Abstract
The vaccinia virus (VV) I3L gene product is a single-stranded DNA-binding protein made early in infection that localizes to the cytoplasmic sites of viral DNA replication (S. C. Rochester and P. Traktman, J. Virol. 72:2917-2926, 1998). Surprisingly, when replication was blocked, the protein localized to distinct cytoplasmic spots (A. Domi and G. Beaud, J. Gen. Virol. 81:1231-1235, 2000). Here these I3L-positive spots were characterized in more detail. By using an anti-I3L peptide antibody we confirmed that the protein localized to the cytoplasmic sites of viral DNA replication by both immunofluorescence and electron microscopy (EM). Before replication had started or when replication was inhibited with hydroxyurea or cytosine arabinoside, I3L localized to distinct cytoplasmic punctate structures of homogenous size. We show that these structures are not incoming cores or cytoplasmic sites of VV early mRNA accumulation. Instead, morphological and quantitative data indicate that they are specialized sites where the parental DNA accumulates after its release from incoming viral cores. By EM, these sites appeared as complex, electron-dense structures that were intimately associated with the cellular endoplasmic reticulum (ER). By double labeling of cryosections we show that they contain DNA and a viral early protein, the gene product of E8R. Since E8R is a membrane protein that is able to bind to DNA, the localization of this protein to the I3L puncta suggests that they are composed of membranes. The results are discussed in relation to our previous data showing that the process of viral DNA replication also occurs in close association with the ER.
Vaccinia virus (VV) is the prototype member of the Poxviridae, a family of large DNA viruses. They are characterized by a complex life cycle that occurs entirely in the cytoplasm of the infected host cell, including viral transcription and DNA replication (15). Infection is initiated upon entry and delivery of the core into the cytoplasm. From this core, which contains the viral genome and enzymes to carry out the process of early transcription, a set of early mRNAs is transcribed (11, 16). The translation of the early messengers is required to uncoat the core and to release the parental DNA from the core and for the subsequent process of DNA replication. The latter process initiates the transcription of late genes, which encode for the late proteins that are required for the assembly of new virions (15). Virion assembly results in the production of two infectious forms, the intracellular mature virus that obtains its membranes from the intermediate compartment or smooth ER and the extracellular enveloped virus, which acquires an additional membrane from the trans-Golgi network or from endosomes (21, 26; reviewed in reference 23).
A number of recent studies were aimed at understanding how some of the early steps of viral morphogenesis relate to each other and to determine if and how these steps interact with the host cell. We could show, for instance, that the viral early mRNAs that are made inside the core and then extruded from these cores organize into distinct cytoplasmic structures. Surprisingly, the latter localized some distance away from the cores from which they were derived. Since both cores and the early mRNAs appeared to be associated with microtubules (MTs), we proposed that the core-extruded mRNAs moved along MTs to collect into translation-competent, MT-bound structures (14).
A long-standing dogma in the poxvirus field was that protein synthesis early in infection is required for the subsequent process of core uncoating and release of the parental DNA into the cytoplasm (7, 10, 11, 20). In a recent study this was confirmed by EM. By labeling cryosections with antibodies to DNA, it was shown that the viral genome remained inside the core when cells were treated with cycloheximide (CX), while the DNA labeling was gradually lost from the cores in the absence of the inhibitor (13). In another study it was shown that if core uncoating was prevented (by blocking protein synthesis), cores were often seen to accumulate close to the endoplasmic reticulum (ER) (17). Subsequent electron microscopy (EM) observations provided a possible explanation for this accumulation close to the ER, since EM images strongly suggested that the parental DNA also associated with the cytosolic side of the ER upon its release from incoming cores (13). Moreover, the putative association of the parental DNA with the ER was consistent with other data showing that the process of DNA replication also occurred in close association with this organelle. Over the time of infection the replication sites became entirely surrounded by membranes of the ER, a process that facilitated efficient viral DNA synthesis (25). It was thus proposed that after entry, intracellular cores associated with both MTs and ER, cellular structures that are well known to intimately interact with each other (1). Upon core uncoating the parental DNA was then delivered to the cytosolic side of the ER, where the subsequent process of DNA replication occurred (13).
In another set of experiments a quantitative relationship between incoming cores, the sites of VV early mRNA accumulation and DNA replication sites, was established. More than 40 years ago a study by Cairns (3) showed that each infectious particle is able to establish a replication site. In contrast, about 4 years later biochemical experiments suggested that in HeLa cells only 50% of the incoming cores uncoat their parental DNA (9). This observation indirectly implied that in HeLa cells 50% of incoming particles are apparently infectious and are consequently able to establish a replication site (9). A more recent study with HeLa cells showed that the amount of intracellular cores increased linearly with increasing multiplicities of infection (MOI). By quantifying incoming cores and replication sites by light microscopy, we could show that at relatively low MOI one core resulted in the formation of one replication site, consistent with the results of Cairns (3). These data implied, in contrast to those obtained by Joklik (9), that in HeLa cells all intracellular cores uncoat their genome, since each core formed one replication site (13). At relatively high MOI, however, we observed a different relationship between the number of incoming cores and replication sites; although the amount of cores still increased with increasing MOI, the number of replication sites remained constant. We therefore proposed that there is a limit to the amount of cytoplasmic replication sites that can be made in HeLa cells. Since the low-MOI experiments demonstrated that all intracellular cores are infectious, these data furthermore suggested that at high MOI more than one core (genome) contributed to one replication site (13). This was supported by the observation that although the number of replication sites remained constant at higher MOI, both the average size of the replication sites and the extent of replication did increase linearly with increasing MOI. Apparently, if more than one genome contributed to one replication site at higher MOIs, the latter increased in size because the availability of more parental DNA resulted in an increase in the extent of viral DNA replication (13).
The VV gene product of I3L has recently been identified as an abundant single-stranded DNA-binding protein (18). The protein is made at early and intermediate times postinfection, acquires serine/threonine phosphorylation, and is associated with the viral replication sites (18). Furthermore, when replication was blocked, I3L localized to distinct cytoplasmic spots (6, 18). Domi and Beaud (6) suggested that the latter are the precursors of DNA replication, since they incorporated BrdU upon washout of a viral DNA replication inhibitor.
The aim of the present study was to characterize in more detail the I3L-positive structures as they occur in the absence of replication. We confirm that in the absence of DNA replication I3L associates with cytoplasmic structures of homogenous size. We show by immunofluorescence and EM that these contain DNA and provide quantitative data to support the notion that this is viral parental DNA. Finally, we show by EM that the I3L- and DNA-containing structures are electron-dense aggregates that are closely associated with the cytosolic side of the ER, consistent with previous data suggesting that core uncoating and the release of the parental DNA occurs close to these same membranes.
MATERIALS AND METHODS
Virus, cells, and antibodies.
HeLa cells, obtained from the American Type Tissue Collection, were grown as described previously (24). The WR strain of VV was propagated and semipurified as described by Pedersen et al. (17) or was purified on sucrose gradients according to Jensen et al. (8). Virus stocks were plaque titrated on BSC-40 cells, and the amount of purified virus was quantified by optical density at 260 nm measurements as described previously (8). The following antibodies were used throughout this study. A peptide corresponding to amino acids 1 to 16 of the I3L sequence was made, and antibodies were raised in rabbits by Neosystems (Strassbourg, France) according to the protocol of the manufacturer. The antibodies to H5R, to VV cores, to A14L, and to E8R have been described previously (17, 19, 25). An antibody to DNA (monoclonal 1D12) was kindly provided by Robert Rubin (University of Albuquerque, N. Mex.), and the antibody to A10L was a kind gift of Denis Hruby (29). Hoechst and 4′,6′-diamidino-2-phenylindole (DAPI) were from Sigma, and the rat monoclonal antibody to BrU was purchased from Harlan Seralab (Loughborough, United Kingdom). Affinity purification of the I3L antibody by using the peptide to which the antibody was raised was performed as described previously (25). Secondary antibodies, anti-rabbit, anti-mouse, or anti-rat coupled to fluorescein isothiocyanate (FITC) or rhodamine, were from Dianova (Jackson Immunochemicals, Hamburg, Germany).
Western blotting and NP-40-DTT treatment of purified virus.
HeLa cells were infected for 3 or 6 h with 5 mM hydroxyurea (HU; Sigma) or were mock infected. The cells were rinsed with phosphate-buffered saline (PBS) and were scraped from the dish in PBS. The total amount of cells in suspension was determined in a Neubauer cell counter. The cells were collected by centrifugation, and the pelleted cells were resuspended in sample buffer. The volume of the sample buffer was adjusted such that all samples contained an equal amount of cells per volume. Equal volumes of sample were then loaded on a 12.5% polyacrylamide gel; after electrophoresis the gel was blotted onto polyvinylidene difluoride (PVDF) membrane, and the I3L protein was detected by using the crude antibody followed by goat anti-rabbit horseradish peroxidase (Bio-Rad) at a 1:2,000 dilution and ECL. Nonidet-P40-dithiothreitol (DTT) treatment of sucrose gradient-purified virus was performed essentially as described previously (8).
Immunofluorescence, infections, and drug treatment.
The immunofluorescence procedure was essentially as described previously (4). HeLa cells grown on coverslips were infected for 30 min at 37°C with WR and were sonicated for 1 min before infection at the indicated MOI. Cells were washed with PBS and were incubated alone or with 5 mM HU or 50 μM cytosine arabinoside (araC; Sigma) and were fixed at the indicated times postinfection with 3% paraformaldehyde in PBS. As described before (13), HU was always prepared fresh as a 1 M stock at the day of the infection and was subsequently discarded. Under these conditions the drug efficiently and consistently blocks viral replication. For the HU washout experiment HU was added at 2 mM immediately after infection, and at 2 h 45 min CX (Sigma) was added at 25 μg/ml. At 3 h postinfection cells were washed at least 10 times with PBS before further incubation in the presence of CX. For the quantitation of intracellular cores, I3L structures, and replication sites, cells were infected as described above in the presence of CX (25 μg/ml) or HU or were mock infected and were fixed at 3 h postinfection. Fixed cells were labeled with the anti-core or anti-I3L antibodies, and cores, I3L structures, and replication sites were counted in 30 cells. For the measurement of the average size of cores and I3L structures (accumulated in the presence of CX or HU, respectively), pictures were taken with a Zeiss Axiovert 200 microscope with fixed integration times. The relative size of labeled structures was evaluated by applying iMAQVisionBuilder (National Instruments) software to the pictures to allow the estimation of the area (but not the real size) occupied by the labeled structure (I3L spot or intracellular core). The average relative sizes (the labeled area) were calculated from 80 I3L spots or cores.
BrUTP transfection and BrdU incubations.
BrUTP transfection to visualize viral early mRNAs was done as described previously (14). Briefly, cells were infected for 15 min in the presence of 1 μg of actinomycin D/ml and then were transfected with BrUTP (Sigma) for 1 h in the presence of the drug. Cells were then washed extensively to remove the drug, at which time HU was added to allow viral transcription and a synchronized incorporation of BrUTP into the viral early transcripts. Cells were fixed at 3 h after actinomycin D washout and were double labeled with anti-BrU and anti-I3L. To test for viral replication, cells were infected for 30 min as described above and then were incubated with 25 μM BrdU (Sigma) in the presence or absence of 5 mM HU or 50 μM araC. Cells were fixed at 3 h postinfection by incubation for 4 min at −20°C in methanol. They were then double labeled with anti-BrU and anti-I3L antibodies.
EM.
For EM, HeLa cells were grown in 6-cm-diameter dishes, infected as described above at the indicated MOI, and then incubated with or without 5 mM HU. The cells were fixed at 3 h postinfection, and cryosections were prepared as described previously (28). Thawed sections were labeled with the affinity-purified anti-I3L antibody at a dilution of 1:12. Double labeling was carried out according to the guidelines of Slot et al. (22).
RESULTS
Generation and characterization of an antibody to I3L.
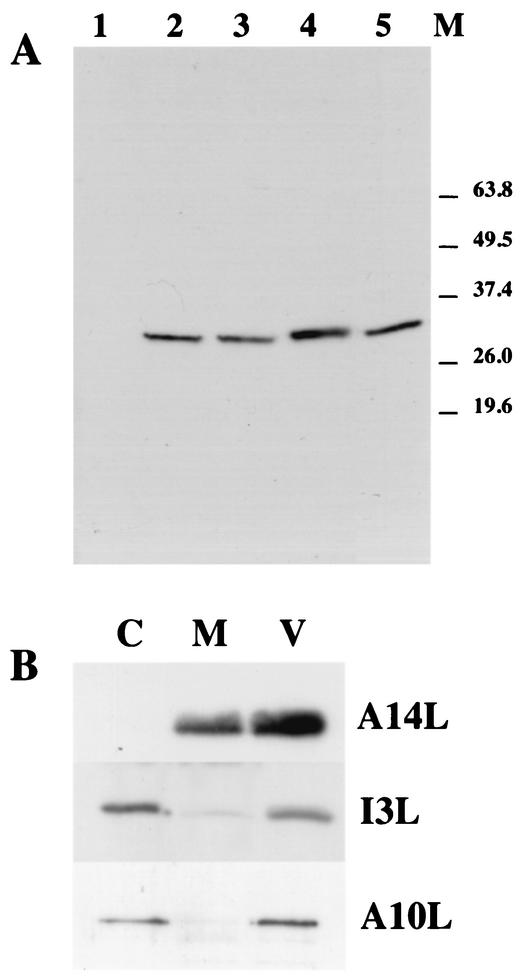
In order to characterize the I3L gene product (hereafter referred to as I3L) a peptide antibody was raised against the extreme N terminus of the protein (amino acids 1 to 16). The specificity of the antibody was first tested by Western blots. Cells were mock infected or were infected with HU for 3 h to block viral replication or for 6 h without inhibitor. Cell lysates as well as purified virus were analyzed by Western blotting by using the anti-I3L antibody. As shown in Fig. 1A, a single band migrating around 35 kDa was detected in infected but not uninfected cells. Although the predicted molecular size of I3L is 29.9 kDa, Tseng et al. (27) and Rochester and Traktman (18) showed that in sodium dodecyl sulfate-polyacrylamide gel electrophoresis the protein migrates around 34 kDa, consistent with our observations. The protein was also synthesized in the presence of HU, indicating that it is an early protein (see also reference 18). I3L also appeared to be packaged into virions as seen by the presence of the same band in purified virion preparations (Fig. 1A). To determine the subvirus localization of the protein, purified virus was treated with NP-40 and DTT and the NP-40-DTT-soluble membrane proteins were separated from the insoluble core fraction by centrifugation. By this treatment the I3L protein was found predominantly in the core fraction (Fig. 1B), indicating that the protein is a viral core protein.
FIG. 1.
Characterization of the I3L antibody by Western blots. (A) HeLa cells grown in 10-cm-diameter dishes were mock infected (lane 1) or were infected at an MOI of 10 in the presence (lane 3) or absence (lane 2 and 4) of 5 mM HU. Cells were scraped from the dish at 3 (lanes 2 and 3) or 6 h (lane 4) postinfection and were resuspended in PBS. The amount of cells in the suspension was determined. The cells were collected by centrifugation and were resuspended in sample buffer. The volume of sample buffer was adjusted such that all samples contained the same amount of cells per volume. Lane 5 depicts 0.66 μg (protein concentration) of sucrose gradient-purified virus. The samples were separated on a 12.5% polyacrylamide gel blotted onto PVDF membrane, and the I3L protein was detected by Western blotting and ECL. The position of the molecular size marker proteins (M) is indicated on the right. (B) Sucrose gradient-purified virus was treated with NP-40 and DTT to separate membrane (M) from core (C) proteins. Membrane and core fractions and purified virus (V) were run on 12.5% polyacrylamide gels and were blotted onto PVDF membrane. I3L was detected by Western blotting followed by ECL. As a control the PVDF membrane was also probed with antibodies to A14L and A10L, two major virus-associated membrane and core proteins, respectively. For I3L and A10L the same amounts of virus (0.66 μg of protein concentration per lane) or core/membrane fraction was used, while for A14L 5 times less material was loaded onto the gel.
Thus, the antibody recognized a protein with a molecular weight expected of the I3L protein that was made early in infection and packaged into virions.
By immunofluorescence microscopy I3L localizes to the replication sites, but in the absence of replication it localized to discrete punctate structures.
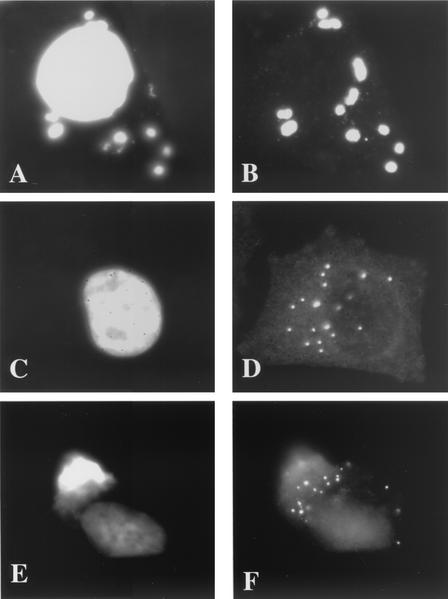
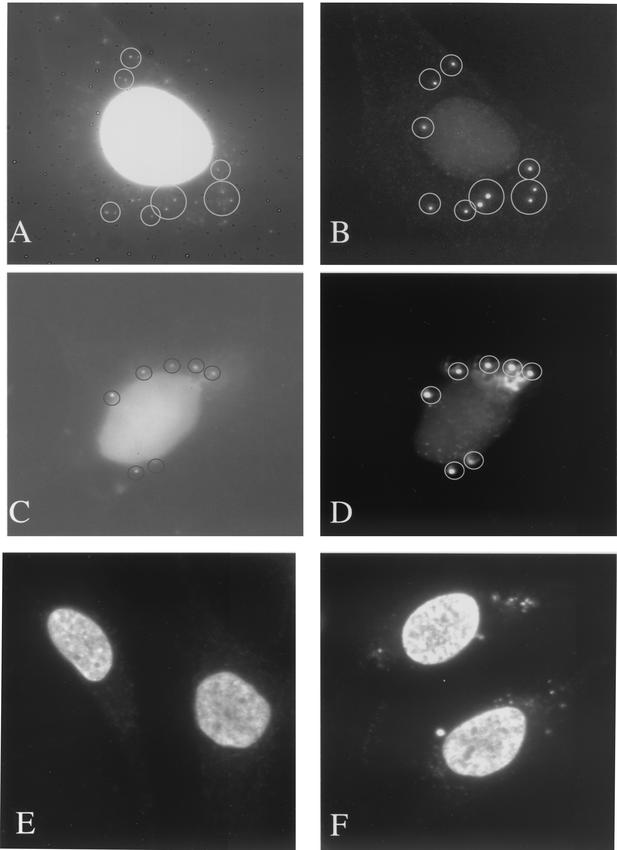
The localization of I3L was subsequently analyzed by immunofluorescence microscopy. Because the antibody showed some background labeling on uninfected cells, it was first affinity purified by using the peptide to which it was raised. No background labeling could be detected when uninfected cells were labeled with the purified antibody (data not shown). As before, I3L colocalized with the viral replication sites at 3 h postinfection (Fig. 2A and B). In the presence of HU, however, the protein appeared to collect in typical punctate structures (Fig. 2C and D) that were evidently not labeled with Hoechst (see below). Similar Hoechst-negative I3L punctate structures were also seen at 90 min postinfection, before replication had started, in the absence of HU, indicating that they are not artifacts of HU treatment (Fig. 2E and F). In the presence of cytosine araC, another inhibitor of viral DNA synthesis, I3L accumulated in similar punctate structures that were not appreciably labeled with Hoechst (Fig. 3A and B). The I3L labeling seen in the presence of either HU or araC significantly differed from the pattern of anti-H5R labeling (Fig. 3C and D). Whereas the H5R protein associates with replication sites as soon as replication is initiated (25), in the absence of replication we found that this protein showed a general cytoplasmic pattern and did not concentrate into typical punctate structures. It should be noted that the image in Fig. 3C and D was chosen to show that although the H5R pattern was mostly cytoplasmic, in some cells the antibody also faintly labeled some spots that resembled the I3L spots.
FIG. 2.
By immunofluorescence the I3L antibody labels distinct structures in the absence of viral replication. HeLa cells were infected at an MOI of 10 and were fixed at 3 h postinfection, except for those shown in panels E and F, which were fixed at 90 min postinfection. Cells shown in panels A, B, E, and F were untreated, and those depicted in panels C and D were treated with 5 mM HU. Fixed cells were double labeled with Hoechst (A, C, and E) and anti-I3L antibody (B, D, and F) followed by anti-rabbit antibody coupled to FITC.
FIG. 3.
I3L accumulates in similar structures in the presence of araC while H5R shows a different pattern. HeLa cells were infected at an MOI of 10 and were treated with either 50 μM araC (A and B) or 5 mM HU (C and D). Cells were double labeled with anti-I3L (B) or anti-H5R (D) antibody and Hoechst (A and C). Note that the cell shown in panels C and D was chosen to show that although H5R gives a general cytoplasmic labeling in the presence of HU in most cells, in some cells a subset of the protein also localizes to discrete spots.
In summary, the data suggest that in the absence of viral replication I3L collects into distinct cytoplasmic structures.
The I3L labeling in the presence of HU does not correspond to incoming cores or to the cytoplasmic sites of early mRNA accumulation.
Similar punctate structures that accumulate in the presence of inhibitors of DNA replication have been described before (6). In the latter study, the punctate structures were shown to contain the gene products of B1R, I3L, and H5R and were suggested to be the precursors of viral replication. Since the I3L structures accumulated in the presence of inhibitors of viral replication and were formed before replication had started (see above), they were likely to be a structure of the VV life cycle that precedes replication. A number of assays that enabled researchers to follow the early VV structures by light microscopy and EM were recently described. An antibody to VV cores was generated that recognizes incoming cores, but not intact particles, by immunofluorescence microscopy (12). Furthermore, an assay to follow the fate of the viral early mRNAs once released from the core was previously established (14).
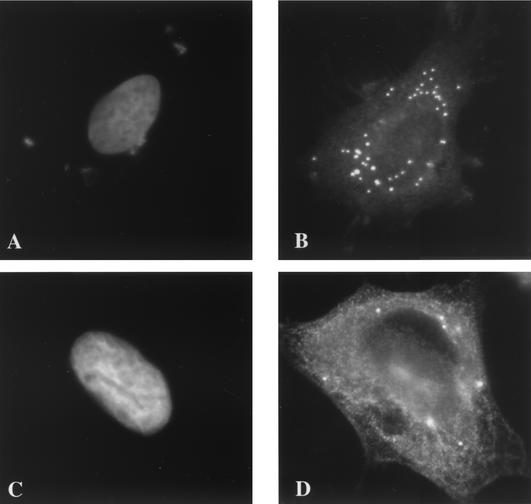
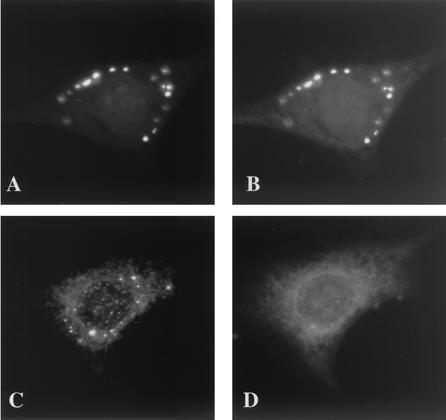
We therefore tested next whether the I3L structures corresponded to either incoming cores or to the sites where the viral early mRNAs accumulate by light microscopy.
Since the anti-core and -I3L antibodies are both raised in rabbits, it was not possible to localize these structures by double-labeling experiments. Therefore, cells were infected in the presence of CX to accumulate intracellular cores or with HU to accumulate the I3L structures. HU is known to block replication but not the core uncoating and degradation that occur prior to this process (see reference 13 and references therein). Cells were infected for 3 h in the presence of either CX or HU, fixed, and labeled with anti-core or with anti-I3L antibodies and the patterns were compared. As expected, in cells infected in the presence of CX, many cores could be detected upon labeling with the anti-core antibody (Fig. 4A). In contrast, no cores (or very few; see the legend to Fig. 4B) were seen in the presence of HU, indicating that, as expected, under these conditions the cores were degraded (Fig. 4B). The reverse pattern was observed with the anti-I3L antibody. No I3L-positive spots were seen in the presence of CX (Fig. 4C), while they were easily discernible in the presence of HU (Fig. 4D). The latter data also suggested that although I3L is associated with the NP-40-DTT-insoluble core fraction of isolated virus (see Fig. 1B), either the antibody did not recognize intracellular cores or I3L was not associated with incoming viral cores.
FIG. 4.
The I3L pattern seen in the presence of HU does not correspond to incoming cores or to the sites of viral early mRNA accumulation. (A through D) Cells were infected for 3 h at an MOI of 10 in the presence of either 25 μg of CX/ml (A and C) or 5 mM HU (B and D). Cells shown in panels A and B are labeled with the anti-core (red channel) antibody, and those shown in panels C and D were labeled with anti-I3L (red channel) antibody. The blue channel is Hoechst labeling showing the absence of labeling in the cytoplasm of infected cells. The cell in panel B (with HU and without CX) was chosen to show that under these conditions occasionally a core can be detected (one red spot that is visible in the cytoplasm) that is not yet degraded. Cells shown in panels E to G were infected in the presence of 1 μg of actinomycin D/ml and were lipofected with 10 mM BrUTP. After 1 h of transfection the cells were washed 10 times with PBS and were incubated for another 3 h in the presence of 5 mM HU to allow the incorporation of BrUTP into viral early mRNAs. The cells were fixed and triple labeled with anti-I3L (E), anti-BrU (F), and Hoechst (blue channel in panel G) antibodies followed by anti-rabbit antibody coupled to rhodamine and anti-rat antibody coupled to FITC. Panel G shows the merge.
Having established that the I3L structures were not incoming cores, we next investigated whether they represented VV early mRNAs. For this we used a recently described assay in which infected cells are transfected with BrUTP in the presence of HU. Under these conditions BrUTP is incorporated into discrete cytoplasmic spots that correspond to VV early mRNAs (14) and that showed some resemblance to the I3L structures. Cells were infected and transfected with BrUTP in the presence of HU as described before and were double labeled with anti-BrU and anti-I3L antibodies. Many labeled punctate structures were seen in infected and/or transfected cells, but in none of the puncta did the anti-BrU and anti-I3L labeling colocalize (Fig. 4E to G). It should be noted that the BrUTP transfection protocol used to visualize VV mRNAs apparently inhibited the formation of the I3L puncta, since fewer I3L spots were seen in infected and/or transfected cells. Nevertheless, since the BrU and I3L labeling did not overlap, the I3L puncta did not appear to be sites where the VV early mRNAs accumulate.
The combined data show that the I3L spots do not correspond to incoming cores or to the viral early mRNAs.
The I3L structures contain DNA which is not derived from newly synthesized DNA.
Since the I3L structures did not colocalize with the early mRNAs or represent incoming cores, we next considered the possibility that they represented specialized cytoplasmic sites where the parental DNA accumulates before replication. This idea was supported by the fact that upon close inspection of the Hoechst labeling a faint signal was sometimes seen in the I3L structures. To better visualize the putative DNA inside the I3L spots, cells infected for 3 h in the presence of HU were double labeled with anti-I3L and either DAPI or with an anti-DNA antibody. DAPI, another DNA dye, generally stains DNA more brightly than Hoechst. For routine studies we prefer to use Hoechst, since Hoechst dye labels the viral replication sites only in the cytoplasm of infected cells while DAPI also stains additional cytoplasmic structures, such as (most likely) mitochondria (our unpublished observations).
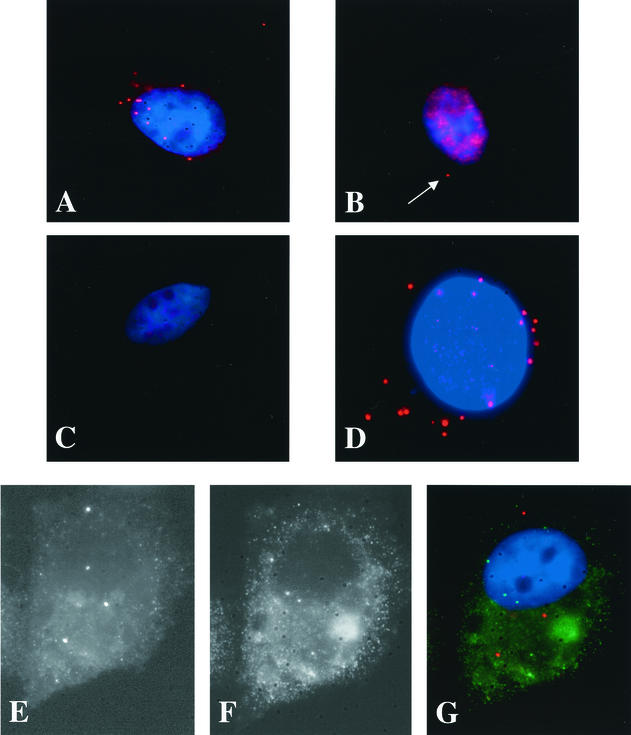
Upon double labeling of fixed cells with DAPI and anti-I3L antibody, we observed that the I3L structures were faintly but clearly labeled with DAPI (Fig. 5A and B). As before, the latter dye also labeled cytoplasmic structures that were not I3L positive. More convincing colocalization was obtained with I3L and an anti-DNA antibody. The anti-DNA labeling was not only brighter than that with DAPI, but also no other cytoplasmic structures beside the I3L spots were seen to label with the antibody by light microscopy (Fig. 5C and D). The cytoplasmic spots that were double labeled with anti-DNA and anti-I3L were also seen upon single labeling with the anti-DNA antibody. Whereas at the antibody dilution used in uninfected cells no cytoplasmic labeling was seen with the latter antibody, we could readily observe anti-DNA-positive spots in infected cells (Fig. 5E and F). Because these spots were not seen in uninfected cells, we suggest that they corresponded to the I3L structures (Fig. 5E and F). Identical results were obtained with araC, showing that the I3L spots were specifically and faintly labeled with anti-DNA antibody or DAPI (data not shown).
FIG. 5.
I3L structures contain DNA. Cells were infected at an MOI of 10 in the presence of 5 mM HU and were fixed at 3 h postinfection. They were then double labeled with anti-I3L (B and D) and anti-DNA (A) antibodies or DAPI (C). The images show that all I3L structures (encircled) are also (weakly) labeled with DAPI or anti-DNA. For panels E and F cells were mock infected (E) or were infected (F) at an MOI of 10 in the presence of HU and were fixed at 3 h postinfection. Fixed cells were single labeled with anti-DNA antibody. Both images were taken with the same exposure time.
To ascertain that the small amounts of DNA that were apparently contained in the I3L spots did not result from an incomplete inhibition of DNA synthesis, cells were mock infected or were infected in the presence of HU or araC (data not shown) and incubated for 2.5 h with BrdU, a compound that is readily membrane permeable and efficiently incorporated into newly synthesized (viral) DNA. Cells were fixed at 3 h postinfection, and DNA synthesis was subsequently detected by immunofluorescence by using antibodies to BrU. In untreated infected cells, prominent anti-BrU-positive structures that colocalized with I3L and that represented viral DNA replication sites were observed (Fig. 6A and B). In HU-treated cells, however, the anti-BrU labeling showed a faint cytoplasmic pattern and did not seem to concentrate in or colocalize with the I3L spots (Fig. 6C and D). Identical results were obtained if araC was used instead of HU (data not shown).
FIG. 6.
I3L structures do not contain newly synthesized DNA. HeLa cells were infected at an MOI of 10 for 30 min. After absorption the cells were incubated with 25 μM BrdU with (C and D) or without (A and B) 5 mM HU. Cells were fixed at 3 h postinfection by incubation for 4 min at −20°C in methanol (the anti-BrU labeling does not work on paraformaldehyde-fixed cells). Cells were then double labeled with anti-I3L (A and C) and anti-BrU (B and D) followed by anti-rabbit rhodamine and anti-rat FITC antibodies. Note that the methanol fixation leads to a poorer preservation of the I3L spots.
Since the I3L structures contain DNA in the presence of either HU or araC but did not incorporate BrdU even after long BrdU incubation times, the data strongly suggest that the I3L structures contain parental DNA that accumulates in the cytoplasm after core uncoating. The next set of experiments was aimed at further testing this hypothesis.
The amount of I3L structures is related to the amount of incoming cores and shows a direct relationship with the amount of DNA replication sites.
If the I3L structures represented the viral parental DNA, we expected a quantitative relationship between the number of I3L spots and that of incoming cores that carry the viral genome. Furthermore, we predicted a relationship between the amount of I3L structures and replication sites. Indeed, Domi and Beaud (6) showed that the amount of punctate B1R structures that possibly represent the same structures as the I3L spots we observed increased linearly with increasing MOI. The quantitative relationship between MOI, the amount of incoming cores, and the number of replication sites was recently studied in detail (13). It was shown that the number of incoming cores, detected by using the anti-core antibody (see above), increased linearly with increasing MOI. Moreover, at lower MOIs we observed a one-to-one relationship between intracellular cores and replication sites. At higher MOIs, however, the amount of replication sites did not further increase (see Introduction).
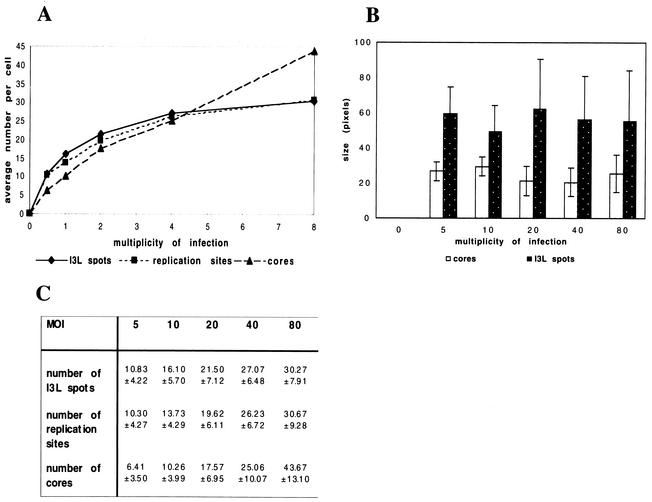
Here, we asked whether a similar relationship existed between incoming cores and I3L spots and between the I3L spots and viral replication sites. For this, cells were infected with increasing MOI under three different conditions: first, in the presence of CX to accumulate intracellular cores; second, in the presence HU to accumulate I3L spots; and finally, without CX or HU to allow the formation of replication sites. Cells were fixed at 3 h postinfection and were labeled with the anti-core antibody or with anti-I3L antibody, and the amount of intracellular cores, I3L spots, and replication sites were counted in 30 cells. The number of intracellular cores increased linearly with increasing MOI, consistent with recent data (Fig. 7A and C). Strikingly, the amounts of I3L spots and replication sites showed a pattern that completely overlapped (Fig. 7A and C); at lower MOI the numbers of both structures increased linearly, and both showed an approximately one-to-one relationship with the amount of cores (see above). At an MOI resulting in more than 30 cores per cell, the curves for both structures then leveled off and the amount of both the I3L spots and the replication sites did not significantly increase anymore (Fig. 7A and C). Thus, at all MOIs there was a direct, one-to-one relationship between I3L structures and replication sites.
FIG. 7.
Relationship between incoming cores, I3L structures, and DNA replication sites. (A and C) Cells were infected with increasing MOI (indicated) in the presence of 25 μg of CX/ml and in the presence or absence of 5 mM HU and were fixed at 3 h postinfection. Cells treated with CX were labeled with the anti-core antibody, and intracellular cores were counted. The cells treated with or without HU were labeled with anti-I3L antibody, and the I3L-positive structures or replication sites were counted. The graph in panel A represents the average amount of cores, I3L structures, or replication sites in 30 cells as a function of the MOI. Because the standard deviations added to the graph affected the graphic presentation of the data, the average amounts and standard deviations are shown in the table in panel C. (B) Infected cells were infected at different MOIs in the presence of CX (to accumulate intracellular cores) or HU (to accumulate I3L structures), fixed at 3 h postinfection, and labeled with the anti-core or anti-I3L antibody, respectively. The average fluorescently labeled area (size in pixels) occupied by 80 intracellular cores or I3L structures was measured at different MOI (see Materials and Methods). It shows that at all MOI the average labeled area (representing the relative size) of both the cores and the I3L structures remains constant.
Since the number of I3L structures did not increase at higher MOI, the data suggested that under these conditions more than one viral genome contributed to the formation of one I3L structure. We thus expected the I3L spots to be larger at higher MOI, since they were formed from more viral genomes. Upon measuring the average relative size of the I3L spots at increasing MOI, however, we observed that their size did not increase but instead remained constant (Fig. 7B). Our subjective impression was, however, that at higher MOI the DAPI or anti-DNA labeling of the I3L spots became brighter at higher amounts of virus, suggesting that they contained more DNA. Attempts to quantify the anti-DNA signal inside the I3L spots at different MOI were hampered by the relatively low anti-DNA signal, making it difficult to reliably measure the fluorescence intensity (data not shown).
In conclusion, although the I3L spots apparently organized into structures of homogenous size, they are likely to contain more DNA at higher MOI, suggesting that at higher MOI more than one genome contributes to one I3L spot as a consequence to the formation of a replication site.
Newly synthesized DNA is initiated at the sites of I3L accumulation.
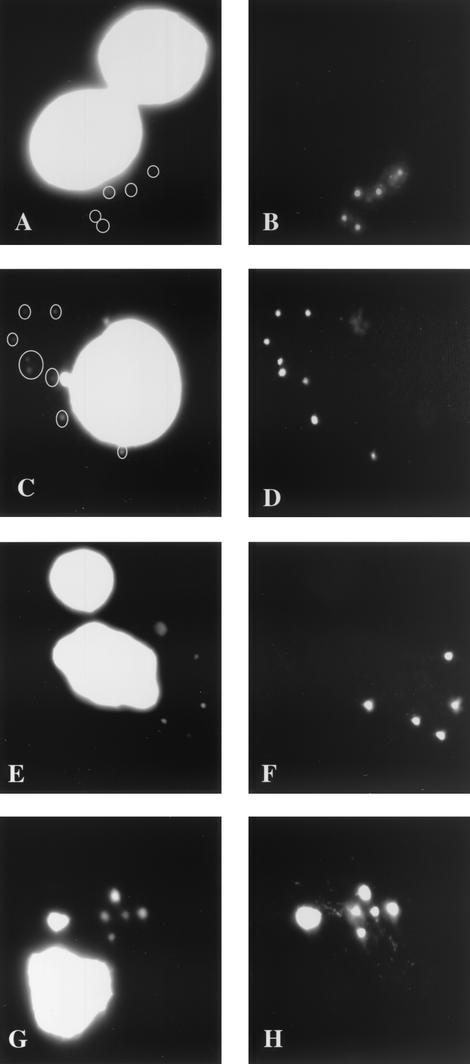
The above experiments indicated that the I3L structures contained the parental DNA that had been released into the cytoplasm after core uncoating. If correct, DNA replication should start from these sites after HU washout. To test this, cells were infected with HU for 3 h and then were extensively washed to remove HU. CX was then added to block further protein synthesis. It is known that the addition of CX after HU washout permits viral replication to some extent (2). The cells were double labeled with anti-I3L antibody and Hoechst. At 0 min postwashout no (or very faint) Hoechst labeling was detected, confirming that HU efficiently blocked replication (Fig. 8A and B). From 30 min postwashout, however, the Hoechst labeling could readily be detected and completely overlapped with the I3L labeling (Fig. 8C and D). The DNA replication sites were more clearly seen at 60 and 120 min postwashout because they had grown in size and were more brightly labeled with Hoechst (Fig. 8E to H). Importantly, the Hoechst labeling entirely colocalized with the I3L pattern at all times after HU washout.
FIG. 8.
DNA replication starts at the sites of I3L accumulation. Cells were infected for 3 h at an MOI of 10 in the presence of 2 mM HU. At 2 h 45 min CX was added at 25 μg/ml to inhibit protein synthesis. Fifteen minutes later the cells were washed 10 times with PBS to remove HU, and they were incubated further in medium with CX. They were then fixed at 0 (A and B), 30 (C and D), 60 (E and F), and 120 (G and H) min post-HU washout. Cells were double labeled with Hoechst (A, C, E, and G) and anti-I3L antibody (B, D, F, and H).
These data strongly suggest that replication starts at the sites of I3L accumulation, providing another indication for the idea that they contain the parental DNA.
EM reveals a complex structure that contains DNA and is associated with the ER.
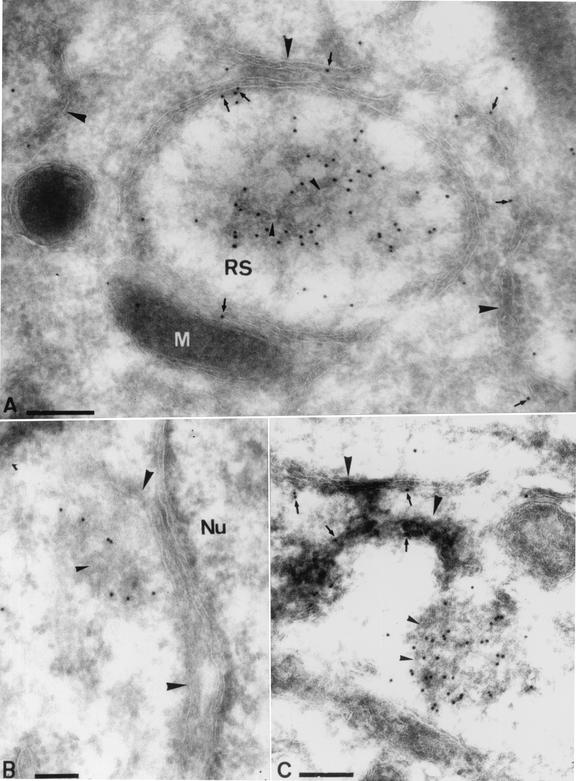
We next wanted to study the morphology of the I3L structures at the ultrastructural level by EM. We first confirmed by EM that I3L localized to the replication sites at 3 h postinfection without HU, as shown by immunofluorescence (Fig. 2A and B). At this time of infection the replication sites could readily be detected as structures with a different electron density than that of the surrounding cytoplasm and that they were almost completely enwrapped by rough ER membranes, consistent with previous results (25) (Fig. 9A). As expected for a soluble DNA-binding protein, the anti-I3L antibody labeled distinct patches in the central part of the replication site. Upon closer inspection, however, we also noticed labeling that was closely associated with the ER (Fig. 9A).
FIG. 9.
EM characterization of the I3L structures. (A) Cells were infected at an MOI of 10 and were fixed at 3 h postinfection. (B and C) Cells were infected at an MOI of 80 in the presence of HU to accumulate the I3L structures. Cryosections were labeled with affinity-purified anti-I3L antibody. Panel A shows a replication site (RS) surrounded by ER membranes (large arrowheads). Other ER membranes not surrounding the replication sites are found close by (large arrowheads). Labeling for I3L is found on the central part of the replication site as well as on ER membranes (small arrows). Note that the I3L labeling inside the replication site is associated with a specific structure that shows white lines typical of membranes (small arrowheads). Panels B and C show the typical I3L structures as they accumulate in the presence of HU. Note that in both images the abundantly labeled I3L structures seem attached to membranes of the ER (large arrowheads). I3L labeling can also be found on the ER itself (small arrows in panel C). Close inspection of the I3L-labeled structures reveals white lines typical of membranes inside the structures (small arrowheads). M, mitochondrion; Nu, nucleus. Bars, 200 nm.
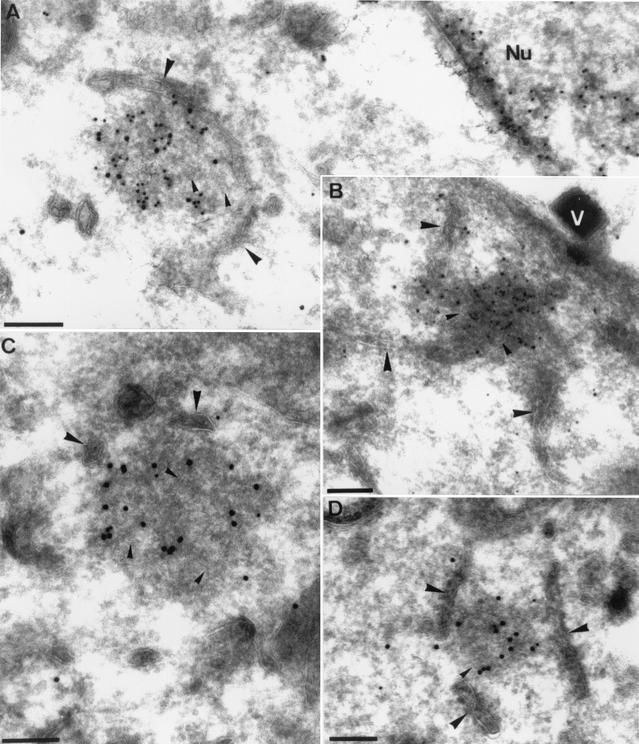
Low ER labeling was also observed if cells were infected in the presence of HU (Fig. 9C). The I3L spots that were prominently seen by light microscopy appeared as abundantly labeled electron-dense structures. These electron-dense I3L-positive structures were always closely associated with membranes of the ER (Fig. 9B and C and 10A to D). Double labeling of I3L and protein disulfide isomerase, a marker of the ER, confirmed that the membranes to which the I3L structures were attached were derived from the ER (data not shown).
FIG. 10.
Double labeling of the I3L structures with anti-I3L antibody and either anti-DNA or anti-E8R antibody. Cells were infected at an MOI of 80 in the presence of HU and were fixed at 3 h postinfection. (A and B) Sections were double labeled with anti-I3L (15-nm gold particles) and anti-DNA (10-nm gold particles) antibodies. (C and D) Sections were double labeled with anti-I3L (15-nm gold particles) and anti-E8R (10-nm gold particles) antibodies. In all images the ER can be found close by or attached to the typical I3L structures (large arrowheads). Putative tubular membranes on the inside of the I3L structures are indicated with small arrowheads. Nu, nucleus that is abundantly labeled for anti-DNA antibody. V, a virion attached to the plasma membrane. Bars, 200 nm.
To confirm that the I3L structures contained (parental) DNA, cryosections of cells infected for 3 h in the presence of HU were double labeled with anti-I3L and anti-DNA antibodies. Indeed, the electron-dense I3L structures were also abundantly labeled with anti-DNA antibody (Fig. 10A and B). In the cytoplasm of cells infected in the presence of HU they represented the only structures besides mitochondria that were labeled with the anti-DNA antibody.
Since the I3L structures appeared to be closely associated with the ER, we asked whether a viral membrane protein could be involved in this putative membrane association. The gene product of E8R, a viral early membrane protein with a putative role in the wrapping of the ER around the replication sites, was recently identified and characterized (5, 25). This protein localized to the ER around the replication sites, and in the absence of replication E8R accumulated in the ER and on vesicular structures on one side of the Golgi complex (5). Sections of cells infected in the presence of HU were therefore double labeled with I3L and anti-E8R. We expected E8R to localize to the ER membranes to which the I3L structures were attached but not to the structures themselves. Surprisingly, rather than labeling the ER cisterna with which the I3L structures were associated the anti-E8R antibody showed low but specific labeling of the structures themselves, as they were clearly double labeled for both E8R and I3L (Fig. 10C and D). Since E8R is a membrane protein, these results suggested that the I3L structures may contain membranes. Indeed, upon close inspection we obtained the impression that they might contain convoluted tubular membranes (Fig. 9B and C and 10A to C).
In conclusion, the collective data strongly suggest that upon release from the core the parental DNA becomes organized into specialized I3L-containing structures that are associated with the cytoplasmic side of the ER and that may contain membranes.
DISCUSSION
The I3L antibody highlights specific cytoplasmic spots.
By using antibodies to I3L, the present study characterizes in detail a VV-induced cytoplasmic structure that accumulates when viral replication is blocked. As expected, I3L colocalized with the DNA replication sites in infected cells. In the absence of replication the protein appeared to localize to discrete organized spots in the cytoplasm. Similar structures that accumulate in the absence of viral replication have been described by others (6, 18). Domi and Beaud (6) described punctate structures that accumulated in the presence of araC and that were labeled with antibodies to I3L as well as B1R and H5R. They suggested these to be the precursors of the viral replication sites, since they incorporated BrdU upon HU washout. Our study confirms and extends the observations made by Domi and Beaud. We show that the I3L structures are not incoming cores or the sites where VV early mRNAs accumulate. Their formation was blocked by an inhibitor of protein synthesis and core uncoating. Although the structures did not incorporate BrdU in the presence of HU or araC, we show by immunofluorescence and EM that they contain small amounts of DNA. The amount of I3L spots increased linearly with increasing amounts of cores at moderate MOI. Interestingly, the number of I3L structures showed a direct one-to-one relationship with the amount of cytoplasmic replication sites at all MOI tested. Finally, EM showed that the I3L structures were intimately associated with the ER (see below). Together, our data argue that the I3L structures represent specialized sites where the parental DNA accumulates upon its release from incoming cores.
I3L highlights a complex structure that is closely associated with the ER.
The I3L puncta seen by immunofluorescence appeared as a remarkable electron-dense structure by EM that was always in close proximity or was attached to the ER. A reasonable assumption is that this complex structure may serve at least three purposes. First, to prepare and organize the parental DNA, together with a complex of proteins, for the subsequent process of DNA replication. Second, by coating the parental DNA with a putative protein complex, to prevent the DNA from unwanted degradation and disintegration. Finally, to anchor the parental DNA to a specific cellular membrane of the ER, the same membranes where replication subsequently occurs (see below).
The notion that the parental DNA becomes organized into a complex structure upon core uncoating is interesting in light of results obtained almost 40 years ago by Joklik (9). As mentioned in the introduction, this study concluded that in HeLa cells only 50% of the incoming cores are able to uncoat the parental DNA. This conclusion was based on an assay in which HeLa cells were infected with [3H]thymidine-labeled viral particles. Uncoating was then quantified by the gradual acquisition of the 3H-labeled viral DNA of sensitivity to digestion with DNase. Since about 50% of the [3H]thymidine label eventually became sensitive to this enzyme, Joklik concluded that only 50% of the incoming cores were able to uncoat the parental DNA in HeLa cells. Our combined studies, however, suggest that in HeLa cells all intracellular cores are infectious (see Introduction) and that all are able to uncoat their genome (13 and this study). Moreover, our unpublished observations suggest that all intracellular cores eventually become degraded in HeLa cells. The apparent discrepancy between our results and those obtained by Joklik can now be explained by the observation that the viral genome, once released from the incoming core, becomes tightly associated with the I3L-containing complex described in the present study. It is conceivable that within this ER-associated complex the DNA is (partially) protected from DNase digestion because it is coated with proteins and because it is possibly associated with membranes.
By EM the I3L protein could be detected in aggregates in association with the parental DNA in discrete structures closely associated with the ER. Some of this protein could also be localized more sparsely to the ER, suggesting that it may form a scaffold for the replication machinery in the process to become organized. The electron-dense regions of I3L accumulation were apparently enriched in membranes, since they labeled with an antibody to E8R, a membrane protein that binds to DNA (5) and that were previously localized to the ER membranes surrounding the replication sites (25). This notion was supported by our EM images strongly suggesting that the I3L structures might contain highly tubulated membranes. Thus, we suggest that these structures are likely to comprise a mesh of convoluted tubules that are directly connected to the ER. However, biochemical evidence supporting the idea that the I3L spots contain membranes is presently lacking, and further studies are needed to address this point in more detail.
Multiple roles of the ER in the VV life cycle.
The present data are entirely consistent with and extend previous observations. It was shown previously that the process of DNA replication occurs in close association with the rough ER. The replication sites become almost completely surrounded by these cellular membranes, a process that facilitates replication (25). Moreover, in two other studies it was found that incoming cores accumulate close to the ER, and upon core uncoating the parental DNA was apparently being delivered to these same membranes (13, 17). The present study now extends these results by showing that the parental DNA becomes organized into a complex, ER-associated structure and that the I3L protein may have a role in organizing this structure. Thus, our data now show that the parental DNA associates with the cytoplasmic face of the same membrane organelle from which replication is subsequently initiated. The challenge will be to find what mediates the targeting of the parental DNA to the ER and which molecules are involved.
Acknowledgments
We would like to thank Bob Rubin (University of Albuquerque) for his generous gift of anti-DNA antibodies, without which this report would have been much less convincing. We thank Gareth Griffiths for critical reading of the manuscript and Birgit Schramm for help with microscopy.
Part of this work was supported by an EU TMR grant (to L.D.).
REFERENCES
- 1.Allan, V. 1996. Role of motor proteins in organizing the endoplasmic reticulum and Golgi apparatus. Semin. Cell Dev. Biol. 7:335-342. [Google Scholar]
- 2.Beaud, G., and R. Beaud. 1997. Preferential virosomal location of underphosphorylated H5R protein synthesized in vaccinia virus-infected cells. J. Gen. Virol. 78:3297-3302. [DOI] [PubMed] [Google Scholar]
- 3.Cairns, H. J. F. 1960. The initiation of vaccinia infection. Virology 11:603-623. [DOI] [PubMed] [Google Scholar]
- 4.Den Boon, J. A., E. J. Snijder, J. Krijnse Locker, M. C. Horzinek, and P. J. M. Rottier. 1991. Another triple-spanning envelope protein among intracellular budding RNA viruses: the torovirus E protein. Virology 182:655-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doglio, L., A. De Marco, S. Schleich, N. Roos, and J. Krijnse Locker. 2002. The Vaccinia Virus E8R gene product; a viral membrane protein that is made early in infection and packaged into the virions' core. J. Virol. 76:9773-9786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Domi, A., and G. Beaud. 2000. The punctate sites of accumulation of vaccinia virus early proteins are precursors of sites of viral DNA synthesis. J. Gen. Virol. 81:1231-1235. [DOI] [PubMed] [Google Scholar]
- 7.Holowczak, J. A. 1972. Uncoating of poxviruses. I. Detection and characterization of subviral particles in the uncoating process. Virology 50:216-232. [DOI] [PubMed] [Google Scholar]
- 8.Jensen, O. N., T. Houthaeve, A. Shevchenko, S. Cudmore, M. Mann, G. Griffiths, and J. Krijnse Locker. 1996. Identification of the major membrane and core proteins of vaccinia virus by two-dimensional electrophoresis. J. Virol. 70:7485-7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Joklik, W. K. 1964. The intracellular uncoating of poxvirus DNA. I. The fate of radioactively-labeled rabbitpox virus. J. Mol. Biol. 8:263-276. [DOI] [PubMed] [Google Scholar]
- 10.Joklik, W. K. 1964. The intracellular uncoating of poxvirus DNA. II The molecular basis of the uncoating process. J. Mol. Biol. 8:277-288. [DOI] [PubMed] [Google Scholar]
- 11.Kates, J. R., and B. R. McAuslan. 1967. Messenger RNA synthesis by a 'coated' viral genome. Proc. Natl. Acad. Sci. 57:314-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krijnse Locker, J., A. Kuehn, S. Schleich, G. Rutter, H. Hohenberg, R. Wepf, and G. Griffiths. 2000. Entry of the two infectious forms of vaccinia virus at the plasma membrane is signaling-dependent for the IMV but not the EEV. Mol. Biol. Cell 11:2497-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mallardo, M., E. Leithe, S. Schleich, N. Roos, L. Doglio, and J. Krijnse Locker. 2002. On the relationship between intracellular cores, early mRNA and DNA-replication sites. J. Virol. 76:5167-5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mallardo, M., S. Schleich, and J. Krijnse Locker. 2001. Microtubule-dependent organization of vaccinia virus core-derived early mRNAs to distinct cytoplasmic structures. Mol. Biol. Cell 12:3875-3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moss, B. 1996. Poxviridae: the viruses and their replication, p. 2637-2671. In B. N. Fields, D. M. Knipe, R. M. Chanock, M. S. Hirsch, J. L. Melnick, T. P. Monath, and B. Roizman (ed.), Fields virology, 3rd ed. Lippincott-Raven Press, Philadelphia, Pa.
- 16.Moss, B. 1990. Regulation of vaccinia virus transcription. Annu. Rev. Biochem. 59:661-688. [DOI] [PubMed] [Google Scholar]
- 17.Pedersen, K., E. J. Snijder, S. Schleich, N. Roos, G. Griffiths, and J. Krijnse Locker. 2000. Characterization of vaccinia virus intracellular cores: implications for viral uncoating and core structure. J. Virol. 74:3525-3536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rochester, S. C., and P. Traktman. 1998. Characterization of the single-stranded DNA binding protein encoded by the vaccinia virus I3 gene. J. Virol. 72:2917-2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salmons, T., A. Kuhn, F. Wylie, S. Schleich, J. R. Rodriguez, D. Rodriguez, M. Esteban, G. Griffiths, and J. Krijnse Locker. 1997. Vaccinia virus membrane proteins p8 and p16 are cotranslationally inserted into the rough ER and retained in the intermediate compartment. J. Virol. 71:7404-7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarov, I., and W. K. Joklik. 1972c. Characterization of intermediates in the uncoating of vaccinia virus DNA. Virology 50:593-602. [DOI] [PubMed] [Google Scholar]
- 21.Schmelz, M., B. Sodeik, M. Ericsson, E. J. Wolffe, H. Shida, G. Hiller, and G. Griffiths. 1994. Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans Golgi network. J. Virol. 68:130-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Slot, J. W., H. J. Geuze, S. Gigengack, G. E. Lienhard, and D. E. James. 1991. Immuno-localization of the insulin regulatable glucose transporter in brown adipose tissue of the rat. J. Cell Biol. 113:123-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sodeik, B., and J. Krijnse Locker. 2002. Assembly of vaccinia virus revisited: de novo membrane synthesis or acquisition from the host? Trends Microbiol. 10:15-24. [DOI] [PubMed] [Google Scholar]
- 24.Sodeik, B., R. W. Doms, M. Ericsson, G. Hiller, C. E. Machamer, W. van't Hof, G. van Meer, B. Moss, and G. Griffiths. 1993. Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J. Cell Biol. 121:521-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tolonen, N., L. Doglio, S. Schleich, and J. Krijnse Locker. 2001. Vaccinia virus DNA replication occurs in ER-enclosed cytoplasmic mini-nuclei. Mol. Biol. Cell 12:2031-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tooze, J., M. Hollinshead, B. Reis, K. Radsak, and H. Kern. 1993. Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur. J. Cell Biol. 60:163-178. [PubMed] [Google Scholar]
- 27.Tseng, M., N. Palaniyar, W. Zhang, and D. H. Evans. 1999. DNA binding and aggregation properties of the vaccinia virus I3L gene product. J. Biol. Chem. 274:21637-21644. [DOI] [PubMed] [Google Scholar]
- 28.van der Meer, Y., E. J. Snijder, J. C. Dobbe, S. Schleich, M. R. Denison, W. J. M. Spaan, and J. Krijnse Locker. 1999. The localization of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J. Virol. 73:7641-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Slyke, J. K., S. S. Whitehead, E. M. Wilson, and D. E. Hruby. 1991. The multistep proteolytic maturation pathway utilized by vaccinia virus P4a protein: a degenerate conserved cleavage motif within core proteins. Virology 183:467-478. [DOI] [PubMed] [Google Scholar]