Abstract
Higher-order structures in the 5′ untranslated region (UTR) of plus-strand RNA viruses are known in many cases to function as cis-acting elements in RNA translation, replication, or transcription. Here we describe evidence supporting the structure and a cis-acting function in defective interfering (DI) RNA replication of stem-loop III, the third of four predicted higher-order structures mapping within the 210-nucleotide (nt) 5′ UTR of the 32-kb bovine coronavirus (BCoV) genome. Stem-loop III maps at nt 97 through 116, has a calculated free energy of −9.1 kcal/mol in the positive strand and −3.0 kcal/mol in the negative strand, and has associated with it beginning at nt 100 an open reading frame (ORF) potentially encoding an 8-amino-acid peptide. Stem-loop III is presumed to function in the positive strand, but its strand of action has not been established. Stem-loop III (i) shows phylogenetic conservation among group 2 coronaviruses and appears to have a homolog in coronavirus groups 1 and 3, (ii) has in all coronaviruses for which sequence is known a closely associated short, AUG-initiated intra-5′ UTR ORF, (iii) is supported by enzyme structure-probing evidence in BCoV RNA, (iv) must maintain stem integrity for DI RNA replication in BCoV DI RNA, and (v) shows a positive correlation between maintenance of the short ORF and maximal DI RNA accumulation in BCoV DI RNA. These results indicate that stem-loop III in the BCoV 5′ UTR is a cis-acting element for DI RNA replication and that its associated intra-5′ UTR ORF may function to enhance replication. It is postulated that these two elements function similarly in the virus genome.
The 5′ and 3′ untranslated regions (UTR) in the coronavirus RNA genome and their counterparts in the antigenome are presumed to carry primary and higher-order structural elements important for genome replication, possibly as sites for the binding of viral RNA-dependent RNA polymerase (RdRp) and associated proteins in formation of the replication complex. For bovine coronavirus (BCoV), one molecule being used for the identification of UTR-mapping cis-acting RNA replication elements is a cloned, naturally occurring 2.13-kb defective interfering (DI) RNA that undergoes replication in the presence of BCoV helper virus (5-7). The BCoV DI RNA is composed of the fused termini of the virus genome [the 5′-terminal 496 nucleotides (nt) and the 3′-terminal 1,637 nt exclusive of the poly(A) tail, which is fused at the third nucleotide upstream of the N start codon, and a poly(A) tail of 68 nt]. To facilitate quantitation of replication by Northern assays, a 30-nt in-frame reporter sequence has been inserted at a unique BglII site at position 1093 to form construct pDrep1 (6). To date, with the use of this and DI RNA molecules from the mouse hepatitis coronavirus (MHV), another group 2 coronavirus, five putative UTR-mapping, cis-acting replication elements have been experimentally identified. Four of these show phylogenetic conservation and map within the 3′ UTR. They include the 3′-terminal poly(A) tail (46), a long-range stem interaction between nt 65 to 71 and 124 to 130 from the base of the poly(A) tail in BCoV (nt 68 through 74 and 136 through 142 in MHV-JHM) (30), a hairpin-type pseudoknot consisting of nt 173 through 226 in BCoV (nt 185 through 238 in MHV-A59) (53), and a 5′-proximal extended bulged stem-loop consisting of nt 222 through 289 in BCoV (nt 234 through 301 in MHV-A59) mapping immediately downstream of the N gene stop codon and overlapping the pseudoknot by 5 nt (21, 22). The importance of the 5′-proximal intra-3′ UTR bulged stem-loop in RNA replication has been confirmed in the context of the intact MHV genome (21). Within BCoV DI RNA, the 65-nt leader, comprising all of an identified stem-loop I and part of an identified stem-loop II, has been shown to function in DI RNA replication (6). The identity of 5′ and 3′ UTR-mapping cis-acting elements is consistent with results of deletion analyses showing dependence of DI RNA replication on coronavirus terminal sequences (6, 10, 23, 24, 29).
Here we extend earlier observations on a potential 5′ UTR-mapping stem-loop III (7) and examine its function in DI RNA replication. We describe its phylogenetic conservation among group 2 coronaviruses, compare its properties with a potential homolog in coronavirus groups 1 and 3, test its existence in BCoV RNA by further enzyme structure probing, document by mutation analyses the requirement of the stem for replication, and identify a potentially important role for the associated intra-5′ UTR open reading frame (ORF) in the replication of BCoV DI RNA.
MATERIALS AND METHODS
Virus and cells.
A DI RNA-free stock of the Mebus strain of BCoV at 4.5 × 108 PFU/ml was prepared and used as a helper virus as previously described (5-7, 53). The human rectal tumor cell line HRT-18 (49) was used as previously described (5-7, 16-19, 37, 53).
Plasmid constructs.
Construction of pGEM3Zf(−)-based pDrep1 (Promega) has been previously described (6). To construct the plasmid derivatives of pDrep1, overlap PCR mutagenesis (20) was done by using oligonucleotides listed in Table 1. The strategy used for constructing each of the derivatives was the same as that for pSLIII-mut1R except for the differences noted. For making pSLIII-mut1R, NdeI(+), SL3R(−), and pDrep1 DNA were used in the first PCR, HpaI(+), GEM3Zf(−)NdeI(−), and pDrep1 DNA were used in the second reaction, and GEM3Zf(−)NdeI(−) and NdeI(+) and the products of the first two reactions were used in the third reaction to make a 1,183-nt product that was trimmed to 693 nt with restriction endonucleases NgoMIV and AvrII and was cloned into NgoMIV/AvrII-linearized pDrep1. For making pSLIII-mut1L, SL3L(−) replaced SL3R(−) in the first reaction. For making pSLIII-mut1L/R, SL3L(−) replaced SL3R(−) and pSLIII-mut1R DNA replaced pDrep1 DNA in the first reaction, and pSLIII-mut1R DNA replaced pDrep1 DNA in the second reaction. For making pSLIII-mut2L, SRST3L(−) replaced SL3R(−) in the first reaction, and SRST3L(+) replaced HpaI(+) in the second reaction. For making pSLIII-mut2R, SRST3R(×) replaced SL3R(-) in the first reaction, and SRST3R(+) replaced HpaI(+) in the second reaction. For making pSLIII-mut2L/R, SRST3double(−) replaced SL3R(−) and pSLIII-2R DNA replaced pDrep1 DNA in the first reaction, and SRST3double(+) replaced HpaI(+) and pSLIII-mut2R DNA replaced pDrep1 DNA in the second reaction. For making pSLIII-mutG109→A, SRG109T(−) replaced SL3R(−) in the first reaction, and SRG109T(+) replaced HpaI(+) in the second reaction. For making pSLIII-mutG110→A, SRG110T(−) replaced SL3R(−) in the first reaction, and SRG110T(+) replaced HpaI(+) in the second reaction. For making pSLIII-mut5, SRAU99(−) replaced SL3R(−) in the first reaction, and SRAU99(+) replaced HpaI(+) in the second reaction. For making pSLIII-mut6, SR81(−) replaced SL3R(−) in the first reaction, and SR81(+) replaced HpaI(+) in the second reaction. For making pSLIII-mut3L, SRT101C(−) replaced SL3R(−) in the first reaction, and SRT101C(+) replaced HpaI(+) in the second reaction. For making pSLIII-mut4L/R, SRUTRSTOP(−) replaced SL3R(−) in the first reaction, and SRUTRSTOP(+) replaced HpaI(+) in the second reaction.
TABLE 1.
Oligonucleotides used in this study
| Oligonucleotidea | Polarity | Sequence (5′-3′)b | Binding region in pDrep1 |
|---|---|---|---|
| GEM3Zf(−)Nde1(−) | + | GAGAGTGCACCATATGCGGTGT | 2498-2519c |
| Hpa1(+) | − | GGCTGAAAGCTGTTAACAGCAGAAATG | 140-166 |
| Nde1(+) | − | CCTCCAAATCATATGGACGTGTATTC | 456-481 |
| SL3L(−) | + | CCACTCCCTGTATTgatgGCTTGTGGGC | 84-111 |
| SL3R(−) | + | GCTTGTGGGCtatcATTTTTCATAGTGG | 102-129 |
| SR81(−) | + | CATCCACTCCCTGTATagaATGCTTGTGGGaGTAGATTTTTCATAGTGG | 81-129 |
| SR81(+) | − | CCACTATGAAAAATCTACtCCCACAAGCATtctATACATTTAGTGGATG | 81-129 |
| SRAU99(−) | + | CATCCACTCCCTGTATTCaATGCTTGTGGGCGTtGATTTTTCATAGTGG | 81-129 |
| SRAU99(+) | − | CCACTATGAAAAATCaACGCCCACAAGCATtGAATACAGGGAGTGGATG | 81-129 |
| SRDM(+) | − | GTTGATCTTCGACATTGTGACC | 204-225 |
| SRG109T(−) | + | CTATGCTTGTGtGCGTAGATTTTTCATAG | 98-126 |
| SRG109T(+) | − | CTATGAAAAATCTACGCaCACAAGCATAG | 98-126 |
| SRG110T(−) | + | CTATGCTTGTGGtCGTAGATTTTTCATAG | 98-126 |
| SRG110T(+) | − | CTATGAAAAATCTACGaCCACAAGCATAG | 98-126 |
| SRleader(−) | + | GATTGTGAGCGATTTGCGTGCGTGC | 1-25 |
| SRST3double(−) | + | CACTCCCTGTATagaATGggTGTGtcCGTtct | 85-116 |
| SRST3double(+) | − | agaACGgaCACAccCATtctATACAGGGAGTG | 85-186 |
| SRST3L(−) | + | CACTCCCTGTATagaATGggTGTGGGCGTAGA | 85-116 |
| SRST3L(+) | − | TCTACGCCCACAccCATtctATACAGGGAGTG | 85-116 |
| SRST3R(−) | + | GTATTCTATGCTTGTGtcCGTtctTTTTTCATAGTGG | 93-129 |
| SRST3R(+) | − | CCACTATGAAAAAagaACGgaCACAAGCATAGAATAC | 93-129 |
| SRT101C(−) | + | CCACTCCCTGTATTCTAcGCTTGTGGGCG | 84-112 |
| SRT101C(+) | − | CGCCCACAAGCgTAGAATACAGGGAGTGG | 84-112 |
| SRUTRSTOP(−) | + | CCACTCCCTGTATTCTtaGCTTGTGGGCtaAGATTTTTCATAGTG | 84-128 |
| SRUTRSTOP(+) | − | CACTATGAAAAATCTtaGCCCACAAGCtaAGAATACAGGGAGTGG | 84-128 |
| TGEV8(+) | − | CATGGCACCATCCTTGGCAACCCAGA | 1098-1123 |
| TGEV8b(+) | − | CATGGCACCATCCTTGGCA | 1105-1123 |
The positive and negative symbols in the oligonucleotide names indicate the polarity of the nucleic acid to which the oligonucleotide anneals.
Lowercase bases represent mutated sites.
Indicates nucleotide positions in the pGEM3Zf(−) vector.
Enzyme structure probing of RNA.
Enzyme structure probing of RNA was carried out as previously described (37). Briefly, for in vitro synthesis of RNA, 10 μg of XbaI-linearized mung bean nuclease-blunt-ended pDrep1 DNA in a 200-μl reaction volume was transcribed at 37°C for 1 h by using 80 U of T7 RNA polymerase (Promega) to yield approximately 40 μg of a 595-nt-long vector-free transcript. RNA was treated with 10 U of RNase-free DNase (Promega) at 37°C for 30 min, extracted with phenol-chloroform and chloroform, chromatographed through a Biospin 6 column (Bio-Rad), spectrophotometrically quantitated, and stored in water at −20°C. For RNase treatments, 40 μg of RNA was heat denatured at 65°C for 3 min and renatured by slow cooling (0.5 h) to 35°C in a 400-μl reaction volume containing 30 mM Tris HCl (pH 7.5)-20 mM MgCl2-300 mM KCl, and aliquots containing 2 μg of sample RNA and 5 μg of yeast tRNA were incubated in 100 μl of the same buffer and 0.05, 0.5, 1, 2, or 4 U of RNase CV1 (Pharmacia), 0.05, 0.5, 1, 5, or 10 U of RNase T1 (GIBCO), or 0.05, 1, 2, 3, or 4 U of RNase T2 (GIBCO). RNase digestion was carried out at 25°C for 15 min and was terminated by the addition of 150 μl of 0.5 M sodium acetate, after which the RNA was extracted with phenol-chloroform and chloroform and was ethanol precipitated. Digested RNA preparations were used in primer extension reactions with 5′-end-labeled plus-strand-binding oligonucleotide SRDM(+) (Fig. 1). Undigested RNA was used with the same primer in dideoxynucleotidyl sequencing reactions to generate a sequence marker. Products were analyzed on a DNA sequencing gel of 6% polyacrylamide.
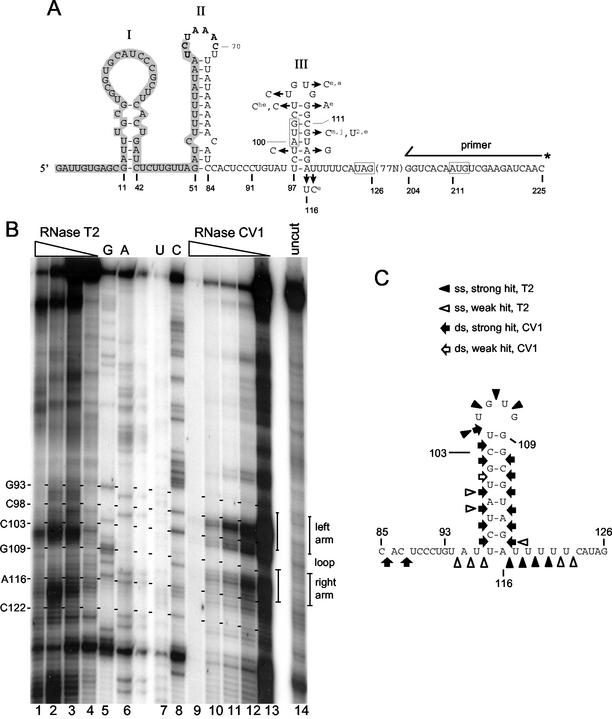
FIG. 1.
Phylogenetic and enzyme structure-probing evidence for stem-loop III. (A) Predicted secondary structures within the first 126 nt of the 210-nt 5′ UTR of the BCoV genome and the site from which 5′-end-labeled primer was extended for stem-loop III structure determination. The 65-nt BCoV leader sequence is shaded, and the leader-associated UCUAAAC intergenic sequence (nt 64 to 70) found within loop II is identified in bold. The start and stop codons for the intra-5′ UTR ORF and the start codon for ORF 1a are boxed. Previously described stem-loops I and II are identified. Stem-loop III is shown along with the variant bases found among the eight sequenced members of group 2 coronaviruses. Bases named with no superscript are found in all MHV strains (A59, 2, and JHM). Superscripts: 2, MHV-2; a, MHV-A59; j, MHV-JHM; he, HECoV-4408; e, ECoV. Stem-loop III is identical in BCoV, HEV, and HCoV-OC43. (B) Enzyme probing data for stem-loop III. Depicted is a DNA sequencing gel showing separation of primer extension products. RNase digestion conditions for the single-strand-digesting RNase T2 and double-strand-digesting RNase CV1 are described in the text. Lanes 5 to 8 show the products of an RNA sequencing reaction done on undigested RNA to identify the base positions. (C) Summary of enzyme probing data shown in panel B.
Northern assay for DI RNA replication.
The Northern assay was performed essentially as previously described (5-7, 19, 53). Briefly, cells at 80% confluency (∼106 cells per dish) in 35-mm-diameter dishes were infected with BCoV at a multiplicity of 10 PFU per cell and were transfected with 500 ng of DI RNA transcript in Lipofectin (Invitrogen) at 1 h postinfection (hpi). For passage of progeny virus, supernatant fluids were harvested at 48 hpi and 500 μl was used to infect freshly confluent cells in a 35-mm-diameter dish. RNA was harvested at the designated times and at 48 h after virus passages 1 and 2 (VP1 and -2, respectively). Total RNA extracted by the NP-40-proteinase K method (yielding approximately 10 μg per dish) was stored as an ethanol precipitate, and precisely one fourth of the preparation was used per lane for formaldehyde-agarose gel electrophoresis. Approximately 1 ng of transcript was loaded per lane as a size marker. Northern blots were probed with reporter-specific oligonucleotide porcine transmissible gastroenteritis virus (TGEV) 8(+) 5′-end labeled with 32P to specific activities of 1.5 × 105 to 3.5 × 105 cpm/pmol (Cerenkov counts). Blots were read with a Packard InstantImager Autoradiography System for quantitation and were exposed to Kodak XAR-5 film for 6 h to 5 days at −80°C for imaging. Images were recorded by digital scanning of the film. In every experiment in which the replication of mutants of pDrep1 RNA was examined, wild-type (wt) pDrep1 RNA was examined in parallel.
Sequence analysis of the stem-loop III region in progeny DI RNAs.
For direct sequencing of asymmetrically amplified cDNA, the procedure described by Hofmann et al. (17) for analysis of plus-strand RNA was used. For this sequencing, oligonucleotides TGEV8b(+) and SRleader(−) were used for reverse transcription-PCR (RT-PCR) with RNA extracted at 48 hpi from cells infected with the VP1 and, in some cases, with VP2, and 5′end-radiolabeled oligonucleotide SRDM(+), HpaI(+), or SRleader(−) was used for sequencing. When mixed sequences were revealed by this method, RT-PCR fragments were cloned with the TOPO XL PCR cloning kit (Invitrogen) and were sequenced.
Synthetic oligonucleotides and accession numbers.
Oligonucleotides used in this study are described in Table 1, and GenBank accession numbers for the sequences studied are M62375, M16620, and M30612 for BCoV-Mebus, AF523843 for human coronavirus (HCoV)-OC43, AF523844 for human enteric coronavirus (HECoV)-4408, AF523845 for porcine hemagglutinating encephalitis virus (HEV)-67N, AF523846 for equine coronavirus (ECoV)-NC99, NC001846 for mouse hepatitis virus (MHV)-2, M27198 for MHV-A59, M55148 for MHV-JHM, 234093 for TGEV-Purdue 116, 69721 for HCoV-229E, and M42 for avian infectious bronchitis virus (IBV)-Beaudette.
RESULTS
A predicted stem-loop III with an associated short ORF in the BCoV 5′ UTR shows phylogenetic conservation among group 2 coronaviruses.
Earlier studies that used the Tinoco algorithm (48) had predicted a potential stem-loop III with a helix of 7 bp and a loop of 6 nt mapping at nt 97 through 116 in the 210-nt 5′ UTR of BCoV, although enzyme structure probing had indicated the stem was probably 8 bp and the loop was 4 nt in length (7). An AUG start codon for an intra-5′ UTR ORF of 24 nt was found within the stem beginning at base 100. In the same study, stem-loops I and II were identified as components of a required 5′-terminal cis-acting element for BCoV DI RNA replication. Structure predictions, however, have failed to show a consistent pattern for stem-loops I and II among the group 2 coronaviruses (7 and data not shown), suggesting that perhaps a specific sequence rather than higher-order structures in the 5′-terminal leader region is important for this cis-acting function. To further evaluate the higher-order nature of stem-loop III in BCoV, the genomic 5′-terminal sequences were analyzed by the mfold program of Zuker (http://bioinfo.math.rpi.edu/∼zukerm/) (32, 54), and a stem-loop with a stem of 8 bp, a loop of 4 nt, and a free energy value of −9.1 kcal/mol was predicted (Fig. 1A). In the minus strand, a stem-loop with an internal loop, a terminal hexaloop, and a free energy of −3.0 kcal/mol was also predicted (Fig. 2). The UGUG loop in the positive strand does not belong to any identified family of especially stable tetraloops (UNCG, GNRA, CUNG) (52), suggesting that it may not contribute extraordinarily to stem-loop stability.
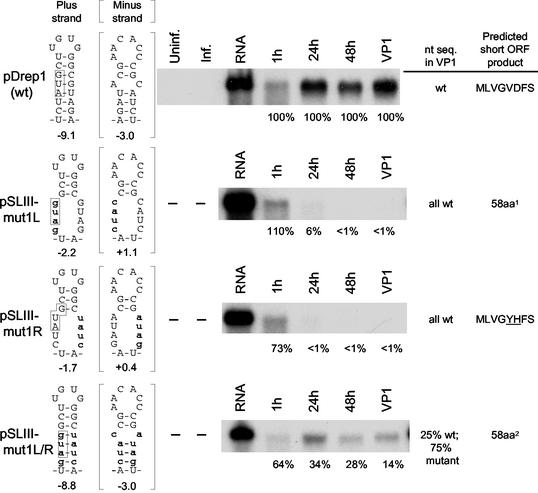
FIG. 2.
Integrity of stem III is required for DI RNA replication as determined by analysis of mutant 1 variants of pDrep1. The mfold-predicted structures of wt and mutant transcripts of pDrep1 (pSLIII-mut1L, pSLIII-mut1R, and pSLIII-mut1L/R) in the positive and negative strands and their calculated free energies in kilocalories per mole are shown. The start codon for the intra-5′ UTR ORF is boxed (note that this moves 1 base position upstream in pSLIII-mut1L and pSLIII-mut1L/R relative to that of wt). Mutated bases are shown as lowercase letters. For Northern analyses, T7 RNA polymerase-generated transcripts of linearized plasmids were transfected into BCoV-infected cells, and RNA was extracted at the indicated times posttransfection and probed with a reporter-specific radiolabeled oligonucleotide. Accumulation of mutant molecules was determined by quantitative blotting of RNA harvested at the times indicated and at 48 h postinfection with VP1. Accumulation of pDrep1 progeny was considered to be 100%. The sequence of the RNA in the progeny replicons was determined by either bulk RT-PCR sequencing by using pDrep1-specific primers as described in the text or by first cloning and sequencing plasmid DNA from isolated colonies. Lanes labels: Uninf., mock-infected, nontransfected cells; Inf., infected, nontransfected cells; RNA, sample of the transcript. Superscript 1 indicates the sequence MACGRRFFIVVSIFISAVNSFQPGTCCILGSGPPIGHNVEDQQIRSRTTLGSRISMDV. Superscript 2 indicates that the sequence is identical to the amino acid sequence predicted for mutant pSLIII-L except that LG replace RR at positions 5 and 6.
5′ UTR sequences for 12 coronaviruses have now been reported, enabling comparisons of predicted higher-order structures (see references associated with the GenBank accession numbers given above). For the eight members in group 2, BCoV-Mebus, HCoV-OC43, HECoV-4408, HEV-67N, ECoV-NC99, MHV-2, MHV-A59, and MHV-JHM, an mfold-predicted stem-loop III with small variations was found (stem-loop III is identical in BCoV, HCoV-OC43, and HEV) (Fig. 1A and Table 2). That is, there was predicted (i) a stem-loop III in the positive strand with a stem (with or without an internal loop) of 7 or 8 nt, a terminal tetraloop, and a net negative free energy within the range of −11.5 to −5.1 kcal/mol, (ii) a stem-loop in the negative strand with a net negative free energy within the range of −9.3 to −3.0 kcal/mol, and (iii) an AUG start codon for an ORF of 24 nt beginning at the third or fourth base position in the stem of the positive strand. The stem begins at base 96 for ECoV, 97 for MHV-A59 and MHV-2, and 101 for MHV-JHM, and an internal loop occurs at the fourth base position in the stem for all MHV strains and at the fifth base in ECoV. Although there is no evidence for a conserved primary sequence among coronavirus groups 1 to 3 in the region of stem-loop III, mfold analyses of groups 1 and 3 revealed a possible stem-loop III homolog in the same relative position within the 5′ UTR (see Discussion). Interestingly, all possess a closely associated AUG start codon for a short intra-5′ UTR ORF (Table 2). Thus, from folding predictions and evidence of phylogenetic conservation it was postulated that stem-loop III in BCoV DI RNA is a higher-order structure with possible function in RNA replication.
TABLE 2.
Properties of predicted coronavirus intra-5′ UTR stem-loop III, stem-loop III-associated ORF, and ORF-encoded oligonucleotide
| Coronavirus | Free energy of stem-loop III in positive strand (kcal/mol)a | Free energy of stem-loop III in negative strand (kcal/mol)a | Kozak context of short ORF start codon | Predicted no. of amino acids encoded by short ORF | Amino acid sequence of short ORF product |
|---|---|---|---|---|---|
| BCoV-Mebus | −9.1 | −3.0 | UCUaugC | 8 | MLVGVDFS |
| HCoV-OC43 | −9.1 | −3.0 | UCUaugC | 8 | MLVGVDFS |
| HECoV-4408 | −11.5 | −6.1 | UCUaugC | 8 | MPVGVDFS |
| HEV-67N | −9.1 | −3.0 | UCUaugC | 8 | MLVGVDFS |
| ECoV-NC99 | −5.1 | −4.2 | UCUaugC | 8 | MLASLDFS |
| MHV-A59 | −9.0 | −9.3 | UCCaugC | 8 | MPAGLVLS |
| MHV-JHM | −9.0 | −9.3 | UCCaugC | 8 | MPVGLVLS |
| MHV-2 | −9.0 | −8.6 | UCCaugC | 8 | MPVGLVLS |
| TGEV-Purdue | −12.9b | −9.0b | UCUaugA | 3 | MKS |
| HCoV-229E | −17.4b | −6.7b | GCUaugG | 11c | MAGIFDAGVVVc |
| IBV-Beau M42 | −19.9b | −18.4b | UGGaugG | 11 | MAPGHLSGFCY |
As determined by the Zuker algorithm.
For TGEV, HCoV-229E, and IBV, the Zuker algorithm predicts a larger stem-loop than for the group 2 coronaviruses.
A second ORF beginning 16 nt downstream from the first encodes the amino acid sequence MLES.
Enzyme structure probing supports the existence of stem-loop III in BCoV RNA.
Previous enzyme structure probing of the 5′-terminal 262-nt region of the BCoV genome had revealed, along with stem-loops I (nt 11 through 42) and II (nt 51 through 84), a stem-loop III with an 8-base stem and a tetraloop (7). Since RNA secondary structures can be significantly influenced by flanking sequences, evidence for stem-loop III was reexamined in the context of a 595-nt transcript. For this, 5′-terminal transcripts of XbaI-linearized pDrep1 DNA were made and were subjected to digestion with either single-strand-specific RNase T1 (data not shown) or T2 or with double-strand-specific RNase CV1, and the products were analyzed by reverse transcriptase extensions of 5′-end-labeled primer SRDM(+) (Fig. 1B, lanes 1 to 4 and 9 to 13). Radiolabeled products of an RNA sequencing reaction were analyzed in parallel to determine sites of RNase digestion (Fig. 1B, lanes 5 to 8). The results show single- and double-stranded regions in agreement with an mfold-predicted structure (Fig. 1C).
Integrity of stem-loop III is required for BCoV DI RNA replication.
To examine the importance of the helical region in stem-loop III for DI RNA replication, site-directed mutations in pDrep1 predicted to disrupt and then restore base pairing with compensatory changes were made, and transcripts were tested for replication. A stem was deemed required for replication if helix-disrupting mutations led to loss of accumulation (as determined by the absence of mutant progeny molecules in VP1 RNA) and structure-restoring compensatory mutations led to some level of restored accumulation (as determined by the presence of the double mutant progeny in VP1 RNA). Analyses focused on DI RNA progeny in VP1, since (i) passaged (i.e., packaged) molecules should be free of long-lived transfected mutant transcripts that would otherwise appear as progeny in RT-PCR assays and (ii) Northern assays alone do not discriminate between replicating mutant and replicating (recombination-derived revertant) wt molecules. Here it was assumed that stem-loop III does not function as a packaging signal (see Discussion). In the first of two sets of mutants examined, pSLIII-mut1L and pSLIII-mut1R were made in which four of the eight base pairs were disrupted in each of the left and right arms leading to stem-loop III free energy values of −2.2 and −1.7 kcal/mol in the positive strands, respectively, and no predicted structures in the negative strands (Fig. 2). By Northern analysis, 6% or less of wt levels of RNA had accumulated by 24 hpi and less than 1% by 48 hpt and by 48 hpi in VP1, but sequence analysis of asymmetrically amplified cDNA from VP1 RNA showed the products of both mutants to be revertants to wt, and thus the mutants had not replicated. By contrast, accumulation of progeny from double mutant pSLIII-mut1L/R for which the free energy value had become −8.8 kcal/mol in the positive strand and −3.0 kcal/mol in the negative strand was 14% of wt levels in VP1, and asymmetrically amplified cDNA from progeny molecules in VP1 as well as in cDNA clones of these showed progeny to be a mixture of wt and double mutant sequences (Fig. 2). Of eight clones sequenced, two were wt and six were the SLIII-mut1L/R mutant. cDNA from VP2 also showed a mixture of wt and mutant sequences, indicating continued replication of the double mutant through a second virus passage. These results together indicate that the helical region of stem-loop III either in the positive or negative strand or both is a requirement for DI RNA replication. A lower rate of accumulation in the double mutant than in wt, however, suggested that factors other than the helical nature of the stem, perhaps the stability of the stem, the nucleotide sequence within the stem, the size and location of the stem-loop-associated ORF, or the ORF product, might also be important for maximal accumulation.
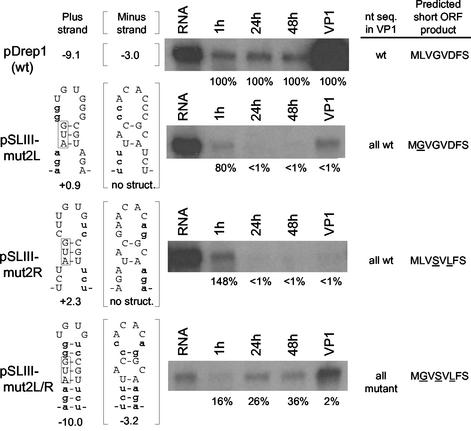
To test for the requirement of the stem under conditions that would minimally disrupt the short ORF, a second set of three mutants was examined in which the short ORF product differed from the wt by no more than three amino acids (Fig. 3). In pSLIII-mut2L, 5 of the 8 bases in the left arm were changed, leaving only those in the wt 100AUG codon unchanged and increasing the free energy value in the positive strand to +0.9 kcal/mol and leading to no predicted structure in the negative strand. The changes altered only the second amino acid (L→G), yielding a potential oligopeptide product of MGVGVDFS. In pSLIII-mut2R, 5 nt in the right arm were changed, maintaining the 111CGU base pairing partner with 100AUG and increasing the free energy in the positive strand to +2.3 kcal/mol and also leading to no predicted structure in the negative strand and potentially encoding an oligopeptide product of MLVSVLFS. In the double mutant pSLIII-mut2L/R, the calculated stem-loop III free energy was −10.0 kcal/mol in the positive strand and −3.2 kcal/mol in the negative strand, and the potential ORF product was MGVSVLFS. Northern analyses of progeny from pSLIII-mut2L and pSLIII-mut2R showed an accumulation of <1% of wt levels at 24 and 48 hpt and in VP1, and sequence analyses of asymmetrically amplified cDNAs showed progeny to be all revertant wt molecules, indicating that the mutants had not replicated. On the other hand, accumulation of progeny from the double mutant pSLIII-mut2L/R was >26% of wt levels at 24 and 48 hpt and >2% in VP1, and all progeny in VP1 had retained the mutant sequences. Thus, these results are consistent with those of the experiment described in Fig. 2 and suggest that stem-loop III either in the positive or negative strand or both is required for replication. A less-than-wt level of accumulation for the double mutant, however, would suggest again that perhaps the nucleotide sequence within the stem or possibly the amino acid sequence of the short ORF product were not optimal for maximal accumulation.
FIG. 3.
Integrity of stem III is required for DI RNA replication as determined by analysis of mutant 2 variants of pDrep1. The assays depicted were carried out as described in the legend to Fig. 2, except that mutant variants (pSLIII-mut2L, pSLIII-mut2R, and pSLIII-2L/R) were used.
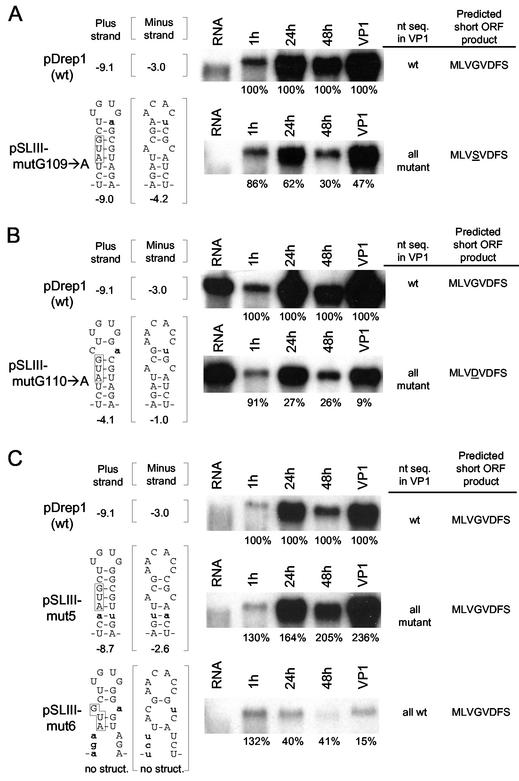
The notion that the nucleotide sequence within the stem or the amino acid sequence of the ORF product are important factors in accumulation in the context of an otherwise stable stem was supported by results from mutants in which single base changes were made in the upper part of the stem. In mutant pSLIII-mutG109→A, a stem of 8 nt was predicted for which the free energy of stem-loop III became −9.0 kcal/mol in the positive strand and −4.2 kcal/mol in the negative strand, and the potential product became MLVSVDFS; in pSLIII-mutG110→A, a stem of 7 nt was predicted for which the free energy of stem-loop III became −4.1 kcal/mole in the positive strand and −1.0 in the negative strand, and the potential ORF product became MLVDVDFS (Fig. 4A and B). For these, although only mutant sequences were found in VP1 DI RNA, accumulation was still less than maximal in that only 47 and 9% of wt levels, respectively, were found in VP1. Thus, to evaluate the role of stem-loop III integrity in mutants that contained nucleotide changes but still produced a potential wt short ORF product, double mutants were made by changing bases in codon wobble positions such that a wt ORF product was retained but that the stem was either maintained or disrupted. For these, pSLIII-mut5 was made in which stem III was maintained in both the positive (−8.71 kcal/mol) and negative (−2.6 kcal/mol) strands, and pSLIII-mut6 was made in which no structure was predicted in either strand of stem-loop III (Fig. 4C). Accumulation of pSLIII-mut5 progeny was no less than that of wt DI RNA at all time points, and the sequences of VP1 progeny were all mutant molecules, indicating that the stable stem mutants potentially producing a wt ORF product replicated at least as well as wt stem-loop III. On the other hand, accumulation of pSLIII-mut6 in which no stem-loop III structure was predicted was at less than wt levels, and only 15% of wt levels were produced in VP1 for which all progeny had wt sequence. These results together demonstrate that the integrity of stem-loop III is important for replication independent of any effect of the short ORF product and that at least some nucleotide changes in the stem are well tolerated.
FIG. 4.
Importance of stem III for DI RNA replication independent of the potential short ORF product. The importance of stem III in the presence of little (A and B) or no (C) change in the potential short ORF product was measured by assays described in the legend to Fig. 2. (A) Accumulation and sequence of progeny from wt or mutant transcripts bearing a mutation at base 109 (pSLIII-mutG109→A). (B) Accumulation and sequence of progeny from wt or mutant transcripts bearing a mutation at base 110 (pSLII0-mutG110→A). (C) Accumulation and sequence of progeny of wt or mutants with changes at codon wobble positions that maintain the wt ORF product MLVGVDFS. Stem III was maintained in pSLIII-mut5 but was disrupted in transcripts of pSLIII-mut6. Assays were carried out as described in the legend to Fig. 2.
Replication of BCoV DI RNA shows a strong preference for the presence of a stem-loop III-associated short ORF.
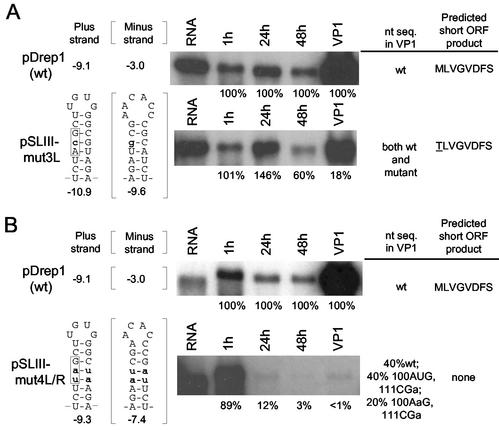
An association of an AUG-initiated short ORF with stem-loop III (or its presumed homolog) in all coronaviruses (see Discussion) led us to hypothesize that the short ORF plays a role as a cis-acting element in replication. To test whether it does, the DI RNA was modified in ways that were predicted to either diminish or prevent translation of the ORF but minimally perturb the integrity of stem-loop III. In the first modification, pSLIII-mut3L was made in which 100AUG was changed to 100AcG, a threonine codon known to function inefficiently as a translation start codon (38) (Fig. 5A). In this construct it was predicted by mfold that base pairing in the positive strand would be maintained between the AcG codon and the cognate wt triplet 111CGU and that the negative free energy of stem-loop III would increase to −10.9 kcal/mol in the positive strand and increase approximately threefold to −9.6 kcal/mol in the negative strand. By Northern analysis, accumulation of transcripts in VP1 was 18% of wt, and the sequence of the replicating progeny in VP1 was a mixture of both wt and mutant, indicating that some of these mutants had replicated. Progeny in VP2 were all wt. These results indicated that transcripts with an AcG-initiated short ORF can be replicated but with less efficiency than wt DI RNA and that an AUG-initiated ORF is preferred.
FIG. 5.
Stem-loop III-associated short ORF correlates with maximal DI RNA accumulation. (A) Accumulation and sequence of progeny from DI RNA in which the stem-loop III-associated short ORF begins with an AcG threonine codon (pSLIII-mut3L). The sequence of the progeny in VP1 was determined to be a mixture by sequencing the asymmetrically amplified RT-PCR product. (B) Accumulation and sequence of mutant transcripts in which the stem-loop III-associated short ORF begins with a uaG stop codon (pSLIII-4L/R). The sequence of progeny molecules in VP1 was determined from clones of the RT-PCR product. Assays were carried out as described in the legend to Fig. 2.
To test the importance of the short ORF by another approach, pSLIII-mut4L/R was made in which the 100AUG was changed to a 100uaG stop codon and its cognate base pairing triplet was changed from 111CGU to 111Cua to maintain full base pairing in the stem (Fig. 5B). In pSLIII-mut4L/R, the mfold-predicted stem-loop III had a free energy of −9.3 kcal/mol in the positive strand and −7.4 kcal/mol in the negative strand, and it had no predicted ORF product. Whereas Northern analysis showed an accumulation of <1% of wt after VP1, sequence analysis of asymmetrically amplified cDNA of progeny at VP1 revealed a mixture of 100A(U,a)G and 111C(G,u)(U,a). Thus, the cDNA products were cloned and sequenced to resolve the makeup of progeny molecules. Of 13 clones, 5 were wt sequence, 3 showed 100AUG but with a cognate base-pairing 111CGa triplet (potentially encoding the peptide MLVGEDFS), and 5 showed 100AaG (known not to function as a start codon [38]) with a cognate 111CGa triplet. No clones had the sequence of the original mutations of 100uaG or 111Cua. All progeny in VP2 were wt as determined from sequencing asymmetrically amplified cDNA. These results, therefore, established that molecules with the non-start codon AaG had replicated poorly and those with a uaG non-start codon had not replicated, indicating, as did the experimental results with mutant pSLIII-mut3L (Fig. 5A), that there is a preference by the DI RNA for a 100AUG-initiated short ORF in replicating molecules.
DISCUSSION
In this study we present evidence that supports the existence of a predicted stem-loop III mapping at nt 97 through 116 in the 210-nt-long 5′ UTR of the BCoV genome and show through mutagenesis analyses that the stem of this structure is required for the replication of BCoV DI RNA. Stem-loop III has an associated AUG that begins an intra-5′ UTR ORF of 24 nt, and mutagenesis studies of this suggested its maintenance is preferred by the DI RNA for replication. The short ORF or its product, therefore, may act to enhance replication. Comparison of 5′ UTR sequences of eight group 2 coronaviruses revealed that stem-loop III and the associated ORF are largely conserved in position and structure. Is a stem-loop III also found in the other coronavirus groups? mfold analyses of the 5′ UTR sequences of the group 1 coronaviruses HCoV-229E and TGEV and the group 3 coronavirus IBV indicated that a bulged stem-loop III analog, if not homolog, of approximately twice the size with an associated intra-5′ UTR ORF is positioned similarly within the 5′ UTR (Fig. 6). Except for HCoV-229E, the predicted loop is also a tetraloop (in HCoV-229E it is a 3-base loop), and in all cases the AUG codon for the associated intra-5′ UTR ORF is within the stem (Fig. 6 and Table 2). ORF-encoded oligopeptides range from 3 amino acids in length in TGEV to 11 amino acids in IBV and appear to be dissimilar in sequence (Table 2).
FIG. 6.
Potential stem-loop III homologs in coronavirus groups 1 and 3 are predicted by the Zuker mfold algorithm. AUG codons for potential stem-loop-associated ORFs are boxed. In HCoV-229E the second AUG is in the +1 frame relative to the first ORF.
From experiments described here it was not established how stem-loop III or the ORF function mechanistically in DI RNA replication. However, several questions regarding mechanism are raised. (i) Does stem-loop III function in the positive or negative strand, or both? Since base pair covariations among group 2 members maintain stem-loop III in the negative strand (free energy range, −9.3 to −3.0 kcal/mol) as well as in the positive strand (−11.5 to −5.1 kcal/mol) (Table 2), the possibility exists that stem-loop III functions in either strand or in both strands. The question was not answered by our experiments, since net negative free energy values were maintained in both the negative (−4.2 to −1.0 kcal/mol)- and positive (−10.9 to −4.1 kcal/mol)-strand stem-loop III structures in all mutants that replicated. (ii) What trans-acting factors, if any, bind stem-loop III? Precedents in other plus-strand RNA viruses (3, 11, 14, 15, 47, and references therein) lead us to speculate that stem-loop III most likely binds viral or cellular protein(s) in the positive strand. This would be consistent with studies showing translation of an ORF attached to the genomic 5′ UTR of BCoV to be regulated differently in infected and uninfected cells (42). However, a cellular protein of 55 kDa has been found to bind to nt 56 through 112 in the MHV-JHM 5′ genome terminus, and cellular proteins of 35 and 38 kDa have been found to bind to the minus-strand counterpart of this sequence (12). Since this sequence represents the upstream half of stem-loop III, these proteins become candidates for playing a role in stem-loop III function. Preliminary experiments in our laboratory have indicated that viral and cellular proteins bind the positive-strand form of stem-loop III, whereas only cellular proteins bind the negative-strand form (S. Raman and D. Brian, unpublished data). Thus, the interactions of stem-loop III with trans-acting elements of either protein or RNA need to be further examined. (iii) Do the bases in the loop of stem-loop III serve a specific function? Inasmuch as loops (terminal and internal) and bulges within RNA often function as protein binding sites (36), the function of the terminal loop sequence will need to be evaluated. (iv) Does the intra-5′ UTR ORF affect replication through a cis-acting influence on translation? Several short upstream ORFs have now been shown to profoundly influence the translation of downstream ORFs in eukaryotic and viral RNAs through a variety of mechanisms (13, 33, 35). Inasmuch as the replication of some group 2 DI RNAs show a cis preference for translation (5, 50), a parallel may exist in genome replication. If so, the influence of a genomic upstream ORF may significantly affect genome replication. Certainly, a short intraleader ORF in a mutant BCoV genome that arose during persistent infection in cell culture had a measurable effect on the translation from an ORF 1 start codon in vitro (18). Curiously, none of the stem-loop III-associated ORFs begin within a highly favorable Kozak context (25 and Table 2), suggesting that translation from this ORF may not need to be efficient to fulfill a translational requirement. In addition, our results showing that at least some molecules with a totally foreign short ORF (i.e., the ORF beginning at nt 99 in pSLIII-mut1L/R shown in Fig. 2) can replicate indicates that perhaps any ORF and not necessarily only the wt ORF can suffice. In this regard, it would seem possible that an inefficient start codon such as CUG (38) at position 91 (Fig. 1), which is in frame with the wt short ORF, could suffice for replication, and maybe this is what initiates the ORF function for the replicating progeny of mutant pSLIII-mut4L/R in which a non-start AaG (lys) codon is found at position 100 (Fig. 5B). If this is true, then perhaps the function of the short ORF in replication is necessary and not just preferred for replication. In this regard, it is also curious that a short 5′ UTR-positioned ORF in the equine arteritis virus appeared to have no effect on the replication of this nidovirus in cell culture (34). It is a remote possibility, suggested by the results of experiments with the AcG codon, that a weak start codon such as GUG (38) used in the equine arteritis virus experiments (34) could have sufficed to initiate a required translation function of the short ORF. (v) Does stem-loop III function as a packaging signal? Inasmuch as our assay measured the character of VP1 (packaged) molecules, our results would have been influenced by a packaging function of stem-loop III. Thus, despite the mapping of a BCoV packaging signal within ORF 1b at genome nucleotide positions 20,049 to 20,117 (8, 9, and K. Nixon and D. Brian, unpublished), a sequence not present in the BCoV DI RNA studied here, the role of stem-loop III in DI RNA packaging should be examined. It should be noted, however, that with no mutant did we see abundant accumulation at 24 and 48 hpt and a low accumulation in VP1, making it unlikely that a disrupted packaging signal explains the low levels in VP1 observed in Fig. 1 and 2.
The question of what constitutes the cis-acting elements for coronavirus RNA replication is especially relevant to an earlier postulate that coronavirus subgenomic mRNAs (sgmRNAs) behave as replicons in the presence of trans-acting viral RdRp (45). Whereas the postulate is consistent with the observations that (i) the termini on sgmRNAs are theoretically large enough to serve as RdRp promoters for replication (a 5′-terminal region of 65 to 95 nt and a 3′-terminal region of ∼300 nt [4, 26, 51]) (both of which greatly exceed the sizes of alphavirus and myxovirus replication promoters [27, 28]), (ii) coronavirus sgmRNAs have minus-strand counterparts (19, 44, 45) capable of forming mRNA-length, double-stranded (apparent) replicative intermediates (1, 2, 19, 39-41, 43-45), and (iii) sgmRNAs accumulate faster than genome at early times after infection (19, 45), the hypothesis has not been supported by experiments that directly measure sgmRNA accumulation following transfection into helper-virus-infected cells (6, 31). For BCoV, the replicating DI RNA differs from sgmRNA 7 by only 421 nt, the first 137 of which come from nt 74 through 210 in the genomic 5′ UTR and the second 284 nt of which come from the 5′ end of ORF 1a (6). Assuming mRNAs do not replicate as indicated by the results of transfection experiments (6, 31), elements mapping within the 5′-proximal 421 nt in BCoV DI RNA but absent in subgenomic mRNAs must be providing at least part of what is needed for replication. Stem-loop III described here appears to be one of these elements.
Acknowledgments
We thank Cary Brown, Kimberley Nixon, Aykut Ozdarendeli, and Hung-Yi Wu for valuable discussions.
This work was supported by Public Health Service grant AI14267 from the National Institute of Allergy and Infectious Diseases, grant 92-37204-8046 from the U.S. Department of Agriculture, and funds from the University of Tennessee, College of Veterinary Medicine, Center of Excellence Program for Livestock Diseases and Human Health.
REFERENCES
- 1.Baric, R. S., C.-K. Shieh, S. A. Stohlman, and M. M. C. Lai. 1987. Analysis of intracellular small RNAs of mouse hepatitis virus: evidence for discontinuous transcription. Virology 156:342-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baric, R. S., and B. Yount. 2000. Subgenomic negative-strand RNAs function during mouse hepatitis virus infection. J. Virol. 74:4039-4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barton, D. J., B. J. O. Donnell, and J. B. Flanegan. 2001. 5′ Cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 20:1439-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavanagh, D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142:629-633. [PubMed] [Google Scholar]
- 5.Chang, R.-Y, and D. A. Brian. 1996. cis requirement for N-specific protein sequence in bovine coronavirus defective interfering RNA replication. J. Virol. 70:2201-2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, R.-Y., M. A. Hofmann, P. B. Sethna, and D. A. Brian. 1994. A cis-acting function for the coronavirus leader in defective interfering RNA replication. J. Virol. 68:8223-8231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, R.-Y., R. Krishnan, and D. A. Brian. 1996. The UCUAAAC promoter motif is not required for high-frequency leader recombination in bovine coronavirus defective interfering RNA. J. Virol. 70:2720-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chouljenko, V. N., X. Q. Lin, J. Storz, K. G. Kousoulas, and A. E. Gorbalenya. 2001. Comparison of genomic and predicted amino acid sequences of respiratory and enteric bovine coronaviruses isolated from the same animal with fatal shipping pneumonia. J. Gen. Virol. 82:2927-2933. [DOI] [PubMed] [Google Scholar]
- 9.Cologna, R., and B. G. Hogue. 2000. Identification of a bovine coronavirus packaging signal. J. Virol. 74:580-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dalton, K., R. Casais, K. Shaw, K. Stirrups, S. Evans, P. Britton, T. D. K. Brown, and D. Cavanagh. 2001. cis-acting sequences required for coronavirus infectious bronchitis virus defective-RNA replication and packaging. J. Virol. 75:125-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frolov, I., R. Hardy, and C. M. Rice. 2001. cis-acting RNA elements at the 5′ end of Sindbis virus genome RNA regulate minus- and plus-strand synthesis. RNA 7:1638-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furuya, T., and M. M. C. Lai. 1993. Three different cellular proteins bind to complementary sites on the 5′-end-positive and 3′-end-negative strands of mouse hepatitis virus RNA. J. Virol. 67:7215-7222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gabelle, A. P. 1996. Translational control mediated by upstream AUG codons, p. 173-197. In J. W. B. Hershey, M. B. Mathews, and N. Sonenberg (ed.), Translational control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, N.Y.
- 14.Gamarnik, A. V., and R. Andino. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 12:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herold, J., and R. Andino. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 7:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hofmann, M. A., and D. A. Brian. 1991. The 5′ end of coronavirus minus-strand RNAs contains a short poly(U) tract. J. Virol. 65:6331-6333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofmann, M. A., R.-Y. Chang, S. Ku, and D. A. Brian. 1993. Leader-mRNA junction sequences are unique for each subgenomic mRNA species in the bovine coronavirus and remain so throughout persistent infection. Virology 196:163-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hofmann, M. A., S. D. Senanayake, and D. A. Brian. 1993. A translation-attenuating intraleader open reading frame is selected on coronavirus mRNAs during persistent infection. Proc. Natl. Acad. Sci. USA 90:11733-11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hofmann, M. A., P. B. Sethna, and D. A. Brian. 1990. Bovine coronavirus mRNA replication continues throughout persistent infection in cell culture. J. Virol. 64:4108-4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horton, R. M., C. Zeling, N. H. Steffan, and L. R. Pease. 1990. Gene splicing by overlap extension: tailor-made genes using polymerase chain reaction. BioTechniques 8:528-535. [PubMed] [Google Scholar]
- 21.Hsue, B., T. Hartshorne, and P. S. Masters. 2000. Characterization of an essential RNA secondary structure in the 3′ untranslated region of murin coronavirus genome. J. Virol. 74:6911-6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsue, B., and P. S. Masters. 1997. A bulged stem-loop structure in the 3′ untranslated region of the coronavirus mouse hepatitis virus genome is essential for replication. J. Virol. 71:7567-7578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Izeta, A., C. Smerdou, S. Alonso, Z. Penzes, A. Mendez, J. Plana-Duran, and L. Enjuanes. 1999. Replication and packaging of transmissible gastroenteritis coronavirus-derived synthetic minigenomes. J. Virol. 73:1535-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, Y. N., Y. S. Jeong, and S. Makino. 1993. Analysis of cis-acting sequences essential for coronavirus defective interfering RNA replication. Virology 197:53-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozak, M. 1994. Determinants of translational fidelity and efficiency in vertebrate mRNAs. Biochimie 76:815-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lai, M. M. C., and D. Cavanagh. 1997. The molecular biology of coronaviruses. Adv. Virus Res. 48:1-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levis, R., B. G. Weiss, M. Tsiang, H. Huang, and S. Schlesinger. 1986. Deletion mapping of Sindbis virus DI RNAs derived from cDNAs defines the sequences essential for replication and packaging. Cell 44:137-145. [DOI] [PubMed] [Google Scholar]
- 28.Li, X., and P. Palese. 1992. Mutational analysis of the promoter required for influenza virus virion RNA synthesis. J. Virol. 66:4331-4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin, Y. J., C. L. Liao, and M. M. C. Lai. 1994. Identification of the cis-acting signal for minus-strand RNA synthesis of a murine coronavirus: implications for the role of minus-strand RNA in RNA replication and transcription. J. Virol. 68:8131-8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu, Q., R. F. Johnson, and J. L. Leibowitz. 2001. Secondary structural elements within the 3′ untranslated region of mouse hepatitis virus strain JHM genomic RNA. J. Virol. 75:12105-12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Makino, S., M. Joo, and J. K. Makino. 1991. A system for study of coronavirus mRNA synthesis: a regulated, expressed subgenomic defective interfering RNA results from intergenic site insertion. J. Virol. 65:6031-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 33.Mathews, M. B. 1996. Interactions between viruses and the cellular machinery for protein synthesis, p. 500-548. In J. W. B. Hershey, M. B. Mathews, and N. Sonenberg (ed.), Translational control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, N.Y.
- 34.Molenkamp, R., H. van Tol, B. C. D. Rozier, Y. van der Meer, W. J. M. Spann, and E. J. Snijder. 2000. The arterivirus replicase is the only viral protein required for genome replication and subgenomic mRNA transcription. J. Gen. Virol. 81:2491-2496. [DOI] [PubMed] [Google Scholar]
- 35.Morris, D. R., and A. P. Geballe. 2000. Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol. 20:8635-8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nowakowski, J., and I. Tinoco. 1997. RNA structure and stability. Semin. Virol. 8:153-165. [Google Scholar]
- 37.Ozdarendeli, A., S. Ku, S. Rochat, G. D. Williams, S. D. Senanayake, and D. A. Brian. 2001. Downstream sequences influence the choice between a naturally occurring noncanonical and closely positioned upstream canonical heptameric fusion motif during bovine coronavirus subgenomic mRNA synthesis. J. Virol. 75:7362-7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peabody, D. S. 1989. Translation initiation at non-AUG triplets in mammalian cells. J. Biol. Chem. 264:5031-5035. [PubMed] [Google Scholar]
- 39.Sawicki, S. G., and D. L. Sawicki. 1990. Coronavirus transcription: subgenomic mouse hepatitis virus replicative intermediates function in mRNA synthesis. J. Virol. 64:1050-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sawicki, S. G., and D. L. Sawicki. 1995. Coronaviruses use discontinuous extension for synthesis of subgenome-length negative strands. Adv. Exp. Med. Biol. 380:499-506. [DOI] [PubMed] [Google Scholar]
- 41.Sawicki, D. L., T. Wang, and S. G. Sawicki. 2001. The RNA structures engaged in replication and transcription of the A59 strain of mouse hepatitis virus. J. Gen. Virol. 82:385-396. [DOI] [PubMed] [Google Scholar]
- 42.Senanayake, S. D., and D. A. Brian. 1999. Translation from the 5′ UTR of mRNA 1 is repressed, but that from the 5′ UTR of mRNA 7 is stimulated in coronavirus-infected cells. J. Virol. 73:8003-8009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sethna, P. B., and D. A. Brian. 1997. Coronavirus subgenomic and genomic minus-strand RNAs are found in N protein-deficient, membrane-protected replication complexes. J. Virol. 71:7744-7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sethna, P. B., M. A. Hofmann, and D. A. Brian. 1991. Minus-strand copies of replicating coronavirus mRNAs contain antileaders. J. Virol. 65:320-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sethna, P. B., S.-L. Hung, and D. A. Brian. 1989. Coronavirus subgenomic minus-strand RNA and the potential for mRNA replicons. Proc. Natl. Acad. Sci. USA 86:5626-5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spagnolo, J. F., and B. G. Hogue. 2000. Host protein interactions with the 3′ end of bovine coronavirus RNA and the requirement of the poly(A) tail for coronavirus defective genome replication. J. Virol. 74:5053-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teterina, N. L., D. Egger, K. Bienz, D. M. Brown, B. L. Semler, and E. Ehrenfeld. 2001. Requirements for assembly of poliovirus replication complexes and negative-strand RNA synthesis. J. Virol. 75:3841-3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tinoco, I., P. N. Borer, B. Dengler, M. D. Levine, O. C. Uhlenbeck, D. M. Crothers, and J. Gralla. 1973. Improved estimation of secondary structure in ribonucleic acids. Nat. New Biol. 246:40-41. [DOI] [PubMed] [Google Scholar]
- 49.Tompkins, W. A. F., A. M. Watrach, J. D. Schmale, R. M. Schulze, and J. A. Harris. 1974. Cultural and antigenic properties of newly established cell strains derived from adenocarcinomas of the human colon and rectum. J. Natl. Cancer Inst. 52:1101-1106. [DOI] [PubMed] [Google Scholar]
- 50.van der Most, R. G., W. Luytjes, S. Rutjes, and W. J. M. Spaan. 1995. Translation but not the encoded sequence is essential for the efficient propagation of the defective interfering RNAs of the coronavirus mouse hepatitis virus. J. Virol. 69:3744-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van der Most, R. G., and W. J. M. Spaan. 1995. Coronavirus replication, transcription, and RNA recombination, p. 11-31. In S. G. Siddell (ed.), The Coronaviridae. Plenum Press, New York. N.Y.
- 52.Varani, G. 1995. Exceptionally stable nucleic acid hairpins. Annu. Rev. Biophys. Biomol. Struct. 24:379-404. [DOI] [PubMed] [Google Scholar]
- 53.Williams, G. D., R.-Y. Chang, and D. A. Brian. 1999. A phylogenetically conserved hairpin-type 3′ untranslated region pseudoknot functions in coronavirus RNA replication. J. Virol. 73:8349-8355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zuker, M., D. H. Mathews, and D. H. Turner. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide in RNA biochemistry and biotechnology, p. 11-43. In J. Barciszewski and B. F. C. Clark (ed.), NATO ASI Series, Kluwer Academic Publishers, Dordrecht, The Netherlands.