Abstract
Chromatin is suppressive in nature to cellular enzymes that metabolize DNA, mainly due to the inherent inaccessibility of the DNA template. Despite extensive understanding of the involvement of chromatin-modifying factors in transcription, roles of related activities in DNA replication remain largely elusive. Here, we show that the heterodimeric transcriptional elongation factor FACT (facilitates chromatin transcription) is functionally linked to DNA synthesis. Its involvement in DNA replication is partly mediated by the stable association with the replicative helicase complex, MCM, and further by the coexistence with MCM on replication origin. Furthermore, relying on its nucleosome-reorganizing activity, FACT can facilitate chromatin unwinding by the MCM complex, which is otherwise inert on the nucleosomal template. As a consequence, the physical and functional interaction between FACT and MCM is an important determinant in the proper initiation of DNA replication and S phase in vivo. Together, our findings identify FACT as an integral and conserved component of the endogenous replication machinery, and support a model in which the concerted action of helicase and chromatin-modifying activities promotes chromosome replication.
Keywords: chromatin, DNA replication, FACT, MCM helicase, S phase
Introduction
The FACT (facilitates chromatin transcription) complex, a heterodimer of hSpt16 and SSRP1 proteins, represents a class of chromatin structure modulator known to reorganize nucleosomal structure presumably by removal and/or reassembly of the histone H2A-H2B dimers (LeRoy et al, 1998; Orphanides et al, 1998, 1999; Belotserkovskaya et al, 2003). The pleiotropic nature of FACT's functions is illustrated by its involvement in replication, transcription and even DNA repair (Keller et al, 2001; Keller and Lu, 2002; Saunders et al, 2003; Shimojima et al, 2003). These findings support the notion that FACT acts as a core activity that, when in conjunction with pathway-specific cofactors, mediates chromatin loosening during the progression of different DNA-metabolizing events.
The role of FACT in DNA replication was previously documented by genetic and biochemical studies on the yeast (Wittmeyer and Formosa, 1997; Wittmeyer et al, 1999; Schlesinger and Formosa, 2000; Gambus et al, 2006) and Xenopus (Okuhara et al, 1999) FACT homologues (yFACT and DUF, respectively). However, the molecular mechanism underlying the replicative functions of DUF and yFACT as well as the functional link between human FACT and DNA replication have not been substantiated. Conversely, despite extensive studies on dissecting the modular organization and temporal regulation of DNA replication, the role of chromatin in this process has not been fully addressed (Bell and Dutta, 2002; Varga-Weisz, 2005).
In the present study, we show that like its yeast counterpart, human FACT is an important DNA replication factor. We identified the MCM complex as novel interacting partner of the FACT heterodimer, thus providing a basis for FACT's involvement in DNA replication. We further analyzed the physical and functional interactions between FACT and MCM. Our data demonstrate that the conserved replicative role of FACT potentially lies in its ability to facilitate chromatin unwinding by the MCM complex.
Results
Identification of the MCM complex as a novel interacting partner of FACT
Using a proteomic approach described previously (Tan and Lee, 2004), which entails anti-SSRP1 monoclonal antibody (clone 10D1) immunoprecipitation and mass spectrometric analysis, we identified three subunits of the replicative helicase complex MCM, MCM4, MCM6 and MCM7, as novel constituents of the FACT-associated complexes in the HeLa cell (Figure 1A and see Supplementary Table S1). Subsequent Western blot analysis verified these associations and also uncovered an additional subunit, MCM2, in this particular immunocomplex (Figure 1B and G). Additional immunoprecipitation experiment using an anti-SSRP1 polyclonal antibody further revealed coprecipitation of MCM2–6 (Figure 1C). Conversely, endogenous FACT heterodimer was specifically detected in the MCM complexes pulled down by an anti-MCM4 antibody (Figure 1D). These results demonstrate that FACT interacts with the hexameric MCM complex in vivo, as well as a discrete MCM subcomplex that includes MCM2/4/6/7. Moreover, the interaction between FACT and MCM is genuinely DNA-independent, as indicated by the intactness of the 10D1 immunocomplexes under nuclease treatment or in the presence of ethidium bromide (see Supplementary Figure S1A, and data not shown).
Figure 1.

Replicative helicase MCM is a novel interacting protein of FACT. (A) FACT-associated complexes were immunoprecipitated from HeLa nuclear extracts (derived from 2 × 107 cells) using αSSRP1 mAb 10D1, resolved by and shown on a silver-stained gel (also see Tan and Lee, 2004). Specific SSRP1-interacting protein bands that are absent in the control immunoprecipitates (not shown) were identified as MCM4 and MCM6 (∼100 kDa), and MCM7 (80 kDa). The 120-kDa band represents a previously identified associated protein, Nek9 (Tan and Lee, 2004). (B) Western blot analysis of the different immunocomplexes targeted by control (2B12) and αSSRP1 (10D1) mAbs. Immunoblotting was carried out using the indicated antibodies against pan-MCM or individual subunits. The positions of MCM2, MCM4/6 and MCM7 in the IP are indicated by the black, gray and white arrowheads (middle panel), respectively. The amount of the input (Input) is equivalent to 1/40 the IP. The identity of the protein band, marked by the asterisk, is unknown. (C) Western blot analysis of the immunoprecipitates targeted by αSSRP1 polyclonal antibodies (PI, preimmune serum). Immunoblotting was carried out using antibodies against the indicated MCM subunits. The amount of the Input is equivalent to 1/30 of the IP. (D) Western blot analysis of the immunoprecipitates targeted by preimmune serum (PI) or αMCM4 antibodies. Immunoblotting was carried out using the specified antibodies. The amount of the Input is equivalent to 1/50 of the IP. (E) Recombinant FACT heterodimer (FLAG-hSpt16/His6-SSRP1, left panel) and MCM subcomplexes (MCM2/4/6/7, MCM2/3/5, MCM2–7 and MCM4/6/7, as indicated) were isolated as described in Materials and methods and visualized by silver-stained gel. In vitro pull-down assay was carried out using the anti-FLAG M2 agarose, with the indicated combinations of protein complexes. The presence of specifically bound proteins was detected by the indicated antisera (bottom two panels). (F) Bacterially expressed GST and GST-fused MCMs were used as baits in the in vitro pull-down assay. The presence and purity of the immobilized proteins were seen on Coomassie blue-stained gels. The presence of bound FLAG-hSpt16 or His6-SSRP1 protein was detected by immunoblotting with M2 or αSSRP1 antibodies, respectively. The schematic representation shown below the panels depicts the domain organization of MCM4 as well as the presence or absence of interaction between various deleted constructs of MCM4 and the FACT subunits (input is 1/15 of IP). (G) Ablation of endogenous MCM3 and MCM4 expression was achieved by RNAi. Equal loadings of whole-cell extracts (‘Input') derived from HeLa cells transiently harboring control-, MCM3- or MCM4-targeting dsRNA (for 54 h) were blotted with the indicated antibodies. Lysates (30 × of input) were immunoprecipitated with 10D1 (bottom four panels, ‘IP: 10D1') and subsequently probed with the indicated antibodies. (H) In vitro pull-down assay was performed as in (E), using FACT (10 nM) and MCM2–7 (10 nM). GST (15 nM) or GST-fused full-length or deletion constructs of MCM4 (GST-MCM4120–250 GST-MCM4250–380) was added to the binding mixture. Precipitated rFACT as well as co-precipitated MCMs were probed with the indicated antibodies. (I) Extracts were prepared from HeLa cells ectopically expressing empty vector, HA-MCM4120–250, HA-MCM4250–380 or HA-MCM4-FL (full-length), and subsequently subjected to immunoprecipitation using the HA antibody. Immunoprecipitates (IP: HA) and lysate input (Input) were probed for HA, MCM or FACT, as indicated.
To test a direct association of these two complexes, purified preparations of the MCM2/4/6/7 heterotetramer, MCM2/3/5 heterotrimer or MCM2/3/4/5/6/7 hexamer, and the recombinant FLAG-hSpt16/His6-SSRP1 heterodimer (Figure 1E, top panels; see Materials and methods) were subjected to in vitro binding assay using the anti-FLAG M2 agarose. We found that the MCM2/4/6/7 and MCM2/3/4/5/6/7 complexes could be specifically detected in the co-precipitates (Figure 1E, bottom), indicating a direct interaction between these complexes. On the other hand, MCM2/3/5 did not bind FACT. Next, to identify the MCM subunit(s) that confer the specific interaction with FACT, we used GST pull-down assay with GST-fused recombinant MCM subunit proteins, and subsequently found that both FLAG-hSpt16p and His6-SSRP1 can individually bind to GST-MCM4, but not the other MCM subunits (Figure 1F). Such interaction with both subunits of FACT contributes to a tighter association of MCM4, as illustrated by a greater degree of MCM4 interaction with heterodimeric FACT than with either single subunit (Supplementary Figure S1B). Accordingly, downregulation of MCM4 (but not MCM3) expression by RNAi specifically led to disruption of the FACT–MCM immunocomplexes in vivo (Figure 1G and Supplementary Figure S1D). Moreover, this failure of FACT to co-precipitate the MCM complex is attributed primarily to the reduced MCM4 expression but not to the altered cell cycle as a consequence of MCM4 downregulation (Supplementary Figure S1D and F). Consistent with the interaction domain mapping analysis (Figure 1F and Supplementary Figure S1C), we found that co-incubation with interaction domain-containing MCM4 fragment (GST-MCM4120–250, amino acids 120–250) compromises the FACT–MCM interaction in vitro (Figure 1H and Supplementary Figure S1E). Conversely, GST-fused constructs of full-length MCM4 (GST-MCM4-FL) and a control MCM4 fragment (GST-MCM4250–380) did not cause discernible effect (Figure 1H). Furthermore, we found through co-immunoprecipitation experiment that exogenous MCM4120–250 could interact with FACT in vivo but did not associate with the endogenous MCM complex (Figure 1I). Taken together, these data reveal a specific physical interaction between FACT and distinct MCM complexes and that the complex formation is mediated by direct contact of MCM4 with both FACT subunits.
FACT coexists with MCM on the chromosomal replication origin
MCM is a key component of the prereplicative complex that has been shown indispensable during the initiation and elongation steps of DNA replication (Aparicio et al, 1997; Labib et al, 2000; Schwacha and Bell, 2001). Our observation of the interaction between FACT and MCM may thus indicate a functional link between the FACT heterodimer and DNA replication. To directly assess this possibility, we first examined whether FACT is present in vivo at a region of known replication origin, namely the replicator associated with the human lamin B2 gene (Abdurashidova et al, 2000; Paixao et al, 2004). Using the chromatin immunoprecipitation (ChIP) assay, chromatin prepared from cells synchronized at different cell cycle stages was precipitated with the 10D1 monoclonal antibody (Figure 2A and B and Supplementary Figure S2A). Subsequent PCR reactions using specific sets of primers were performed to assess the presence of the lamin B2 origin sequence (Figure 2A, lanes 1–6). As shown in Figure 2A and B, at equal loads of chromatin DNA preparations, we were able to demonstrate specific occupancy of FACT in the lamin B2 origin region in asynchronously growing cells (Figure 2A, compare lanes 1 and 4, and Figure 2B). As a control, sequences of a distant non-transcribed region (Figure 2A, lanes 7–9 and B) and a lamin B2-proximal, non-origin region (Figure 2B) were not significantly enriched in the immunoprecipitates. Additional quantitative analysis indicated that a further enrichment of such origin binding was detectable during the G1/S transition (Figure 2A, compare lanes 1 and 2, and Figure 2B). Conversely, significantly less FACT occupied the origin during S and G2/M phase. Mitotic dissociation of FACT from origin is consistent with our observation that FACT dissociated from condensed chromatin during mitosis (Tan and Lee, 2004). Interestingly, MCM4 underwent an analogous kinetics of origin occupancy (Figure 2B), which also agrees with the previous observations on the origin-association of the pre-RC components (Abdurashidova et al, 2003). This cell cycle-dependent binding pattern of FACT was specific to the origin, as no such change could be seen for FACT's association with the transcribed region of the γ-actin gene (Figure 2A, lanes 10 and 11, and B).
Figure 2.
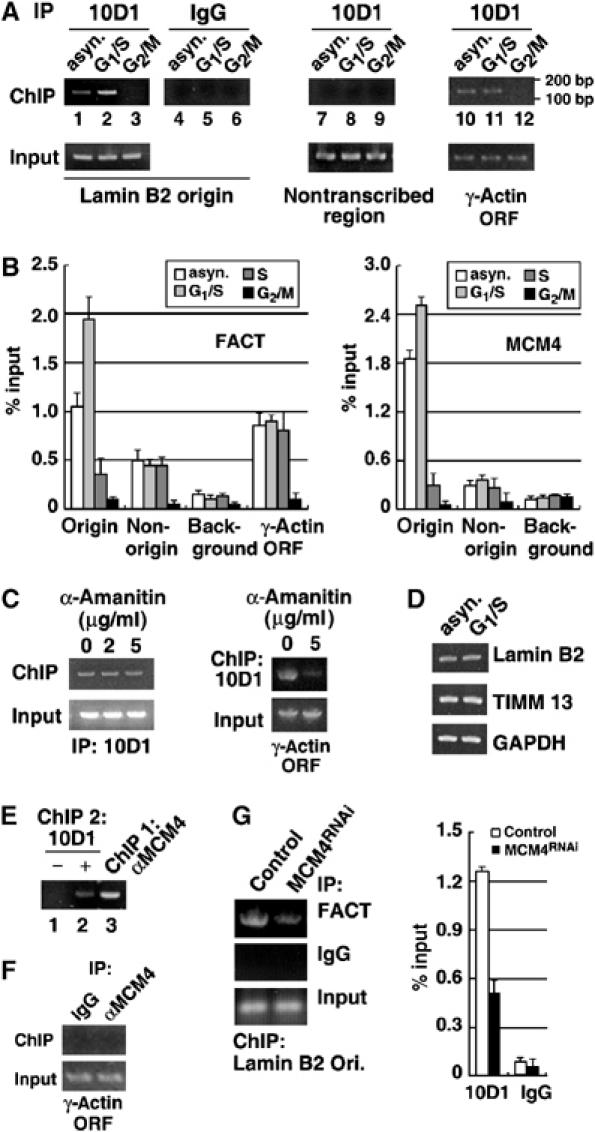
FACT coexists with MCM on the chromosomal replication origin. (A) ChIP was performed as described in Materials and methods. Chromatin fragments were prepared from cells at different stages: asynchronous (lanes 1, 4, 7 and 10), G1/S (lanes 2, 5, 8 abd 11) and G2/M (lanes 3, 6, 9 and 12). Immunoprecipitation was performed with either control (IgG, lanes 4–6) or 10D1 (lanes 1–3, 7–9 and 10–12) antibody. Products (∼160 bp) from final PCR analysis using primers specific to lamin B2 origin (lanes 1–6), to a non-transcribed region (lanes 7–9), or to the γ-actin gene, were resolved by 1.5% agarose gel (positions of the size marker are denoted on the right). Precipitated products are shown in the upper panels, DNA input in the lower. (B) Chromatin samples were immunoprecipitated with 10D1 (FACT, left) or anti-MCM4 (MCM4, right) antibody. The charts represent the quantitative determination of bound DNA, as determined by quantitative RT–PCR. Regions correspond to the lamin B2 origin, a non-origin region, a region of no annotated genes (background), and γ-actin ORF were analyzed (indicated on the x-axis). Data presented are normalized to input DNA (shown in percentage), and the mean±s.d. values from at least four experiments are shown. (C) HeLa cells were treated with various concentrations of α-amanitin for 12 h to inhibit RNA polII transcription. Binding of FACT to the lamin B2 origin (left), or to the non-origin, transcribed region of the γ-actin gene (right), under such treatment was monitored by the ChIP assay using the 10D1 antibody, as in (A). (D) The mRNA level of the lamin B2 (upper panel) and TIMM3 (middle) genes in asynchronous or double thymidine-arrested (G1/S) HeLa cells were examined by RT–PCR. Expression level of GAPDH (lower panel) was used as a loading control. (E) Coexistence of FACT and MCM on origin was demonstrated by a sequential ChIP experiment. Chromatin was first immunoprecipitated by MCM4 antibody (lane 3). A second round of ChIP was performed using control (lane 1) or 10D1 (lane 2) antibody. The presence of DNA fragments corresponding to the lamin B2 origin in the second ChIP was assessed by PCR. (F) Chromatin was prepared from asynchronous cells. ChIP was performed using the preimmune (IgG) or MCM4-specific (αMCM4) antibodies, and was subsequently probed for the presence of the non-origin, transcribed region of the γ-actin gene fragment. (G) Chromatin was prepared from the control and MCM4RNAi cells. ChIP was performed as above to probe for the presence of lamin B2 origin DNA fragment in the FACT immunocomplexes (left). Chart on the right represents quantification of bound lamin B2 origin region and shows the percent of starting material immunoprecipitated by 10D1 or control IgG (noted on the x-axis), as determined by RT–PCR. The mean±s.d. values from at least three experiments are shown.
Since FACT is generally known as a transcription elongation factor, we next asked whether the above observation is a result of transcription-coupled recruitment. As shown by the ChIP result, inhibition of mRNA transcription by α-amanitin, a specific inhibitor of RNA polymerase II, did not alter the binding of FACT to the lamin B2 origin (Figure 2C, left panels). At the same concentration of drug treatment, however, FACT's association with the γ-actin gene locus was significantly blocked (right panels). In addition, we found that levels of RNA transcripts for the two gene loci that flank the lamin B2 origin, lamin B2 and TIMM 13, remained relatively constant during G1/S transition (Figure 2D and Supplementary Figure S2B). We therefore postulate that the increased association of FACT with the origin upon S-phase entry (Figure 2A, lanes 1 and 2) is independent of the transcription activity of neighboring genes or FACT's own transcriptional function. Taken together, occupancy of FACT in lamin B2 origin area may be associated primarily with DNA replication.
Despite the contact of FACT with an endogenous origin region in a chromatin context, it remains a distinct possibility that the FACT–MCM complex may not exist in such chromatin-bound form. To address the issue of whether both MCM and FACT simultaneously assemble in this origin region, a two-step, sequential ChIP assay was performed (Figure 2E). First, sonicated chromatin fragments were immunopurified by anti-MCM4 antibodies. The specific presence of DNA fragments corresponding to lamin B2 origin in this precipitate, but not those of non-origin region, was confirmed by PCR (Figure 2E, lane 3 and F). We next used the FACT antibody to perform a second round of ChIP on chromatin recovered from the first round. The presence of lamin B2 origin chromatin fragments in the 10D1 but not the control precipitate (Figure 2E, compare lanes 1 and 2) demonstrates the coexistence of FACT and MCM molecules in a replication origin region on chromatin. Interestingly, reduced level of MCM4 expression by siRNA treatment (Figure 2G) as well as disruption of the FACT–MCM complex (Figure 4C) both led to weaker association of FACT with the origin. These observations confirm the origin co-occupancy of FACT and MCM and further suggest that such coexistence at the origin is partly dependent on MCM4.
Figure 4.
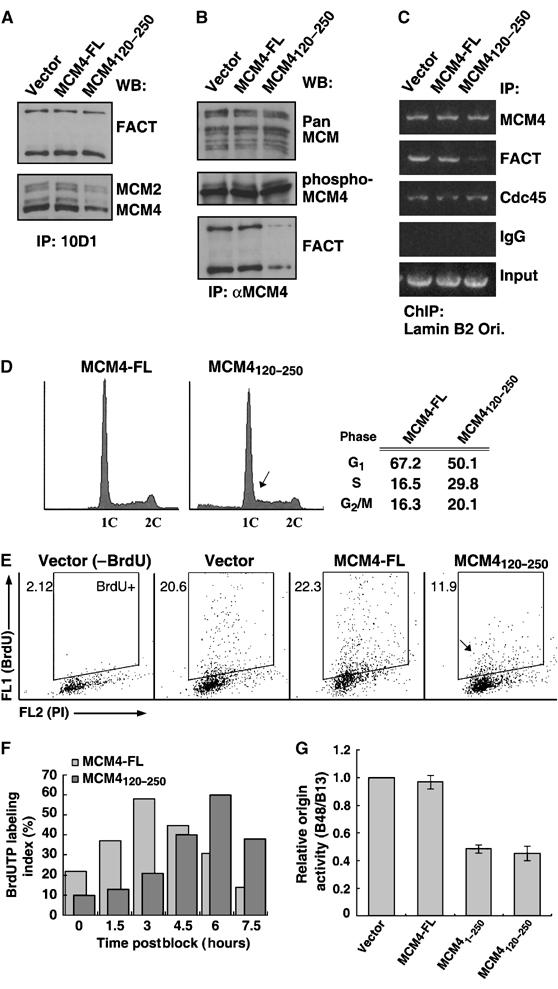
Disruption of the FACT–MCM complex triggers delayed DNA replication initiation. Stable clones of HeLa cells ectopically expressing empty vector, HA-MCM4-FL (full-length) or HA-MCM4120–250 were established as described in the Materials and methods. Mixed clones of each line were subjected to analysis. (A) Extracts were subjected to immunoprecipitation using the 10D1 antibody. Immunoprecipitates were probed for FACT or MCM2/4, as indicated. (B) Anti-MCM4 immunocomplexes isolated from these extracts were subjected to immunoblotting using the pan MCM, phospho-MCM4 (Thr110) and FACT antibodies, as indicated. (C) Using the indicated antibodies, ChIP was performed as in Figure 2 to probe for the presence of lamin B2 origin DNA fragment in the corresponding immunocomplexes. DNA input is shown on the bottom. (D) Cell cycle profiles of the MCM4-FL and MCM4120–250 cells. Cells were subjected to FACS for measurement of DNA content. Cells in the G0/G1, S and G2/M phases were defined by gating. Percentages of gated events are summarized on the right. Arrowhead indicates a major difference in phase distribution (see text). (E) BrdU-incorporation of the indicated cell lines at the G1/S junction. Upon release from thymidine treatment (at time 0), cells were pulse-labeled with BrdUTP. Analog incorporation (FL1) was compared with DNA content (FL2-PI, y-axis). BrdU-stained cells were defined by electronic gating (‘BrdU+'). Percentages of gated events are shown (in the upper left corner of each panel). The ‘Vector (−BrdU)' panel represents the distribution of cells in the absence of BrdU labeling, and thus serves as a control for the gating of stained cells. The arrow indicates the distribution of BrdU+ cells of the MCM4120–250 line (see text). (F) MCM4-FL, MCM41–250 and MCM4120–250 cells were synchronized and released at the G1/S junction and, at the indicated time points, pulse-labeled with BrdU (30 min) before being harvested. Cell cycle profiles and degree of BrdU incorporation were monitored by FACS. The labeling index quantitatively corresponds to the percentage (%) of BrdU-positive cells in a particular cell population. (G) Origin function of the lamin B2 replicator in the three indicated cell lines was assessed as described in Materials and methods (also see Supplementary Figure S5). The bar graph is a quantitative representation of the origin activity, with the activity in the control cells represented as 1. Data are averaged±s.d. of three independent experiments.
FACT promotes DNA unwinding activity of the MCM helicase on nucleosomal template
FACT was initially isolated based on its ability to facilitate RNA polymerase passage on a nucleosomal template. We thus speculated that the involvement of FACT in the process of DNA replication might potentially be mediated by a similar mechanism wherein the chromatin unwinding (or passage) activity of MCM, through interacting with FACT, is upregulated. To test this hypothesis, we first generated a forked, linear DNA template of ∼200 bp onto which mononucleosomal particles can be assembled for assaying the MCM unwinding activity (Figure 3A and B, and Supplementary Figure S3A). FACT interacts with two complexes of MCM, MCM2–7 and MCM4/6/7 (Figure 1). While the former has been regarded as the physiologically relevant form of the helicase, only the latter has been reported as the enzymatically active assembly in vitro (Ishimi, 1997; Lee and Hurwitz, 2001), and is therefore used in our study. As shown in Figure 3C, purified MCM4/6/7 complex possessed unwinding activity toward naked DNA substrate (Figure 3C, lanes 2 and 3). Conversely, the MCM2/3/5 heterotrimer (lanes 7 and 8) and recombinant FACT heterodimer did not (see Supplementary Figure S3B). The helicase activity of the active MCMs was reduced significantly, however, when the nucleosome was introduced onto the DNA template, suggesting that the MCM helicase might be minimally active, or even inert, on chromatin in vivo (Figure 3D, lanes 1 and 2). Strikingly, the addition of the recombinant FACT heterodimer moderately relieved the nucleosome-dependent inhibition on the MCM helicase activity (Figure 3D, lanes 3 and 4). Pretreatment of the nucleosomal template with FACT before addition of the helicase slightly augmented the MCM-mediated chromatin unwinding (Figure 3F, lane 3), further signifying that relaxed nucleosomal DNA serves as a more appropriate substrate for MCM. This effect of FACT on MCM is nucleosome-dependent, as FACT did not alter the MCM helicase activity in the context of naked DNA (Figure 3C, lane 4). Lack of nucleosomal DNA unwinding by FACT alone also indicates that such DNA strand separation was specifically induced by MCM but not contaminating activities in the FACT fraction (see Supplementary Figure S3C).
Figure 3.
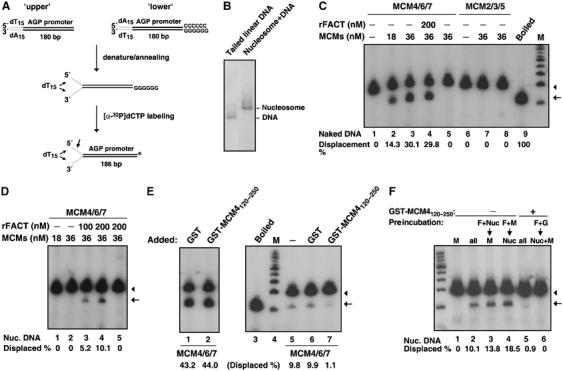
FACT promotes the DNA unwinding activity of the MCM helicase on nucleosomal template. (A) Schematic representation of the strategy for generating a linear, tailed DNA template by PCR. The presence of 5′ dT (15-mer) tail is known to increase the helicase activity of MCM (You et al, 2003). Asterisk marks the site of labeling. See Materials and methods for details. (B) Reconstitution of nucleosome cores onto the ∼200-bp, end-labeled DNA fragment. Aliquot of the transfer reactions were analyzed and bands corresponding to reconstituted nucleosomes and free DNA are indicated. (C) Helicase assay was performed with the indicated amounts of isolated MCM4/6/7 (lanes 2–5) or MCM2/3/5 (lanes 7 and 8), or in combination with purified recombinant FACT (lane 4). Reactions were performed on the free DNA substrate. Lane 5 represents unwinding reactions done in the absence of ATP. Deprotienated reaction products were resolved by native gel electrophoresis. M, 100 bp ladder DNA marker. Denaturation of the DNA substrate by heat is also shown (‘Boiled', lane 9). Degree of DNA unwinding/displacement for each reaction was calculated based on the quantified signal ratio of ss- to ds-DNA bands. It is expressed in percentage (%) and shown on the bottom of the figure. (D) DNA helicase assay was carried out as in (C), except the use of the reconstituted nucleosomal template. Unwinding of the nucleosomal DNA by various combinations of MCM4/6/7 and/or FACT complexes (amounts indicated on top) was monitored by autoradiography. Lane 5 represents unwinding reaction carried out in the absence of ATP. Degree of DNA unwinding/displacement is determined as above. (E) DNA helicase activity of the FACT–MCMs complex was assayed on the naked DNA (lanes 1 and 2) or the nucleosomal (lanes 3–7) template, as described above. MCM4/6/7 (36 nM) (lanes 1–2 and 5–7) was assayed in combination with 200 nM of purified recombinant FACT. Before reaction, the complexes were preincubated with GST (50 nM, lanes 1 and 6), GST-fused deletion construct of MCM4 (GST-MCM4120–250, 50 nM, lanes 2 and 7) or buffer (lane 5). Percentage of displaced DNA is calculated as above and shown below the panel. M, DNA marker (lane 4). Heat denaturation of the DNA is also shown (‘Boiled', lane 3). (F) DNA helicase activity was assayed on the nucleosomal template, as described above. Preincubation experiments were performed with the indicated combinations of proteins or template (‘Preincubation:'), for a reaction length of 20 min, before adding the remaining components. Reactions were also performed without (lanes 1–4) or with (lanes 5 and 6) GST-MCM120–250. M, MCM4/6/7; F, rFACT; Nuc, nuclesomal DNA; G, GST-MCM120–250; all, all components without any preincubation. Lane 1: MCM4/6/7 alone. In (C–F), the positions of the double- and single-stranded fragments are indicated by arrowheads and arrows, respectively.
Next, to unequivocally elucidate whether such functional cooperation between FACT and MCM depends on their physical interaction, we performed the helicase assay again with the addition of GST-MCM4120–250, the protein fragment that was shown to weaken the FACT–MCM association (Figure 1H). As a control, the GST-MCM4120–250 fragment did not alter the intrinsic helicase activity of the MCM complexes on the naked DNA substrate (Figure 3E, lanes 1 and 2). When the FACT–MCM complexes were partially disrupted by GST-MCM4120–250, their catalytic activity on the nucleosomal template correspondingly dropped (Figure 3E, compare lanes 5 and 7). On the other hand, the control GST did not exhibit such effect (lane 6). Interestingly, by preincubating FACT with the GST-MCM4120–250 fragment before helicase reaction, we observed a yet stronger, almost complete inhibition of the unwinding activity (Figure 3F, lane 6). Conversely, preincubation of FACT and MCM, which allows complex formation before adding substrate, consequently facilitated a greater extent of chromatin unwinding as compared to the coincubation reaction (Figure 3F, lane 4). Together, these results strongly support the notion that the FACT–MCM interaction is a critical determinant for their functional cooperation in unwinding nuclesomal DNA. Such functional interaction implies a positive regulatory role of FACT on the MCM-mediated DNA unwinding during chromatin replication.
The FACT–MCM interaction is important for the proper initiation of DNA replication
Having demonstrated the functional cooperation between FACT and MCM in chromatin unwinding, we next sought to address the physiological significance of this interaction. In an attempt to disrupt the interaction between FACT and MCM, we first examined the effect of expressing the MCM4 N-terminal fragment (MCM4120–250) in HeLa cells. Similar to what was observed in the above in vitro interaction assay, ectopic expression of MCM4120–250 fragment led to a partially reduced complex formation between FACT and MCM in vivo (Figure 4A and B). On the other hand, in cells those stably harbor the control or the full-length MCM4-enconding vectors, the FACT–MCM immunocomplexes remained intact (Figure 4A and B, see control and MCM4-FL). The interaction-interference effect of MCM4120–250 is specific, as overexpression of this protein fragment did not disturb various attributes of the MCM complex, such as protein expression (Figure 1I), complex composition (Figure 4B), origin binding (Figure 4C), phosphorylation of MCM4 (Figure 4B) and DNA helicase activity (data no shown). Moreover, the presence of this fragment did not affect origin binding of Cdc45 (Figure 4C), a replication initiation factor whose recruitment follows that of the MCM complex (Bell and Dutta, 2002). Additional control experiment also excluded the possibility that the effect of MCM4120–250 arises from altered FACT-mediated gene transcription (Supplementary Figure S4). Intriguingly, disruption of the FACT–MCM complex renders FACT less engaged at the origin (Figure 4C), implying an important role of MCM in targeting FACT to the replication origin.
When we subjected the stable cell lines (control and MCM4 derivatives) to cell cycle kinetics analysis, we found a greater distribution of cells in the S phase in the MCM4120–250-expressing line, as compared to the MCM4-FL line (30 versus 17%), suggestive of a delayed S phase progression (Figure 4D). In particular, there was a greater number of G1/S- and early S-phase cells (indicated by an arrow in Figure 4D). To obtain more detailed information about this potential replicative defect, an analysis of BrdU incorporation during S phase progression was performed. Figure 4E shows that, while control and MCM4-FL cells exhibited normal ongoing DNA synthesis at the G1/S junction (∼20% of the total population is BrdU-positive), the MCM4120–250 line had about only half of the actively replicating cell. Moreover, closer examination of the BrdU-incorporation profile of the MCM4120–250 cells revealed that even the BrdU+ cells had relatively low level of analog incorporation (indicated by an arrow in Figure 4E), signifying an inefficient DNA synthesis.
By quantitatively monitoring the degree of BrdU incorporation (or ‘labeling index') as cells progress through S phase, replication efficiency of a certain culture can be assessed. This analysis subsequently revealed that in the presence of MCM4120–250, cells exhibited markedly delayed DNA synthesis at the onset stage (Figure 4F). Based on the physical and functional association of FACT with replication origin, we next sought to determine whether this defective replication initiation is a consequence of weaker origin firing. Using a quantitative origin mapping procedure devised by Giacca, Pelizon and Falaschi (Giacca et al, 1997), which entails isolation of nascent DNA and quantification of specific origin DNA fragments by a competitive PCR technique (see Materials and methods), activity of the lamin B2 replicator can be assessed (see Supplementary Figure S5). We showed using this method that knockdown of MCM4 expression by RNAi resulted in a nearly complete abolishment of origin activity (<10%), while downregulation of FACT also caused a significant reduction (30% of the control) (see Supplementary Figure S5). Importantly, we in fact found that a destabilized FACT–MCM complex led to a reduced origin activity of the lamin B2 replicator (Figure 4G, compare MCM41–250 and MCM4120–250 to control and MCM4-FL). Collectively, these findings are consistent with our model that FACT and MCM functionally cooperate in chromatin unwinding (Figure 3), and further indicate that such interaction may be physiologically important for promoting the initiation of DNA replication.
Discussion
An important discovery of our work is that FACT contributes to chromatin DNA replication regulation through its physical and functional interaction with the active forms of the MCM complexes: MCM4/6/7 (Ishimi, 1997; Schwacha and Bell, 2001) and MCM2–7 (Labib et al, 2000). A very recent report has demonstrated that the yeast MCM helicase, with the assistance of the GINS complex, interacts with the replisome progression complexes (RPCs) (Gambus et al, 2006). Interestingly, constituents of RPC include, among various factors, the yeast FACT heterodimer. This finding corroborates the reported interaction between FACT and MCM, and, together with our observation of analogous interaction between the fly counterparts (BC Tan and SC Lee, unpublished observations), substantiates the evolutionary conservation and functional significance of this complex. While the study on the yeast RPC did not further dissect the FACT–MCM association, our present work revealed several mechanistic and functional attributes underlying this interaction. In addition to its direct association with the replicative helicase complex, the involvement of FACT in DNA replication was further strengthened by its coexistence with MCM on the replication origin as well as its functional cooperation with MCM in promoting initiation of DNA synthesis. Moreover, we demonstrated that a potential mechanism of FACT's replicative function lies in conferring upon the MCM complex an ability to unwind DNA in a nucleosomal context. Taken together, our results directly implicate the highly conserved role of FACT in DNA replication and further outline a model of the collaborative function of the FACT and MCM complexes in chromatin unwinding.
It is noteworthy that we did not identify proteins in the FACT immunocomplexes that correspond to the other components of the yeast replisome complex (Gambus et al, 2006). One of the probable explanations for this different outcome may lie in the methodologies of isolating the targeted complexes (i.e. antibody or cell extraction condition). Also, we presently cannot rule out the possibility that there exists more than one subassembly of the replisome complexes. In this regard, FACT–MCM may be one of the stable subcomplexes. Furthermore, it is likely that there is an inter-species distinction in the formation of replisome or replication-associated multiprotein complexes. Nevertheless, these recent findings collectively strengthen the conserved replicative role of the FACT–MCM interaction.
Chromatin is inhibitory in nature to various DNA transactions, generally by rendering the DNA template structurally inaccessible. Analogous to transcription, the progression of DNA replication must overcome the structural hindrance imposed by the nucleosomal template. Indeed, based on our observation, nucleosomes efficiently reduces the DNA helicase activity of MCM (Figure 3D), an effect similarly observed in the case of RNA Pol II-mediated transcription (Orphanides et al, 1998). It is likely that, as accompanied by the MCM-mediated recruitment of FACT, the local chromatin structure around the origin undergoes replication-associated changes. In that capacity, FACT's unique mode of action may be through destabilizing the structure of nucleosome without altering its epigenetic status (Belotserkovskaya et al, 2003), consequently generating a more appropriate substrate template for MCM's unwinding activity. Our identification of FACT as a replication factor thus clearly underscores the importance of chromatin structure in origin firing and replication progression and, more importantly, an intrinsic requirement of a chromatin remodeling/loosening mechanism. Indeed, putative involvement of chromatin modifiers, such as CHRAC (Alexiadis et al, 1998), WICH (Bozhenok et al, 2002; Poot et al, 2004), HBO1 (Burke et al, 2001), Sir2p (Pappas et al, 2004) and Rpd3 (Aggarwal and Calvi, 2004), in DNA replication was recently identified. New findings linking histone hyperacetylation to active origins within the terminal repeats of the Kaposi's sarcoma-associated herpevirus genome (Stedman et al, 2004) and the chorion locus in Drosophila follicle cells (Aggarwal and Calvi, 2004) lend further support to the notion that DNA replication is under epigenetic control. Collectively, involvement of these factors emphasizes the direct role of chromatin context in DNA replication.
Existence of replication-associated chromatin-modulating factors other than FACT is further evident in our results. Downregulation of the FACT–MCM interaction led to considerable, but not complete, loss of replication origin function (Figure 3H) and defective DNA replication initiation (Figure 3E–G), suggesting the partially redundant nature of FACT's activity. Moreover, results from the in vitro helicase assay indicate that, even in the presence of FACT, the nucleosomal duplex unwinding by MCM could not be fully restored to a level comparable to that on the naked DNA template (Figure 3D). We thus speculate that the establishment of an optimal nucleosomal structure in which the template is readily accessible to MCM may require multiple activities (histone modification, for instance), with FACT being an important component. Together, these observations point to a regulatory function of FACT's activity, and further suggest that, as in transcription, multiple factors/complexes, coordinately or independently, may constitute the prevalent chromatin regulatory mechanism during replication events.
Materials and methods
Preparation of recombinant proteins and MCM complex
FLAG-hSpt16p and His6-SSRP1 were expressed by baculovirus-infected insect cells (Sf9) and purified by anti-FLAG (M2) immunoaffinity column and Ni-NTA agarose (Qiagen), respectively, according to the manufacturer's instructions and procedures described previously (Tan and Lee, 2004). For the chromatin unwinding (Figure 3) assays, the expression and purification of recombinant FACT heterodimer were carried out according to the procedures outlined elsewhere (Belotserkovskaya et al, 2003). Purification of the MCM4/6/7 and MCM2/4/6/7 complexes was carried out based on a previously established protocol (Ishimi, 1997). Bacterially expressed GST-fused MCM2, 3, 4, 5, 6 and 7 were obtained from the BL21(Lys) strain. MCM proteins were cleaved from the GST fusion using the appropriate protease, and purified based on a protocol provided by the manufacturer (Amersham). Preparation of MCM2/3/5 and MCM2–7 was carried out by mixing the corresponding recombinant proteins in equimolar ratio and subsequently purifying through Sephacryl S-400 gel filtration chromatography. For the GST-fused MCM31–250, MCM41–250, MCM4120–250 and MCM4250–380 constructs, cDNA fragments corresponding to the first 250 amino acids (750 bp) or amino acids 120–250 or 250–380 (393 bp) of these proteins were first amplified by PCR and subsequently ligated to the pGEX-4T-1 vector (Amersham Pharmasia). Bacterial expression of these proteins and purification by the Glutathione Sepharose 4B (Amersham Pharmasia) were based on the manufacturer's protocols.
Antibodies and Western blot analysis
Generation of monoclonal antibodies against SSRP1 (2B12/control and 10D1) and hSpt16p was described previously (Tan and Lee, 2004). Anti-SSRP1 rabbit polyclonal antibodies were produced and purified using the full-length recombinant protein. Polyclonal antibodies against human pan-MCM were purchased from BD biosciences. Anti-Cdc45 rabbit polyclonal antibodes were purchased from Santa Cruz Biotechnology. Monoclonal antibody against Cdc6 and rabbit antisera against MCM3, MCM4, MCM5 and MCM6 were produced with the following peptide antigens and affinity purified by Dagene (Taiwan). MCM3: SDTEEEMPQVHTPKTAD; MCM4: SRRGRATPAQTPRSED; MCM5: KEVADEVTRPRPSGE; MCM6: KYLQLAEELIRPERNT. Polyclonal antibody against MCM2 was generated using a recombinant protein fragment of MCM2 (a.a. 792–892). Immunizing peptide for the phopho-MCM4 (Thr110) specific antibody was SGVRGpTPVRQRPD (pT being the phosphorylation site). Western blot analysis was performed after electrophoretic separation of polypeptides by 7.5 or 10% SDS–PAGE and trasfer to Hybond-C membranes. Blots were probed with the indicated primary and appropriate secondary antibodies, and detected by ECL chemiluminescence (Amersham).
Immunoprecipitation and in vitro pull-down assay
HeLa cells were extracted using a buffer containing: 20 mM HEPES (pH 7.4), 0.2 M NaCl, 0.5% TX-100, 5% glycerol, 1 mM EDTA, 1 mM EGTA, 10 mM β-glycerophosphate, 2 mM Na3VO4, 1 mM NaF, 1 mM DTT, 10 mM PMSF, 1 μg/ml leupeptin and 1μg/ml pepstatin A. For preparation of nuclear extracts, HeLa nuclei were isolated and lysed in nuclear extraction buffer (10 mM HEPES with pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 0.1% TX-100, 0.4 M NaCl, 10% glycerol, 1 mM DTT, 10 mM PMSF, 1 μg/ml leupeptin and 1μg/ml pepstatin A). All immunoprecipitations were performed with the indicated antibodies prebound to protein G-Sepharose (Amersham), and washed in the cell lysis buffer. For the in vitro pull-down assay, purified and bound GST-MCMs were independently incubated with eluted FLAG-hSpt16p or His6-SSRP1 in the cell lysis buffer. For FACT–MCM complex interaction assay, bound FACT was incubated with purified MCM subcomplexes. Protein-bound beads were then washed four times in the same buffer.
Plasmid-based dsRNAi
To establish a plasmid-based dsRNAi system targeting endogenous SSRP1, MCM3 or MCM4, annealed oligonucleotides corresponding to partial sequence were designed and ligated to the pSuper.neo+GFP (OligoEngine) according to the manufacturer's instructions. The cDNA sequence of the targeted mRNA region for different genes is as follows: SSRP1: 5′-TGGCAAGACCTTTGACTAC-3′ (nucleotides 677–695); MCM3: 5′-AAACGAGAAGAGGGCTAAC-3′ (nucleotides 171–189); MCM4: 5′-GACACCACACACAGTTATC-3′ (nucleotides 1095–1113). The same sequence in the inverted orientation was used as the nonspecific dsRNAi control.
Cell culture and transfection
All HeLa cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 100 U/ml penicillin and streptomycin. Cells were transfected using Lipofectamine (GIBCO) according to the manufacturer's instructions. Transient transfection was done for 48–54 h prior to cell harvest, unless otherwise noted. Generation of cell lines stably harboring MCM4 variants (Figure 4) was performed by first transfection of the corresponding plasmid and subsequent clone selection in the presence of 1 mg/ml G418.
Cell cycle synchronization and analysis
Cells were synchronized at the G1/S boundary by double thymidine block. Subconfluent culture of HeLa cells was blocked with 2 mM thymidine (final) and incubated for 19 h. Cells were washed three times with phosphate-buffered saline (PBS) and then incubated with fresh serum-rich medium for 10 h. Subsequently, the second block with 2 mM thymidine lasted for 15 h. To obtain S-phase cells, double thymidine-arrested cells were washed three times with PBS and released into S-phase in complete medium for 4 h. Mitotic cells were harvested by first treating subconfluent cells with 100 ng/ml nocodazole for 14 h, and subsequent detachment by mechanical shake-off. Synchronization was followed by flow-cytometric analysis (Supplementary Figure S2A). DNA content-based profiling of cell cycle (Figure 4D) as well as kinetics of DNA synthesis (via BrdU labeling) (Figure 4E and F) were also performed by FACS analysis, as described previously (Tan and Lee, 2004). Percentages of BrdU-positive cells and the ‘labeling index' were quantitatively determined by the CellQuest software (Becton Dickinson). α-Amanitin (Sigma) was dissolved in PBS. Treatment of drug (of the indicated concentrations) was performed for 12–14 h in a 37°C cell culture incubator.
RT–PCR
First-strand cDNA synthesis was performed with the SuperScript II Reverse Transcriptase (Invitrogen), according to the manufacturer's instructions. Sequences of the primers used to PCR-amplify the lamin B2, TIMM 13 or GADPH transcripts are as follows:

PCR reactions were performed for 25–27 cycles. For quantitative determination by real-time PCR, target transcripts were analyzed using the LightCycler system and LightCycler-FastStart DNA Master SYBR Green I dye (Roche Diagnostics). Triplicate PCRs were performed. Lamin B2 and TIMM13 mRNA abundance was analyzed using the above primer sets. Results were normalized to GAPDH values of the respective sample.
Chromatin immunoprecipitation
ChIP assays were modified from previously described methods (Su et al, 2003; Paixao et al, 2004). Briefly, HeLa cells (exponentially growing or synchronized) were crosslinked with 1% formaldehyde for 10 min at 37°C. The nuclei were isolated and sonicated into oligonucleosomes of ∼500–600 bp in length. The sheared chromatin was immunoprecipitated overnight with protein G-agarose previously bound with the 10D1, anti-MCM4 or control antibody. After extensive washes, the immunoprecipitates were subjected to deproteination and crosslinking reversal. For the sequential ChIP experiment, precipitate from the first round (anti-MCM4 IP) was recovered by an elution solution (1% SDS and 0.1 M NaHCO3) and used to perform a second round of ChIP using the 10D1 or control antibody. The presence of genomic DNA in the precipitates was detected by PCR with the B48 primer set and a background primer set. The background primers anneal to a region with no annotated genes, 30 kb upstream of the lamine B2 origin sequence on chromosome 19, and have the following sequences: 5′-CTATGCCAAGCCCATTCTAGGTCCT-3′ (sense), 5′-GCAGGGAAACTGTGCACAGCAAGAG-3′ (antisense). The primer pair corresponding to the coding region of the γ-actin gene locus has the following sequences: 5′-GCTGTTCCAGGCTCTGTTCC-3′ (sense), 5-ATGCTCACACGCCACAACATGC-3′ (antisense). Upon amplification for 27–30 cycles, the products were resolved by 2% agarose gels and visualized with ethidium bromide staining.
Real-time PCR
DNA samples from ChIP preparations were quantified by real-time PCR using LightCycler system and LightCycler FastStart TaqMan Probe Mix (Roche Diagnostics) with dual labeled probes selected from the human Universal ProbeLibraray Set (Roche Diagnostics). The probes are labeled with a reporter dye at 5′-end and a quencher at the 3′-end. The primers and the probes were: B48 (origin): forward primer, 5′-TGTACAACACTCCAATAAACATTTTG-3′; reverse primer, 5′-GAGCTTCCCCTCAGGAATAAA-3′; probe, #34; B13 (non-origin): forward primer, 5′-GCCAGCTGGGTGGTGATA-3′; reverse primer, 5′-GAGGCGTGTTTTCCTCCTC-3′; probe, #70; background: forward primer, 5′-AAACGTGACCTCAGACAGAGC-3′; reverse primer, 5′-CTGGCAGGTCTGGGACTATG-3′; probe, #7. Triplicate PCRs for each sample were carried out. The results are given as percentages of inputs and represent the mean±s.d. of at least three independent experiments. Control ChIP assays with nonspecific antisera were performed in each ChIP experiment.
Competitive PCR-based measurement of origin activity
Genomic DNA was isolated from 5 × 107 exponentially growing HeLa (control or dsRNAi) cells based on the protocol described elsewhere (Paixao et al, 2004). Upon fractionation by sucrose gradient centrifugation, single-stranded (nascent) DNA fragments of the length 0.7–1.5 kb were isolated and concentrated by ethanol precipitation. Competitive PCR analysis was performed with the B48 and B13 primer sets (Dx/Sx), sequences of which were detailed in the above publication. To generate a competitor template, a DNA segment that carries the tandem sequences of the B48 and B13 primers at two ends separated by a 180-bp linker DNA sequence in middle was amplified by PCR. Thus, as the result of competitive PCR, the length of competitor products would be ∼240 bp, as opposed to 160 bp of the genomic DNA products. Competitive PCR amplification was performed with a constant volume of nascent DNA and decreasing amounts of the competitor fragment, at the conditions of 95°C for 30 s, 56°C for 25 s and 72°C for 25 s (40 cycles). PCR products were resolved on agarose gel (2%) and stained with ethidium bromide. Intensities of DNA bands on the UV-illuminated images were quantitatively determined by a Fujifilm Luminescent Image Analyzer LAS-1000plus and the software Image Gauge. To determine the amount of target genomic molecules in the nascent DNA sample, linear relationship between the competitor/target genomic DNA (C/T) ratios and the concentrations of the input competitor was first plotted and deduced. Based on the equation, the target DNA concentration was then calculated as the amount of competitor DNA at C/T=1. For each type of cell line, the ratio of the DNA products amplified by B48 and B13 primer pairs was subsequently evaluated. The capacity of the endogenous origin in promoting DNA replication initiation was then compared between control and SSRP1-knockdown cells.
Chromatin unwinding assay
A PCR-based strategy was applied to generate the linear, tailed DNA substrate for the nucleosome reconstitution and the helicase assay (Figure 3A). The template backbone is a 180 bp-long sequence covering part of the mouse AGP gene promoter (Chang et al, 1990). Two sets of primer pairs were designed to generate from PCR amplification two types of DNA segments, ‘upper' and ‘lower'. The sequences of the primers are:

Both PCR products were purified and mixed equally and then subjected to denaturing (95°C for 5 min) and renaturing (65°C for 15 min, 37°C for 1 h). Only one of the four likely renatured intermediates (25% of the final products) can subsequently be isotopically labeled by T4 DNA polymerase in the presence of [α-32P]dCTP. Nucleosome cores were prepared from HeLa cells (Mizzen et al, 1999) and reconstituted onto the 200-bp, end-labeled DNA fragment by octamer transfer method (Studitsky et al, 1995; Utley et al, 1996). Helicase assay was performed on the naked DNA or nucleosomal templates with the indicated amounts of factors under the conditions as described previously (You et al, 2003). Preincubation experiments (Figure 3F) were performed with the indicated proteins or template, for a reaction length of 20 min, before adding the remaining components. The reactions were terminated and deproteinated by the addition of EDTA (20 mM), SDS (1%) and 5 μg of proteinase K. Degree of unwinding was observed as in the DNA helicase assay (7.5% native PAGE/TBE).
Supplementary Material
Supplementary Table and Figures
Acknowledgments
We thank Hiroshi Nojima for the MCM plasmids. We are especially grateful to Drs Ming-Jer Tsai, Ruey-Hwa Chen, Ching-Jin Chang and Margaret S Ho for critical reading of the manuscript, and members of the SC Lee and CT Chien labs for technical assistance. This work was supported by National Science Council Grants NSC93-2320-B002-109 and the Institute of Biological Chemistry, Academia Sinica (to SCL), and National Health Research Institute (NHRI) postdoctoral fellowship award PD9302 (to BCT).
References
- Abdurashidova G, Danailov MB, Ochem A, Triolo G, Djeliova V, Radulescu S, Vindigni A, Riva S, Falaschi A (2003) Localization of proteins bound to a replication origin of human DNA along the cell cycle. EMBO J 22: 4294–4303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdurashidova G, Deganuto M, Klima R, Riva S, Biamonti G, Giacca M, Falaschi A (2000) Start sites of bidirectional DNA synthesis at the human lamin B2 origin. Science 287: 2023–2026 [DOI] [PubMed] [Google Scholar]
- Aggarwal BD, Calvi BR (2004) Chromatin regulates origin activity in Drosophila follicle cells. Nature 430: 372–376 [DOI] [PubMed] [Google Scholar]
- Alexiadis V, Varga-Weisz PD, Bonte E, Becker PB, Gruss C (1998) In vitro chromatin remodelling by chromatin accessibility complex (CHRAC) at the SV40 origin of DNA replication. Embo J 17: 3428–3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio OM, Weinstein DM, Bell SP (1997) Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell 91: 59–69 [DOI] [PubMed] [Google Scholar]
- Bell SP, Dutta A (2002) DNA replication in eukaryotic cells. Annu Rev Biochem 71: 333–374 [DOI] [PubMed] [Google Scholar]
- Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D (2003) FACT facilitates transcription-dependent nucleosome alteration. Science 301: 1090–1093 [DOI] [PubMed] [Google Scholar]
- Bozhenok L, Wade PA, Varga-Weisz P (2002) WSTF-ISWI chromatin remodeling complex targets heterochromatic replication foci. EMBO J 21: 2231–2241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke TW, Cook JG, Asano M, Nevins JR (2001) Replication factors MCM2 and ORC1 interact with the histone acetyltransferase HBO1. J Biol Chem 276: 15397–15408 [DOI] [PubMed] [Google Scholar]
- Chang CJ, Chen TT, Lei HY, Chen DS, Lee SC (1990) Molecular cloning of a transcription factor, AGP/EBP, that belongs to members of the C/EBP family. Mol Cell Biol 10: 6642–6653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K (2006) GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol 8: 358–366 [DOI] [PubMed] [Google Scholar]
- Giacca M, Pelizon C, Falaschi A (1997) Mapping replication origins by quantifying relative abundance of nascent DNA strands using competitive polymerase chain reaction. Methods 13: 301–312 [DOI] [PubMed] [Google Scholar]
- Ishimi Y (1997) A DNA helicase activity is associated with an MCM4, -6, and -7 protein complex. J Biol Chem 272: 24508–24513 [DOI] [PubMed] [Google Scholar]
- Keller DM, Lu H (2002) p53 serine 392 phosphorylation increases after UV through induction of the assembly of the CK2.hSPT16.SSRP1 complex. J Biol Chem 277: 50206–50213 [DOI] [PubMed] [Google Scholar]
- Keller DM, Zeng X, Wang Y, Zhang QH, Kapoor M, Shu H, Goodman R, Lozano G, Zhao Y, Lu H (2001) A DNA damage-induced p53 serine 392 kinase complex contains CK2, hSpt16, and SSRP1. Mol Cell 7: 283–292 [DOI] [PubMed] [Google Scholar]
- Labib K, Tercero JA, Diffley JF (2000) Uninterrupted MCM2–7 function required for DNA replication fork progression. Science 288: 1643–1647 [DOI] [PubMed] [Google Scholar]
- Lee JK, Hurwitz J (2001) Processive DNA helicase activity of the minichromosome maintenance proteins 4, 6, and 7 complex requires forked DNA structures. Proc Natl Acad Sci USA 98: 54–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeRoy G, Orphanides G, Lane WS, Reinberg D (1998) Requirement of RSF and FACT for transcription of chromatin templates in vitro. Science 282: 1900–1904 [DOI] [PubMed] [Google Scholar]
- Mizzen CA, Brownell JE, Cook RG, Allis CD (1999) Histone acetyltransferases: preparation of substrates and assay procedures. Methods Enzymol 304: 675–696 [DOI] [PubMed] [Google Scholar]
- Okuhara K, Ohta K, Seo H, Shioda M, Yamada T, Tanaka Y, Dohmae N, Seyama Y, Shibata T, Murofushi H (1999) A DNA unwinding factor involved in DNA replication in cell-free extracts of Xenopus eggs. Curr Biol 9: 341–350 [DOI] [PubMed] [Google Scholar]
- Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D (1998) FACT, a factor that facilitates transcript elongation through nucleosomes. Cell 92: 105–116 [DOI] [PubMed] [Google Scholar]
- Orphanides G, Wu WH, Lane WS, Hampsey M, Reinberg D (1999) The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature 400: 284–288 [DOI] [PubMed] [Google Scholar]
- Paixao S, Colaluca IN, Cubells M, Peverali FA, Destro A, Giadrossi S, Giacca M, Falaschi A, Riva S, Biamonti G (2004) Modular structure of the human lamin B2 replicator. Mol Cell Biol 24: 2958–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas DL Jr, Frisch R, Weinreich M (2004) The NAD(+)-dependent Sir2p histone deacetylase is a negative regulator of chromosomal DNA replication. Genes Dev 18: 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poot RA, Bozhenok L, van den Berg DL, Steffensen S, Ferreira F, Grimaldi M, Gilbert N, Ferreira J, Varga-Weisz PD (2004) The Williams syndrome transcription factor interacts with PCNA to target chromatin remodelling by ISWI to replication foci. Nat Cell Biol 6: 1236–1244 [DOI] [PubMed] [Google Scholar]
- Saunders A, Werner J, Andrulis ED, Nakayama T, Hirose S, Reinberg D, Lis JT (2003) Tracking FACT and the RNA polymerase II elongation complex through chromatin in vivo. Science 301: 1094–1096 [DOI] [PubMed] [Google Scholar]
- Schlesinger MB, Formosa T (2000) POB3 is required for both transcription and replication in the yeast Saccharomyces cerevisiae. Genetics 155: 1593–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacha A, Bell SP (2001) Interactions between two catalytically distinct MCM subgroups are essential for coordinated ATP hydrolysis and DNA replication. Mol Cell 8: 1093–1104 [DOI] [PubMed] [Google Scholar]
- Shimojima T, Okada M, Nakayama T, Ueda H, Okawa K, Iwamatsu A, Handa H, Hirose S (2003) Drosophila FACT contributes to Hox gene expression through physical and functional interactions with GAGA factor. Genes Dev 17: 1605–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedman W, Deng Z, Lu F, Lieberman PM (2004) ORC, MCM, and histone hyperacetylation at the Kaposi's sarcoma-associated herpesvirus latent replication origin. J Virol 78: 12566–12575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studitsky VM, Clark DJ, Felsenfeld G (1995) Overcoming a nucleosomal barrier to transcription. Cell 83: 19–27 [DOI] [PubMed] [Google Scholar]
- Su WC, Chou HY, Chang CJ, Lee YM, Chen WH, Huang KH, Lee MY, Lee SC (2003) Differential activation of a C/EBP beta isoform by a novel redox switch may confer the lipopolysaccharide-inducible expression of interleukin-6 gene. J Biol Chem 278: 51150–51158 [DOI] [PubMed] [Google Scholar]
- Tan BC, Lee SC (2004) Nek9, a novel FACT-associated protein, modulates interphase progression. J Biol Chem 279: 9321–9330 [DOI] [PubMed] [Google Scholar]
- Utley RT, Owen-Hughes TA, Juan LJ, Cote J, Adams CC, Workman JL (1996) In vitro analysis of transcription factor binding to nucleosomes and nucleosome disruption/displacement. Methods Enzymol 274: 276–291 [DOI] [PubMed] [Google Scholar]
- Varga-Weisz P (2005) Chromatin remodeling factors and DNA replication. Prog Mol Subcell Biol 38: 1–30 [DOI] [PubMed] [Google Scholar]
- Wittmeyer J, Formosa T (1997) The Saccharomyces cerevisiae DNA polymerase alpha catalytic subunit interacts with Cdc68/Spt16 and with Pob3, a protein similar to an HMG1-like protein. Mol Cell Biol 17: 4178–4190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmeyer J, Joss L, Formosa T (1999) Spt16 and Pob3 of Saccharomyces cerevisiae form an essential, abundant heterodimer that is nuclear, chromatin-associated, and copurifies with DNA polymerase alpha. Biochemistry 38: 8961–8971 [DOI] [PubMed] [Google Scholar]
- You Z, Ishimi Y, Mizuno T, Sugasawa K, Hanaoka F, Masai H (2003) Thymine-rich single-stranded DNA activates Mcm4/6/7 helicase on Y-fork and bubble-like substrates. EMBO J 22: 6148–6160 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table and Figures


