimeras, the ALY protein was divided into three domains: the two N- and C-terminal domains containing variable, nonconserved sequences and a more conserved central domain containing the RNA-binding motif (RRM) (Fig. 4). Eight different GFP-tagged chimeras, resulting from all the possible permutations between the three domains of these two ALYs were analyzed by infiltration in N. benthamiana plants either with or without P19, followed by confocal microscopy.
FIG. 4.
Chimeras between AtALY2 and AtALY3 reveal domains required for relocalization by P19 and nucleolar localization. The left panel of each pair shows the nuclear distribution of GFP-tagged ALY chimeras expressed in the absence of P19. The right panel of each pair shows a field view of GFP-derived fluorescence from coexpression of ALY-GFP and P19-mRFP. The chimera name denotes the origin of the three domains (N-terminal, RRM, and C-terminal domains in that order). The relocation of ALY-GFP to the cytoplasm by coexpression with P19-mRFP (+) and the absence of relocalization (−) are indicated. The bars are 10 μm for the nucleus panel and 100 μm for the field panel.
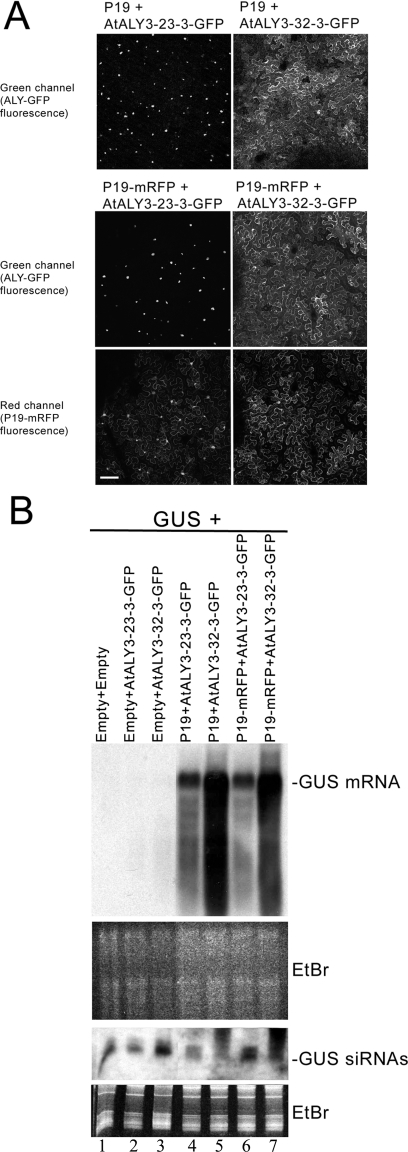
While all the chimeras were nuclear when expressed alone, some displayed the nucleus-to-cytoplasm delocalization phenotype when coexpressed with P19. This phenotype was determined solely by the presence of the central RRM domain of AtALY2 (Fig. 4, field panels). Further mapping of the delocalization motif was made using additional, intra-RRM chimeras, revealing that the AtALY3 chimera carrying the C-terminal RRM fragment from AtALY2 (ALY3-32-3-GFP) relocated to the cytoplasm in the presence of P19 (or P19-mRFP), whereas the AtALY3 chimera carrying the N-terminal RRM fragment from AtALY2 (ALY3-23-3-GFP) remained nuclear (Fig. 5A). Thus, the C-terminal half of the AtALY2 RRM domain was solely responsible for the delocalization phenotype induced by P19.
FIG. 5.
(A) The localization of ALY to the nucleus or cytoplasm in the presence of P19 maps to the C-terminal part of the central RRM motif. An AtALY3 chimera carrying the N-terminal RRM portion from AtALY2 (ALY3-23-3-GFP) remained nuclear in the presence of P19 or P19-mRFP (top and middle left panels). In contrast, an AtALY3 chimera carrying the C-terminal RRM fragment from AtAL2 (ALY3-32-3-GFP) relocalized to the cytoplasm in the presence of either P19 or P19-mRFP (top and middle right panels). Bar, 100 μm. (B) The subcellular location of ALY affects the suppressor activity of P19. Northern blot analysis of RNAs extracted from plants probed for GUS coinfiltrated with the intra-RRM chimeras shown in panel A. The blots show GUS mRNA and GUS siRNAs, with ethidium bromide (EtBr) staining as a loading control. All plants infiltrated with a mixture of Agrobacterium cultures containing the GUS gene together with empty binary vector alone (lane 1) or empty binary vector plus AtALY3-23-3-GFP (lanes 2, 4, and 6) or AtALY3-32-3-GFP (lanes 3, 5, and 7). Lanes 1 to 3, no P19; lanes 4 and 5, P19, lanes 6 and 7, P19-mRFP. The suppressor activity of P19 decreased in the presence of the chimera (AtALY3-23-3-GFP) that remained nuclear (lanes 4 and 6 versus lanes 5 and 7).
Nucleolar targeting of AtALY3 did not map to a single domain. While the presence of the C-terminal domain from AtALY3 in a chimera (e.g., AtALY2-2-3-GFP) was sufficient for it to be targeted to the nucleolus, replacing the C-terminal domain of AtALY3 with that of AtALY2 (ALY3-3-2-GFP) did not prevent accumulation of this protein in the nucleolus (Fig. 4, nucleus panels). Replacing both the RRM and C-terminal domains (AtALY3-2-2-GFP) did prevent nucleolar accumulation. In addition, chimera AtALY2-3-2-GFP displayed a mixed phenotype, with nonnucleolar targeting in the majority of epidermal nuclei (Fig. 4, nucleus panel), but some nucleolar targeting in the nuclei of occasional epidermal cells was also observed (data not shown).
Effects of ALY RRM domain swaps on P19 suppressor function.
The experiments described above showed that the four Arabidopsis and N. benthamiana ALY proteins that reduced P19 suppressor function in an agroinfiltration assay were those that remained nuclear during coexpression experiments with P19 (this work and earlier work [23]) and that relocated the P19 protein to the nucleus (AtALY1, AtALY3, NbALY615, and NbALY1693). Conversely, those ALY proteins that relocalized to the cytoplasm together with P19 (AtALY2, AtALY4, NbALY617, and NbALY916) did not interfere with P19 suppressor activity. These data show that the accumulation of P19 in the nucleus mediated by ALY had an inhibitory effect on silencing suppression by P19. To dissect the roles of the different ALY domains on the inhibition of P19 suppressor function, the AtALY2 and AtALY3 RRM domain chimeras described above were analyzed in the GUS mRNA silencing/suppression assay.
As was shown before, AtALY3-GFP, which remains nuclear in the presence of P19 or P19-mRFP, reduced the suppressor activity of P19 (Fig. 1A, lane 9 versus lane 6). Similarly, the AtALY3 RRM chimera carrying the N-terminal RRM fragment from AtALY2 (ALY3-23-3-GFP), which also remains nuclear in the presence of P19 or P19-mRFP, reduced the activity of the suppressor (Fig. 5B, lanes 4 and 6 versus lanes 5 and 7). However, the chimera carrying the C-terminal RRM fragment from AtALY2 (ALY3-32-3-GFP), which relocalizes to the cytoplasm, did not have an inhibitory effect on the suppression function (Fig. 5B, lanes 5 and 7 versus lanes 4 and 6). These results demonstrate that the effect of ALY on the suppressor activity of P19 is linked to the nuclear versus cytoplasmic localization phenotype that maps to the C-terminal domain of the central RRM motif of the ALY protein.
Localization of the site of P19-ALY interaction.
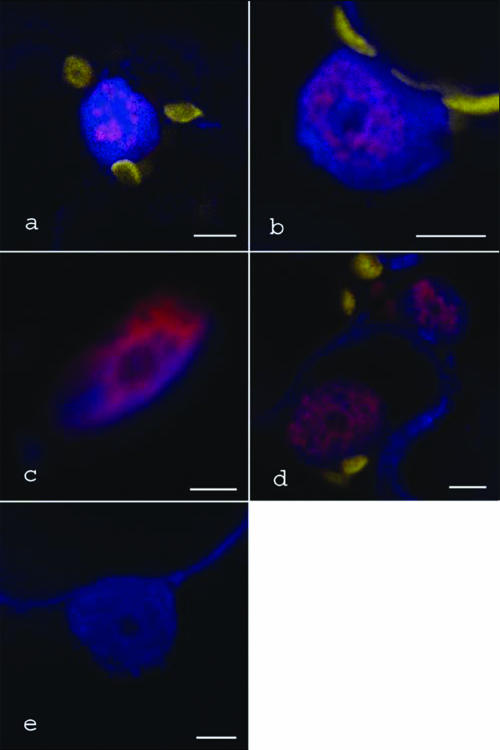
The results so far revealed that in some combinations P19 and ALY both localized to the nucleus. To confirm that the colocalization reflected actual physical interaction between these proteins, we used the technique of bimolecular fluorescence complementation (BiFC) whereby one of the proteins under study is fused to a nonfunctional truncated fluorescent reporter, while the second assay protein is fused to a complementing fragment of the same reporter (9). Our studies examined plant cells coinfiltrated to express P19 fused at the C terminus to an N-terminal fragment of mRFP and each of the four Arabidopsis ALY proteins fused at the C terminus to a C-terminal fragment of mRFP. For P19 with AtALY1, AtALY2, or AtALY4, we observed diffuse patterns of mRFP fluorescence in the nuclei of cells, the location of which was confirmed by counterstaining with DAPI (Fig. 6a to d). Cells coexpressing the P19 and AtALY3 BiFC constructs appeared to show mRFP fluorescence in nuclei, but these nuclei were distorted in shape and difficult to counterstain with DAPI. Nevertheless, these results suggest that P19 and all four Arabidopsis ALY proteins interact in the nucleus but not, to any detectable extent, in the cytoplasm. As a control, plants were infiltrated with BiFC constructs of P19 and an Arabidopsis 19S proteosome subunit protein (At1g29150), which is located in the nucleus and cytoplasm and is functional in other, unrelated BiFC experiments (data not shown). No mRFP fluorescence was detected with this combination of proteins (Fig. 6e).
FIG. 6.
BiFC images of ALY-P19 interaction in nuclei. Nuclei are DAPI stained and appear blue, and chloroplasts appear yellow. Pink areas show interaction between the split-mRFP-tagged ALY and P19 proteins. (a) AtALY1; (b) AtALY2; (c) AtALY3; (d) AtALY4; (e) lack of mRFP complementation between P19 and an Arabidopsis proteosome subunit protein. Bars, 5 μm.
DISCUSSION
The TBSV P19 protein has a well-studied function as a suppressor of the plant RNA silencing pathway and is, thus, a key determinant of virus pathogenicity. Depending on the host plant species, defects in P19 expression result in impairment of TBSV short-distance or long-distance movement together with a reduction in disease symptom production. On the basis of its ability to bind ds siRNA molecules, it has been proposed that P19 fulfils its silencing suppression function by sequestration of siRNAs, which prevents their incorporation in the RISC complex and inhibits the spread of a systemic silencing signal (10, 24, 28). However, it is not well understood what might be the molecular functions of P19 in its diverse roles as a pathogenicity determinant, whether or how these functions are related to its siRNA binding and silencing suppression activity, and whether additional host factors are involved.
P19 has been shown previously to interact specifically with a family of plant ALY proteins (13, 23). Here we provide evidence that the interaction with particular ALY proteins alters the subcellular localization of P19 and has an impact on its silencing suppression activity.
Antiviral RNA silencing is thought to occur in the cytoplasm, and this correlates well with the proposed role of P19 as a scavenger of siRNAs in the cytoplasm (20). We, and others found that GFP-tagged P19 also is predominantly a cytoplasmic protein (12, 23), and Western blot analysis of extracts of plants infected by TBSV detected the majority of P19 protein in the soluble (cytoplasmic, S30) protein fraction (17).
We found that in infiltration experiments tagging of P19 with mRFP did not inhibit its suppressor activity, making it a suitable reporter for subsequent studies.
While it was shown previously that some of the ALY proteins are relocalized from the nucleus to the cytoplasm by the presence of P19 (23), we have now found that conversely, the other members of the ALY protein family change the intracellular localization of the P19 protein. When P19-mRFP was expressed by itself, it localized predominantly to the cytoplasm but was also found in the nucleus. However, P19-mRFP did not localize to the nucleolus. When P19-mRFP was coexpressed with the Arabidopsis ALY protein AtALY1 or AtALY3 or with the N. benthamiana ALY protein NbALY615 or NbALY1693, P19-mRFP was located mostly in the nucleus and particularly in the nucleolus.
In these coexpression studies, silencing suppression activity of P19 was significantly decreased, indicating that interaction with these particular ALY proteins in conjunction with a delocalization of P19 to the nucleus/nucleolus interferes with P19 function. Although a direct involvement of ALY RNA export factors in the RNA silencing process has not been demonstrated so far, the effects of some of the ALYs on P19 function hint at a specific role of these proteins, possibly in the context of a plant defense strategy counteracting the viral suppression of silencing. In a TBSV-infected plant, the precise pattern of P19 intracellular distribution probably would depend on the relative expression levels of the different ALY proteins in a particular cell or tissue type, and this might influence the suppression activity of P19. Indeed, it was found previously that targeting P19 to the nucleus by introducing a nuclear localization signal into the protein altered the accumulation of siRNAs and of miRNAs, which are themselves processed in the nucleus (12). Northern blot analysis showed that all of the ALY genes are expressed in leaves, whereas analysis of Arabidopsis ALY promoters using transcriptional fusions with GUS showed that there are differences in the expression level and tissue distribution of the ALY mRNAs (data not shown). Thus, we suggest the interaction of P19 with ALY could indeed have different consequences for suppression of silencing in different parts of the plant. Attempts to silence expression of each of the ALY genes in N. benthamiana using transient expression of inverted-repeat constructs were not successful, and because Arabidopsis thaliana is not a host for TBSV infection, we are not able to investigate ALY mutants in this plant. Consequently, direct evidence of the biological role of ALY in P19 function has not yet been obtained.
By constructing chimeras between two Arabidopsis ALY proteins with different abilities to delocalize P19, we demonstrated that the C-terminal part of the ALY RRM domain influences whether the ALY protein is relocalized in the presence of P19 from the nucleus to the cytoplasm, whether the P19 protein similarly is localized to the cytoplasm or nucleus, and whether the P19 protein efficiently suppresses RNA silencing. The RRM domain is conserved in all ALY proteins and has been shown previously to be necessary and sufficient for the P19-ALY interaction (23). It remains to be shown whether the protein interactions and RNA-binding capacities of the ALY RRM domain are directly relevant for P19 siRNA binding and silencing suppression activity or whether the ALY proteins affect P19 function primarily by altering it subcellular location.
When we used the BiFC technique to examine the physical interaction between P19 and the Arabidopsis ALY proteins in vivo, some surprising results were obtained. The BiFC interaction signal for P19 and AtALY1 or AtALY3 was located in the nucleus and nucleolus, which correlates with the location of these proteins when individually tagged and coexpressed. However, the BiFC interaction signal for P19 and AtALY2 or AtALY4 was also located in the nucleus, even though these proteins individually are localized to the cytoplasm when coexpressed. Our previous model of the ALY-P19 interaction proposed that for AtALY2 and AtALY4, these proteins interacted with P19 in the cytoplasm, which prevented their shuttling back into the nucleus. The BiFC data do not support this model but suggest a more complex pattern of movement.
Because we did not detect ALY-P19 interaction in the cytoplasm, we suggest that on formation this complex is moved very rapidly into the nucleus. Animal ALY proteins shuttle between the cytoplasm and nucleus, and we expect at least some of the plant ALYs to behave in the same way. We hypothesize that for P19 with the nonrelocalized ALYs (AtALY1, AtALY3, NbALY615, and NbALY1693), the complex cannot, because of physical masking or modification by P19, engage with the nuclear export pathway, and thus, each of the proteins remains in the nucleus. However, for P19 and AtALY2, AtALY4, NbALY617, or NbALY916, the complex is able to be reexported into the cytoplasm, perhaps involving dissociation of P19 and ALY. However, these ALYs must be modified during this process or incorporated into a different complex so that they cannot shuttle back into the nucleus. Perhaps the relocalizable (AtALY2 and AtALY4) and nonrelocalizable (AtALY1 and AtALY3) ALY proteins exist in different complexes in the nucleus and/or use different pathways to exit the nucleus, which could be affected differently by P19 binding. It may also be pertinent that P19 forms a homodimer when binding siRNAs and that ALY proteins also dimerize with themselves and with each other (data not shown). These interactions may complicate the ALY-P19 interaction that is seen in the BiFC data and may influence the partitioning of interacting complexes between the nucleus and cytoplasm.
We previously suggested that because the normal, nuclear distribution of the relocalizable ALY proteins was altered by P19, this subset of proteins is the “true” target for modification during TBSV infection. However, as we have now shown that the nonrelocalized ALY proteins sequester P19 in the nucleus and interfere with its silencing suppression activity, this subset of ALY proteins may also have an active role in modulating the virus pathogenicity.
Acknowledgments
The Scottish Crop Research Institute is supported by grants from the Scottish Executive Environment and Rural Affairs Department.
REFERENCES
- 1.Baumberger, N., and D. C. Baulcombe. 2005. Arabidopsis ARGONAUTE1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl. Acad. Sci. USA 102:11928-11933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bracha-Drori, K., K. Shichrur, A. Katz, M. Oliva, R. Angelovici, S. Yalovsky, and N. Ohad. 2004. Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J. 40:419-427. [DOI] [PubMed] [Google Scholar]
- 3.Campbell, R. E., O. Tour, A. E. Palmer, P. A. Steinbach, G. S. Baird, D. A. Zacharias, and R. Y. Tsien. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99:7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Canto, T., F. Cillo, and P. Palukaitis. 2002. Generation of siRNAs by T-DNA sequences does not require active transcription or homology to sequences in the plant. Mol. Plant-Microbe Interact. 15:1137-1146. [DOI] [PubMed] [Google Scholar]
- 5.Dunoyer, P., C. H. Lecellier, E. A. Parizotto, C. Himber, and O. Voinnet. 2004. Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. Plant Cell 16:1235-1250. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Fricker, M. D., A. Parsons, M. Tlalka, E. Blancaflor, S. Gilroy, A. J. Meyer, and C. Plieth. 2001. Fluorescent probes for living plant cells, p. 35-84. In C. Hawes and B. Satiat-Jeunemaitre (ed.), Plant cell biology. Oxford University Press, Oxford, United Kingdom.
- 7.Gatfield, D., and E. Izaurralde. 2002. REF1/Aly and the additional exon junction complex proteins are dispensable for nuclear mRNA export. J. Cell Biol. 159:579-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Havelda, Z., C. Hornyik, A. Crescenzi, and J. Burgyan. 2003. In situ characterization of Cymbidium ringspot tombusvirus infection-induced posttranscriptional gene silencing in Nicotiana benthamiana. J. Virol. 77:6082-6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu, C. D., and T. K. Kerppola. 2003. Simultaneous visualization of multiple protein interactions in living cells using multicolour fluorescence complementation analysis. Nat. Biotechnol. 21:539-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lakatos, L., G. Szittya, D. Silhavy, and J. Burgyan. 2004. Molecular mechanism of RNA silencing suppression mediated by P19 protein of tombusvirus. EMBO J. 23:876-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Molnar, A., T. Csorba, L. Lakatos, E. Varallyay, C. Lacomme, and J. Burgyan. 2005. Plant virus-derived small interfering RNAs originate predominantly from highly structured single-stranded viral RNAs. J. Virol. 79:7812-7818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Papp, I., M. F. Mette, W. Aufsatz, L. Daxinger, S. E. Schauer, A. Ray, J. van der Winden, M. Matzke, and A. J. M. Matzke. 2003. Evidence for nuclear processing of plant micro RNA and short interfering RNA precursors. Plant Physiol. 132:1382-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park, J. W., S. Faure-Rabasse, M. A. Robinson, B. Desvoyes, and H. B. Scholthof. 2004. The multifunctional plant viral suppressor of gene silencing P19 interacts with itself and an RNA binding host protein. Virology 323:49-58. [DOI] [PubMed] [Google Scholar]
- 14.Qiu, W., J. W. Park, and H. B. Scholthof. 2002. Tombusvirus P19-mediated suppression of virus-induced gene silencing is controlled by genetic and dosage features that influence pathenogenicity. Mol. Plant-Microbe Interact. 15:269-280. [DOI] [PubMed] [Google Scholar]
- 15.Qu, F., and T. J. Morris. 2002. Efficient infection of Nicotiana benthamiana by Tomato bushy stunt virus is facilitated by the coat protein and maintained by p19 through suppression of silencing. Mol. Plant-Microbe Interact. 15:193-202. [DOI] [PubMed] [Google Scholar]
- 16.Qu, F., X. Ye, G. Hou, S. Sato, T. E. Clemente, and T. J. Morris. 2005. RDR6 has a broad-spectrum but temperature-dependent antiviral defense role in Nicotiana benthamiana. J. Virol. 79:15209-15217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scholthof, H. B., K. B. G. Scholthof, M. Kikkert, and A. O. Jackson. 1995. Tomato bushy stunt virus spread is regulated by two nested genes that function in cell-to-cell movement and host-dependent systemic invasion. Virology 213:425-438. [DOI] [PubMed] [Google Scholar]
- 18.Scholthof, H. B., T. J. Morris, and A. O. Jackson. 1995. Identification of tomato bushy stunt virus host-specific symptom determinants by expression of individual genes from a potato virus X vector. Plant Cell 7:1157-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwach, F., F. E. Vaistij, L. Jones, and D. C. Baulcombe. 2005. An RNA-dependent RNA polymerase prevents meristem invasion by Potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 138:1842-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silhavy, D., A. Molnar, A. Lucioli, G. Szittya, C. Hornyik, M. Tabazza, and J. Burgyan. 2002. A viral protein suppresses silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J. 21:3070-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Storozhenko, S., D. Inze, M. Van Montagu, and S. Kushnir. 2001. Arabidopsis coactivator ALY-like proteins, DIP1 and DIP2, interact physically with the DNA-binding domain of the Zn-finger poly(ADP-ribose) polymerase. J. Exp. Bot. 52:1375-1380. [PubMed] [Google Scholar]
- 22.Turina, M., R. Omarov, J. F. Murphy, C. Bazaldua-Hernandez, B. Desvoyes, and H. B. Scholthof. 2003. A newly identified role for Tomato bushy stunt virus P19 in short distance spread. Mol. Plant Pathol. 4:67-72. [DOI] [PubMed] [Google Scholar]
- 23.Uhrig, J., T. Canto, D. Marshall, and S. A. MacFarlane. 2004. Relocalization of nuclear ALY proteins to the cytoplasm by the Tomato bushy stunt virus p19 pathogenicity protein. Plant Physiol. 135:2411-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vargason, J. M., G. Szittya, J. Burgyan, and T. M. Tanaka Hall. 2003. Size selective recognition of siRNA by an RNA silencing suppressor. Cell 115:799-811. [DOI] [PubMed] [Google Scholar]
- 25.Virbasius, C. M. A., S. Wagner, and M. R. Green. 1999. A human nuclear-localized chaperone that regulates dimerization, DNA binding, and transcriptional activity of bZIP proteins. Mol. Cell 4:219-228. [DOI] [PubMed] [Google Scholar]
- 26.Voinnet, O. 2005. Induction and suppression of RNA silencing: insights from viral infections. Nat. Rev. Genet. 6:206-220. [DOI] [PubMed] [Google Scholar]
- 27.Voinnet, O., C. Lederer, and D. C. Baulcombe. 2000. A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103:157-167. [DOI] [PubMed] [Google Scholar]
- 28.Ye, K., L. Malinina, and D. J. Patel. 2003. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature 426:874-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou, Z., M. J. Luo, K. Straesser, J. Katahira, E. Hurt, and R. Reed. 2000. The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature 407:401-405. [DOI] [PubMed] [Google Scholar]