Abstract
Spitz (Spi) is a TGFα homolog that is a cardinal ligand for the Drosophila EGF receptor throughout development. Cleavage of the ubiquitously expressed transmembrane form of Spi (mSpi) precedes EGF receptor activation. We show that the Star and Rhomboid (Rho) proteins are necessary for Spi cleavage in Drosophila cells. Complexes between the Spi and Star proteins, as well as between the Star and Rho proteins were identified, but no Spi–Star–Rho triple complex was detected. This observation suggests a sequential activity of Star and Rho in mSpi processing. The interactions between Spi and Star regulate the intracellular trafficking of Spi. The Spi precursor is retained in the periphery of the nucleus. Coexpression of Star promotes translocation of Spi to a compartment where Rho is present both in cells and in embryos. A Star deletion construct that maintains binding to Spi and Rho, but is unable to facilitate Spi translocation, lost biological activity. These results underscore the importance of regulated intracellular trafficking in processing of a TGFα family ligand.
Keywords: Spitz, Star, Rhomboid, EGF receptor, intracellular trafficking, ligand cleavage
The EGF receptor (DER) pathway in Drosophila fulfills multiple roles at all stages of development. Most notable is the specification of cell fates in the follicle cells, the embryo, and the imaginal discs. Other roles include control of cell division, suppression of apoptosis, and regulation of cell migration (for review, see Schweitzer and Shilo 1997; Nilson and Schupbach 1999). The wide spectrum of DER functions necessitates a tight control of receptor activation in terms of the timing of induction as well as the spatial domains of activation. This regulation resides at the level of the ligands.
Flexible regulation of receptor activation is achieved by a battery of ligands with distinct properties that are suitable for the biological contexts in which they function. Gurken, a TGFα homolog, is restricted to the activation of DER in the follicle cells of the ovary (Neuman-Silberberg and Schupbach 1993; Gonzalez-Reyes et al. 1995; Roth et al. 1995). Vein, a secreted ligand, possesses an inherently weaker activation capacity and is used in tissues where low activation levels are required, for example, as a positive feedback reinforcement to the initial activation of the receptor by other ligands (Schnepp et al. 1996, 1998; Wasserman and Freeman 1998; Golembo et al. 1999). Vein also induces the muscle attachment cell fate, following its accumulation at the receiving cell (Yarnitzky et al. 1997). Argos is a secreted ligand that binds the receptor but inhibits activation by competing with the activating ligands (Freeman et al. 1992; Schweitzer et al. 1995a; Jin et al. 2000). It is induced as a negative feedback response and plays a major role in restricting the activation range of the activating ligands (Golembo et al. 1996b). Finally, the cardinal activating ligand is Spitz (Spi), which is responsible for DER activation in most tissues (Rutledge et al. 1992).
Spi is produced as an inactive transmembrane precursor (mSpi) that is broadly expressed, but is inactive as such. Even after ectopic expression of the Spi precursor, no activation of DER is observed. Only when cleaved does Spi become a potent ligand of DER, as can be assayed by the induction of target genes and the accumulation of activated MAP kinase (dpERK) (Schweitzer et al. 1995b; Gabay et al. 1996, 1997). Regulated cleavage of Spi precursor is thus the key to DER induction. Elements participating in the regulation of Spi have been identified by genetic screens. The cuticle phenotype of spi mutant embryos is characterized, most notably, by reduction in the width of the denticle bands and occasional fusion of denticle bands between adjacent segments. Several other mutants with similar phenotypes have been classed as the spitz group (Mayer and Nusslein-Volhard 1988). Most notable are rhomboid (rho), encoding a protein with seven transmembrane domains (Bier et al. 1990), and Star coding for a type II single transmembrane domain protein (Kolodkin et al. 1994). Rho homologs were identified in Caenorhaditis elegans and vertebrates (Pascall and Brown 1998), and several additional members of the family are also present in the Drosophila genome (Guichard et al. 2000; Wasserman et al. 2000). Rho protein is concentrated in punctate dots under the plasma membrane (Sturtevant et al. 1996). Star, which is a novel type II transmembrane domain protein, is mostly in the periphery of the nucleus, consistent with rough endoplasmic reticulum (ER) staining (Pickup and Banerjee 1999).
Several experimental approaches have placed Rho and Star upstream of Spi activity. The ability to trigger DER ectopically by expression of secreted Spi (sSpi) provided a way to carry out epistasis tests. Expression of sSpi can induce ectopic phenotypes in embryos mutant for rho or Star, thus bypassing the requirement for these genes (Schweitzer et al. 1995b). Second, rho or Star is able to function nonautonomously in the embryo and the wing. When the normal gene products of Rho or Star were provided only in the midline cells in embryos deficient for the respective gene product, rescue of EGF receptor activation in the adjacent ectodermal cells was observed (Golembo et al. 1996a). This finding is consistent with the capacity of Rho and Star to promote production of a secreted ligand in the midline cells. Similarly rho or Star mutant clones in the wing show nonautonomous defects in the formation of veins (Guichard et al. 1999). Finally, examination of Spi processing in a heterologous Xenopus system showed that both Rho and Star must be expressed in the same cells as mSpi, for the production of sSpi (Bang and Kintner 2000). The above results imply that Rho and Star regulate the cleavage of mSpi.
Both DER and Spi are ubiquitously expressed (Zak et al. 1990; Rutledge et al. 1992). As a rule, the dynamic activation pattern of the DER pathway follows expression of rho (Gabay et al. 1997). Accordingly, ectopic expression of Rho triggers DER activation in all tissues (Noll et al. 1994; Golembo et al. 1996a). In some cases, rho itself becomes a target gene for the DER pathway, for example, during oogenesis and eye development (Sapir et al. 1998; Wasserman and Freeman 1998). This feedback allows multiple cycles of activation of the pathway, as in every cell where Rho is expressed, a new round of ligand processing ensues. In contrast, the Star protein is expressed ubiquitously (Kolodkin et al. 1994). However, its levels may be limiting, as Star mutants were consistently isolated in many screens for dosage-sensitive suppressors of the pathway. This feature is true especially in the eye, where Star has a haplo-insufficient, dominant phenotype due to the reduced activation of DER.
Although the involvement of Star and Rho in Spi processing has been established, their mechanism of action was not known. We show that Star and Rho can promote mSpi cleavage in Drosophila S2 cells. This finding provides a cell culture system to dissect the mechanism of processing. Protein–protein interactions between the different components were identified. mSpi–Star or Star–Rho complexes were detected, but no triple complex was found. This result is in accordance with a sequential activity of Star and Rho in mSpi processing. Intracellular trafficking of mSpi is regulated and crucial for cleavage. mSpi is retained in the periphery of the nucleus and extends into the cytoplasm in a manner consistent with an ER localization pattern. Star alters the trafficking of mSpi, targeting it to a compartment containing Rho, where cleavage can take place. A deletion construct of Star that binds mSpi but does not translocate it, is biologically inactive. These results highlight the role of Star in promoting intracellular trafficking of mSpi, to allow processing.
Results
Star and Rho are required for Spi cleavage in S2 cells
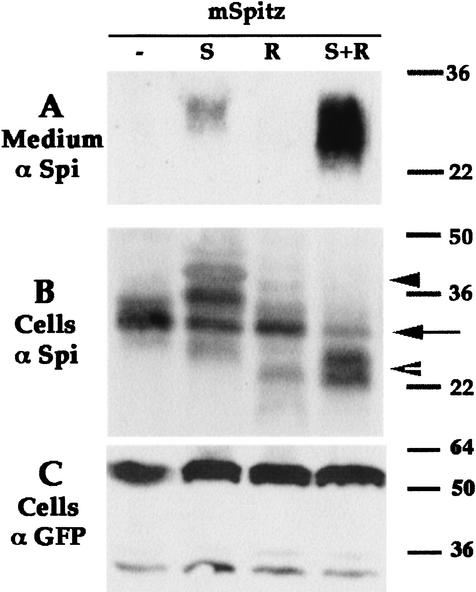
Star and Rho were implicated in Spi processing on the basis of genetic data and epistasis experiments. However, their direct role in cleavage has not been demonstrated in Drosophila. We explored the possibility of establishing a cleavage assay for Spi in Drosophila Schneider S2 cells. Previous experiments have shown that expression of mSpi in S2 cells does not lead to the appearance of sSpi in the medium. In contrast, expression of sSpi in these cells results in accumulation of biologically active Spi in the medium (Schweitzer et al. 1995b; Fig. 1A). Endogenous Star or Rho proteins could not be detected in S2 cells with the available antibodies (Schweitzer et al. 1995b). The effects of Rho or Star on processing of mSpi could thus be tested.
Figure 1.
Star and Rho are required for mSpi processing in S2 cells. Schneider S2 cells were transiently transfected with combinations of different expression constructs. mSpi alone did not undergo cleavage, coexpression with Star gave rise to low levels of sSpi in the medium, whereas coexpression with Star and Rho generated high levels of sSpi. Note that in the cell lysates higher molecular weight forms of mSpi were observed in the mSpi+Star cells (arrowhead), whereas lower molecular weight forms were detected in cells expressing mSpi+Star+Rho (open arrowhead), and the levels of original mSpi (arrow) were reduced. (A) Medium probed with anti-Spi. (B) Cell lysates probed with anti-Spi. (C) Cell lysates probed with anti-GFP, to detect cotransfected UAS-CD8–GFP show similar transfection efficiencies. In this experiment we used mSpi, Star-HA (S), and Rho-HA (R).
Coexpression of mSpi and Rho in a transient transfection did not yield sSpi in the medium. In contrast, coexpression of mSpi and Star resulted in accumulation of low levels of sSpi in the medium (Fig. 1A). The low levels of sSpi detected after Star coexpression could reflect a residual activity of the endogenous Rho expressed by the S2 cells or Rho-independent processing. To examine the possible role of Rho in processing, cells coexpressing mSpi, Star, and Rho were generated. Large quantities of sSpi were detected in the medium of these cells (Fig. 1A). Activity of the cleaved protein was tested on S2 cells expressing DER. Accumulation of dpERK in these cells was indeed observed (data not shown).
Star and Rho also affected the level and processing of mSpi in the producing cells. In extracts of cells expressing the three proteins, the level of mSpi was reduced, and lower molecular weight forms appeared, which may correspond to the cleaved Spi before it has been secreted to the medium. In contrast, extracts of cells expressing mSpi and Star displayed higher molecular weight forms of Spi (Fig. 1B). These forms were reduced in size following treatment with PNGase F, which cleaves high mannose and complex N-linked oligosaccharides, typical to the Golgi (data not shown). These results show that Star and Rho are required for high level cleavage of Spi. The mSpi cleavage assay in S2 cells provided the basis for examining the mechanism of Spi processing.
Dissection of functional Star domains
To identify the domain(s) of Star required for its biological activity, we wanted to generate deletion constructs. Because Star is a novel protein with no defined domains (except for the transmembrane domain), no clues were available for the generation of such constructs. Searching the database, we identified a Bombyx mori cDNA sharing homology with Star. Complete sequence of this cDNA identified a type II transmembrane protein of 315 amino acids, which we termed BmS. Notably, the amino-terminal cytoplasmic domain of this protein is only 74 residues long, and the major boxes of homology are located immediately carboxy-terminal to the transmembrane domain (Fig. 2A). This pattern of homology raised the possibility that most of the amino-terminal domain of Star may be dispensable for its function, and prompted us to examine constructs of Star missing most of the amino- or carboxy-terminal domains. Two truncated Star constructs were generated. NTM contains the entire amino-terminal and transmembrane domains, and is truncated 16 residues after the transmembrane domain. Conversely, in TMC, the amino-terminal 259 residues were deleted, and an initiator methionine was added (schematized in Fig. 2B).
Figure 2.
B. mori Star homolog and Star deletion constructs. (A) Alignment of the BmS and Star protein sequences. Note the shorter amino-terminal domain of BmS, and the homology to Star at the carboxyl terminus. The transmembrane domain is underlined, and the amino or carboxyl terminus of TMC or NTM are shown by filled or open arrowheads, respectively. (B) Schematic representation of the Star deletion constructs and BmS. The carboxy-terminal domain is extracellular, as Star and BmS are type II transmembrane proteins (BmS GenBank accession no. AF455272).
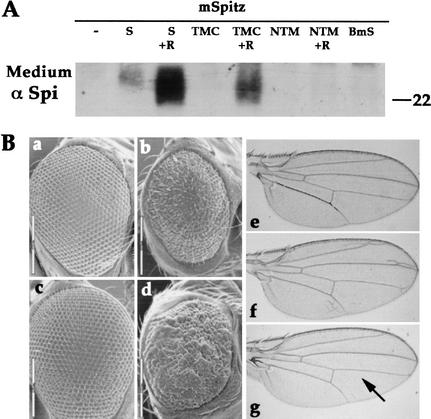
The biological activity of the constructs was tested in S2 cells. Only the TMC construct was capable of promoting mSpi cleavage, but at a lower efficiency than full-length Star. sSpi could be detected in the medium of cells coexpressing mSpi, TMC and Rho, but not in the medium of cells expressing only mSpi and TMC (Fig. 3A). Stable transfected lines expressing mSpi and TMC provided a more sensitive assay and produced low levels of sSpi (data not shown). The lower levels of sSpi, however, indicated that this construct was less potent than the full-length Star protein. The NTM and BmS constructs were not active alone or even in the presence of Rho (Fig. 3A and data not shown). In addition, after sequencing the Star54 null allele, we identified a termination codon at position Q387 in the extracellular domain. This protein is 67 residues longer than NTM and yet is inactive, indicating that the carboxy-terminal region of Star is essential for its biological function.
Figure 3.
Biological activity of Star deletion constructs and BmS. (A) The capacity of Star constructs to promote mSpi cleavage in S2 cells was tested alone or in combination with Rho. The medium was probed with anti-Spi. sSpi was detected only in the medium of cells transfected with mSpi, TMC, and Rho. Thus, only TMC showed biological activity, which, however, is lower than that of full-length Star. (B) The activity of the constructs was also tested in transgenic flies, for their capacity to rescue Star haplo-insufficiency in the eye, and induce ectopic eye and wing phenotypes. The NTM and BmS did not show any activity (data not shown), whereas the TMC construct displayed a weak activity and was thus capable of rescuing the haplo-insufficiency, but did not elicit ectopic phenotypes. (a) wild-type; (b) Star −/+; (c) Star −/+; GMR-Gal4/UAS-TMC; (d) Star −/+; GMR-Gal4/UAS-Star; (e) MS1096-Gal4; (f) MS1096-Gal4/UAS-Star; (g) MS1096-Gal4/UAS-TMC. (Arrow) Ectopic wing veins. The TMC construct normally gave rise to wild-type wings and only rarely to small extra veins.
The TMC, NTM, and BmS constructs were also tested in flies heterozygous for the Star mutation, which display rough eyes due to haplo-insufficiency. Again, only ectopic expression of the TMC construct in eye discs (by GMR-Gal4) was capable of rescuing the phenotype, giving rise to wild-type eyes (Fig. 3Bc and data not shown). High levels of expression of full-length Star (in Star heterozygous flies, as well as in wild-type flies) led to the appearance of rough eyes and to the formation of extra wing veins, consistent with DER hyperactivation (Fig. 3B, panels d and f). The reduced biological activity of TMC was evident, as it was not capable of eliciting these hyperactivation phenotypes (Fig. 3B, panels c and g). We conclude that, in cells and in flies, only the Star construct containing the carboxy-terminal domain retained the essential activities of Star, but was less potent than full-length Star.
Star forms a complex with Spi
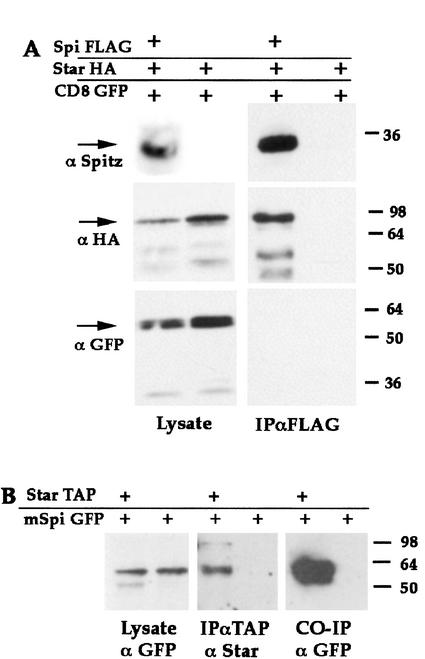
Having shown that Star is essential for mSpi cleavage, we asked whether it interacts directly with the mSpi protein. We examined the capacity of Star to form protein complexes with mSpi. Cells were cotransfected with constructs expressing mSpi–FLAG, Star–HA, and, as a negative control, CD8–GFP. Expression of the three proteins in cell lysates was verified by probing with anti-Spi, anti-HA, and anti-GFP, respectively. mSpi was immunoprecipitated by anti-FLAG, and coprecipitation was followed by anti-HA and anti-GFP immunoblotting (Fig. 4A). Indeed, immunoprecipitation of mSpi gave rise to significant coprecipitation of Star. Absence of CD8–GFP coprecipitation indicated that the observed interactions were specific. In all subsequent coimmunoprecipitation experiments, CD8–GFP was included as a control for the specificity of coimmunoprecipitation, but will not be shown. The robust interaction between mSpi and Star was also demonstrated in the reciprocal experiment, where immunoprecipitation of Star–TAP resulted in coprecipitation of mSpi–GFP (Fig. 4B).
Figure 4.
mSpi–Star protein interactions. (A) mSpi-FLAG was expressed in S2 cells with Star–HA and CD8–GFP. Immunoprecipitation of mSpi–FLAG coprecipitated Star–HA (detected by anti-HA), but not CD8–GFP (detected by anti-GFP). The presence of mSpi–GFP was verified by anti-Spi. (B) In the reciprocal experiment, mSpi–GFP was expressed with Star–TAP. Immunoprecipitation of Star–TAP led to coprecipitation of mSpi.
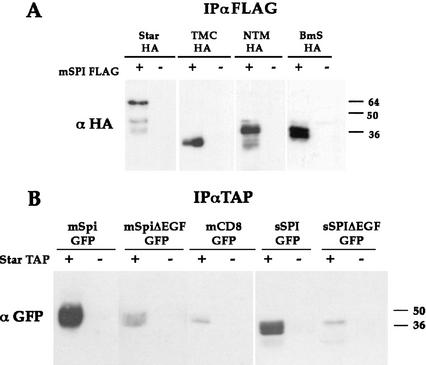
In view of the association between Spi and Star, we examined whether the biological activity of the Star deletion constructs correlates with the capacity to bind Spi. The NTM, TMC, or BmS constructs were coexpressed with mSpi in S2 cells. Although only the TMC construct, containing an intact carboxy-terminal domain, was active in cells and flies (Fig. 3), all three proteins coprecipitated mSpi with similar efficiencies (Fig. 5A). The basis for loss of Star biological activity, in spite of the ability to bind Spi, will be described below.
Figure 5.
Dissection of mSpi–Star interaction domains. (A) The capacity of the Star constructs to interact with mSpi was analyzed by coexpression of mSpi–FLAG, immunoprecipitation of mSpi–FLAG, and monitoring of coprecipitation of Star. All constructs tested retained the binding to mSpi. (B) The domains of mSpi required for interaction with Star were dissected by expression of Star–TAP together with Spi deletion constructs fused to GFP. Following immunoprecipitation of Star–TAP, coprecipitation of Spi was tested by anti-GFP. The EGF domain of Spi was shown to be essential for binding to Star.
To identify the domain(s) of mSpi mediating the interaction with Star, mSpi–GFP was constructed. GFP was inserted amino-terminal to the EGF domain. This construct retained the capacity to undergo Star- and Rho-dependent cleavage in S2 cells. It was also biologically active in flies, as coexpressed mSpi–GFP and Star in wing discs gave rise to abnormal, wrinkled wings, in accordance with previous experiments (Pickup and Banerjee 1999; data not shown). mSpi–GFP and several deletion constructs were tested for their ability to interact with Star. A secreted Spi protein lacking the cytoplasmic and transmembrane domains retained the capacity to bind Star, pointing to the extracellular domain of Spi as the mediator of binding. In accordance with this conclusion, a mSpi protein lacking the EGF domain showed only residual levels of binding to Star. Further deletions showed that the EGF domain of Spi is necessary for the interaction, as secreted constructs lacking this domain showed only background association with Star (Fig. 5B). Thus, the EGF domain of Spi mediates the interaction with Star.
Star associates with Rho
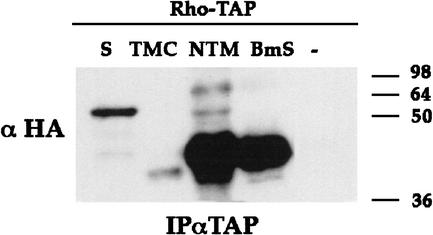
The interaction between Star and mSpi prompted us to examine whether Star also associates with Rho. Star–HA was coexpressed with Rho–TAP in S2 cells. Immunoprecipitation of Rho–TAP showed coprecipitation of Star (Fig. 6). In the reciprocal experiment, immunoprecipitation of Star–HA or Star–TAP coprecipitated Rho (data not shown). To identify the regions of Star necessary for this interaction, the experiment was repeated with the three Star constructs. The NTM and BmS constructs retained the interaction with Rho, whereas the TMC construct showed no detectable interaction or only weak interaction (Fig. 6). It appears that sequences within the amino-terminal cytoplasmic domain of Star that are not present in TMC mediate the binding to Rho.
Figure 6.
Star–Rho protein interactions. Interactions between Star and Rho were demonstrated by expression of Rho–TAP and Star–HA, immunoprecipitation of Rho–TAP, and detection of coprecipitating Star–HA. Examination of Star deletion constructs showed that NTM–HA and BmS–HA retained strong binding to Rho, whereas TMC–HA showed weak, borderline interaction. The cytoplasmic domain of Star is thus mediating the interaction with Rho.
Sequential binding of Star to Spi and Rho
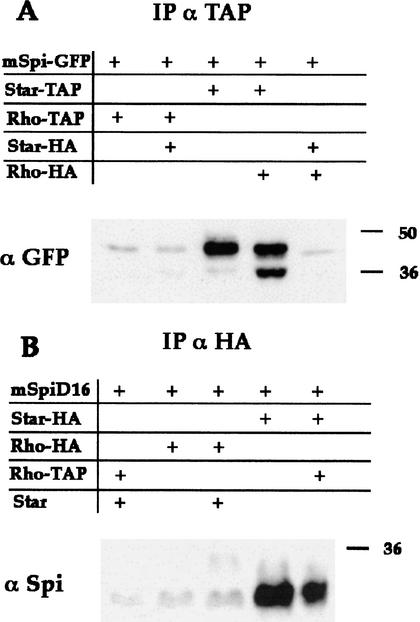
The capacity of Star to bind both Spi and Rho, raised the possibility that Star may function as a scaffold protein, to form a trimeric protein complex. Examination of possible direct interactions between mSpi and Rho failed to detect any significant coprecipitation (Fig. 7A). If a trimeric complex is formed, we would expect Star to promote coprecipitation of mSpi by Rho. However, we did not detect any elevation in mSpi coprecipitation with Rho when Star was also expressed. Under these conditions, Star retained the capacity to bind mSpi, regardless of the expression of Rho (Fig. 7A). Therefore, it appears that whereas mSpi–Star or Star–Rho complexes are formed in the cells, no trimeric complex is present.
Figure 7.
Sequential activity of Star and Rho in mSpi processing. The presence of mSpi–Star–Rho triple complexes was examined. (A) Rho–TAP is unable to precipitate detectable levels of mSpi. It was thus possible to ask whether coexpression of Star, which forms mSpi–Star and Star–Rho complexes, can promote the formation of a triple complex and allow Rho–TAP to coimmunoprecipitate mSpi–GFP, as detected by anti-GFP. The results were negative. Coprecipitation of mSpi–GFP with Star–TAP was also detected by anti-GFP, in the presence or absence of Rho. (B) To examine the possibility of a short-lived triple complex due to mSpi cleavage, a construct of mSpi devoid of a site necessary for cleavage was used (mSpiΔ16) and its coprecipitation was monitored by anti-Spi. Rho–HA or Star–HA were immunoprecipitated by anti-HA. Again, no triple complex was detected, arguing against the presence of a short-lived triple complex.
Because coexpression of Rho and Star significantly promoted processing of mSpi, it is possible that short-lived trimeric complexes are formed as a result of cleavage of mSpi. To circumvent this problem, we examined the formation of complexes with a noncleavable mSpi precursor. A mSpi protein in which 16 amino acids between the EGF and transmembrane domains were deleted, does not undergo cleavage in S2 cells (data not shown). A similar construct failed to be cleaved in the Xenopus assays (Bang and Kintner 2000). Like mSpi, the deleted mSpi protein readily formed complexes with Star, but not with Rho (Fig. 7B). Again, even in the presence of Star, Rho was not capable of coprecipitating this uncleavable Spi construct (Fig. 7B), arguing against the presence of even a transient triple complex. This experiment suggests that Star and Rho function sequentially in mSpi processing.
mSpi, Star, and Rho intracellular localization
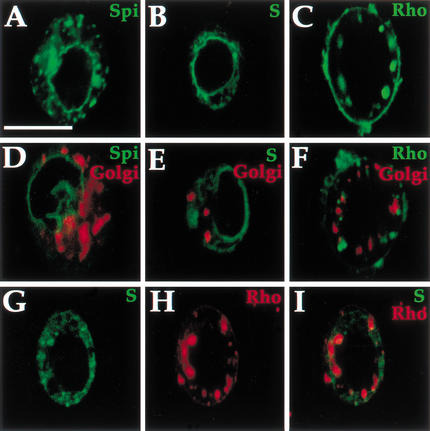
In view of the apparent sequential roles of Star and Rho, we sought to identify the cellular compartments in which they are active. The subcellular localization of Star and Rho may provide a clue as to their order of activity. Star protein was reported to be in the periphery of the nucleus, consistent with an ER localization (Pickup and Banerjee 1999). Immunostaining of S2 cells to follow Star overexpression confirmed this localization pattern (Fig. 8B,E). Immunostaining of Rho in S2 cells overexpressing Rho showed a punctate distribution that did not colocalize with a 120-kD integral Golgi membrane protein (Stanley et al. 1997), as well as plasma membrane staining (Fig. 8C,F). This pattern is in accordance with previous observations in embryos showing punctate endogenous Rho distribution. Following overexpression, plasma membrane staining was also detected (Sturtevant et al. 1996). Expression of Star and Rho showed only a restricted overlap (Fig. 8G–I). The distinct cellular distribution of these proteins suggests that a key feature in the regulation of mSpi processing may be its cellular trafficking. This possibility is in accord with the sequential activities of Star and Rho implied by the coprecipitation results.
Figure 8.
Intracellular localization of mSpi, Star, and Rho. Localization was monitored in live cells following transfection (using GFP), or in fixed cells following antibody staining. (A,D) mSpi–GFP shows a predominantly peripheral nuclear distribution (green), that does not colocalize with the Golgi marker (red); (B,E) Similarly, Star–HA shows a peripheral nuclear distribution (green) with no Golgi colocalization; (C,F) Rho–HA (green) is found in a punctate distribution, which is mostly nonoverlapping with the Golgi. (G–I) Double staining of Star and Rho–HA (anti-Star green, anti-HA red) shows the distinct distributions with minimal overlap. Scale bar, 20 μm.
The distribution of mSpi–GFP was followed in cells. mSpi–GFP showed a peripheral nuclear localization in S2 cells, as well as diffuse cytosolic staining, but it was not detected in the Golgi or plasma membrane (Fig. 8A,D). Therefore, it appears that the mSpi protein is retained in the ER, a compartment in which it cannot undergo cleavage.
To identify the domain(s) of mSpi required for its ER retention, a construct containing intact extracellular and transmembrane domains, but lacking the intracellular domain was examined. This protein showed only residual levels of peripheral nuclear localization and was detected primarily in a punctate pattern, consistent with exit from the ER (Fig. 9B). Therefore, the cytoplasmic domain of mSpi is necessary for its ER retention.
Figure 9.
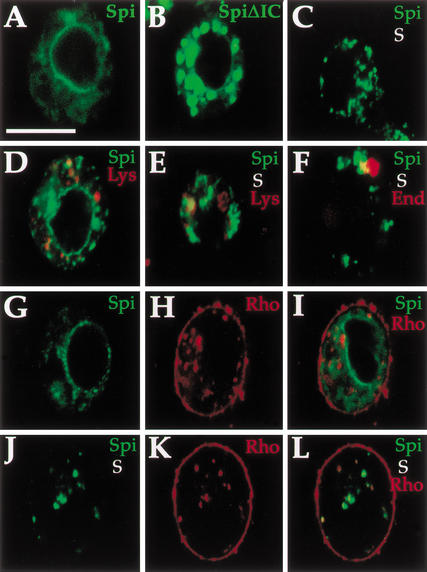
Star translocates mSpi from the ER in S2 cells. The effects of Star and Rho on mSpi intracellular distribution were examined in S2 cells. (A) mSpi–GFP showed a peripheral nuclear distribution. (B) mSpiΔIC–GFP lacking the intracellular domain was not retained and predominantly localized to plaques. (C) Following cotransfection with Star, the peripheral nuclear mSpi disappeared, and instead a punctate distribution was observed (mSpi–GFP, green, Star–HA, red). (D) mSpi–GFP showed only limited colocalization with lysosomes. (E) Similarly, the punctate distribution in mSpi–GFP+Star showed only some overlap with lysosomes (red), or endosomes (red) (F). (G–I) Coexpression of mSpi–GFP and Rho did not alter the distribution of mSpi or Rho (mSpi–GFP green, Rho–HA red). (J–L) Coexpression of mSpi–GFP, Star, and Rho–HA gave rise to a punctate mSpi–GFP distribution that was weaker in intensity than the transfections of mSpi+Star, because of a high level of cleavage and secretion. The mSpi–GFP vesicles predominantly colocalized with the Rho staining, indicating that Star targeted mSpi to a compartment containing Rho and consistent with sequential activity (mSpi–GFP, green; Rho–HA, red). Bar, 20 μm.
Star alters mSpi distribution
The peripheral nuclear distribution of Star and mSpi appears similar, whereas the punctate localization of Rho suggests that it is present in a more advanced compartment along the secretory network. Because Rho is required for efficient mSpi cleavage, how does mSpi reach the Rho compartment? One option is that Star forms a complex with mSpi and alters its intracellular localization. The distribution of mSpi–GFP was examined following coexpression with Star. Indeed, the original peripheral nuclear distribution of mSpi–GFP disappeared, and instead it was found in a prominent punctate pattern (Fig. 9C), which does not correspond to the Golgi and only partly overlaps with the lysosomes and endosomes (Fig. 9E,F). The alteration in mSpi distribution in the presence of Star was observed in all cells.
In contrast, expression of Rho with mSpi–GFP did not alter its distribution, in agreement with a lack of coprecipitation (Fig. 9G–I). However, coexpression of Rho with mSpi–GFP and Star reduced dramatically the levels of mSpi–GFP observed in the cells, in accordance with the results obtained in anti-Spi blots (Fig. 1B). The residual mSpi–GFP was found in a punctate staining colocalizing with Rho (Fig. 9J–L). These results showed that Rho can alter the levels of mSpi in the cells, but only in the presence of Star. They are consistent with sequential activity of Star and Rho, where Star is required first to transport mSpi from the ER to the compartment containing Rho, and Rho subsequently facilitates the cleavage and secretion.
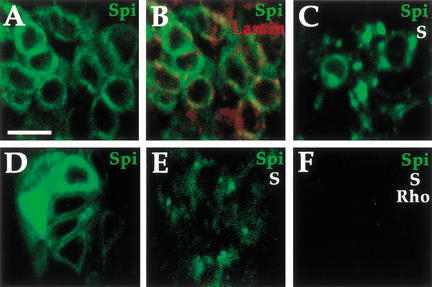
To test the distribution of mSpi and the effect o,,f Star in embryos, the mSpi–GFP construct was expressed in embryos. After crossing several independent transgenes of the UAS-mSpi–GFP construct to embryonic Gal4 drivers, only very weak fluorescence was seen, in contrast with other GFP constructs such as UAS-CD8–GFP. This result may indicate a high turnover and low steady-state levels of the noncleaved, mSpi precursor. Only in a small percentage of embryos could the mSpi–GFP be visualized. Embryos expressing mSpi–GFP showed a peripheral nuclear distribution colocalizing with the nuclear membrane protein Lamin. In contrast, embryos expressing mSpi–GFP and Star showed a punctate distribution (Fig. 10A–C). Although Star is normally expressed in all embryonic cells, we assume that the overexpression of mSpi–GFP alone generated an excess of this protein and thus essentially represents the mSpi distribution in the absence of Star.
Figure 10.
Star translocates mSpi from the ER in embryos. The effect of Star on mSpi–GFP localization was tested in embryos. (A) actin–Gal4/UAS-mSpi–GFP embryos show a peripheral nuclear localization. (B) Merge with anti-Lamin (red) marking the nuclear membrane. (C) actin–Gal4/UAS-mSpi–GFP/UAS–Star embryos show only residual peripheral nuclear Spi, and primarily a punctate localization. (D) Injection of UAS-mSpi–GFP into actin–Gal4 cyncytial embryos gave rise at stage 10 to a peripheral nuclear distribution. (E) Injection of UAS-mSpi–GFP and UAS-Star into prd–Gal4 embryos gave rise to the punctate distribution of mSpi–GFP. Similar results were obtained when UAS-mSpi–GFP was injected into prd–Gal4/UAS-Star embryos (data not shown). (F) Injection of UAS-mSpi–GFP+UAS-rho into prd–Gal4/UAS–Star embryos retained the punctate distribution of mSpi–GFP, but the intensity was lower, presumably due to secretion. Bar, 20 μm.
In an alternative way to circumvent the high mSpi turnover, we followed the distribution of mSpi–GFP after transient injection of the UAS-mSpi–GFP construct to embryos carrying prd–Gal4 or actin–Gal4. Monitoring fluorescence several hours after injection, a bright peripheral nuclear distribution of mSpi–GFP was seen (Fig. 10D). Coinjection of mSpi–GFP and Star constructs eliminated this distribution and led to the accumulation of mSpi–GFP in a punctate pattern, similar to the distribution in S2 cells (Fig. 10E). Conversely, coinjection of mSpi–GFP and Rho did not alter the mSpi peripheral nuclear distribution (data not shown). Finally, coinjection of mSpi–GFP, Star, and Rho also gave rise to the mSpi–GFP plaques, but their intensity was decreased (Fig. 10F). We conclude that the effect of Star on mSpi localization we observed in S2 cells also occurs in embryos and is likely to reflect the essential role of Star in mSpi intracellular trafficking.
Biological activity of Star constructs correlates with mSpi trafficking
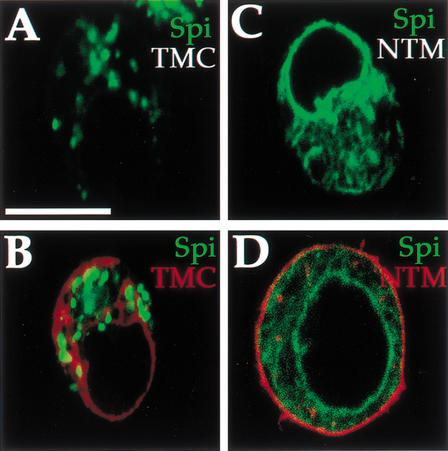
We wanted to test whether the biological activity of Star indeed corresponds to the capacity to promote mSpi trafficking. Although the two Star deletion constructs retained the ability to bind mSpi (Fig. 5A), only the TMC construct containing an intact carboxy-terminal domain was biologically active in cells and flies (Fig. 3). Each of the two Star constructs was coexpressed with the mSpi–GFP construct. In the TMC expressing cells, mSpi–GFP displayed the same punctate distribution observed when coexpressed with full-length Star (Fig. 11A,B). In contrast, mSpi–GFP remained in the peripheral nuclear pattern in the presence of NTM and BmS (Fig. 11C,D and data not shown). These results imply that the carboxy-terminal domain is required for the ability of Star to target mSpi to the proper compartment. Without this property, Star is not biologically active.
Figure 11.
Biological activity of Star correlates with mSpi trafficking. (A,B) The TMC construct, which is biologically active, albeit less than full-length Star, retained the ability to translocate mSpi–GFP from the ER (mSpi–GFP, green, TMC–HA, red). (C,D) The NTM construct, which binds mSpi but is not biologically active, failed to alter the peripheral nuclear distribution of mSpi (mSpi–GFP, green, NTM–HA, red). Interestingly, instead of the peripheral nuclear distribution of Star and TMC, NTM was found weakly in vesicles and predominantly on the plasma membrane, suggesting that the cellular distribution of Star itself is regulated and important for mSpi shuttling. Bar, 20 μm.
How is the cellular distribution of Star itself regulated? In cells, Star is found in a peripheral nuclear distribution (Fig. 8B). Examination of the distribution of the Star deletion constructs showed that TMC retained the same distribution as full-length Star (Fig. 11B). In contrast, NTM was not seen in the ER and was detected instead in a weak punctate and prominent membrane staining (Fig. 11D). Thus, the cellular distribution of Star itself is regulated and residues in the carboxy-terminal domain may promote interactions with other proteins that regulate the peripheral nuclear localization of Star.
Discussion
The DER pathway functions multiple times during Drosophila development to trigger diverse cell fates. Spi is the prominent ligand, which is used repeatedly, alone or in combination with other ligands. Processing of mSpi is the major limiting step in the activation of the DER pathway during development, and its regulation determines the precise activation pattern of DER. Whereas the Spi precursor is inactive, the secreted form is extremely potent. The mechanisms restricting the activity of mSpi on the one hand, while precisely triggering cleavage on the other, are thus at the heart of DER pathway regulation.
Several experimental results implicated the Rho and Star proteins in the mSpi cleavage process. Analysis of mutants in spi, rho, or Star showed similar phenotypes. Epistasis and rescue experiments placed Rho and Star in the cleavage process of Spi. By establishing a Drosophila cell culture assay for mSpi processing that requires the expression of Star and Rho, we explored the mechanism of cleavage. On the basis of protein–protein interactions and cellular localization studies in cells as well as in flies, we propose a sequential requirement for Star and Rho in the regulation of mSpi cleavage. The emerging steps will be outlined below.
Star as a trafficking molecule of mSpi
Although mSpi contains a signal peptide and transmembrane domain, the protein does not reach the plasma membrane, even in cases where it does not undergo cleavage. Rather, mSpi is found in a peripheral nuclear distribution extending into the cytoplasm, consistent with an ER compartment (Fig. 8). We do not know whether this ER retention relies on retrograde transport of mSpi from the Golgi to the ER, or on failure of mSpi to be transported from the ER. The end result, however, is that mSpi is normally sequestered in a compartment in which it cannot undergo cleavage. This sequestering provides a pool of inactive precursor that can be recruited for cleavage. Analysis of mSpi deletion constructs showed that the cytoplasmic domain of mSpi is required for ER retention (Fig. 9B). It is possible that an RR pair positioned four residues before the carboxyl terminus of mSpi may provide the retrieval signal common to ER membrane proteins (Mellman and Warren 2000).
Star is emerging as a critical modulator of mSpi intracellular distribution by forming a tight association with mSpi (Fig. 4). Star can alter the localization of mSpi and divert it from the original peripheral nuclear position (Figs. 9 and 10). The simplest hypothesis is that Star functions as a cargo receptor, necessary for transporting mSpi out of the ER (Ellgaard et al. 1999). During coexpression of mSpi and Star, the majority of mSpi protein accumulates in a punctate distribution in cells and in embryos. These bodies do not colocalize with the Golgi and only partially colocalize with lysosomes or endosomes. The appearance of higher molecular weight forms of mSpi in the presence of Star that are PNGase sensitive may reflect additional modifications of mSpi taking place en route to this compartment.
We demonstrated that the EGF domain of mSpi mediates the primary interaction with Star. We assume that binding between these proteins is a direct one, because we observed efficient coprecipitation of Spi–Star complexes after overexpression. This result is in agreement with findings of Hsiung et al. (2001), who identified the EGF domain of Spi as the crucial domain for binding to Star, using a GST pull-down assay. Interestingly, Hsiung et al. (2001) observed low endogenous levels of mSpi in the eye disc and identified the capacity of Star to stabilize Spi and elevate the levels of detectable Spi protein, similar to the rapid turnover of mSpi–GFP we identified in embryos.
The interaction with Star appears to be specific to Spi, as other ligands interacting with the EGF receptor, such as Vein–GFP or Argos–GFP, were not retained in the ER, and their intracellular localization was not modified by coexpression of Star. In addition, these ligands did not show any significant binding to Star (data not shown). It will be interesting to examine whether Star can modify the distribution of Gurken, which, like Spi, contains a transmembrane domain.
The importance of mSpi trafficking by Star was demonstrated by examination of the Star deletion constructs and the B. mori homolog. All three proteins bind Spi, but only the TMC construct is biologically active (Figs. 3 and 5). The correlation between the biological activity of the Star constructs and their capacity to export mSpi highlights the significance of mSpi intracellular trafficking by Star. Only the TMC construct could transport mSpi from the peripheral nuclear position, and retain biological activity (Figs. 3 and 11). Furthermore, the aberrant localization of the NTM construct in weak punctate plaques and in the plasma membrane suggests that the cellular localization of Star itself is regulated through interaction of unknown proteins with its carboxy-terminal domain.
The role of Star in trafficking mSpi to the Rho compartment, to allow mSpi cleavage, has also been demonstrated in a recent paper (Lee et al. 2001). This work showed that trafficking and cleavage of mSpi can be promoted efficiently in COS cells coexpressing the Drosophila Star and Rho proteins. Star- and Rho-mediated cleavage of mSpi was also demonstrated previously in Xenopus oocytes (Bang and Kintner 2000). The capacity of the trafficking and cleavage machinery of mSpi to function in vertebrate cells again argues for a direct interaction between these proteins. The tight regulation of retention and degradation of mSpi, suggest that additional proteins, which may not be dedicated only to mSpi, participate in the process. Localization of Star itself may also be regulated by protein interactions mediated by the carboxy-terminal domain.
Cellular compartmentalization is also crucial for regulation of other developmental signaling pathways. The Pipe protein, essential for dorso-ventral patterning of the follicle cells, is retained in the ER and requires translocation to the Golgi mediated by Windbeutel (Sen et al. 2000). The Rab23 protein involved in vesicle trafficking was recently shown to specifically down-regulate signaling of the Hedgehog pathway in mice (Eggenschwiler et al. 2001).
Rho promotes Spi cleavage
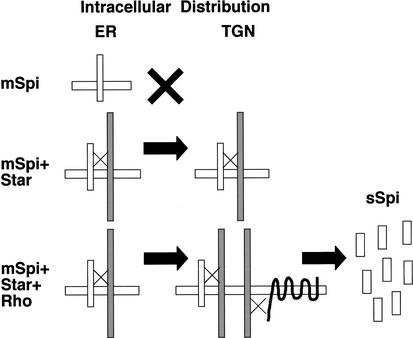
Although both Star and Rho are necessary for mSpi processing, several observations indicate that they function sequentially, rather than simultaneously. First, no triple complex was found between Spi, Star, and Rho. Rather, only complexes of Spi–Star or Star–Rho were detected (Figs. 6 and 7). Second, the cellular localization of Star and Rho is mostly nonoverlapping. Star is predominantly in a peripheral nuclear distribution, consistent with ER localization, whereas Rho shows a punctate staining, as well as membrane localization when overexpressed (Fig. 8). Finally, Star alters the peripheral nuclear localization of mSpi, whereas Rho has no effect on the intracellular distribution of mSpi. In cells expressing mSpi, Star, and Rho, the punctate localization of mSpi was shown to colocalize with Rho (Fig. 9). Thus, Star seems to act first, to transport mSpi to a compartment where Rho is located. A scheme for the sequential activities of Star and Rho in mSpi processing is shown (Fig. 12).
Figure 12.
Scheme for the sequential activities of Star and Rho in mSpi processing. mSpi is retained in a peripheral nuclear distribution, consistent with the ER. Expression of Star promotes binding to mSpi and shuttling to a compartment that does not colocalize with the Golgi and is possibly the trans Golgi network (TGN). In the absence of Rho, efficient cleavage does not take place. When mSpi, Star and Rho are expressed, mSpi is shuttled by Star to a TGN compartment containing Rho, thus leading to efficient cleavage and secretion of Spi.
What is this compartment? It does not colocalize with an integral Golgi marker, with endosomes, or with lysosomes. We assume it reflects a late compartment in the trans Golgi network. The cleavage apparatus of other secreted proteins was shown to be located in such a compartment. For example, Kex2 is required for cleavage of the yeast alpha factor pheromone and is localized in the trans Golgi network (Redding et al. 1996). Cleavage of pro-opiomelanocortin was shown to take place in the immature secretory granules, following sulfation in the trans Golgi network (Fernandez et al. 1997).
How does the interaction between the Star and Rho proteins impinge on the cleavage mechanism? We believe that the association of Rho with Star is significant for their biological activity. Indeed, the TMC construct, which did not show a detectable Rho association, was less potent than full-length Star (Figs. 3 and 6). It is tempting to speculate that the association between Star (carrying mSpi) and Rho, may target mSpi efficiently to the cleavage apparatus. In addition, Rho binding to Star may release mSpi from the mSpi–Star complex. A recent paper provides indications for the activity of Rho as a serine protease that mediates regulated intramembrane proteolysis of Spi (Urban et al. 2001).
Both mSpi and Star are broadly expressed throughout Drosophila development. What prevents the Star–mSpi complexes that reach the Golgi from releasing mSpi and allowing its cleavage by nonspecific proteases in cells that do not express Rho? We would like to propose that the tight association between Star and mSpi and the possible release of mSpi only after Star–Rho interaction, provides an additional mechanism for regulation of mSpi cleavage, after it has been shuttled to the Golgi.
Is Rho absolutely essential for cleavage, or can residual cleavage take place in the absence of Rho? Normally, during all stages of development, Rho is indeed essential for efficient activation of DER by Spi. It was shown, however, that uniform overexpression of mSpi and Star in the wing disc can lead to a phenotype consistent with ubiquitous, low level activation of DER (Pickup and Banerjee 1999). This activation appears to be Rho-independent, as Rho is expressed only in a restricted pattern prefiguring the veins. Thus, high levels of mSpi in the TGN may allow cleavage by other mechanisms, which are less efficient. Similarly, in S2 cells, Star was able to promote low level cleavage of mSpi, without coexpression of Rho (Fig. 1). However, in this case, it is not possible to rule out the contribution of residual endogenous Rho levels.
The results of this work open a set of questions regarding mSpi processing, in particular, and cleavage of TGFα-like ligands, in general. Several ligands of the ErbB family receptors in vertebrates (e.g., TGFα, HB EGF, and Neuregulin) are produced as transmembrane precursors, but can efficiently activate the receptor only in their cleaved form. In view of the essential role of Star, it is surprising that no Star homologs have been identified to date in vertebrates. Because the Star domain mediating interaction with mSpi may be restricted, it is possible that different proteins that retain this domain fulfill a similar role to Star in vertebrates. Alternatively, other intracellular carrier systems, such as PDZ domain proteins, may transport the ligand precursor (Fernandez-Larrea et al. 1999; Kuo et al. 2000).
In conclusion, this work shows that the intracellular localization and trafficking of mSpi is crucial for its regulated cleavage. mSpi is normally retained in a peripheral nuclear compartment where it does not undergo cleavage and may be rapidly degraded. Star, which is also enriched in this compartment, associates with mSpi and translocates it to a compartment in which Rho is enriched, to allow cleavage. The association between Star and Rho may allow Star to efficiently deliver mSpi. The cleavage process thus entails orchestration of trafficking and protein–protein associations to ensure tight regulation of mSpi processing and, hence, DER activation.
Materials and methods
DNA constructs
All constructs were generated by PCR, checked by sequencing and inserted into the pUAST vector. UAS-mSpi was described previously (Schweitzer et al. 1995b), UAS-mSpi–GFP was generated by insertion of pEGFP-C1 (Clontech) after mSpi residue Thr 54. The biological activity of the construct was verified by the S2 cleavage assay and by expression with Star in wing discs. UAS-sSpi–GFP was generated by insertion of a termination codon after P128, 11 residues before the transmembrane domain of the above construct. Biological activity was demonstrated by expression in embryos and discs. UAS-mSpiΔEGF–GFP and UAS-sSpiΔEGF–GFP were generated by removal of the EGF domain (residues C78–C117 in Spi). UAS-mSpi–FLAG was generated by insertion of the FLAG epitope sequence after mSpi residue M59. UAS-mSpiΔ16 was generated by removal of mSpi residues C108–D123. This protein is inactive in cells and in flies. UAS-mSpiΔIC–GFP was generated by insertion of a termination codon after L173 in Spi.
UAS-Star was described previously (Golembo et al. 1996a). UAS-Star–HA was generated by insertion of the HA tag at the carboxyl terminus and displayed biological activity in the S2 cleavage assay. UAS-NTM–HA was generated by use of residues 1–322 from Star, fused at the carboxyl terminus to the HA tag. The UAS-TMC–HA construct was produced by insertion of an ATG codon before Star residue Ser 259, and addition of HA at the carboxyl terminus. For the UAS-BmS–HA, we used B. mori cDNA clone WS00200 (provided by K. Mita, Utsunomiya University, Tochigi, Japan) and fused HA to the carboxyl terminus. UAS-Star–TAP was generated by fusion of the TAP tag (provided by B. Seraphin, EMBL, Heidelberg, Germany) to the carboxyl terminus. The activity of this construct was verified by the S2 cleavage assay and by the induction of a rough eye phenotype following a cross to GMR–Gal4 flies.
UAS-rho–HA; UAS-rho–FLAG, and UAS-rho–TAP were generated by insertion of the tags at the carboxyl terminus and displayed biological activity following expression in the wing disc induced by MS1096-Gal4. UAS-mouse CD8–GFP was provided by L. Luo (Stanford University, CA), and actin–Gal4 by S. Cohen (EMBL, Heidelberg, Germany).
Sequence of the S54allele was derived from PCR products of the exons, obtained from a heterozygous line. The mutated codon was recognized by a double peak.
Antibodies and dyes
Rat monoclonal anti-Spi was described previously (Schweitzer et al. 1995b). Rat polyclonal anti-Star was generated against the entire carboxy-terminal domain proceeding the transmembrane domain. Mouse anti-GFP (Roche) was used for blotting at 1/1,000 (of 0.4 mg/mL). Mouse monoclonal anti-HA (BAbCO) was used at 1/1,000 for blotting. Mouse anti-Golgi (Calbiochem; Stanley et al. 1997) was used for staining at 1/100. LysoTracker Red DND99 (Molecular Probes) was used on live cells at 50 nM (2 h incubation at 25°C). Early and late endosomes were identified by a 30-min incubation with 1 mg/mL Dextran, rhodamine B (70,000 MW) from Molecular Probes. Rabbit anti-Lamin was used at 1/200. HRP-conjugated secondary antibodies (1/10,000), and fluorescent secondary antibodies (1/300; Jackson ImmunoResearch) were used.
Transient transfection of S2 cells
For transfection of cells, calcium phosphate precipitates were generated (Chen and Okayama 1987), by use of 5–30 μg of total DNA. Expression of all pUAST constructs was driven by cotransfection with an actin–Gal4 plasmid. Typically, 5 × 106 cells were transfected and grown in 5 mL of Schneider medium (Beit Haemek) and 10% FCS. Medium, cells, or cell lysates were harvested 48–72 h after transfection.
Flies
Transgenic flies carrying UAS constructs of NTM–HA,TMC–HA, and BmS–HA were generated. UAS-Star was described previously (Golembo et al. 1996a). Activity of these constructs was checked in StarIIN heterozygous flies.
For expression of constructs in eye or wing discs, we used GMR-Gal4 or MS1096-Gal4, respectively. prd–Gal4 flies containing the insert on chromosome 3 and actin–Gal4 flies carrying the insert on chromosome 2 were used for the mSpi–GFP localization studies in embryos. For transient embryo injections, 0.3 mg/mL of each DNA was suspended in standard injection buffer, and injected into syncitial embryos. Embryos were covered in Halocarbon oil, incubated at 25°C and visualized by confocal microscopy 6–7 h after egg lay. Variable numbers of cells expressing the injected constructs were observed and were more prominent in the posterior part of the embryo, closer to the site of injection. The intracellular pattern, however, was reproducible in each case.
Immunoprecipitation and blotting
For detection of sSpi, 45 μL of medium was analyzed. Cells were lysed for 1.5 h on ice in 0.5 mL of 20 mM Tris (pH 8.0), 150 mM NaCl, 10% glycerol, 2 mM EDTA, 1 mM Na3VO4, 10 mM CHAPS (Sigma), and Complete protein inhibitors (Roche). The lysate was cleared by a 30-min centrifugation at 13,000 RPM, and the supernatant was subsequently used. For each lane, 30 μL of the lysate was used for assaying a lysate gel sample. For immunoprecipitation, the same lysate was precleared by shaking with the appropriate beads without antibody for 2 h at 4°C. The lysate was incubated for 2 h with beads conjugated to the antibody, or incubated for 2 h with the antibody followed by a 2-h incubation with the appropriate beads. The beads were washed 5 times in the lysis buffer, followed by resuspension of the beads in sample buffer. For immunoprecipitation of FLAG, anti-FLAG M2 affinity gel (Sigma) was used, and,for immunoprecipitation of HA, we used HA antibodies (BAbCO) followed by protein A–Sepharose CL-4B (Amersham Pharmacia Biotech). Immunoprecipitation of TAP and elusion by TEV protease were carried out according to Rigaut et al. (1999). SDS-PAGE gels (15%) were semi-dry blotted (Biometra). Primary antibody binding was detected by SuperSignal (Pierce). PNGase F (New England Biolabs) was used according to the manufacturer's instructions.
Acknowledgments
We thank S. Cohen, L. Luo, K. Mita, and B. Seraphin for reagents, K. Moses for open discussions, the Shilo group for discussions and support, and Jeff Gerst, Lilly Glazer, Zeev Paroush, Dalia Rosin, and Talila Volk for critical reading of the manuscript. This work was supported by the U.S.–Israel Binational Fund to B.S., who is an incumbent of the Hilda and Cecil Lewis Professorial chair in Molecular Genetics.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Benny.Shilo@Weizmann.ac.il; FAX 972-8-9344108.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.214202.
References
- Bang AG, Kintner C. Rhomboid and Star facilitate presentation and processing of the Drosophila TGFα homolog Spitz. Genes & Dev. 2000;14:177–186. [PMC free article] [PubMed] [Google Scholar]
- Bier E, Jan LY, Jan YN. rhomboid, a gene required for dorsoventral axis establishment and peripheral nervous system development in Drosophila melanogaster. Genes & Dev. 1990;4:190–203. doi: 10.1101/gad.4.2.190. [DOI] [PubMed] [Google Scholar]
- Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggenschwiler JT, Espinoza E, Anderson KV. Rab23 is an essential negative regulator of the mouse Sonic hedgehog signalling pathway. Nature. 2001;412:194–198. doi: 10.1038/35084089. [DOI] [PubMed] [Google Scholar]
- Ellgaard L, Molinari M, Helenius A. Setting the standards: Quality control in the secretory pathway. Science. 1999;286:1882–1888. doi: 10.1126/science.286.5446.1882. [DOI] [PubMed] [Google Scholar]
- Fernandez CJ, Haugwitz M, Eaton B, Moore HP. Distinct molecular events during secretory granule biogenesis revealed by sensitivities to brefeldin A. Mol Biol Cell. 1997;8:2171–2185. doi: 10.1091/mbc.8.11.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Larrea J, Merlos-Suarez A, Urena JM, Baselga J, Arribas J. A role for a PDZ protein in the early secretory pathway for the targeting of proTGF-alpha to the cell surface. Mol Cell. 1999;3:423–433. doi: 10.1016/s1097-2765(00)80470-0. [DOI] [PubMed] [Google Scholar]
- Freeman M, Klambt C, Goodman CS, Rubin GM. The argos gene encodes a diffusible factor that regulates cell fate decisions in the Drosophila eye. Cell. 1992;69:963–975. doi: 10.1016/0092-8674(92)90615-j. [DOI] [PubMed] [Google Scholar]
- Gabay L, Scholz H, Golembo M, Klaes A, Shilo BZ, Klambt C. EGF receptor signaling induces pointed P1 transcription and inactivates Yan protein in the Drosophila embryonic ventral ectoderm. Development. 1996;122:3355–3362. doi: 10.1242/dev.122.11.3355. [DOI] [PubMed] [Google Scholar]
- Gabay L, Seger R, Shilo BZ. In situ activation pattern of Drosophila EGF receptor pathway during development. Science. 1997;277:1103–1106. doi: 10.1126/science.277.5329.1103. [DOI] [PubMed] [Google Scholar]
- Golembo M, Raz E, Shilo BZ. The Drosophila embryonic midline is the site of Spitz processing, and induces activation of the EGF receptor in the ventral ectoderm. Development. 1996a;122:3363–3370. doi: 10.1242/dev.122.11.3363. [DOI] [PubMed] [Google Scholar]
- Golembo M, Schweitzer R, Freeman M, Shilo BZ. Argos transcription is induced by the Drosophila EGF receptor pathway to form an inhibitory feedback loop. Development. 1996b;122:223–230. doi: 10.1242/dev.122.1.223. [DOI] [PubMed] [Google Scholar]
- Golembo M, Yarnitzky T, Volk T, Shilo BZ. Vein expression is induced by the EGF receptor pathway to provide a positive feedback loop in patterning the Drosophila embryonic ventral ectoderm. Genes & Dev. 1999;13:158–162. doi: 10.1101/gad.13.2.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Reyes A, Elliott H, St. Johnston D. Polarization of both major body axes in Drosophila by gurken-torpedo signalling. Nature. 1995;375:654–658. doi: 10.1038/375654a0. [DOI] [PubMed] [Google Scholar]
- Guichard A, Biehs B, Sturtevant MA, Wickline L, Chacko J, Howard K, Bier E. rhomboid and Star interact synergistically to promote EGFR/MAPK signaling during Drosophila wing vein development. Development. 1999;126:2663–2676. doi: 10.1242/dev.126.12.2663. [DOI] [PubMed] [Google Scholar]
- Guichard A, Roark M, Ronshaugen M, Bier E. brother of rhomboid, a rhomboid-related gene expressed during early Drosophila oogenesis, promotes EGF-R/MAPK signaling. Dev Biol. 2000;226:255–266. doi: 10.1006/dbio.2000.9851. [DOI] [PubMed] [Google Scholar]
- Hsiung F, Griffis ER, Pickup A, Powers MA, Moses K. Function of the Drosophila TGF-α homolog Spitz is controlled by Star and interacts directly with Star. Mech Dev. 2001;107:13–23. doi: 10.1016/s0925-4773(01)00432-4. [DOI] [PubMed] [Google Scholar]
- Jin MH, Sawamoto K, Ito M, Okano H. The interaction between the Drosophila secreted protein argos and the epidermal growth factor receptor inhibits dimerization of the receptor and binding of secreted spitz to the receptor. Mol Cell Biol. 2000;20:2098–2107. doi: 10.1128/mcb.20.6.2098-2107.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodkin AL, Pickup AT, Lin DM, Goodman CS, Banerjee U. Characterization of Star and its interactions with sevenless and EGF receptor during photoreceptor cell development in Drosophila. Development. 1994;120:1731–1745. doi: 10.1242/dev.120.7.1731. [DOI] [PubMed] [Google Scholar]
- Kuo A, Zhong C, Lane WS, Derynck R. Transmembrane transforming growth factor alpha tethers to the PDZ domain-containing, Golgi membrane-associated protein p59/GRASP55. EMBO J. 2000;19:6427–6439. doi: 10.1093/emboj/19.23.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JR, Urban S, Freeman M. Regulated intracellular ligand transport and proteolysis control EGF signal activation in Drosophila. Cell. 2001;107:161–171. doi: 10.1016/s0092-8674(01)00526-8. [DOI] [PubMed] [Google Scholar]
- Mayer U, Nusslein-Volhard C. A group of genes required for pattern formation in the ventral ectoderm of the Drosophila embryo. Genes & Dev. 1988;2:1496–1511. doi: 10.1101/gad.2.11.1496. [DOI] [PubMed] [Google Scholar]
- Mellman I, Warren G. The road taken: Past and future foundations of membrane traffic. Cell. 2000;100:99–112. doi: 10.1016/s0092-8674(00)81687-6. [DOI] [PubMed] [Google Scholar]
- Neuman-Silberberg FS, Schupbach T. The Drosophila dorsoventral patterning gene gurken produces a dorsally localized RNA and encodes a TGFα-like protein. Cell. 1993;75:165–174. [PubMed] [Google Scholar]
- Nilson LA, Schupbach T. EGF receptor signaling in Drosophila oogenesis. Curr Top Dev Biol. 1999;44:203–243. doi: 10.1016/s0070-2153(08)60471-8. [DOI] [PubMed] [Google Scholar]
- Noll R, Sturtevant MA, Gollapudi RR, Bier E. New functions of the Drosophila rhomboid gene during embryonic and adult development are revealed by a novel genetic method, enhancer piracy. Development. 1994;120:2329–2338. doi: 10.1242/dev.120.8.2329. [DOI] [PubMed] [Google Scholar]
- Pascall JC, Brown KD. Characterization of a mammalian cDNA encoding a protein with high sequence similarity to the Drosophila regulatory protein Rhomboid. FEBS Lett. 1998;429:337–340. doi: 10.1016/s0014-5793(98)00622-x. [DOI] [PubMed] [Google Scholar]
- Pickup AT, Banerjee U. The role of star in the production of an activated ligand for the EGF receptor signaling pathway. Dev Biol. 1999;205:254–259. doi: 10.1006/dbio.1998.9119. [DOI] [PubMed] [Google Scholar]
- Redding K, Seeger M, Payne GS, Fuller RS. The effects of clathrin inactivation on localization of Kex2 protease are independent of the TGN localization signal in the cytosolic tail of Kex2p. Mol Biol Cell. 1996;7:1667–1677. doi: 10.1091/mbc.7.11.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nature Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- Roth S, Neuman-Silberberg FS, Barcelo G, Schupbach T. cornichon and the EGF receptor signaling process are necessary for both anterior-posterior and dorsal-ventral pattern formation in Drosophila. Cell. 1995;81:967–978. doi: 10.1016/0092-8674(95)90016-0. [DOI] [PubMed] [Google Scholar]
- Rutledge BJ, Zhang K, Bier E, Jan YN, Perrimon N. The Drosophila spitz gene encodes a putative EGF-like growth factor involved in dorsal-ventral axis formation and neurogenesis. Genes & Dev. 1992;6:1503–1517. doi: 10.1101/gad.6.8.1503. [DOI] [PubMed] [Google Scholar]
- Sapir A, Schweitzer R, Shilo BZ. Sequential activation of the EGF receptor pathway during Drosophila oogenesis establishes the dorsoventral axis. Development. 1998;125:191–200. doi: 10.1242/dev.125.2.191. [DOI] [PubMed] [Google Scholar]
- Schnepp B, Grumbling G, Donaldson T, Simcox A. Vein is a novel component in the Drosophila epidermal growth factor receptor pathway with similarity to the neuregulins. Genes & Dev. 1996;10:2302–2313. doi: 10.1101/gad.10.18.2302. [DOI] [PubMed] [Google Scholar]
- Schnepp B, Donaldson T, Grumbling G, Ostrowski S, Schweitzer R, Shilo BZ, Simcox A. EGF domain swap converts a Drosophila EGF receptor activator into an inhibitor. Genes & Dev. 1998;12:908–913. doi: 10.1101/gad.12.7.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer R, Shilo BZ. A thousand and one roles for the Drosophila EGF receptor. Trends Genet. 1997;13:191–196. doi: 10.1016/s0168-9525(97)01091-3. [DOI] [PubMed] [Google Scholar]
- Schweitzer R, Howes R, Smith R, Shilo BZ, Freeman M. Inhibition of Drosophila EGF receptor activation by the secreted protein Argos. Nature. 1995a;376:699–702. doi: 10.1038/376699a0. [DOI] [PubMed] [Google Scholar]
- Schweitzer R, Shaharabany M, Seger R, Shilo BZ. Secreted Spitz triggers the DER signaling pathway and is a limiting component in embryonic ventral ectoderm determination. Genes & Dev. 1995b;9:1518–1529. doi: 10.1101/gad.9.12.1518. [DOI] [PubMed] [Google Scholar]
- Sen J, Goltz JS, Konsolaki M, Schupbach T, Stein D. Windbeutel is required for function and correct subcellular localization of the Drosophila patterning protein Pipe. Development. 2000;127:5541–5550. doi: 10.1242/dev.127.24.5541. [DOI] [PubMed] [Google Scholar]
- Stanley H, Botas J, Malhotra V. The mechanism of Golgi segregation during mitosis is cell type specific. Proc Natl Acad Sci. 1997;94:14467–14470. doi: 10.1073/pnas.94.26.14467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturtevant MA, Roark M, O'Neill JW, Biehs B, Colley N, Bier E. The Drosophila rhomboid protein is concentrated in patches at the apical cell surface. Dev Biol. 1996;174:298–309. doi: 10.1006/dbio.1996.0075. [DOI] [PubMed] [Google Scholar]
- Urban S, Lee JR, Freeman M. Drosophila Rhomboid-1 defines a family of putative intramembrane serine proteases. Cell. 2001;107:173–182. doi: 10.1016/s0092-8674(01)00525-6. [DOI] [PubMed] [Google Scholar]
- Wasserman JD, Freeman M. An autoregulatory cascade of EGF receptor signaling patterns the Drosophila egg. Cell. 1998;95:355–364. doi: 10.1016/s0092-8674(00)81767-5. [DOI] [PubMed] [Google Scholar]
- Wasserman JD, Urban S, Freeman M. A family of rhomboid-like genes: Drosophila rhomboid-1 and roughoid/rhomboid-3 cooperate to activate EGF receptor signaling. Genes & Dev. 2000;14:1651–1663. [PMC free article] [PubMed] [Google Scholar]
- Yarnitzky T, Min L, Volk T. The Drosophila neuregulin homolog Vein mediates inductive interactions between myotubes and their epidermal attachment cells. Genes & Dev. 1997;11:2691–2700. doi: 10.1101/gad.11.20.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zak NB, Wides RJ, Schejter ED, Raz E, Shilo BZ. Localization of the DER/flb protein in embryos: Implications on the faint little ball lethal phenotype. Development. 1990;109:865–874. doi: 10.1242/dev.109.4.865. [DOI] [PubMed] [Google Scholar]