Abstract
Polycomb group (PcG) proteins are required to maintain stable repression of the homeotic genes and others throughout development. The PcG proteins ESC and E(Z) are present in a prominent 600-kDa complex as well as in a number of higher-molecular-mass complexes. Here we identify and characterize a 1-MDa ESC/E(Z) complex that is distinguished from the 600-kDa complex by the presence of the PcG protein Polycomblike (PCL) and the histone deacetylase RPD3. In addition, the 1-MDa complex shares with the 600-kDa complex the histone binding protein p55 and the PcG protein SU(Z)12. Coimmunoprecipitation assays performed on embryo extracts and gel filtration column fractions indicate that, during embryogenesis E(Z), SU(Z)12, and p55 are present in all ESC complexes, while PCL and RPD3 are associated with ESC, E(Z), SU(Z)12, and p55 only in the 1-MDa complex. Glutathione transferase pulldown assays demonstrate that RPD3 binds directly to PCL via the conserved PHD fingers of PCL and the N terminus of RPD3. PCL and E(Z) colocalize virtually completely on polytene chromosomes and are associated with a subset of RPD3 sites. As previously shown for E(Z) and RPD3, PCL and SU(Z)12 are also recruited to the insertion site of a minimal Ubx Polycomb response element transgene in vivo. Consistent with these biochemical and cytological results, Rpd3 mutations enhance the phenotypes of Pcl mutants, further indicating that RPD3 is required for PcG silencing and possibly for PCL function. These results suggest that there may be multiple ESC/E(Z) complexes with distinct functions in vivo.
Polycomb group (PcG) and Trithorax group (trxG) proteins are required for long-term maintenance of “active” and “inactive” chromatin states, respectively. PcG proteins form complexes that act through Polycomb response elements (PREs) to create “silent” chromatin states, while trxG proteins create “active” chromatin states. Two distinct PcG complexes have been reported, a 2-MDa Polycomb Repressive Complex 1 (PRC1) (39) and a 600-kDa ESC/E(Z) complex (33, 47). Many of the previously identified PcG proteins have been found to be components of one or the other of these two complexes. PRC1 contains the PcG proteins PC, PH, PSC, and SCM (39) and associates directly with components of the transcriptional machinery, including TBP and TAFs (37), suggesting that its repressive effects are ultimately exerted directly at promoters.
In addition to ESC and E(Z) the 600-kDa ESC/E(Z) complex contains the histone binding protein p55 (47) and the recently identified PcG protein SU(Z)12 (6, 10, 15, 32). The presence of a SET domain in E(Z) strongly suggested that it functions as a histone methyltransferase, and this has recently been demonstrated (10, 15, 32). It was previously suggested that the PRC1 and ESC/E(Z) complexes function cooperatively, enzymatic modification of histones by the ESC/E(Z) complex being a prerequisite for the recruitment of PRC1 (47). This is supported by recent evidence that the two complexes are transiently associated during early embryogenesis (36) and by evidence that methylation of Ubx PRE-associated nucleosomes is required for stable association of PRC1 with the Ubx PRE (10, 15). The histone deacetylase RPD3 is also associated with ESC/E(Z) complexes (15, 47) and the homologous mammalian EED/EZH2 complexes (47, 50). However, RPD3 may not be stably associated with the 600-kDa complex but with one or more larger ESC/E(Z) complexes also detected during early embryogenesis (22).
A number of previously identified PcG proteins have not been found in either the PRC1 or the 600-kDa ESC/E(Z) complex, suggesting that there are likely to be additional PcG complexes. The PcG protein PCL was recently reported to interact with E(Z) but does not appear to be a component of the 600-kDa complex (34).
Here we report the identification an ∼1-MDa ESC/E(Z) complex that is distinct from the previously characterized 600-kDa ESC/E(Z) complex. Both complexes contain the PcG proteins ESC, E(Z), and SU(Z)12 as well as the histone binding protein p55. In addition, the 1-MDa complex also contains the PcG protein PCL and the histone deacetylase RPD3. RPD3 appears to be predominantly if not exclusively associated with the 1-MDa complex and not the 600-kDa complex. Direct binding of RPD3 to PCL in vitro suggests that PCL may be required for stable association of RPD3 with ESC/E(Z) complexes. The 1-MDa ESC/E(Z) complex does not contain PC or PSC, indicating that it is also distinct from the PRC1 complex. PCL and E(Z) binding sites on polytene chromosomes coincide and correspond to a subset of the more numerous RPD3 sites. As previously shown for E(Z) and RPD3, PCL and SU(Z)12 are recruited to the Ubx PRE in vivo, further suggesting that these proteins act in concert directly at the PRE. Genetic interactions between Pcl and Rpd3 mutations also suggest that RPD3 is required for PCL function. We discuss the possible functional relationship between the these different ESC/E(Z) complexes.
MATERIALS AND METHODS
Protein expression constructs.
The pGEX3-PCL and pBluescript-SK-PCL constructs were gifts from Robert Saint (University of Adelaide, Adelaide, Australia). The cDNA constructs encoding PCL residues 1 to 139, 140 to 858, 423 to 858, 573 to 858, and 838 to 1043 were generated by PCR and inserted into pGEM-T vector (Promega) by TA cloning. Then all PCL-coding sequence fragments were inserted into the pET-15 vector at NdeI and BamHI sites, respectively. These constructs were used to produce 35S-labeled PCL proteins by using the TNT T7 Quick Coupled Transcription/Translation System (Promega). pET-H6-PCL140-541 and pET-H6-PCL423-609 were derived from pET-H6-PCL140-858 and pET-H6-PCL423-858 by excision of the NsiI-BamHI and XhoI-BamHI fragments, respectively, followed by recircularization by blunt end ligation after end fill-in treatment with Klenow DNA polymerase. The BamHI-EcoRI fragment (encoding PCL residues 423 to 567) from pGEM-PCL423-858 was inserted into pGEX-2T vector to yield pGEX-PCL423-567. The NdeI-NsiI fragment from pGEM-PCL838-1043 was inserted into modified pGEX-2T vector to yield pGEX-PCL838-1043. GST fusion constructs containing full-length ESC, E(Z), and RPD3 were described previously (47). The pGEX-RPD3 (1-148) was derived from pGEX-RPD3 full-length by excision of an EcoRI fragment and recircularization. The BamHI-NotI cut fragment of RPD3 was inserted into pGEX-4T-1 to yield a construct of pGEX-RPD3 (114-442). The pGEX-RPD3 (148-442) was derived from pGEX-RPD3 (114-442) by deletion of BamHI-EcoRI fragment, end-blunt by the Klenow DNA polymerase, and recircularization. The BalI-EcoRI fragment encoding RPD3 residues 24 to 148 was inserted into the pGEX-2T vector at SmaI and EcoRI sites. pGEX-RPD3 (24-129) and pGEX-RPD3 (114-148) were derived from the pGEX-RPD3 (24-148) by excision of the BglII-EcoRI and BamHI-BamHI fragments, respectively, blunt ending by the Klenow DNA polymerase, and recircularization. The N- terminally truncated form of RPD3 was generated by inserting PCR fragment encoding amino acids 346 to 521 of RPD3 into a modified pGEX-2T vector at NdeI and NsiI. Further details of all construction can be obtained upon request. Glutathione Sepharose 4B (Amersham Pharmacia) and Ni-nitrilotriacetic acid agarose (Qiagen) were used for purification of glutathione transferase (GST) fusion and six-histidine-tagged proteins, respectively.
Antibody production and purification.
Rabbit anti-PCL antibodies were raised against a recombinant PCL protein containing the first 139 residues of PCL and an N-terminal six-histidine tag. Six-histidine-PCL1-139 protein was expressed in Escherichia coli and purified on Ni-nitrilotriacetic acid agarose (Qiagen). For affinity purification of anti-PCL antibodies from serum, purified six-histidine-PCL1-139 protein was coupled to CNBr-activated Sepharose 4 Fast Flow (Amersham Pharmacia) according to the manufacturer's instructions. Anti-PCL serum was loaded onto the six-histidine-PCL 1-139-coupled Sepharose affinity column, and bound antibodies were eluted with 0.1 M glycine (pH 2.7) after extensive washing. Purified antibodies were examined by Coomassie blue staining after sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and were assayed by Western blotting with goat anti-rabbit immunoglobulin G (IgG)-horseradish peroxidase secondary antibody. According to the revised Pcl sequence (2), a single PCL protein containing 1,043 amino acid residues is predicted (revised from 857), with a calculated molecular mass of 115 kDa. On Western blots of 0- to 16-h embryo nuclear extracts, affinity-purified anti-PCL antibodies recognize only two very closely spaced bands (Fig. 1A) that are the same size as in vitro translated PCL (see Fig. 3D, top panel), with an apparent molecular mass on SDS-PAGE of approximately 145 kDa. Rabbit anti-ESC antibodies were raised against the N terminus of ESC and affinity purified; its characterization will be described elsewhere. Mouse and rabbit anti-E(Z) antibodies were generated as previously described (46, 47) and were antigen affinity purified. Polyclonal anti-SU(Z)12 antibodies were raised in chicken against a synthetic peptide (MAPAKKREKDSNPDG-C) corresponding to the N-terminal 15 residues of the SU(Z)12 protein, conjugated to keyhole limpet hemocyanin via a C-terminal cysteine residue by using a maleimide cross-linker (Agrisera, Vännäs, Sweden). The chicken IgY was affinity purified on an UltraLink Iodoacetyl column as described by the manufacturer (Pierce). It recognizes a protein, on Western blots of embryo extracts, that is the same size as in vitro translated SU(Z)12. Additional guinea pig polyclonal anti-SU(Z)12 antibodies were raised against recombinant SU(Z)12 (residues 624 to 900) and were purified on a HiTrap Protein A column (Amersham Pharmacia). They recognize a single SU(Z)12 band on a Western blot of embryonic extract. Anti-PSC monoclonal antibody is from Paul Adler. Affinity-purified rabbit anti-RPD3, p55, and p105 antibodies were generous gifts from Jim Kadonaga and Jessica Tyler.
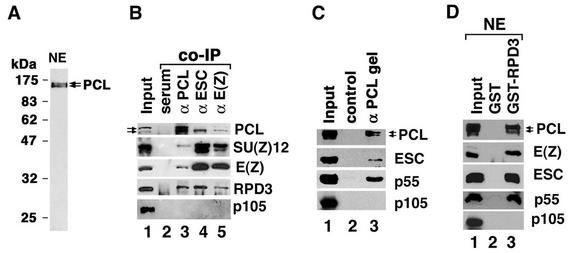
FIG. 1.
PCL is physically associated with RPD3, SU(Z)12, E(Z), ESC and p55 in vivo. (A) Zero- to 16-h embryo nuclear extracts (NE) were separated by SDS-11% PAGE, and PCL was detected by Western with affinity purified anti-PCL antibodies. No signal was detected with the rabbit preimmune serum. (B) Co-IP of PCL, ESC, and E(Z) with SU(Z)12 and RPD3 from 0- to 16-h whole-embryo extracts by use of protein G Sepharose and purified rabbit anti-PCL (lane 3), anti-ESC (lane 4), and anti-E(Z) antibodies (lane 5). Preimmune rabbit serum was used in lane 2 for negative control. SU(Z)12 degradation product (lower band in the second panel) was detected in whole-embryo extracts but not in nuclear extracts (Fig. 4A, panel 5). (C) Purified anti-PCL antibodies were coupled to CNBr-activated Sepharose to generate an affinity gel. Embryo extracts (lane 1) were mixed with the affinity gel. After extensive washing, bound proteins were eluted by 0.1 M glycine (pH 2.7) and detected in lane 3. Lane 2 shows a parallel control with preimmune serum-coupled gel. (D) GST pulldown assay with use of GST-RPD3 (lane 3) and 0- to 16-h nuclear extracts (NE). Lanes 1 show 10% (B) and 20% (C and D) of the total sample used. Proteins were detected by Western blotting and were indicated at the right side of each panel. The bottom gels (B, C, and D) show that p105, which is associated with p55 in the abundant ∼600-kDa CAF-1 complex, is not present in immunoprecipitates, indicating that immunoprecipitates are not contaminated with CAF-1.
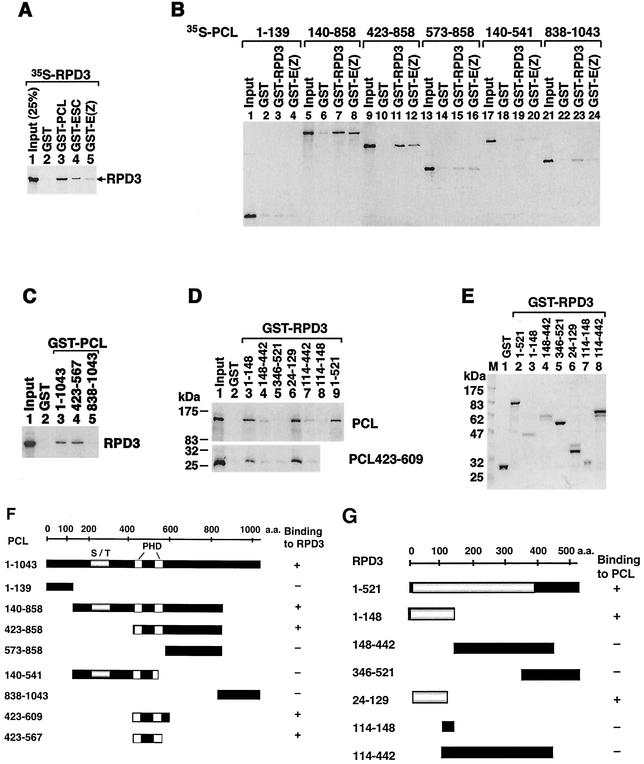
FIG. 3.
In vitro binding of PCL and RPD3. In vitro translated 35S-labeled PCL (full-length and truncated forms) and RPD3 proteins (input lanes in panels A to D), as well as GST fusion proteins (indicated at top of appropriate lanes), were used to test direct interaction of PCL and RPD3 and to map regions responsible for interactions. Lanes with GST alone in panels A through D serve as negative controls and show background binding. Signals that are ≥3-fold higher than background were considered significant specific binding. Proteins were separated by SDS-PAGE and visualized by autoradiography (A to D). Purified GST fusion proteins used for panel D were shown in panel E with Coomassie blue staining. (F and G) Schematic summary of results of in vitro binding experiments and the PCL and RPD3 constructs tested. For PCL (F), the highly conserved PHD fingers are indicated by a white box, and the serine-threonine-rich region (S/T) is indicated by a gray box. For RPD3 (G), the highly conserved N terminus is indicated by a gray box. The in vitro binding result (+ or −) is indicated to the right of each construct. Note that the PCL constructs containing both PHD fingers (PCL423-567) and the RPD3 constructs containing the N terminus (24-129) retain binding activity. a.a., amino acids.
Embryo extract preparation and fractionation.
Whole-embryo extracts and embryo nuclear extracts were prepared and fractionated on a Superose 6 HR 10/30 gel filtration column by using an AKTA Purifier system as previously described (47) with extraction buffer containing 0.3 M NaCl and elution buffer containing 0.2 M NaCl. Fractions (0.5 ml/tube) were collected after elution of the first 5 ml and were stored at −80°C for future use. Proteins in fractions were analyzed by Western blots with the enhanced chemiluminescence detection system (Pierce).
IP and in vitro binding assays.
Coimmunoprecipitation (co-IP) and GST pulldown assays were performed as described previously (46). GST pulldown assays were carried out under modified conditions with binding buffer (20 mM HEPES, pH 7.5, 0.15 M NaCl, 1 mg of bovine serum albumin [BSA]/ml, 10% glycerol, 0.2% NP-40, and 1 mM dithiothreitol). The binding reaction was carried out at 4°C with rotation for 80 min. The beads were then washed twice with 300 μl of binding buffer with 0.3 M NaCl and twice with 300 μl of washing buffer (50 mM Tris-HCl, pH 8.5, 0.3 M NaCl, 10% glycerol, 1 mM dithiothreitol, 0.1% NP-40, and 0.1% Triton X-100). After final washing with 50 mM sodium phosphate, proteins bound to the beads were eluted with 2× SDS sample buffer and were analyzed by Western blotting. Co-IP assays using anti-ESC, anti-SU(Z)12, and anti-PCL antibodies were carried out from gel filtration column fractions in the same way as co-IP from embryo extracts. Protein G-Sepharose (∼15 μl/tube) was preincubated with binding buffer containing 1 mg of BSA/ml and was then incubated with purified antibodies for at least 30 min. After washing of protein G beads to remove any unbound material, 180 μl of sample from each fraction (after prespinning to remove any precipitates) and 100 μl of binding buffer with protease and phosphatase inhibitors were mixed with the beads. The mixture was incubated for 2 h at 4°C with rotation. Beads were washed with modified binding buffer containing 0.3 M NaCl and no BSA, as described above. To prevent DNA-dependent protein associations, embryo extracts and fraction samples were incubated with 80 to 100 μg of ethidium bromide (EtBr)/ml on ice for at least 30 min (26). Precipitates were removed by centrifugation, and supernatants were used for co-IP. To test whether p55 coimmunoprecipitates with ESC from fractions, we used a transgenic line expressing a previously characterized epitope-tagged FLAG-ESC transgene and the anti-FLAG M2 Sepharose gel (46, 47).
Immunostaining of polytene chromosomes.
To determine the extent of colocalization of PCL with RPD3 and of PCL with E(Z), sequential staining of salivary gland polytene chromosomes with rabbit antibodies was carried out as described (49). Chromosomes from third instar wild-type larvae and from the transgenic line containing a minimal Ubx PRE construct were immunostained with anti-PCL and anti-SU(Z)12 antibodies as previously described (47).
Genetic interaction assays.
To determine whether Rpd3 mutations enhance the weak dominant phenotypes of Pcl mutants, Rpd3 mutant females (Rpd3313/Rpd3313 or Rpd3303/TM3 or Rpd3R8/TM3 or Rpd3R24/TM3) were crossed to Pcl11/CyO males or Pcl7/CyO males. Reciprocal crosses were also done. Progeny males heterozygous for both mutations were scored for enhancement of the frequency and severity of the extra sex comb phenotype (transformation of second and third thoracic [T2 and T3] legs to T1 leg identity) and transformation of abdominal segment 4 (A4) to abdominal segment A5, two PcG phenotypes that occur at low frequency in Pcl mutant heterozygotes (19). The Rpd3 alleles used are described by Mottus et al. (31) and have no PcG phenotypes on their own when heterozygous. Rpd3R24 is a null allele. Pcl11 is a putative null allele (23). To assay genetic interactions between Pcl and Su(z)12 mutations, Pcl7/CyO females were crossed to sc z wis; Su(z)121/TM3, Sb males and non-Cy, non-Sb male progeny (heterozygous for both PcG mutations) were scored for enhancement of the frequency and severity of the weak low-penetration extra sex comb phenotype exhibited in flies heterozygous for either of the mutations alone. The number and size of extra sex combs were scored. For controls, Su(z)121/+ and Pcl7/+ females were crossed to sc z wis males to eliminate any effects of the balancers. The Pcl13 allele was also tested in the same manner and yielded similar results.
RESULTS
PCL is associated with RPD3, ESC, E(Z), SU(Z)12, and p55 in vivo.
It was previously observed that, while ESC and E(Z) are present in a prominent 600-kDa complex, they also cofractionate over a broad high-molecular-weight range from 0.8 to 2 MDa, particularly during early embryogenesis (22). To begin to characterize these larger ESC/E(Z) complexes, we first asked whether they might be distinguished from the 600-kDa complex by the presence of additional PcG proteins that had not been previously identified as components of either the 600-kDa ESC/E(Z) complex or the 2-MDa PRC1 complex. Among candidates for such proteins are PCL and RPD3. It was recently shown that, while RPD3 is associated with ESC and E(Z) (47), it is associated with a high-molecular-weight ESC/E(Z) complex(es) and is not stably associated with the 600-kDa complex (22). The PcG protein PCL (34) was recently shown to interact with E(Z) in vitro and to coimmunoprecipitate with E(Z) from embryo extracts, although it has not been identified as a component of the 600-kDa ESC/E(Z) complex. Using affinity-purified antibodies raised against PCL, we confirmed that immunoprecipitates obtained with anti-PCL antibodies contain E(Z) and determined that they also contain SU(Z)12 and RPD3 (Fig. 1B, lane 3). These proteins were not detected in immunoprecipitates obtained with preimmune serum (lane 2). Immunoprecipitates obtained with anti-ESC (Fig. 1B, lane 4) and anti-E(Z) antibodies (lane 5) also contain PCL and RPD3 in addition to SU(Z)12 and E(Z).
We could not readily determine whether p55 and ESC are present in anti-PCL immmunoprecipitates by Western blotting after IP, since the anti-p55 and anti-ESC antibodies were also raised in rabbits and since the IgG heavy chain migrated at approximately the same position as p55 and ESC on SDS-PAGE. To circumvent this problem, we coupled affinity-purified anti-PCL antibody to CNBr-activated Sepharose to yield an anti-PCL affinity gel. Normal rabbit serum coupled to Sepharose serves as a negative control. We observed that the PCL affinity gel specifically bound PCL, p55, and ESC (Fig. 1C, lane 3) as well as E(Z) and SU(Z)12 (data not shown). Taken together, these results indicate that PCL associates with RPD3, p55, ESC, E(Z), and SU(Z)12 in vivo, suggesting the existence of a complex(es) containing them.
The association of PCL with RPD3 in vivo was further tested by GST pulldown from nuclear extract. As shown in Fig. 1D, GST-RPD3 specifically pulls down PCL (lane 3, top panel). As previously shown, GST-RPD3 also pulls down E(Z), ESC, and p55 (lane 3, lower panels), while GST alone does not (lane 2).
Identification of an ESC/E(Z) complex that contains PCL and RPD3.
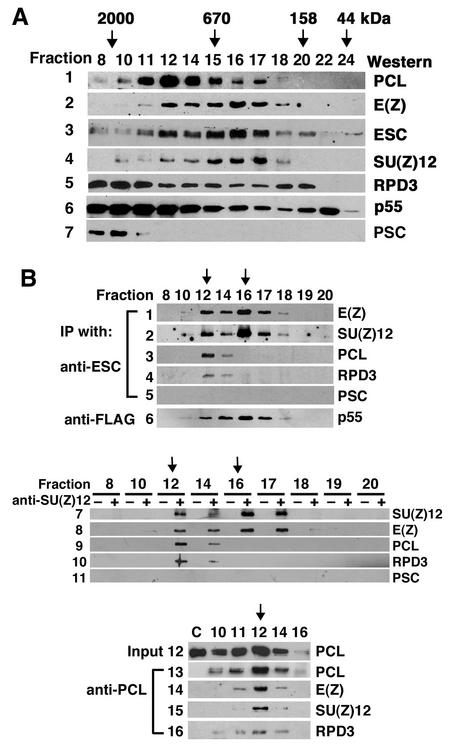
To identify ESC/E(Z) complexes containing PCL and RPD3, we fractionated 0- to 16-h embryo nuclear extracts on a Superose 6 gel filtration column to separate protein complexes by size and then carried out IP from individual fractions to examine the association of PCL with ESC and E(Z) in complexes of different sizes. The fractionation of nuclear extracts, shown in Fig. 2A, suggests that there are multiple PCL-containing complexes in fractions 10 to 17. ESC, E(Z), and SU(Z)12 cofractionate precisely with each other in fractions 11 to 18 with peaks in fraction 12 (∼1 MDa) and 16 (∼600 kDa [panels 2 through 4 in Fig. 2A]). The broad distributions of the abundant RPD3 and p55 proteins (Fig. 2A, panels 5 and 6) reflect their presence in multiple different complexes. The PcG protein PSC, a component of the 2-MDa PRC1, is detected only in fractions 8 to 11 (Fig. 2A, panel 7). To further test the association of PCL and RPD3 with ESC/E(Z) complexes, we determined whether they coimmunoprecipitate with ESC from fractions 8 to 20. Fractions were first treated with EtBr to eliminate DNA-dependent associations of these proteins. As shown in Fig. 2B, anti-ESC antibodies coimmunoprecipitated E(Z) and SU(Z)12 (panels 1 and 2) mainly from fractions 12 to 17 (and weakly from fractions 10 and 18), with the same pattern and peaks at fractions 12 and 16 (indicated by arrows in Fig. 2B). Although PCL and RPD3 are each distributed over a broad range (Fig. 2A, panels 1 and 5), they coimmunoprecipitated only with ESC from fractions 12 and 14, with the same pattern and peak at fraction 12 (Fig. 2B, panels 3 and 4). They did not coimmunoprecipitate from fraction 16 or 17, which contains the 600-kDa ESC/E(Z) complex. These data indicate that PCL and RPD3 are only present in an ∼1-MDa ESC/E(Z) complex but not in the 600-kDa ESC/E(Z) complex, consistent with our previous finding that RPD3 is present in larger ESC complex(es) (22). To test for the presence of p55 in anti-ESC immunoprecipitates from fractions, we fractionated embryo nuclear extracts made from a transgenic line expressing a previously characterized FLAG-ESC transgene (46, 47). As shown in Fig. 2B, panel 6, the anti-FLAG M2 affinity gel also bound p55 from fractions 10 to 18, with a profile similar to that of E(Z) and SU(Z)12 (panels 1 and 2), reflecting the presence of ESC, E(Z), SU(Z)12, and p55 in multiple complexes of different sizes. The PcG protein PSC was not detected in anti-ESC immunoprecipitates from any fraction (panel 5), consistent with its association with the PRC1 complex. The profile of ESC/E(Z) complexes was further confirmed by IP from fractions with anti-SU(Z)12 (Fig. 2B, panels 7 to 11). As seen with anti-ESC above, anti-SU(Z)12 antibodies immunoprecipitated SU(Z)12 itself and E(Z) from fractions 12 to 17 (Fig. 2B, panels 7 and 8), and PCL and RPD3 from fractions 12 and 14 (Fig. 2B, panels 9 and 10) but did not coimmunoprecipitate PSC (Fig. 2B, panel 11).
FIG. 2.
Identification of an ESC/E(Z) complex that contains PCL and RPD3. (A) Freshly made wild-type 0- to 16-h embryo nuclear extracts (NE) (0.2 ml) were fractionated on a Superose 6 HR 10/30 column. The protein in each fraction (fraction number indicated at top of each lane) was analyzed by Western blotting and is indicated at the right side of each panel. The column was calibrated with a series of standard proteins, and mass is indicated at the top with arrows. Fraction 6 is void volume. It is estimated that fraction 10 (or 9) is approximately 2 MDa, based on the presence of PRC1 (PSC) in fractions 8 to 11. (B) EtBr-treated fraction samples from panel A were used for co-IP with anti-ESC (panels 1 to 5), anti-SU(Z)12 (panels 7 to 11), and anti-PCL (panels 13 to 16). Preimmune serum was used for negative controls for IP from fractions and showed no signal (see lanes marked “−” in panels 7 to 11). Forty microliters of NE was diluted four times in the column elution buffer and was used for co-IP in the absence of anti-PCL antibodies, which was also served as negative control (lane C) in panels 13 to 16. Panel 12 shows PCL in each fraction used for IP in panels 13 to 16. Zero- to 16-h nuclear extracts from embryos expressing FLAG-ESC were fractionated as for Fig. 2A. And then EtBr-treated fraction samples were used for co-IP with anti-FLAG M2 gel. p55 eluted from the M2 gel by the FLAG peptide was analyzed by Western blotting for panel 6. Wild-type nuclear extract control on M2 gel was performed as previously described (47), showing no p55 or p105.
To further confirm the presence of PCL in a 1-MDa ESC/E(Z) complex, we carried out similar IP from fractions with anti-PCL antibodies (Fig. 2B, panels 13 to 16). Although E(Z) and SU(Z)12 were present in many fractions, significant amounts of each were present in anti-PCL immunoprecipitates from fraction 12 (and to a much lesser extent from fractions 11 and 14) (Fig. 2B, panels 14 and 15), consistent with the presence of PCL in anti-ESC immunoprecipitates from fraction 12 (and fraction 14) (Fig. 2B, panel 3). RPD3 was also present in anti-PCL immunoprecipitates from fractions 10, 11, 12, and 14 (with a peak at 12) (panel 16 in Fig. 2B), suggesting that RPD3 is physically associated with PCL in additional complexes. These fraction co-IP data indicate that PCL is associated with RPD3, ESC, E(Z), SU(Z)12, and p55 in a 1-MDa complex but not in the 600-kDa ESC/E(Z) complex. Interestingly, while a PCL doublet is immunoprecipitated by anti-PCL antibodies (Fig. 2B, panel 13; also see lane 3 in Fig. 1B and C), only the smaller PCL species was brought down by anti-ESC (Fig. 2B, panel 3; also see lane 4 in Fig. 1B), suggesting that even in fraction 12 not all of the PCL is associated with the 1-MDa ESC/E(Z) complex. Other PCL complexes in fraction 10 (and some in fractions 11 and 12) are evidently not associated with the ESC/E(Z) complex (panels 13 to 15 in Fig. 2B).
PCL binds directly to RPD3 via the PCL PHD fingers and the RPD3 N terminus.
The association of both PCL and RPD3 with a 1-MDa ESC/E(Z) complex but not the 600-kDa complex, as well as their co-IP from other size fractions, raises the possibility that these two proteins bind directly to each other. By use of GST pulldown assays, it was previously shown that in vitro translated RPD3 is pulled down by GST-ESC and GST-E(Z) (47). As shown in Fig. 3A, lane 3, in vitro translated 35S-labeled RPD3 was also pulled down by GST-PCL but not by GST alone (lane 2). Under identical conditions, using similar amounts of GST fusion proteins, GST-PCL pulled down a substantially greater amount of RPD3 than did GST-ESC and GST-E(Z) (compare lane 3 to lanes 4 and 5), suggesting that PCL interacts more strongly with RPD3. Conversely, in vitro translated PCL was also pulled down by GST-RPD3 (see below and Fig. 3D). To map the region of PCL that binds RPD3, we in vitro translated different parts of PCL and tested their ability to bind to GST-RPD3. Since E(Z) binds directly to PCL PHD fingers (34), GST-E(Z) was used as a side-by-side positive control. As shown in Fig. 3B, GST-RPD3 pulled down a significant amount of any PCL protein fragment that contains both PHD fingers (lanes 7 and 11), like GST-E(Z) (lanes 8 and 12). However, PCL140-541 (lane 19), which contains the first PHD finger but lacks the C-terminal half of the second PHD finger, as well as other PCL fragments containing neither PHD finger, was unable to bind efficiently to GST-RPD3 (lanes 3, 15, 19, and 23). Conversely, GST-PCL423-567 (Fig. 3C, lane 4), which contains only the two PHD fingers, retained the full binding activity of PCL (lane 3), while the C terminus of PCL did not (lane 5). Using the same assay and GST-RPD3 fusion proteins containing different regions of RPD3, we mapped the region of RPD3 that interacts with PCL. As shown in Fig. 3D, GST-RPD3 pulled down full-length PCL (lane 9, upper panel). Two N-terminal fragments of RPD3, containing residues 1 to 148 (lane 3) or just residues 24 to 129 (lane 6), bind to PCL (the upper panel) and PCL PHD fingers (the lower panel). All purified GST fusion proteins used for Fig. 3D are shown Coomassie blue stained in Fig. 3E. The amounts of different fusion proteins were adjusted to approximately the same levels in the assays. Results of binding experiments and constructs tested for mapping regions that mediate PCL-RPD3 binding are summarized in Fig. 3F and G.
Association of PCL with the 1-MDa ESC/E(Z) complex is no longer detectable by late embryogenesis.
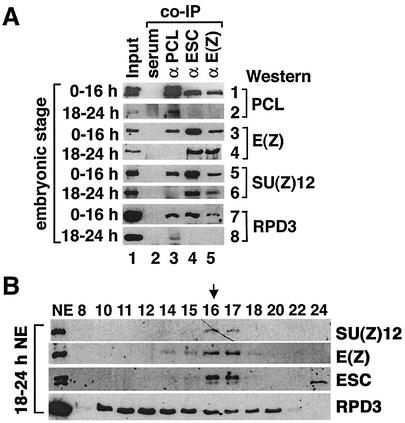
ESC/E(Z) complexes undergo dynamic changes during embryogenesis (22). To determine the temporal profile of the 1-MDa ESC/E(Z) complex, we carried out co-IP assays with staged embryo extracts and found that PCL was associated with RPD3, ESC, E(Z), and SU(Z)12 in 0- to 3-h, 12- to 24-h, and 0- to 16-h embryo nuclear extracts (Fig. 4A, panels 1, 3, 5, and 7). In 18- to 24-h embryo extracts, while RPD3 continues to be associated with PCL (Fig. 4A, lane 3 in panel 8), we can detect no association of RPD3 or PCL with ESC, E(Z), or SU(Z)12 (Fig. 4A, lanes 4 and 5 in panels 2 and 8 and lane 3 in panels 4 and 6). In contrast, ESC, E(Z), and SU(Z)12 continue to be associated with each other (lanes 4 and 5 in panels 4 and 6). Consistent with this, when 18- to 24-h nuclear extract was fractionated on a Superose 6 column (Fig. 4B), ESC/E(Z) complexes were found mostly in fraction 16 and 17 (600 kDa) but not in fraction 12 (∼1 MDa). Comparing the ESC/E(Z) distributions in 18- to 24-h and 0- to 16-h extracts (Fig. 2A, panels 2 and 3), we found that, while the ESC protein level drops in late embryogenesis, the 600-kDa ESC complex is still detectable but that the larger complexes are no longer detectable. This suggests that the 1-MDa complex is not required during late embryogenesis. Whether it reappears later in development remains to be determined. The developmental profile of PCL itself changes during embryogenesis, its level dropping dramatically after 18 h, paralleling the profile of the 1-MDa complex (data not shown).
FIG. 4.
The association of PCL with ESC, E(Z), and SU(Z)12 is no longer detectable by late embryogenesis. (A) EtBr-treated 0- to 16-h and 18- to 24-h embryo nuclear extracts (type of extract indicated at the left side of each panel) were used for co-IP assays with rabbit anti-PCL (lane 3), anti-ESC (lane 4), and anti-E(Z) antibodies (lane 5) and preimmune rabbit serum (lane 2). Other proteins present in immunoprecipitates were detected by Western blotting with rabbit anti-PCL and anti-RPD3 antibodies, mouse anti-E(Z) antibodies, and chicken anti-SU(Z)12 antibodies (antibody type indicated at the right side of each panel). (B) Fractionation of 18- to 24-h nuclear extracts (NE) on a Superose 6 column, same as for Fig. 3A.
Chromosomal colocalization of PCL with RPD3 and E(Z).
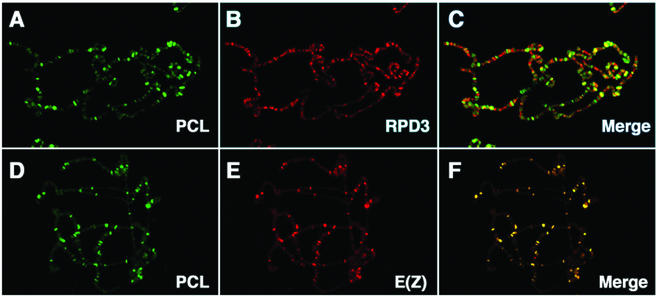
Carrington and Jones originally reported that E(Z) is associated with fewer than half the sites bound by other PcG proteins (12). It was previously shown that E(Z) and ESC are associated with at least 100 specific chromosomal sites and colocalize at virtually all of them (46). PCL was also previously reported to be associated with about 100 specific sites and to colocalize completely with PC (27). To further investigate the association of PCL with RPD3 and E(Z) in vivo, we examined the colocalization of PCL with RPD3 and with E(Z) on polytene chromosomes. As shown in Fig. 5B, RPD3 binds to many sites on the polytene chromosomes (13), consistent with its being an abundant protein associated with many different complexes. PCL, shown in green, is detected at fewer than 100 sites (Fig. 5A) and colocalizes with many but not all RPD3 sites (Fig. 5C). PCL also colocalizes with E(Z) at virtually all sites (Fig. 5F). Moreover, many but not all sites where PCL colocalizes with E(Z) are also RPD3 sites (e.g., note that RPD3 is not found at the 65D landmark PcG site in Fig. 6E and F, confirming a previous report [13]). These data are consistent with the biochemical evidence presented above that PCL is physically associated with RPD3, ESC, and E(Z) in vivo.
FIG. 5.
Colocalization of PCL with RPD3 and E(Z) on salivary gland polytene chromosomes. PCL binding sites on chromosomes were detected with affinity-purified anti-PCL primary antibodies and fluorescein isothiocyanate-labeled secondary antibodies (green [A and D]). RPD3 and E(Z) were detected by sequential immunofluorescence staining by using these antibodies as second primary and Texas red-labeled second secondary antibodies on the same chromosomes. Panels C and F show merged images of panels A and B and panels D and E, respectively. Yellow bands (C and F) indicate sites of colocalization. Control experiments performed in parallel, where the second primary antibodies were omitted, showed no signal due to Texas red-labeled secondary antibody binding to the first primary antibody.
FIG. 6.
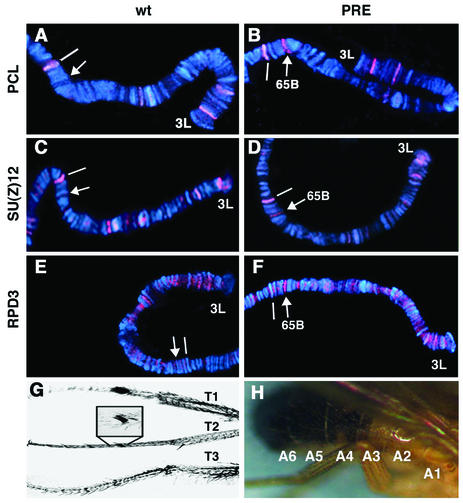
(A to F) Binding of PCL, SU(Z)12, and RPD3 to the Ubx PRE in vivo. The chromosomal binding sites of PCL (red [A and B]), SU(Z)12 (C and D), and RPD3 (E and F) on the distal portion of chromosome arm 3L are shown in a wild-type larva (wt) (A, C, and E) and in a transformant (PRE) (B, D, and F) containing the 670-bp PstI-NdeI fragment of the Ubx PRE inserted at 65B. A landmark site at 65D, which is bound by PCL and other PcG proteins, is indicated by a white line. The arrows indicate new PCL, SU(Z)12, and RPD3 binding sites created at the insertion site of the PRE construct in the transformant (B, D, and F); no band is present at this site in wild-type controls (A, C, and E). The RPD3 binding site at the insertion site of the PRE construct (F) was further confirmed by double staining of PCL and RPD3 (data not shown). (G) Typical transformation of T2 to T1 leg identity seen in over 90% of Pcl11/+; Rpd3303/+ double heterozygotes. (H) Typical A4-to-A5 transformation seen in 35% Pcl7/+; Rpd3303/+ double heterozygotes.
PCL and SU(Z)12 are bound to the Ubx PRE in vivo.
Previously we showed that RPD3 is required for PRE-mediated silencing of a mini-white reporter in transgenic flies and that E(Z) and RPD3 are bound to the insertion site of a transgene containing an isolated 670-bp Ubx PRE in vivo (47). Consistent with the biochemical and cytological data presented above, PCL (Fig. 6A and B) and SU(Z)12 (Fig. 6C and D) are also recruited to the same Ubx PRE in vivo. Figure 6E and F confirm our previous report that RPD3 is also recruited to this PRE transgene when a different anti-RPD3 antibody is used. This suggests that RPD3 and E(Z) and PCL and SU(Z)12 act together directly at the PRE to mediate silencing.
Genetic interaction of Pcl mutations with Su(z)12 and Rpd3 mutations.
Pcl and E(z) mutations have been shown to act as dominant enhancers of esc mutant phenotypes. Indeed Pcl and E(z) mutations have the strongest such enhancing effects of all the PcG mutations tested (9). Conversely, Pcl and esc mutations, like a number of other PcG mutations, also strongly suppress the dominant phenotypes of E(z)Trm, a novel gain-of-function allele of E(z) (5). These data indicate that Pcl is required for the function of ESC and E(Z) and suggest that the physical association of ESC, E(Z), and PCL in the 1-MDa complex is functionally important.
We have also found that Pcl and Su(z)12 mutations interact strongly with each other. One hundred percent of Pcl7/+; Su(z)121/+ double heterozygotes (n = 20 males) exhibit well-developed sex combs on all six legs, i.e., extra sex combs on both mesothoracic and both metathoracic legs. This characteristic PcG phenotype seen in adults is indicative of a transformation of T2 and T3 legs to T1 leg identity. By comparison, Su(z)121/+ heterozygotes (n = 20 males) have an average of only 0.35 ± 0.58 legs, with any evidence of an extra sex comb, usually comprised of just one tooth, indicative of a very weak partial transformation of a T2 or T3 leg to T1 leg identity. In Pcl7/+ heterozygotes (n = 20 males) an average of 2.15 ± 0.81 legs exhibit any evidence of an extra sex comb, typically with only two or three teeth. Similar results were obtained with the Pcl13 allele (data not shown). The strong enhancement of both the frequency and severity of the extra sex comb phenotype observed in Pcl7/+; Su(z)121/+ double heterozygotes is consistent with the observed physical association of PCL with SU(Z)12 in the 1-MDa ESC/E(Z) complex and suggests that this association is functionally important for PcG silencing.
While Rpd3 mutations do not themselves produce homeotic transformations characteristic of PcG mutants (17, 31), they do perturb PRE-mediated reporter silencing (47) and have been shown to act as dominant enhancers of Pc mutant phenotypes (13). This indicates that RPD3 is required for PcG silencing, consistent with an observed physical association of RPD3 with PC-containing complexes (13). We tested for similar genetic interactions between Rpd3 and Pcl mutations. Males heterozygous for either the null Pcl11 or the Pcl7 allele were crossed to females heterozygous for the Rpd3303 allele, and their male progeny were examined for enhancement of the low-penetrance dominant extra sex comb phenotype of Pcl11. Sixty-seven percent (n = 101) of the Pcl11/+; Rpd3303/+ double heterozygotes exhibit an extra sex comb phenotype compared to 7% (n = 71) of their Pcl11/+ siblings (Fig. 6G). Similar enhancement of the Pcl11 extra sex comb phenotype was obtained with the Rpd3313 allele (90%; n = 124) and the null Rpd3R24 allele (data not shown). The extra sex comb phenotype of the Pcl7 allele was similarly enhanced by these Rpd3 mutations (data not shown). Pcl7 and Pcl11 heterozygotes also exhibit another characteristic PcG phenotype with low penetrance: transformation of A4 to A5 identity. This phenotype is also enhanced in Pcl/+; Rpd3/+ double heterozygotes. Figure 6H shows a typical A4-to-A5 transformation seen in 35% (n = 124) of Pcl7/+; Rpd3303/+ double heterozygotes (versus 0% in Pcl7/+ siblings; n = 115). Similar enhancement of this phenotype is observed with the Rpd3R24and Rpd3326 alleles and also in Pcl11/+; Rpd3R24/+ double heterozygotes (data not shown). Taken together, these genetic results further implicate RPD3 in PcG silencing, and together with the biochemical and cytological data presented above, the strong enhancement of Pcl mutant phenotypes is consistent with the possibility that RPD3 is required for the function of PCL in the 1-MDa ESC/E(Z) complex.
DISCUSSION
Association of RPD3 with ESC/E(Z) complexes.
We have identified a ∼1-MDa ESC/E(Z) complex that is distinguished from the previously characterized 600-kDa complex by the presence of the PcG protein PCL and the histone deacetylase RPD3. It was previously shown that RPD3 is associated with a complex containing ESC, E(Z), and p55 and that RPD3 and E(Z) are bound to the Ubx PRE in vivo (47). Histone deacetylase activity was previously reported to be associated with a complex containing mammalian ESC and E(Z) homologs (50) and has recently been detected in a purified ∼600-kDa ESC/E(Z) complex (15). However, these studies did not resolve the 1-MDa complex from the 600-kDa complex. The data presented here indicate that RPD3 is stably associated only with the 1-MDa ESC/E(Z) complex but not the 600-kDa complex. Our previous observation that the 600-kDa ESC/E(Z) complex contains some RPD3 reflects the presence of material from the tail of the distribution of the 1-MDa ESC/E(Z) complex, which overlaps the distribution of the 600-kDa complex in fraction 14 on a Superose 6 column. This fraction had been included in the material subjected to subsequent affinity purification via FLAG epitope-tagged ESC (47). A similar explanation is likely to pertain to the recent confirmation by one group that RPD3 is present in a purified 600-kDa ESC/E(Z) complex (15), while others have not detected it (10, 32).
RPD3 appears to bind more strongly to PCL than to ESC and E(Z) in vitro, consistent with the possibility that the binding of RPD3 to PCL may be essential for stable association of RPD3 with the 1-MDa ESC/E(Z) complex. The respective binding regions of both RPD3 and PCL have been highly conserved in evolution, suggesting that their interaction is likely to be conserved in evolution and functionally important. The enhancement of Pcl mutant phenotypes by Rpd3 and Su(z)12 mutations, together with similar genetic interactions between Pcl, E(z), and esc (9) and the requirement of RPD3 for PRE-mediated silencing of a mini-white reporter (47), are further evidence of the functional importance of the association of PCL and RPD3 with ESC, E(Z), and SU(Z)12.
RPD3 has also been found in a complex with another SET domain-containing histone methyltransferase, SU(VAR)3-9 (16). Genetic evidence indicates that RPD3 is required for SU(VAR)3-9-dependent heterochromatin-mediated repression in vivo, suggesting that deacetylation of acetylated histones by RPD3 may be an essential prerequisite for their methylation by SU(VAR)3-9 (16). The RPD3 present in the 1-MDa ESC/E(Z) complex may play a similar essential role in preparing acetylated histones for methylation by E(Z). We are presently identifying appropriate E(z) and Rpd3 genotypes that will allow us to determine whether RPD3 is required for E(Z) function in vivo.
Role of PCL in ESC/E(Z) complexes.
Genetic studies have revealed that, while the phenotypes associated with loss-of-function mutations in different PcG genes have generally similar phenotypes, they differ in detail, suggesting that individual PcG proteins may have distinct functions and that the requirements for individual PcG proteins can differ depending on tissue, cell type, or target gene (20, 29, 30, 40, 42). Compared to most PcG mutations, which transform all embryonic segments to A8 identity [e.g., esc, E(z), and Pc], embryos homozygous for apparently null Pcl mutations (derived from germ line clones) exhibit only incomplete transformations to more posterior segment identities, thoracic segments appearing almost unaffected (8). Consistent with this, in Pcl mutant embryos only weak derepression of the homeotic genes is observed in the epidermis, while strong derepression occurs in the central nervous system and visceral mesoderm (42). This suggests that PCL is partially dispensable in the epidermis, at least for homeotic gene silencing. These tissue-specific differences in phenotypic effects distinguish Pcl mutants from esc, E(z), and Pc mutants, which exhibit strong derepression of the homeotic genes in all tissues (29, 42). The reason for the apparently lesser requirement for PCL in the epidermis, at least for silencing of the homeotic genes, is unknown. It might reflect differences in the composition of ESC/E(Z) complexes in different tissues or at different target genes, as has been found for other PcG proteins (44). Our evidence is not inconsistent with the possibility that the 600-kDa and 1-MDa complexes represent distinct functional complexes in vivo that are differentially required in different contexts. However, it also remains possible that the 600-kDa complex is simply a stable intermediate in the assembly of a larger, biologically functional complex(es).
The function of PCL in PcG silencing is presently unknown. PCL contains two PHD fingers, a motif found in a wide variety of proteins (1), including the chromatin-associated trxG proteins TRX (43), ASH1 (48), and ASH2 (3) and the histone acetyltransferase CBP, where it is required for histone acetyltransferase activity (25). PHD fingers in TRX and its human homolog MLL mediate homodimerization and binding to the cyclophilin Cyp33 (4, 21). A PHD finger in the KAP-1 corepressor is required for binding of KAP-1 to the Mi-2α subunit of the NuRD complex (38). The PHD fingers in several proteins have recently been shown to possess an intrinsic E3 ubiquitin ligase activity (14, 28). The RING finger motif, which is closely related to the PHD finger in sequence and structure (7, 11, 35), has also been shown to have E3 ubiquitin ligase activity in a number of proteins (7). This raises the possibility that PCL may also function as an E3 ubiquitin ligase. Given its association with an ESC/E(Z) complex, possible substrates might include histones. Histone ubiquitination has been associated with transcriptional silencing in yeast (24, 41), where ubiquitination of histone H2B by a complex containing the E2 and E3 ubiquitin ligases Rad6 and Bre1 has recently been shown to be a prerequisite for methylation of histone H3 by the SET1 protein (18, 45, 51). The association of PCL with E(Z) might reflect a similar coupling of histone ubiquitination and methylation. We are investigating this possibility.
ADDENDUM IN PROOF
Kuzmichev et al. recently presented evidence suggesting that mammalian ESC and E(Z) homologs may also exist in distinct complexes with and without the RPD3 homologs HDAC1 and HDAC2 (A. Kuzmichev, K. Nishioka, H. Erdjument-Bromage, P. Tempst, and D. Reinberg, Genes Dev. 16:2893-2905, 2002.)
Acknowledgments
We thank Robert Saint (Australian National University) for providing pGEX3-PCL and pBluescript-SK-PCL constructs, Jim Kadonaga and Jessica Tyler (University of California at San Diego) for their generous gift of rabbit polyclonal RPD3 and p55 antibodies, and Paul Adler (University of Virginia) for monoclonal PSC antibodies.
This work was supported by a grant from the NIH (GM39255) to P. J. H. and by grants from the J. C. Kempe and Nilsson-Ehle Funds to Å.R.-L.
REFERENCES
- 1.Aasland, R., T. J. Gibson, and A. F. Stewart. 1995. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 20:56-59. [DOI] [PubMed] [Google Scholar]
- 2.Adams, M. D., S. E. Celniker, et al. 2000. The genome sequence of Drosophila melanogaster. Science 287:2185-2195. [DOI] [PubMed] [Google Scholar]
- 3.Adamson, A. L., and A. Shearn. 1996. Molecular genetic analysis of Drosophila ash2, a member of the trithorax group required for imaginal disc pattern formation. Genetics 144:621-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson, M., K. Fair, S. Amero, S. Nelson, P. J. Harte, and M. O. Diaz. 2002. A new family of cyclophilins with an RNA recognition motif that interact with members of the trx/MLL protein family in Drosophila and human cells. Dev. Genes Evol. 212:107-113. [DOI] [PubMed] [Google Scholar]
- 5.Bajusz, I., L. Sipos, Z. Gyorgypal, E. A. Carrington, R. S. Jones, J. Gausz, and H. Gyurkovics. 2001. The Trithorax-mimic allele of Enhancer of zeste renders active domains of target genes accessible to Polycomb-Group-dependent silencing in Drosophila melanogaster. Genetics 159:1135-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Birve, A., A. K. Sengupta, D. Beuchle, J. Larsson, J. A. Kennison, Å. Rasmuson-Lestander, and J. Muller. 2001. Su(z)12, a novel Drosophila Polycomb group gene that is conserved in vertebrates and plants. Development 128:3371-3379. [DOI] [PubMed] [Google Scholar]
- 7.Borden, K. L., and P. S. Freemont. 1996. The RING finger domain: a recent example of a sequence-structure family. Curr. Opin. Struct. Biol. 6:395-401. [DOI] [PubMed] [Google Scholar]
- 8.Breen, T. R., and I. M. Duncan. 1986. Maternal expression of genes that regulate the bithorax complex of Drosophila melanogaster. Dev. Biol. 118:442-456. [DOI] [PubMed] [Google Scholar]
- 9.Campbell, R. B., D. A. Sinclair, M. Couling, and H. W. Brock. 1995. Genetic interactions and dosage effects of Polycomb group genes of Drosophila. Mol. Gen. Genet. 246:291-300. [DOI] [PubMed] [Google Scholar]
- 10.Cao, R., L. Wang, H. Wang, L. Xia, H. Erdjument-Bromage, P. Tempst, R. S. Jones, and Y. Zhang. 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298:1039-1043. [DOI] [PubMed] [Google Scholar]
- 11.Capili, A. D., D. C. Schultz, F. J. Rauscher, and K. L. Borden. 2001. Solution structure of the PHD domain from the KAP-1 corepressor: structural determinants for PHD, RING and LIM zinc-binding domains. EMBO J. 20:165-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carrington, E. A., and R. S. Jones. 1996. The Drosophila Enhancer of zeste gene encodes a chromosomal protein: examination of wild-type and mutant protein distribution. Development 122:4073-4083. [DOI] [PubMed] [Google Scholar]
- 13.Chang, Y. L., Y. H. Peng, I. C. Pan, D. S. Sun, B. King, and D. H. Huang. 2001. Essential role of Drosophila HDAC1 in homeotic gene silencing. Proc. Natl. Acad. Sci. USA 98:9730-9735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coscoy, L., D. J. Sanchez, and D. Ganem. 2001. A novel class of herpesvirus-encoded membrane-bound E3 ubiquitin ligases regulates endocytosis of proteins involved in immune recognition. J. Cell Biol. 155:1265-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Czermin, B., R. Melfi, D. McCabe, V. Seitz, A. Imhof, and V. Pirrotta. 2002. Drosophila Enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111:185-196. [DOI] [PubMed] [Google Scholar]
- 16.Czermin, B., G. Schotta, B. B. Hulsmann, A. Brehm, P. B. Becker, G. Reuter, and A. Imhof. 2001. Physical and functional association of SU(VAR)3-9 and HDAC1 in Drosophila. EMBO Rep. 2:915-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.DeRubertis, F., D. Kadosh, S. Henchoz, D. Pauli, G. Reuter, K. Struhl, and P. Spierer. 1996. The histone deacetylase RPD3 counteracts genomic silencing in Drosophila and yeast. Nature 384:589-591. [DOI] [PubMed] [Google Scholar]
- 18.Dover, J., J. Schneider, M. A. Tawiah-Boateng, A. Wood, K. Dean, M. Johnston, and A. Shilatifard. 2002. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 277:28368-28371. [DOI] [PubMed] [Google Scholar]
- 19.Duncan, I. M. 1982. Polycomblike: a gene that appears to be required for the normal expression of the bithorax and antennapedia gene complexes of Drosophila melanogaster. Genetics 102:49-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dura, J. M., N. B. Randsholt, J. Deatrick, I. Erk, P. Santamaria, J. D. Freeman, S. J. Freeman, D. Weddell, and H. W. Brock. 1987. A complex genetic locus, polyhomeotic, is required for segmental specification and epidermal development in D. melanogaster. Cell 51:829-839. [DOI] [PubMed] [Google Scholar]
- 21.Fair, K., M. Anderson, E. Bulanova, H. Mi, M. Tropschug, and M. O. Diaz. 2001. Protein interactions of the MLL PHD fingers modulate MLL target gene regulation in human cells. Mol. Cell. Biol. 21:3589-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furuyama, T., F. Tie, and P. J. Harte. 2003. The Drosophila Polycomb group proteins ESC and E(Z) are present in multiple distinct complexes that undergo dynamic changes during development. Genesis 35:114-124. [DOI] [PubMed] [Google Scholar]
- 23.Gindhart, J. G., Jr., and T. C. Kaufman. 1995. Identification of Polycomb and trithorax group responsive elements in the regulatory region of the Drosophila homeotic gene Sex combs reduced. Genetics 139:797-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang, H., A. Kahana, D. E. Gottschling, L. Prakash, and S. W. Liebman. 1997. The ubiquitin-conjugating enzyme Rad6 (Ubc2) is required for silencing in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:6693-6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kalkhoven, E., H. Teunissen, A. Houweling, C. P. Verrijzer, and A. Zantema. 2002. The PHD type zinc finger is an integral part of the CBP acetyltransferase domain. Mol. Cell. Biol. 22:1961-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lai, J. S., and W. Herr. 1992. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc. Natl. Acad. Sci. USA 89:6958-6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lonie, A., R. D'Andrea, R. Paro, and R. Saint. 1994. Molecular characterization of the Polycomblike gene of Drosophila melanogaster, a transacting negative regulator of homeotic gene expression. Development 120:2629-2636. [DOI] [PubMed] [Google Scholar]
- 28.Lu, Z., S. Xu, C. Joazeiro, M. H. Cobb, and T. Hunter. 2002. The PHD domain of MEKK1 acts as an E3 ubiquitin ligase and mediates ubiquitination and degradation of ERK1/2. Mol. Cell 9:945-956. [DOI] [PubMed] [Google Scholar]
- 29.McKeon, J., and H. W. Brock. 1991. Interactions of the Polycomb group of genes with homeotic loci of Drosophila. Roux Arch. Dev. Biol. 199:387-396. [DOI] [PubMed] [Google Scholar]
- 30.Moazed, D., and P. H. O'Farrell. 1992. Maintenance of the engrailed expression pattern by Polycomb group genes in Drosophila. Development 116:805-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mottus, R., R. E. Sobel, and T. A. Grigliatti. 2000. Mutational analysis of a histone deacetylase in Drosophila melanogaster: missense mutations suppress gene silencing associated with position effect variegation. Genetics 154:657-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Müller, J., C. M. Hart, N. J. Francis, M. L. Vargas, A. Sengupta, B. Wild, E. L. Miller, M. B. O'Connor, R. E. Kingston, and J. A. Simon. 2002. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111:197-208. [DOI] [PubMed] [Google Scholar]
- 33.Ng, J., C. M. Hart, K. Morgan, and J. A. Simon. 2000. A Drosophila ESC-E(Z) protein complex is distinct from other Polycomb group complexes and contains covalently modified ESC. Mol. Cell. Biol. 20:3069-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Connell, S., L. Wang, S. Roberts, C. A. Jones, R. Saint, and R. S. Jones. 2001. Polycomblike PHD fingers mediate conserved interaction with Enhancer of Zeste protein. J. Biol. Chem. 276:43065-43073. [DOI] [PubMed] [Google Scholar]
- 35.Pascual, J., M. Martinez-Yamout, H. J. Dyson, and P. E. Wright. 2000. Structure of the PHD zinc finger from human Williams-Beuren transcription factor. J. Mol. Biol. 304:723-729. [DOI] [PubMed] [Google Scholar]
- 36.Poux, S., R. Melfi, and V. Pirrotta. 2001. Establishment of Polycomb silencing requires a transient interaction between PC and ESC. Genes Dev. 15:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saurin, A. J., Z. Shao, H. Erdjument-Bromage, P. Tempst, and R. E. Kingston. 2001. A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature 412:655-660. [DOI] [PubMed] [Google Scholar]
- 38.Schultz, D. C., J. R. Friedman, and F. J. Rauscher III. 2001. Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2alpha subunit of NuRD. Genes Dev. 15:428-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shao, Z., F. Raible, R. Mollaaghababa, J. R. Guyon, C. T. Wu, W. Bender, and R. E. Kingston. 1999. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 98:37-46. [DOI] [PubMed] [Google Scholar]
- 40.Simon, J., A. Chiang, and W. Bender. 1992. Ten different Polycomb group genes are required for spatial control of the abd-A and Abd-B homeotic products. Development 114:493-505. [DOI] [PubMed] [Google Scholar]
- 41.Singh, J., V. Goel, and A. J. S. Klar. 1998. A novel function of the DNA repair gene rhp6 in mating-type silencing by chromatin remodeling in fission yeast. Mol. Cell. Biol. 18:5511-5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soto, M. C., T. B. Chou, and W. Bender. 1995. Comparison of germline mosaics of genes in the Polycomb group of Drosophila melanogaster. Genetics 140:231-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stassen, M. J., D. B. Bailey, S. Nelson, V. Chinwalla, and P. J. Harte. 1995. The Drosophila trithorax proteins contain a novel variant of the nuclear receptor type DNA binding domain and an ancient conserved motif found in other chromosomal proteins. Mech. Dev. 52:209-223. [DOI] [PubMed] [Google Scholar]
- 44.Strutt, H., and R. Paro. 1997. The Polycomb group protein complex of Drosophila melanogaster has different compositions at different target genes. Mol. Cell. Biol. 17:6773-6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun, Z. W., and C. D. Allis. 2002. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature 418:104-108. [DOI] [PubMed] [Google Scholar]
- 46.Tie, F., T. Furuyama, and P. J. Harte. 1998. The Drosophila Polycomb-Group proteins ESC and E(Z) bind directly to each other and co-localize at multiple chromosomal sites. Development 125:3483-3496. [DOI] [PubMed] [Google Scholar]
- 47.Tie, F., T. Furuyama, J. Prasad-Sinha, E. P. Jane, and P. J. Harte. 2001. The Drosophila Polycomb Group proteins ESC and E(Z) are present in a complex containing the histone-binding protein p55 and the histone deacetylase RPD3. Development 128:275-286. [DOI] [PubMed] [Google Scholar]
- 48.Tripoulas, N., D. LaJeunesse, J. Gildea, and A. Shearn. 1996. The Drosophila ash1 gene product, which is localized at specific sites on polytene chromosomes, contains a SET domain and a PHD finger. Genetics 143:913-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tyler, J. K., K. A. Collins, J. Prasad-Sinha, E. Amiott, M. Bulger, P. J. Harte, R. Kobayashi, and J. T. Kadonaga. 2001. Interaction between the Drosophila CAF-1 and ASF1 chromatin assembly factors. Mol. Cell. Biol. 21:6574-6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van der Vlag, J., and A. P. Otte. 1999. Transcriptional repression mediated by the human Polycomb-Group protein EED involves histone deacetylation. Nat. Genet. 23:474-478. [DOI] [PubMed] [Google Scholar]
- 51.Wood, A., N. J. Krogan, J. Dover, J. Schneider, J. Heidt, M. A. Boateng, K. Dean, A. Golshani, Y. Zhang, J. F. Greenblatt, M. Johnston, and A. Shilatifard. 2003. Bre1, an e3 ubiquitin ligase required for recruitment and substrate selection of rad6 at a promoter. Mol. Cell 11:267-274. [DOI] [PubMed] [Google Scholar]