Abstract
The p21-activated kinases Ste20p and Cla4p carry out undefined functions that are essential for viability during budding in Saccharomyces cerevisiae. To gain insight into the roles of Ste20p, we have used a synthetic lethal mutant screen to identify additional genes that are required in the absence of Cla4p. Altogether, we identified 65 genes, including genes with roles in cell polarity, mitosis, and cell wall maintenance. Herein, we focus on a set that defines a function carried out by Bni1p and several of its interacting proteins. We found that Bni1p and a group of proteins that complex with Bni1p (Bud6p, Spa2p, and Pea2p) are essential in a cla4Δ mutant background. Bni1p, Bud6p, Spa2, and Pea2p are members of a group of polarity determining proteins referred to as the polarisome. Loss of polarisome proteins from a cla4Δ strain causes cells to form elongated buds that have mislocalized septin rings. In contrast, other proteins that interact with or functionally associate with Bni1p and have roles in nuclear migration and cytokinesis, including Num1p and Hof1p, are not essential in the absence of Cla4p. Finally, we have found that Bni1p is phosphorylated in vivo, and a substantial portion of this phosphorylation is dependent on STE20. Together, these results suggest that one function of Ste20p may be to activate the polarisome complex by phosphorylation of Bni1p.
INTRODUCTION
Yeast cells undergo polarized growth during budding and other morphogenetic events in response to intracellular or extracellular cues (Drubin and Nelson, 1996). Polarized cell growth depends on assembly of a polarized actin cytoskeleton, which then directs transport of secretory vesicles containing cell wall and plasma membrane components to the site of growth (Novick and Botstein, 1985; Mulholland et al., 1994; Ayscough et al., 1997; Pruyne et al., 1998). The p21 GTPase Cdc42p plays a critical role in the establishment of subcellular polarity and the execution of subsequent apical growth by regulating the actin cytoskeletion (Adams et al., 1990; Johnson and Pringle, 1990; Ziman et al., 1993; Li et al., 1995; Richman and Johnson, 2000). Cdc42p is also required for septin ring function and for cytokinesis (Richman et al., 1999; Toenjes et al., 1999). How Cdc42p orchestrates these various activities is poorly understood, but some of its target effectors have been identified. For example, Gic1p and Gic2p can bind activated Cdc42p and are important for polarization of the actin cytoskeleton (Brown et al., 1997; Chen et al., 1997). Two other identified effectors for Cdc42p are the related p21-activated protein kinases, Cla4p and Ste20p, both of which interact with activated Cdc42p and localize to sites of polarized growth (Cvrckova et al., 1995; Peter et al., 1996; Leberer et al., 1997; Holly and Blumer, 1999; Mosch et al., 2001). Each kinase has unique roles in the cell. Ste20p functions in pheromone response and haploid invasive growth, whereas Cla4p promotes normal septin function (Ramer and Davis, 1993; Roberts and Fink, 1994; Benton et al., 1997; Tjandra et al., 1998; Gulli et al., 2000; Bose et al., 2001). A cell lacking both kinases is inviable (Cvrckova et al., 1995), demonstrating that Ste20p is essential in the absence of Cla4p (and vice versa). One interpretation of this relationship is that these two kinases share a function that is essential, though other interpretations are possible. Currently, the only proposed targets of Ste20p and Cla4p are the two myosin I homologs Myo3p and Myo5p. Sites in Myo3p and Myo5p are phosphorylated in vitro by Ste20p and are required for in vivo function (Wu et al., 1997). Myo3p and Myo5p are required for actin patch assembly (Evangelista et al., 2000; Geli et al., 2000; Lechler et al., 2000). However, although an activated allele of Myo3p (MYO3S357D) can rescue the polarity defects of myo3Δ myo5Δ mutants, this mutant failed to compensate for the growth defects in ste20Δ cla4Δ mutants, indicating that there are other targets of Ste20p and Cla4p (Wu et al., 1997). To identify the target(s) and function(s) regulated by Ste20p, we have carried out a search for mutations that are synthetically lethal in a cla4Δ mutant background with the expectation that the genes identified would suggest the nature of the physiological events that have been perturbed.
Herein, we present the results of two independent synthetic lethal mutant screens. One screen was based on random mutagenesis of the genome by using a red/white colony sectoring assay (Kranz and Holm, 1990; Bender and Pringle, 1991). The second screen used a yeast genome-wide deletion set and evaluated the viability of cla4Δ paired with 4672 different viable deletion strains (Tong et al., 2001). From the collection of genes defined by these screens, we chose a subset for more detailed investigation. BNI1 is at the center of this study and encodes a formin homology protein (Zahner et al., 1996) that is required to assemble actin cables (Evangelista et al., 2002; Sagot et al., 2002). These cables seem to guide myosin motors that direct secretion, organelle and mRNA inheritance, and mitotic spindle orientation, thereby establishing cell polarity (Evangelista et al., 2002). Bni1p has an intricate network of interactions involving a number of different groups of proteins. One such group of proteins, Bud6p, Spa2p, and Pea2p, complexes with Bni1p to form the “polarisome,” which is involved in apical growth (Sheu et al., 1998; Pruyne and Bretscher, 2000). We show that Bud6p, Spa2p, and Pea2p are essential in a cla4Δ mutant (Evangelista et al., 1997; Fujiwara et al., 1998; Sheu et al., 2000). Conversely, other proteins that interact with Bni1p, including Hof1p and Num1p (involved in cytokinesis and nuclear migration, respectively), are not essential in cells lacking Cla4p (Kamei et al., 1998; Heil-Chapdelaine et al., 2000; Vallen et al., 2000; Farkasovsky and Kuntzel, 2001). Finally, we further show that Bni1p is a Ste20p-dependent phosphoprotein, suggesting that Bni1p's function may be directly regulated by Ste20p. Together, these results imply that Ste20p may play an important role in activating the polarisome.
MATERIALS AND METHODS
Growth Conditions, Plasmids, and Strains
Yeast and bacterial strains were propagated using standard methods (Sambrook et al., 1989; Rose et al., 1990). YEPD and SD media were prepared as described previously (Rose et al., 1990). Yeast transformations were performed using modifications of the LiOAc method (Chen et al., 1992; Gietz et al., 1995). Bacterial transformations, DNA preparations, and plasmid constructions were performed by standard methods (Sambrook et al., 1989). The plasmids used in this study, YCpHIS3cla4-75, pY39tet1HA-BNI1 (p925), pcla4-75-td, and pRS316ADE8CLA4 have been described elsewhere (Cvrckova et al., 1995; Evangelista et al., 1997; Holly and Blumer, 1999; Mitchell and Sprague, 2001). To ensure that the version cla4-75-td we were using was the same as the allele used in Holly and Blumer (1999), we sequenced the pcla4-75-td and the cla4-75-td alleles rescued from our strains. We found that the cla4-75-td alleles rescued from our strains were identical to that of the pcla4-75-td from the Blumer laboratory. Strains that were used in this study are listed in Table 1. Gene deletions were constructed by polymerase chain reaction (PCR) (Baudin et al., 1993) by using either the pRS (Sikorski and Hieter, 1989) or pFA6a (Longtine et al., 1998) plasmid series as templates. In all cases, the entire coding region was replaced with the indicated marker, and successful replacement was confirmed by PCR and phenotype when applicable. Single step gene deletion plasmids for swe1::LEU2, spa2::URA3 (p210) and pea2::URA3 (pNV44) were provided by I. Herskowitz and D. Lew (Booher et al., 1993; Valtz and Herskowitz, 1996). We used bni1::URA3 (p321), a single step gene deletion plasmid, to delete BNI1 (Evangelista et al., 1997). 5-Fluoroorotic acid (5-FOA) (Biovectra, Oxford, CT) was used to select for uracil auxotrophs. The COOH-terminal deletion mutant bni1-CTΔ1 lacks the coding sequence for amino acids 1749–1953 of Bni1p (Lee et al., 1999). bni1-CTΔ1 was created by amplification of the kanMX6 cassette from pFA6a-kanMX6 together with sequences immediately flanking base pairs 5247–5859 of BNI1 by using the forward primer 5′-ATAAATGAATACAAAAAAGCTCAAGCGCAAAATCTAGCCTGAGGCG -CGCCACTTCTAAA-3′ and the reverse primer 5′-GTTTTGGTAT-TACTGTTGTCATAATTTTTTGGTTTAATATTGAATTCGAGCTC-GTTTAAAC-3′ (the sequences flanking base pairs 5247–5859 of BNI1 are underlined) (Longtine et al., 1998). The amplified fragment was transformed into strains SY3357, SY3362, SY3380, and SY3764; the transformants were plated on YEPD medium and incubated overnight at 30°C. The lawn of cells was then replica-plated onto YEPD containing 200 μg/ml G418/geneticin (Invitrogen, Carlsbad, CA) to select stable G418-resistant transformants. The successful creation of the deletion strain was confirmed by PCR, and the protein function was verified in a bnr1Δ strain, in which Bni1p is essential (our unpublished data) (Ozaki-Kuroda et al., 2001).
Table 1.
Yeast strains used in this study
| Straina | Genotype | Source |
|---|---|---|
| SY3357 | MATa leu2-Δ1 ura3-52 his3-Δ200 trp1-Δ63 ade8Δ ade2-101 mfa2-Δ1∷FUS1-lacZ | Mitchell and Sprague (2001) |
| SY3358 | MATα leu2-Δ1 ura3-52 his3-Δ200 lys2-801 trp1-Δ63 ade8Δ ade2-101 mfa2-Δ1∷FUS1 lacZ | Mitchell and Sprague (2001) |
| SY3362 | SY3357 except cla4Δ∷TRP1 (pRS316ADE8CLA4) | Mitchell and Sprague (2001) |
| SY3364 | SY3357 except cla4Δ∷TRP1 ncs1Δ∷LEU2 (pRS316ADE8CLA4) | Mitchell and Sprague (2001) |
| SY3366 | SY3357 except cla4Δ∷TRP1 ncs2-1 (pRS316ADE8CLA4) | Mitchell and Sprague (2001) |
| SY3367 | SY3357 except cla4Δ∷TRP1 ncs3-1 (pRS316ADE8CLA4) | Mitchell and Sprague (2001) |
| SY3368 | SY3357 except cla4Δ∷TRP1 ncs4-1 (pRS316ADE8CLA4) | Mitchell and Sprague (2001) |
| SY3369 | SY3357 except cla4Δ∷TRP1 ncs5-1 (pRS316ADE8CLA4) | Mitchell and Sprague (2001) |
| SY3370 | SY3357 except cla4Δ∷TRP1 ncs6-1 (pRS316ADE8CLA4) | Mitchell and Sprague (2001) |
| SY3371 | SY3357 except cla4Δ∷TRP1 ncs7-1 (pRS316ADE8CLA4) | Mitchell and Sprague (2001) |
| SY3372 | SY3357 except cla4Δ∷TRP1 ncs8-1 (pRS316ADE8CLA4) | Mitchell and Sprague (2001) |
| SY3373 | SY3357 except cla4Δ∷TRP1 ncs10-1 (pRS316ADE8CLA4) | Mitchell and Sprague (2001) |
| SY3380 | SY3357 except cla4Δ∷TRP1 (YCpHIS3cla4-75) | Mitchell and Sprague (2001) |
| SY3403 | SY3357 except cla4Δ∷TRP1 swe1∷LEU2 ncs1Δ∷HIS3 (pRS316ADE8CLA4) | Mitchell and Sprague (2001) |
| SY3756 | SY3357 except cla4Δ∷TRP1 bni1Δ∷HIS3 (pRS316ADE8CLA4) | This study |
| SY3757 | SY3357 except cla4Δ∷TRP1 bud6Δ∷HIS3 (pRS316ADE8CLA4) | This study |
| SY3758 | SY3358 except cla4Δ∷TRP1 spa2∷ura3 (pRS316ADE8CLA4) | This study |
| SY3759 | SY3357 except cla4Δ∷TRP1 pea2∷ura3 (pRS316ADE8CLA4) | This study |
| SY3760 | SY3357 except cla4Δ∷TRP1 bni1∷URA3 (YCpHIS3cla4-75) | This study |
| SY3761 | SY3357 except cla4Δ∷TRP1 bud6Δ∷URA3 (YCpHIS3cla4-75) | This study |
| SY3762 | SY3357 except cla4Δ∷TRP1 spa2∷URA3 (YCpHIS3cla4-75) | This study |
| SY3763 | SY3357 except cla4Δ∷TRP1 pea2∷URA3 (YCpHIS3cla4-75) | This study |
| SY3764 | SY3357 except cla4Δ∷TRP1 ste20Δ∷URA3 (YCpHIS3cla4-75) | This study |
| SY3766 | SY3357 except cla4Δ∷TRP1 swe1∷LEU2 (pRS316ADE8CLA4) | This study |
| SY3767 | SY3357 except cla4Δ∷TRP1 ste20Δ∷TRP1 swe1∷LEU2 (pRS316ADE8CLA4) | This study |
| SY3768 | SY3357 except cla4Δ∷TRP1 bni1∷ura3 swe1∷LEU2 (pRS316ADE8CLA4) | This study |
| SY3769 | SY3357 except cla4Δ∷TRP1 bud6Δ∷his3 swe1∷LEU2 (pRS316ADE8CLA4) | This study |
| SY3770 | SY3358 except cla4Δ∷TRP1 spa2∷ura3 swe1∷LEU2 (pRS316ADE8CLA4) | This study |
| SY3771 | SY3357 except cla4Δ∷TRP1 pea2∷ura3 swe1∷LEU2 (pRS316ADE8CLA4) | This study |
| SY3772 | SY3357 except cla4Δ∷TRP1 swe1∷LEU2 (YCpHIS3cla4-75) | This study |
| SY3773 | SY3357 except cla4Δ∷TRP1 ste20Δ∷URA3 swe1∷LEU2 (YCpHIS3cla4-75) | This study |
| SY3774 | SY3357 except cla4Δ∷TRP1 bni1∷ura3 swe1∷LEU2 (YCpHIS3cla4-75) | This study |
| SY3775 | SY3357 except cla4Δ∷TRP1 bud6Δ∷ura3 swe1∷LEU2 (YCpHIS3cla4-75) | This study |
| SY3776 | SY3357 except cla4Δ∷TRP1 spa2∷ura3 swe1∷LEU2 (YCpHIS3cla4-75) | This study |
| SY3777 | SY3357 except cla4Δ∷TRP1 pea2∷ura3 swe1∷LEU2 (YCpHIS3cla4-75) | This study |
| SY3778 | SY3358 except bni1∷HIS3 | This study |
| SY3779 | SY3358 except bni1∷HIS3 (pY39tet1HA-BNI1) | This study |
| SY3780 | SY3358 except ste20Δ∷TRP1 bni1∷HIS3 (pY39tet1HA-BNI1) | This study |
| SY3781 | SY3357 except cla4Δ∷TRP1 ste20Δ∷TRP1 bni1∷ura3 (YCpHIS3cla4-75) | This study |
| SY3782 | SY3357 except cla4Δ∷TRP1 ste20Δ∷kanMX6 bud6Δ∷ura3 (YCpHIS3cla4-75) | This study |
| SY3783 | SY3357 except cla4Δ∷TRP1 ste20Δ∷kanMX6 spa2∷ura3 (YCpHIS3cla4-75) | This study |
| SY3784 | SY3357 except cla4Δ∷TRP1 ste20Δ∷kanMX6 pea2∷ura3 (YCpHIS3cla4-75) | This study |
| SY3785 | SY3358 except bni1-CTΔ1∷kanMX6 | This study |
| SY3786 | SY3357 except cla4Δ∷TRP1 bni1-CTΔ1∷kanMX6 (pRS316ADE8CLA4) | This study |
| SY3787 | SY3357 except cla4Δ∷TRP1 ste20Δ∷TRP1 bni1-CTΔ1∷kanMX6 (pRS316ADE8CLA4) | This study |
| SY3788 | SY3357 except cla4Δ∷TRP1 bni1-CTΔ1∷kanMX6 (YCpHIS3cla4-75) | This study |
| SY3789 | SY3357 except cla4Δ∷TRP1 ste20Δ∷TRP1 bni1-CTΔ1∷kanMX6 (YCpHIS3cla4-75) | This study |
| SY3790 | SY3357 except cla4Δ∷TRP1 URA3∷cla4-75-td | This study |
| SY3791 | SY3357 except cla4Δ∷TRP1 bni1∷HIS3 URA3∷cla4-75-td | This study |
| SY3792 | SY3357 except cla4Δ∷TRP1 bud6Δ∷ura3 URA3∷cla4-75-td | This study |
| SY3793 | SY3357 except cla4Δ∷TRP1 spa2∷ura3 URA3∷cla4-75-td | This study |
| SY3794 | SY3357 except cla4Δ∷TRP1 pea2∷ura3 URA3∷cla4-75-td | This study |
| SY3795 | SY3357 except cla4Δ∷TRP1 ste20Δ∷TRP1 URA3∷cla4-75-td | This study |
| DY759 | cry1 ade2-101(am) his3-11 leu2-3,112 ura3-1 | Weiss et al. (2000) |
| DY2060 | DY759 except cla4Δ∷LEU2 ste20Δ∷KanMX URA3∷cla4-75-td | Weiss et al. (2000) |
| Y2454 | MATα mfa1Δ∷MFA1pr-HIS3 can1Δ ura3Δ leu2Δ his3Δ1 lys2Δ | This study |
| Y2851 | Y2454 except cla4Δ∷URA3 | This study |
| Y2928 | Y2851 except cla4Δ∷natR | This study |
All are derivatives S288C except DY759 and DY2060 (W303 derivatives).
Synthetic Lethal Mutant Screens
Two independent methods were used to search for mutations that are lethal in a cla4Δ background. Previously, we described the details of the NCS screen by using the colony sectoring assay (Mitchell and Sprague, 2001). Synthetic genetic array analysis (SGA) was also used to identify genes that were essential in a cla4Δ background as described in Tong et al. (2001). Y2928 (MATα cla4Δ::natR mfa1Δ::MFA1pr-HIS3 can1Δ ura3Δ leu2Δ his3Δ1 lys2Δ) was created in two steps. First, CLA4 was deleted from Y2454 by using PCR-based integration with primers (5′-TTTGGTGTAATAAATCGAACA GTGAAACTGAAACATAAAAGAAATAGTGCAAAATGGAAACAGCTATG ACCATG-3′ and 5′-AGAAAT-ACATAAGATTGTAGTATGTATGATATGCTTATAGAAATAGTTGT-GTGCTGTTGTAAAACGACGGCCAGT-3′), which annealed to URA3 and contained CLA4 sequences (underlined), to generate Y2851 (MATα cla4Δ::URA3 mfa1Δ::MFA1pr-HIS3 can1Δ ura3Δ leu2Δ his3Δ1 lys2Δ). Second, cla4Δ::URA3 was switched to cla4Δ::natR by PCR-based integration with primers (5′-AGTATTCTTAACCCAACTGCACAGAACAAA-AACCTGCAGGAAACGAAGATAAATCATGACCACTCTTGACGA -CACGG-3′ and 5′-TTGAAGCTCTAATTTGTGAGTTTAGTATA-CATGCATTTACTTATAATACAGTTTTCTAGGGGCAGGGCATGC-TCAT-3′), which anneal to natMX4 DNA (Goldstein et al., 1999) and contain URA3 sequences (underlined). We performed SGA on cla4Δ::natR four times. A total of 100 potential positives were identified and 62 were confirmed by tetrad analysis.
Isolation of BNI1, BUD6, and Other NCS Genes
Wild-type NCS8 and NCS5 were identified as BNI1 and BUD6 by complementation of ncs8-1 (SY3372) and ncs5-1 (SY3369) mutants, respectively. For NCS8 isolation, 20,000 library transformants yielded six complementing clones from a yeast genomic library (ATCC no. 77162). An 8.6-kb region shared by all of them was sequenced and found to include BNI1. NCS8 was shown to be BNI1 by deletion and linkage analysis (see below). For BUD6, two clones complementing ncs5-1 were found among 8000 library transformants. An 8-kb fragment shared by both complementing plasmids was sequenced. Deletion and subcloning analysis identified BUD6 as the complementing gene. To isolate NCS2, strain SY3366, which harbors an ncs2 mutation, was transformed with a high copy YEp13 based library (ATCC no. 37323), yielding six complementing clones in 6000 transformants. A 2-kb fragment containing two overlapping open reading frames (ORFs) shared by all complementing plasmids was sequenced. Deletion analysis identified YNL119w/YNL120c as the complementing ORF(s). For NCS3 isolation, 7000 library transformants yielded six complementing clones from a high copy YEp13-based library. A 3.6-kb fragment containing three ORFs shared by all complementing plasmids was sequenced. Deletion and subcloning analysis identified UBA4 as the complementing gene. Because ncs4 mutants had a strong mating defect, it seemed reasonable that members of this complementation group could contain mutations in STE20. Indeed, we found that STE20 on a plasmid complemented these mutants. In the case of NCS6, transformation of SY3370 with yeast genomic library (ATCC no. 77162) yielded two complementing clones from 9000 transformants. A 6.8-kb fragment containing six ORFs shared by all complementing plasmids was sequenced. Deletion and subcloning analysis identified YGL211w as the ORF containing the complementing gene. In the case of NCS10, 16,000 transformants of a yeast genomic library yielded 16 complementing clones. A 4.6-kb fragment containing three ORFs shared by all complementing plasmids was sequenced. Deletion and subcloning analysis identified ELP2 as the complementing gene.
Linkage analysis was performed to verify that the cloned genes represented wild-type versions of the mutant alleles. A HIS3 marker was introduced at the locus of interest in a diploid homozygous for the cla4 mutation and heterozygous for the NCS gene of interest. The strain carried plasmid-borne CLA4 so that segregation of the ncs mutation in tetrads could be scored. After sporulation of the marked strains, the Ncs+ phenotype cosegregated with the HIS3 marker in at least 22 tetrads.
Microscopy
Standard microscopic techniques were used, and cells were examined using an Axioplan 2 fluorescence microscope (Carl Zeiss, Thornwood, NY) fitted with an Orca 100 digital camera (Hamamatsu, Bridgewater, NJ). Methods for staining with rhodamine-phalloidin (Molecular Probes, Eugene, OR) to visualize F-actin was performed essentially as described previously (Pringle et al., 1989). All assays were performed in triplicate. Indirect immunofluorescence was performed to visualize the septins by using an α-Cdc3p antibody (a generous gift from John Pringle) (Roberts et al., 1991). Cells were grown in YEPD at 30°C to 0.7 OD600/ml before fixation. Strains containing plasmids were grown first in selective medium, transferred to YEPD, and then grown for 3–4 h at 37°C. Cells were fixed by adding a final concentration of 3.7% formaldehyde to the culture medium for 1 h. The cells were pelleted by centrifugation, resuspended in 4% paraformaldehyde (final concentration) in 50 mM KPO4, pH 6.5, and incubated for 18 h at room temperature. The fixed cells were then spheroplasted and permeabilized with 5% SDS for 5 min. Antibody incubations were carried out for 1 h at 22°C. The secondary antibody used was Alexa (A594)-conjugated goat anti-rabbit antibody (Molecular Probes).
Mating and Invasive Growth Assays
Quantitative mating assays were done using strain 227 as a tester (Sprague, 1991). For invasive growth assays, cells were transformed with YEplac181-FLO8 to allow the S288c background to manifest the switch from the yeast form to the filamentous form (Gagiano et al., 1999). YEplac181-FLO8 was a generous gift from Isak S. Pretorius (Stellenbosch University, Stellenbosch, South Africa). The plate washing assay was performed as previously described (Roberts and Fink, 1994).
In Vivo Labeling
To label with 32Pi, cultures of yeast strains SY3778, SY3779, and SY3780 were pregrown overnight in synthetic medium lacking leucine. Cells were washed once with phosphate-depleted medium (Rubin, 1975) and transferred into phosphate-depleted medium at an OD600 of 0.2 and grown to an OD600 of 0.5. For labeling with 32Pi, 50 ml of culture was harvested, the pellet was suspended in 15 ml of phosphate-depleted medium, and 1 mCi of 32PO4 (ICN Pharmaceuticals, Costa Mesa, CA) was added. After labeling for 45 min, 10 mM sodium azide was added to the cultures and cells were harvested. Cells were spheroplasted as described previously (Graham et al., 1998). Frozen spheroplasts were resuspended in lysis buffer (50 mM Tris, pH 8.0, 1% NP-40, 50 mM NaCl, 1 mM EDTA) containing a mixture of protease inhibitors (1 mM phenylmethanesulfonyl fluoride, 1 μg/ml leupeptin, 1 μg/ml pepstatin A, 1 μg/ml mg/ml aprotinin [all from Sigma-Aldrich, St. Louis, MO] and one tablet of Roche Diagnostics protease inhibitor mixture Complete/25 ml) and phosphatase inhibitors (5 mM sodium pyrophosphate, 0.1 mM sodium metavanadate, 50 mMNaF; all from Sigma-Aldrich). The spheroplasts were incubated in lysis buffer for 15 min at 4°C and unlysed cells were removed by centrifugation at 13,000 × g for 2 min. A total of 0.5 ml of lysate was precleared with 50 μl of protein A-Sepharose beads and bovine serum albumin (1 mg/ml). Bni1p was immunoprecipitated from these extracts with 3 μl of rabbit anti-hemagglutinin (HA) antiserum (a kind gift from T. Stevens, Institute of Molecular Biology, University of Oregon) for 30 min at 4°C, after which 50 μl of protein A-Sepharose beads was added and incubation continued for another 30 min at 4°C. These pellets were then washed four times with lysis buffer, boiled in 1% SDS before the addition of 9 volumes of lysis buffer, and the immunoprecipitation was repeated. These pellets were washed four times in lysis buffer, resuspended in 30 μl of sample buffer, and the entire sample was run on a 6% SDS-PAGE gel, transferred to nitrocellulose, and visualized on a Storm PhosphorImager. Immunoprecipitated Bni1p was also detected by Western blot analysis with an anti-HA monoclonal antibody followed by a horseradish peroxidase-labeled anti-mouse secondary antibody. Blots were developed with ECLplus (Amersham Biosciences, Piscataway, NJ), visualized by chemiluminescence, and quantified by chemifluorescence on a Storm PhosphorImager (Invitrogen, Sunnyvale, CA) with a wavelength of 450 nm.
RESULTS
Identification of BNI1 and BUD6 by Synthetic Lethal Interactions with a cla4 Null Mutation
In an effort to identify activators and targets of Ste20p, we screened for mutations that are lethal in combination with a cla4 null mutation by using two independent methods. The first method used a random mutagenesis of the genome and identified synthetic lethal mutations via a red/white colony sectoring assay (Table 2). Such a screen can be expected to identify two classes of genes: 1) genes that encode upstream activators and downstream effectors of STE20, and 2) genes that share a function with CLA4 that is independent of STE20. Previously, we described the isolation of NCS1 (Needs CLA4 to Survive), which falls into the second class (Mitchell and Sprague, 2001). NCS1/RRD1 was unique among NCS genes in that ncs1Δ mutants were not defective for any known STE20 function. NCS1 is a phosphotyrosyl phosphatase activator that may share a function with CLA4 required at the G2/M phase transition (Mitchell and Sprague, 2001).
Table 2.
NCS mutants
| Complementation group | Gene or ORF | Isolates | swe1Δ Overcomes synthetic lethality | Invasive growth | Mating competency |
|---|---|---|---|---|---|
| NCS1 | RRD1 | 1 | Yes | No defect | WT |
| NCS2 | YNL119w/YNL120c | 6 | Yes | Defect | WT |
| NCS3 | UBA4 | 6 | Yes | Defect | WT |
| NCS4 | STE20 | 4 | No | Defect | 1000 fold lower than wild type |
| NCS5 | BUD6 | 2 | No | Defect | N/D |
| NCS6 | YGL211w | 3 | N/D | Defect | N/D |
| NCS7 | N/D | 2 | Yes | Defect | WT |
| NCS8 | BNI1 | 5 | No | Defect | ∼3-fold lower than wild type |
| NCS10 | ELP2 | 2 | Yes | Defect | WT |
N/D, not determined; WT, wild type.
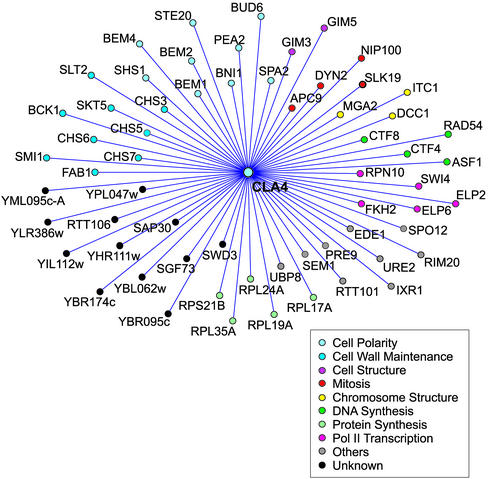
The screen also yielded complementation groups that exhibited some ste20Δ phenotypes; in many cases, a defect in haploid invasive growth and in some cases a partial defect in mating as well. As described in MATERIALS AND METHODS, molecular cloning identified the genes corresponding to these complementation groups (Table 2). In this study, we chose to concentrate on NCS8/BNI1, both because it is required for efficient mating and because a second complementation group, NCS5/BUD6, encodes a protein known to interact with Bni1p (Evangelista et al., 1997). Bni1p (Bud neck involved protein) is a formin homology protein that interacts with a large number of proteins and has many functions attributed to it. These functions include roles in bipolar bud site selection in diploids, cell polarity, cytokinesis, and spindle alignment during nuclear migration and may all stem from its role in actin cable assembly (Zahner et al., 1996; Lee et al., 1999; Miller et al., 1999; Sheu et al., 2000; Vallen et al., 2000; Evangelista et al., 2002). We therefore sought to establish which Bni1p functions were essential in a cla4Δ background and whether Bni1p has a specific link to Ste20p. To this end, we asked whether the loss of other proteins that interact with Bni1p or function in the same processes as Bni1p was lethal in a cla4Δ background. Some of these new double mutants were constructed by transformation with the appropriate gene disruption followed by a genetic cross. Other double mutants were created in the second synthetic lethal screen, which used a systematic method, known as SGA analysis, to construct double mutants (Tong et al., 2001). This screen used a genome-wide deletion set and evaluated the viability of cla4Δ paired with deletion of 4672 open reading frames. The results of this screen can be viewed in Figure 1 and in Supplementary Table 1. The two approaches, random mutagenesis coupled with subsequent directed double mutant construction and the use of the genome-wide deletion set, were complementary. Each identified some unique genes and in cases where the same gene was investigated, the two approaches gave congruent results. The SGA analysis provides a global view of the proteins that become essential in the absence of Cla4p. Such proteins include ones involved in apical growth, bud emergence, cytokinesis, mitosis, and cell wall maintenance. However, although the SGA method enabled us to perform a more complete synthetic lethal analysis, three of the nine complementation groups identified in the random mutagenesis were not identified by the SGA method.
Figure 1.
Genetic interaction network of the synthetic lethal interactions identified by the SGA analysis. The genes that are essential or required for normal growth rate in a cla4Δ are represented as nodes. Each node is color coordinated according to the functional classification of the gene according to YPD (Hodges et al., 1999; Costanzo et al., 2001). For genes that have multiple roles assigned to them, we chose the function that we considered most probable based on a review of published abstracts for the gene of interest.
Polarisome Components Are Essential in a cla4Δ Background
As noted above, loss of either BNI1 or BUD6 is lethal in a cla4Δ strain. Each gene is involved in cell polarity establishment and in bipolar budding (Zahner et al., 1996; Evangelista et al., 1997; Sheu et al., 2000). Moreover, Bni1p and Bud6p have been shown to interact by two-hybrid analysis (Evangelista et al., 1997). We also found that loss of Spa2p, another protein that interacts with Bni1p (Fujiwara et al., 1998), is essential in a cla4Δ mutant background. These three proteins, together with a fourth protein Pea2p, form a 12S complex termed the polarisome that has been suggested to promote polarized morphogenesis (Sheu et al., 1998; Pruyne and Bretscher, 2000). We found that Pea2p is likewise essential in a cla4Δ strain.
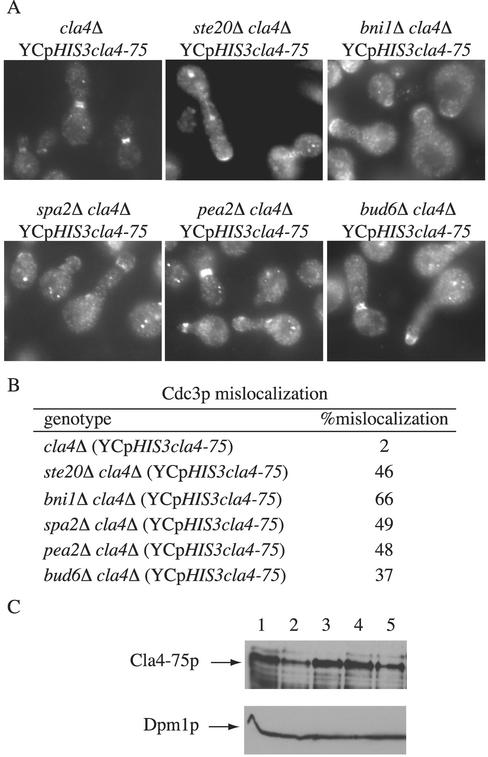
To examine the terminal phenotype of cla4Δ mutants lacking polarisome function, we used a plasmid-borne thermosensitive allele of CLA4 (YCpHIS3cla4-75). A striking phenotype of ste20Δ cla4Δ YCpHIS3cla4-75 mutants is the mislocalization of the septin ring (Cvrckova et al., 1995). We therefore examined septin localization in bni1Δ cla4Δ YCpHIS3cla4-75 and in other polarisome cla4Δ double mutants. The septin phenotype of bni1Δ cla4Δ YCpHIS3cla4-75 mutants at the restrictive temperature resembled that of ste20Δ cla4Δ YCpHIS3cla4-75 mutants. The septin ring was formed at the proper time and location. However, as the bud began to grow, the septin ring frequently localized at the tip of the misshapened bud rather than remaining at the bud neck (Figure 2). These results imply that new growth is on the mother side of the neck rather than the bud side. Similar phenotypes were observed with bud6Δ cla4Δ YCpHIS3cla4-75, spa2Δ cla4Δ YCpHIS3cla4-75, and pea2Δ cla4Δ YCpHIS3cla4-75 mutants (Figure 2). Other aspects of the polarisome cla4 double mutants will be discussed below.
Figure 2.
Morphological phenotypes of bni1Δ cla4Δ, spa2Δ cla4Δ, pea2Δ cla4Δ, and bud6Δ cla4Δ carrying YCpHIS3cla4-75. (A) Exponential cultures of haploid strains SY3380 (cla4Δ), SY3760 (bni1Δ cla4Δ), SY3761 (bud6Δ cla4Δ), SY3762 (spa2Δ cla4Δ), SY3763 (pea2Δ cla4Δ), and SY3764 (ste20Δ cla4Δ) carrying YCpHIS3cla4-75 were grown at 25°C in YEPD, shifted to 37°C for 4 h, fixed, and stained for Cdc3p. (B) Quanitation of Cdc3p mislocalization. For each strain, 250 cells were counted in three independent experiments. (C) Immunoblot analysis by using anti-Cla4p antibodies. Lysates of haploid strains SY3357 (WT; lane 1), SY3380 (cla4Δ; lane 2) SY3764 (ste20Δ cla4Δ; lane 3), SY3760 (bni1Δ cla4Δ; lane4), and SY3761 (bud6Δ cla4Δ; lane 5) carrying YCpHIS3cla4-75 were analyzed by SDS-PAGE and immunoblot analysis by using affinity purified polyclonal anti-Cla4p antibodies (provided by D. Kellogg, Department of Molecular, Cellular, and Developmental Biology, University of California, Santa Cruz) and monoclonal antibodies to Dpm1p (to confirm equal protein loading). The band corresponding to wild-type Cla4p and Cla4-75p are indicated. All strains expressed similar amounts of Cla4-75p.
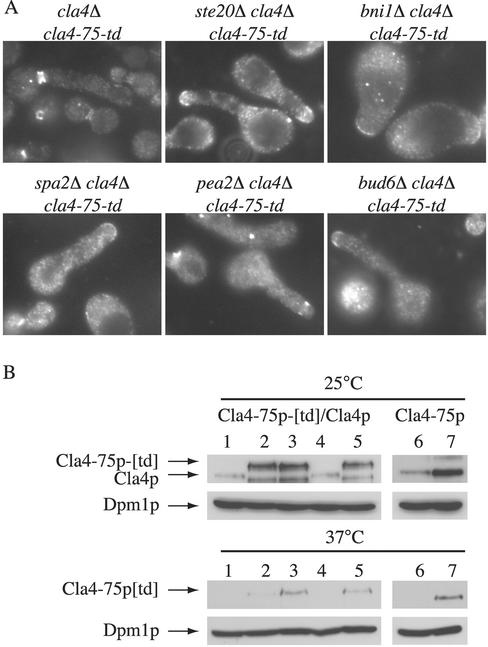
To corroborate the results observed using strains harboring YCpHIS3cla4–75, we also used strains expressing an integrated cla4–75-ts degron construct (cla4–75-td). This version of Cla4p is reported to be degraded rapidly following a shift to the restrictive temperature (Holly and Blumer, 1999); note, however, that wild-type Cla4p and Cla4–75p are also degraded rapidly at 37°C (Figure 3B). The phenotype of bni1Δ cla4Δ cla4–75-td mutants at the restrictive temperature recapitulated that of cells carrying YCpHIS3cla4–75: the septin ring was mislocalized to the tip of the bud (Figure 3A). Together, these results indicate that the polarisome is essential in the absence of Cla4p and further suggest that Ste20p may activate the polarisome.
Figure 3.
Morphological phenotypes of bni1Δ cla4Δ cla4-75-td, spa2Δ cla4Δ cla4-75-td, pea2Δ cla4Δ cla4-75-td, and bud6Δ cla4Δ cla4-75-td. (A) Exponential cultures of haploid strains SY3790 (cla4Δ cla4-75-td), SY3791 (bni1Δ cla4Δ cla4-75-td), SY3792 (bud6Δ cla4Δ cla4-75-td), SY3793 (spa2Δ cla4Δ cla4-75-td), SY3794 (pea2Δ cla4Δ cla4-75-td), and SY3795 (ste20Δ cla4Δ cla4-75-td) were grown at 25°C in YEPD, shifted to 37°C for 4 h, fixed, and stained for Cdc3p. (B) Immunoblot analysis using anti-Cla4p antibodies. Lysates of haploid strains SY3357 (WT; lane 1), SY3790 (cla4Δ cla4-75-td; lane 2), SY3795 (ste20Δ cla4Δ cla4-75-td; lane 3), DY759 (WT; lane4) and DY2060 (ste20Δ cla4Δ cla4-75-td; lane 5), SY3380 (cla4Δ YCpHIS3cla4-75; lane 6), and SY3764 (ste20Δ cla4Δ YCpHIS3cla4-75; lane 7), were grown at 25°C in YEPD, shifted to 37°C for 1 h, and analyzed by SDS-PAGE and immunoblot analysis by using affinity purified polyclonal anti-Cla4p antibodies (provided by D. Kellogg) and monoclonal antibodies to Dpm1p (to confirm equal protein loading). The position of wild-type Cla4p, Cla4-75p, and Cla4-75p[td] are indicated. In lanes 2, 3 and 5, a degradation product of Cla4-75p[td] is visible at 25°C. Lanes 6 and 7 are a darker exposure than lanes 1–5. As previously reported, at 37°C, there is residual Cla4-75p[td] protein present (Holly and Blumer, 1999; Weiss et al., 2000).
Bni1p Is Phosphorylated In Vivo and Phosphorylation Is Partially Dependent on STE20
Because Bni1p and Ste20p colocalize to the bud tip in a Cdc42p-dependent manner (Peter et al., 1996; Leberer et al., 1997; Ozaki-Kuroda et al., 2001) and show similar genetic interactions with CLA4, it seemed plausible that Bni1p and Ste20p might physically interact and that this interaction would be necessary for the essential activity that Bni1p has in the absence Cla4p. To investigate this possibility, we created two-hybrid constructs of full-length, N-terminal, and C-terminal fusions of Ste20p and Bni1p but were unable to detect an interaction. We also failed to detect an interaction using coimmunoprecipitation under a variety of assay conditions. Moreover, the proper localization of Bni1p or Ste20p to the bud tip did not require the presence of the other protein (our unpublished data).
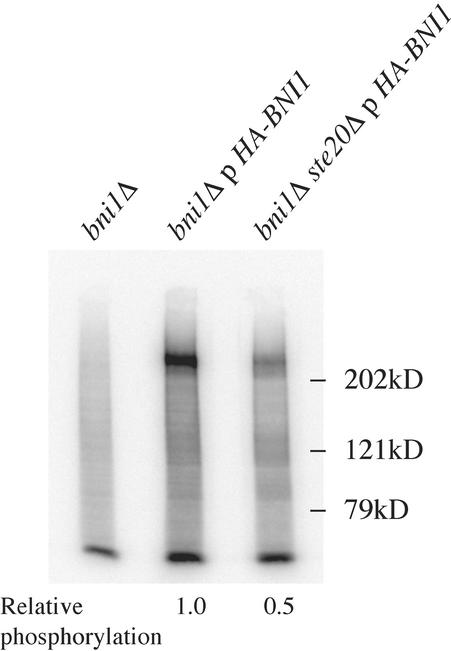
Although we were unable to detect a physical interaction between Ste20p and Bni1p by using the methods described, we considered the possibility that the interaction is transient. In particular, because Ste20p is a protein kinase we asked whether Bni1p is a Ste20p-dependent phosphoprotein. A culture of cells expressing HA-tagged Bni1p was labeled with 32Pi, Bni1p was immunoprecipitated from the labeled extracts with the HA antibody, and radiolabeled proteins in the immune complexes were visualized by a Phosphor-Imager and subsequent immunoblot analysis. Bni1p is indeed a phosphoprotein (Figure 4). Moreover, in cells lacking Ste20p, the amount of phosphorylated Bni1p was twofold less than that found in wild-type cells, suggesting that a substantial portion of Bni1p phosphorylation is dependent on Ste20p in vivo (Figure 4).
Figure 4.
Bni1p is a phosphoprotein. Cultures of SY3779 (bni1Δ) and SY3780 (bni1Δ ste20Δ) carrying pY39tet1HA-BNI1 and SY3778 (bni1Δ) were incubated with 1 mCi of 32PO4. Rabbit anti-HA antibody was used for immunoprecipitation and an anti-HA monoclonal antibody was used for immunoblot analysis. The relative amount of phospho-Bni1p, normalized to total Bni1p, is indicated below each lane.
Septin Ring Mislocalization Is Not the Cause of Lethality in a bni1Δ cla4Δ Cells
Cells lacking CLA4 exhibit a bud morphology that suggests a defect in the apical-to-isotrophic bud transition that occurs in the G2 phase of the cell cycle. This phenotype is reminiscent of the phenotype conferred by misregulation of Cdc28p kinase activity (Lew and Reed, 1995a,b; McMillan et al., 1998; Richman et al., 1999). Indeed, we showed previously that deletion of SWE1, which encodes a protein kinase thought to be part of a morphogenetic checkpoint that negatively regulates Clb1,2p-Cdc28p activity, restores normal bud morphology in cla4Δ mutants (Longtine et al., 2000; Weiss et al., 2000; Mitchell and Sprague, 2001). By bypassing this morphogenetic checkpoint with swe1Δ, we were able to class the NCS genes into two groups, an NCS1-related group and an STE20-related group. Loss of SWE1 restores normal bud morphology and overcomes the synthetic lethality of ncs1Δ cla4Δ YCpHIS3cla4-75 cells. In the case of ste20Δ cla4Δ YCpHIS3cla4-75 cells, however, loss of SWE1 restores the localization of the septin ring to the mother bud junction but does not restore viability (Mitchell and Sprague, 2001). To test whether BNI1 is in the NCS1 group or the STE20 group, we deleted SWE1 in bni1Δ cla4Δ YCpHIS3cla4-75 cells. The loss of SWE1 from bni1Δ cla4Δ YCpHIS3cla4-75 yielded a phenotype similar to that of swe1Δ ste20Δ cla4Δ YCpHIS3cla4-75 cells (Figure 5), suggesting that septin ring mislocalization is not the cause of lethality. Furthermore, this result suggests that BNI1 may facilitate or orchestrate some STE20 functions. Similar results were obtained when SWE1 was deleted from pea2Δ cla4Δ, spa2Δ cla4Δ, and bud6Δ cla4Δ cells carrying YCpHIS3cla4-75 (Figure 5). Moreover, these polarisome genes constitute a group distinct from other NCS genes based on their genetic interactions with SWE1 (Table 2).
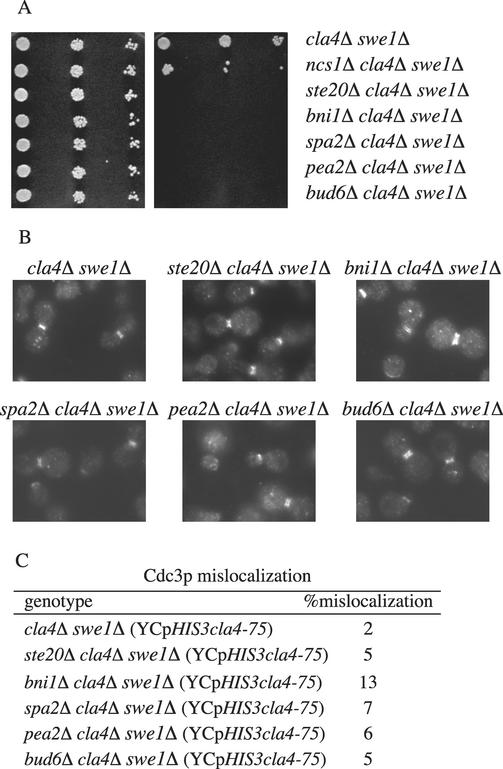
Figure 5.
Loss of SWE1 in bni1Δ cla4Δ restores septin ring localization to the mother-bud junction. (A) Strains SY3766 (cla4Δ swe1Δ), SY3403 (ncs1Δ cla4Δ swe1Δ), SY3767 (ste20Δ cla4Δ swe1Δ), SY3768 (bni1Δ cla4Δ swe1Δ), SY3769 (bud6Δ cla4Δ swe1Δ), SY3770 (spa2Δ cla4Δ swe1Δ), and SY3771 (pea2Δ cla4Δ swe1Δ) carrying pRS316ADE8CLA4 were grown to midlog in YEPD at 30°C. A serial dilution (1/10) was performed starting with 10,000 cells. Cells were spotted onto either YEPD (left) or 5-FOA (right) and grown 3 d at 30°C. (B) Exponential cultures of haploid strains SY3772 (cla4Δ swe1Δ), SY3773 (ste20Δ cla4Δ swe1Δ), SY3774 (bni1Δ cla4Δ swe1Δ), SY3775 (bud6Δ cla4Δ swe1Δ), SY3776 (spa2Δ cla4Δ swe1Δ), and SY3777 (pea2Δ cla4Δ swe1Δ) carrying YCpHIS3cla4-75 were grown at 25°C in YEPD, shifted to 37°C for 4 h, fixed, and stained for Cdc3p. (C) Quantitation of Cdc3p mislocalization. For each strain 250 cells were counted in three independent experiments.
Not All Proteins Involved in Bipolar Budding Pattern Are Essential in cla4Δ Cells
Recent studies have shown that some mutants defective for bipolar budding pattern selection also show a defect in apical growth and that lengthening the apical growth phase enhances the accuracy of bud site selection (Sheu et al., 2000). Ste20p, Bni1p, Pea2p, Spa2p, and Bud6p have been implicated in apical growth and also have roles in bipolar bud site selection (Snyder, 1989; Valtz and Herskowitz, 1996; Zahner et al., 1996; Evangelista et al., 1997; Sheu et al., 2000). To ascertain whether there is a specific connection between their roles in bipolar budding and the essential function they have in the absence of Cla4p, we looked for genetic interactions between CLA4 and other components of the bipolar bud site machinery. Loss of BUD8 was not synthetically lethal with cla4Δ (Figure 1 and Supplementary Table 1). In addition, the SGA screen showed that the loss of other genes that affect the budding pattern in diploids, such as BUD9, BUD14, BUD16-32 was not synthetically lethal with cla4Δ. These results suggest the roles of Bni1p, Spa2p, Pea2p, and Bud6p in bipolar budding pattern are not essential in the absence of Cla4p.
Roles of Bni1p in Spindle Alignment during Nuclear Migration and Cytokinesis Are Not Essential in cla4Δ Cells
In addition to its roles in bipolar budding pattern and apical growth, Bni1p has a role in cytokinesis. Other proteins important for cytokinesis that function with Bni1p are Myo1p, Hof1p, and Bnr1p, a formin homology protein related to Bni1p (Bi et al., 2000). Based on synthetic lethal interactions, BNI1 and MYO1 are believed to be in one functional pathway, whereas HOF1 and BNR1 are in another. Loss of BNI1 is synthetically lethal with bnr1Δ and with hof1Δ. However, none of these genes (except BNI1) is essential in a cla4Δ background (our unpublished data). Based on these results, the role of Bni1p in actomyosin contraction during cytokinesis is not essential in the absence of Cla4p.
Bni1p has also been shown to play a role in positioning the mitotic spindle during nuclear migration (Lee et al., 1999; Miller et al., 1999). We investigated whether the genetic interactions between BNI1 and CLA4 are related to the role of BNI1 in nuclear migration by looking for genetic interactions with KIP3. Kip3p is a kinesin-related protein hypothesized to function with Bni1p to organize and position the mitotic spindle. Loss of KIP3 was not synthetically lethal with cla4Δ (our unpublished data). Furthermore, the loss of NUM1, which encodes a protein that controls interaction of bud-neck cytoskeleton with the nucleus in G2 and also interacts with Bni1p, was not synthetically lethal with cla4Δ (our unpublished data).
Bni1p Has Roles Distinct from Spa2p, Pea2p, Bud6p, and Ste20p in cla4Δ Cells
To gain more insight into the role of Bni1p in cla4Δ cells, we examined the terminal phenotype of bni1Δ cla4Δ YCpHIS3cla4-75 mutants in more detail. With respect to the septin ring localization, the terminal phenotype of bni1Δ cla4Δ YCpHIS3cla4-75 cells was similar to ste20Δ cla4Δ YCpHIS3cla4-75 cells, but with respect to other phenotypes, the two phenotypes were distinct. bni1Δ cla4Δ YCpHIS3cla4-75 cells have both wider bud necks and defects in actin localization compared with ste20Δ cla4Δ YCpHIS3cla4-75 at the restrictive temperature (Figure 6). In particular, bni1Δ cla4Δ YCpHIS3cla4-75 mutants had no visible actin cables and only 6% of the cells had organized patches of actin at the tip of the bud. In contrast, cla4Δ single mutants had no observable defects in actin polarization and only 38% of bni1Δ single mutant cells had defects in actin polarization (Figure 6). Thus, it seems that Bni1p is more critical for actin organization in the absence of Cla4p than in wild-type cells, suggesting that Cla4p may also participate in actin organization but that its role in this process is functionally redundant with that of Bni1p.
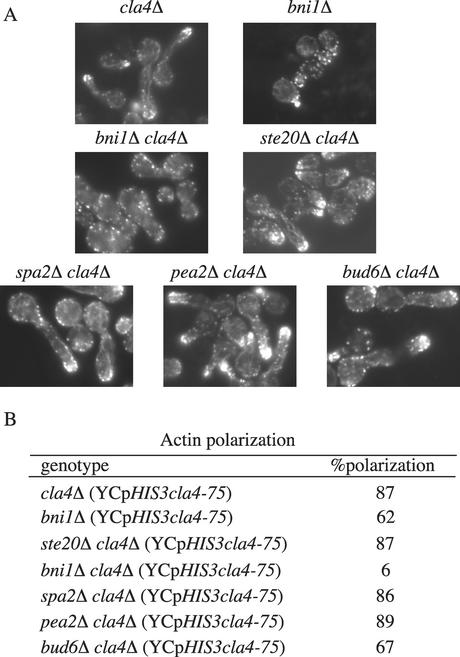
Figure 6.
bni1Δ cla4Δ mutants have a severe actin polarization defect. (A) Strains SY3380 (cla4Δ), SY3778 (bni1Δ), SY3764 (ste20Δ cla4Δ), SY3760 (bni1Δ cla4Δ), SY3761 (bud6Δ cla4Δ), SY3762 (spa2Δ cla4Δ), and SY3763 (pea2Δ cla4Δ) carrying YCpHIS3cla4-75 were grown at 25°C in YEPD, shifted to 37°C for 4 h, fixed, and stained for F-actin (by using rhodamine-phalloidin). (B) Quantitation of actin polarization. For each strain, 250 cells were counted in three independent experiments.
As previously reported, actin polarization in ste20Δ cla4Δ YCpHIS3cla4-75 cells was indistinguishable from that in cla4Δ mutants, with actin cables traversing from mother to bud and actin patches localized toward the bud tip (Cvrckova et al., 1995). Likewise, spa2Δ cla4Δ and pea2Δ cla4Δ mutants carrying the YCpHIS3cla4-75 construct did not seem to have defects in actin organization compared with wild-type cells or ste20Δ cla4Δ YCpHIS3cla4-75 mutants (Figure 6). bud6Δ cla4Δ YCpHIS3cla4-75 mutants had some noticeable actin defects with fewer actin cables and polarized actin patches, yet the defects were not as severe as those of bni1Δ cla4Δ YCpHIS3cla4-75 mutants (Figure 6). Thus, deletion of polarisome genes in cla4Δ cells leads to two broad phenotypic classes, one that includes spa2Δ, pea2Δ, and bud6Δ, with a terminal phenotype resembling that associated with ste20Δ, and another class whose sole member is bni1Δ, associated with a more severe terminal phenotype.
To further test for a functional relationship between Ste20p and the polarisome components, we examined the terminal phenotype of triple mutants containing spa2Δ, pea2Δ, bud6Δ, or bni1Δ in combination with ste20Δ cla4Δ YCpHIS3cla4-75. The terminal phenotypes of spa2Δ, pea2Δ, and bud6Δ triple mutants were similar to that of the ste20Δ cla4Δ YCpHIS3cla4-75 double mutant (Figure 7). Moreover, no synthetic growth defects were observed in strains where the polarisome genes were deleted in combination with ste20Δ. In contrast, the terminal morphology of the bni1Δ triple mutant was more severe than that associated with either the bni1Δ cla4Δ YCpHIS3cla4-75 or the ste20Δ cla4Δ YCpHIS3cla4-75 double mutants. The bni1Δ ste20Δ cla4Δ YCpHIS3cla4-75 cells were large and unbudded with no visible actin cables and mostly unpolarized patches of actin (Figure 7). In these unbudded cells, components of the septin ring still localized to the presumptive bud site, but in 2% of cells, Cdc3p (a septin component) localized to more than one site on the cell (Figure 7). In contrast, the actin defects in ste20Δ bni1Δ mutants were no more severe than that of bni1Δ single mutant cells (62 vs. 59% polarized actin, respectively). Together, these results suggest that Ste20p functions within a Spa2p/Pea2p/Bud6p pathway, but that is also participates in Bni1p-independent functions.
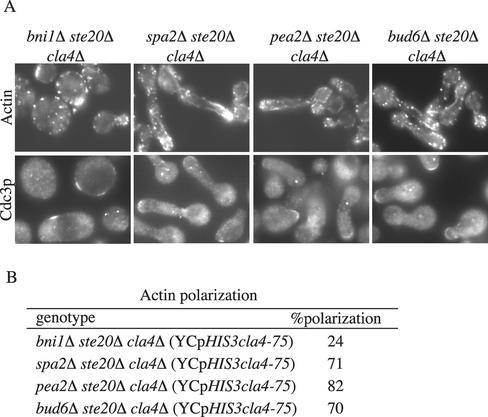
Figure 7.
Terminal phenotype of bni1Δ ste20Δ cla4Δ.. Exponential cultures of SY3781 (bni1Δ cla4Δ ste20Δ), SY3782 (bud6Δ cla4Δ ste20Δ), SY3783 (spa2Δ cla4Δ ste20Δ), and SY3884 (pea2Δ cla4Δ ste20Δ) carrying YCpHIS3cla4-75 were grown at 25°C in YEPD, shifted to 37°C for 4 h, fixed, and stained for F-actin (by using rhodamine-phalloidin) or for Cdc3p. (B) Quantitation of actin polarization. For each strain, 250 cells were counted in three independent experiments.
Disruption of Bni1p-Bud6p Interaction Leads to Synthetic Lethality with cla4Δ
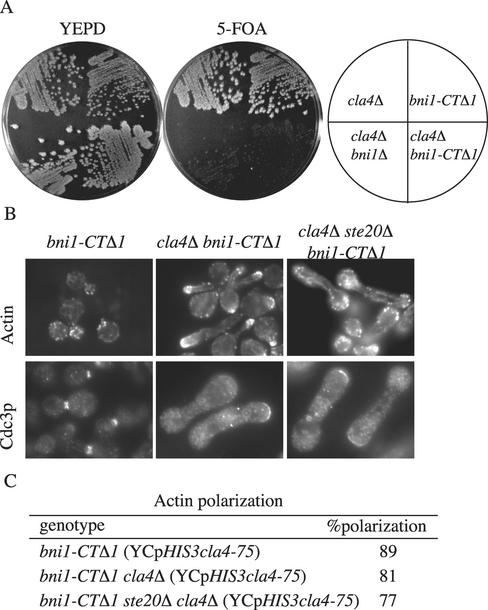
The finding that Bni1p and other polarisome components are essential in the absence of Cla4p, leads to the natural inference the disruption of the interactions between these proteins would lead to synthetic lethality with cla4Δ. To explore this possibility, we examined the phenotype conferred by a version of Bni1p that is lacking the Bud6p binding region but still competent to carry out other Bni1p functions. Amino acid residues 1749–1953 of Bni1p contain the Bud6p binding domain (Evangelista et al., 1997). Accordingly, we constructed an allele of BNI1 that encodes a version of Bni1p truncated at the C terminus (bni1-CTΔ1) (Lee et al., 1999). bni1-CTΔ1 retains some Bni1p functions as it is able to complement the synthetically lethality of a bni1Δ bnr1Δ (Ozaki-Kuroda et al., 2001). In contrast, bni1-CTΔ1 was lethal in a cla4Δ background (Figure 7A). Moreover, the terminal phenotype of bni1-CTΔ cla4Δ YCpHIS3cla4-75 was similar to the ste20Δ cla4Δ YCpHIS3cla4-75 terminal phenotype but not the bni1Δ cla4Δ YCpHIS3cla4-75 phenotype (Figure 8B). The bni1-CTΔ cla4Δ YCpHIS3cla4-75 cells have polarized actin, whereas bni1Δ cla4Δ YCpHIS3cla4-75 cells have no visible cables and unpolarized patches of actin. In addition, the ste20Δ cla4Δ bni1-CTΔ1 YCpHIS3cla4-75 triple mutant phenotype resembled the ste20Δ cla4Δ YCpHIS3cla4-75 and the bni1-CTΔ1 cla4Δ YCpHIS3cla4-75 double mutant phenotypes (Figure 8B). The cells were elongated, with mostly polarized actin, but the septin ring was mislocalized. These results support the idea that the Bni1p C terminus, which interacts with Bud6p, carries out one function that is essential in the absence of Cla4p, whereas the remainder of Bni1p is critical for actin organization.
Figure 8.
bni1-CTΔ1 cla4Δ mutants have the same phenotype as ste20Δ cla4Δ. (A) bni1-CTΔ1 is synthetically lethal with cla4Δ. Strains SY3362 (cla4Δ), SY3756 (bni1Δ cla4Δ), SY3785 (bni1-CTΔ1), and SY3786 (bni1-CTΔ1 cla4Δ) harboring pRS316ADE8CLA4 were streaked on YEPD plates and incubated at 30°C for 2 d. Plates were then replica-plated to 5-FOA and incubated at 30°C for 3 d. (B) Exponential cultures of SY3785 (bni1-CTΔ1), SY3788 (bni1-CTΔ1 cla4Δ) and SY3789 (bni1-CTΔ1 ste20Δ cla4Δ) carrying YCpHIS3cla4-75 were grown at 25°C in YEPD, shifted to 37°C for 4 h, fixed, and stained for F-actin (by using rhodamine-phalloidin) or for Cdc3p. (C) Quantitation of actin polarization. For each strain 250 cells were counted in three independent experiments.
DISCUSSION
The related protein kinases Ste20p and Cla4p have unique activities, an inference made from the distinct phenotypes of strains lacking an individual kinase. However, a strain lacking both kinases is inviable, implying that there is a physiological connection between their activities. One possibility is that Cla4p and Ste20p share an essential activity. A less constrained interpretation is simply that Ste20p carries out a function that is essential in cells lacking Cla4p cells (and vice versa). In an effort to shed light on Ste20p function, we carried out two independent screens for mutations that are lethal in a cla4Δ mutant background. This effort identified a surprisingly large number of genes. Herein, we focus on a group of genes whose products are known to interact. Each of the proteins that form this group, Bni1p, Bud6p, Spa2p, and Pea2p, has been implicated in several different facets of cell biology. For example, Bni1p, a formin homology protein, has been implicated in actin polarization, cytokinesis, nuclear migration, and apical growth. Similarly, Bud6p, Spa2p, and Pea2p are involved in budding pattern determination and apical growth. The common denominator among the functions attributed to this set of proteins is involvement in apical growth, and we therefore suggest that proper regulation of this growth is the essential in the absence of Cla4p. We further suggest that the connection between these proteins and the Ste20p/Cla4p essential function is likely to be direct because Bni1p is a Ste20p-dependent phosphoprotein.
Bni1p Has Roles Distinct from That of Spa2p, Pea2p, Bud6p, or Ste20p in cla4Δ Cells
The lethality of bni1Δ cla4Δ strains may have more than one cause. As summarized above, we think one aspect of Bni1p function that is required in a cla4Δ mutant background is its participation in apical growth. This interpretation is supported by the finding that deletion of the Bni1p C-terminal 200 amino acids, the region of Bni1p that interacts with Bud6p, is lethal in a cla4Δ mutant background. Indeed, the terminal phenotype of bni1-CTΔ1 cla4Δ YCpHIS3cla4-75 is similar to the bud6Δ cla4Δ YCpHIS3cla4-75 terminal phenotype. The C-terminal region is not only the Bud6p interaction domain on Bni1p, but it is also believed to be an autoinhibitory domain. In the case of other formin homology proteins, this autoinhibitory domain has been shown to interact with the Cdc42p binding domain on the same molecule (Alberts, 2001). Perhaps binding of activated Cdc42p releases the autoinhibitory domain and enables Bni1p to interact with Bud6p and the 12S complex. It will be interesting to determine whether the Ste20p-dependent phosphorylation of Bni1p influences interaction of it with Bud6p or other proteins.
Our results suggest that Bni1p has at least one other function that is important in a cla4Δ mutant background. This possibility emerges from the observation that bni1Δ cla4Δ YCpHIS3cla4-75 double mutants have additional phenotypes beyond those seen for the ste20Δ cla4Δ, bni1-CTΔ1 cla4Δ YCpHIS3cla4-75, and bud6Δ cla4Δ YCpHIS3cla4-75 mutant strains. What is this additional important function? In addition to its role in apical growth, Bni1p also has roles in bud site selection, nuclear migration, cytokinesis, and actin polarization. Genetic tests, coupled with careful examination of the terminal phenotype of the bni1Δ cla4Δ YCpHIS3cla4-75 double mutant, point to actin polarization as the likely function. In particular, loss of Hof1p (required for cytokinesis), or Num1p (required for nuclear migration) is not lethal in the absence of Cla4p. However, diminution of Cdc42p activity is lethal in a cla4Δ mutant background (Cvrckova et al., 1995). Cdc42p interacts with Bni1p and is required for polarization of the actin cytoskeleton. Moreover, bni1Δ cla4Δ YCpHIS3cla4-75 mutants contain very few actin cables, whereas ste20Δ cla4Δ, bud6Δ cla4Δ, pea2Δ cla4Δ, and spa2Δ cla4Δ mutants carrying YCpHIS3cla4-75 contain abundant cables. Together, these results support the idea that a Bni1p role in actin polarization is critical in the absence of Cla4p.
CLA4 Synthetic Lethal Universe
The two screens for mutations that are synthetically lethal in a cla4Δ mutant background identified a large number of genes. The number of genes is large in absolute terms, but it is surprisingly large compared with the number of genes identified in a complementary synthetically lethal screen. Specifically, in a preliminary effort to identify mutations synthetically lethal with the absence of STE20 by using the colony-sectoring assay, only the CLA4 gene was identified (Mitchell, Goehring, and Sprague, unpublished data). The functions identified in the CLA4 synthetic screens reported herein cover a wide spectrum of cell biological processes and include bud emergence (BEM1, BEM2, BEM4), cytokinesis (SHS1), nuclear migration (DYN2, NIP100, APC9, SLK19), and cell wall maintenance (GIM5, BCK1, CHS3, SKT5/CHS4, CHS5, CHS6, CHS7, FAB1, SLT2, SMI1). Not all of the proteins, or even a majority of the proteins, involved in a particular process were identified. This finding implies that lethality does not result because an entire process has become essential in the cla4Δ mutant background, but rather implies that a particular activity or role of the protein has become essential.
It is often difficult to identify the targets of signaling proteins. Synthetic lethal screens for genes required in the absence of specific signaling molecules may provide a general means to identify potential downstream targets of the signaling molecule. In this study, we screened for genes that, like STE20, were synthetically lethal with CLA4 and identified the polarisome as a potential target of Ste20p. By extension of this logic, potential downstream targets of Cla4p may be identified in synthetic lethal screens that use query mutations in STE20 or any one of the other genes identified in the CLA4 synthetic lethal universe. For example, because BNI1 is synthetic lethal with CLA4, the set of ∼50 genes that are synthetically lethal with BNI1 (Tong et al., 2001), which includes genes involved in bud emergence, chitin synthase III activity, and the dynein/dynactin spindle orientation pathway, may be Cla4p targets. Candidate targets are then identified by determining which single mutants exhibit phenotypes that resemble facets of the cla4Δ mutant phenotype. Thus, global synthetic lethal networks should be useful for large-scale mapping of functional relationships between signaling molecules and their downstream targets.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Pringle, I. Herskowitz, I. Pretorius, and D. Lew for providing plasmids. We also thank David Rivers, Hilary Kemp, Greg Smith, and Paul Cullen for helpful comments and suggestions. This work was supported by grant GM-30027 (to G.F.S.) from the National Institutes of Health, National Research Service Award GM-18002-03 (to D.A.M.), training grant 5 T32 GM-07759 (to A.S.G.) from the National Institutes of Health, Canadian Institute of Health Research (to C.B.), and operating grant from the National Cancer Institute of Canada (C.B.).
Footnotes
The online version of this article contains supplemental tabular material. The online version of the article is available at www.molbiolcell.org.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–06–0348. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–06–0348.
REFERENCES
- Adams AE, Johnson DI, Longnecker RM, Sloat BF, Pringle JR. CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae. J Cell Biol. 1990;111:131–142. doi: 10.1083/jcb.111.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts AS. Identification of a carboxyl-terminal diaphanous-related formin homology protein autoregulatory domain. J Biol Chem. 2001;276:2824–2830. doi: 10.1074/jbc.M006205200. [DOI] [PubMed] [Google Scholar]
- Ayscough KR, Stryker J, Pokala N, Sanders M, Crews P, Drubin DG. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J Cell Biol. 1997;137:399–416. doi: 10.1083/jcb.137.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender A, Pringle JR. Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:1295–1305. doi: 10.1128/mcb.11.3.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton BK, Tinkelenberg A, Gonzalez I, Cross FR. Cla4p, a Saccharomyces cerevisiae Cdc42p-activated kinase involved in cytokinesis, is activated at mitosis. Mol Cell Biol. 1997;17:5067–5076. doi: 10.1128/mcb.17.9.5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E, Chiavetta JB, Chen H, Chen GC, Chan CS, Pringle JR. Identification of novel, evolutionarily conserved Cdc42p-interacting proteins and of redundant pathways linking Cdc24p and Cdc42p to actin polarization in yeast. Mol Biol Cell. 2000;11:773–793. doi: 10.1091/mbc.11.2.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booher RN, Deshaies RJ, Kirschner MW. Properties of Saccharomyces cerevisiae wee1 and its differential regulation of p34CDC28 in response to G1 and G2 cyclins. EMBO J. 1993;12:3417–3426. doi: 10.1002/j.1460-2075.1993.tb06016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose I, Irazoqui JE, Moskow JJ, Bardes ES, Zyla TR, Lew DJ. Assembly of scaffold-mediated complexes containing Cdc42p, the exchange factor Cdc24p, and the effector Cla4p required for cell cycle-regulated phosphorylation of Cdc24p. J Biol Chem. 2001;276:7176–7186. doi: 10.1074/jbc.M010546200. [DOI] [PubMed] [Google Scholar]
- Brown JL, Jaquenoud M, Gulli MP, Chant J, Peter M. Novel Cdc42-binding proteins Gic1 and Gic2 control cell polarity in yeast. Genes Dev. 1997;11:2972–2982. doi: 10.1101/gad.11.22.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DC, Yang BC, Kuo TT. One-step transformation of yeast in stationary phase. Curr Genet. 1992;21:83–84. doi: 10.1007/BF00318659. [DOI] [PubMed] [Google Scholar]
- Chen GC, Kim YJ, Chan CS. The Cdc42 GTPase-associated proteins Gic1 and Gic2 are required for polarized cell growth in Saccharomyces cerevisiae. Genes Dev. 1997;11:2958–2971. doi: 10.1101/gad.11.22.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo MC, et al. YPD, PombePD, and WormPD. model organism volumes of the BioKnowledge library, an integrated resource for protein information. Nucleic Acids Res. 2001;29:75–79. doi: 10.1093/nar/29.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvrckova F, De Virgilio C, Manser E, Pringle JR, Nasmyth K. Ste20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeast. Genes Dev. 1995;9:1817–1830. doi: 10.1101/gad.9.15.1817. [DOI] [PubMed] [Google Scholar]
- Drubin DG, Nelson WJ. Origins of cell polarity. Cell. 1996;84:335–344. doi: 10.1016/s0092-8674(00)81278-7. [DOI] [PubMed] [Google Scholar]
- Evangelista M, Blundell K, Longtine MS, Chow CJ, Adames N, Pringle JR, Peter M, Boone C. Bni1p, a yeast formin linking Cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- Evangelista M, Klebl BM, Tong AH, Webb BA, Leeuw T, Leberer E, Whiteway M, Thomas DY, Boone C. A role for myosin-I in actin assembly through interactions with Vrp1p, Bee1p, and the Arp2/3 complex. J Cell Biol. 2000;148:353–362. doi: 10.1083/jcb.148.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista M, Pruyne D, Amberg DC, Boone C, Bretscher A. Formins direct Arp2/3-independent actin filament assembly to polarize cell growth in yeast. Nat Cell Biol. 2002;4:32–41. doi: 10.1038/ncb718. [DOI] [PubMed] [Google Scholar]
- Farkasovsky M, Kuntzel H. Cortical Num1p interacts with the dynein intermediate chain PacI1p and cytoplasmic microtubules in budding yeast. J Cell Biol. 2001;152:251–262. doi: 10.1083/jcb.152.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T, Tanaka K, Mino A, Kikyo M, Takahashi K, Shimizu K, Takai Y. Rho1p-Bni1p-Spa2p interactions: implication in localization of Bni1p at the bud site and regulation of the actin cytoskeleton in Saccharomyces cerevisiae. Mol Biol Cell. 1998;9:1221–1233. doi: 10.1091/mbc.9.5.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagiano M, Van Dyk D, Bauer FF, Lambrechts MG, Pretorius IS. Divergent regulation of the evolutionarily closely related promoters of the Saccharomyces cerevisiae STA2 and MUC1 genes. J Bacteriol. 1999;181:6497–6508. doi: 10.1128/jb.181.20.6497-6508.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geli MI, Lombardi R, Schmelzl B, Riezman H. An intact SH3 domain is required for myosin I-induced actin polymerization. EMBO J. 2000;19:4281–4291. doi: 10.1093/emboj/19.16.4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- Goldstein AL, Pan X, McCusker JH. Heterologous URA3MX cassettes for gene replacement in Saccharomyces cerevisiae. Yeast. 1999;15:507–511. doi: 10.1002/(SICI)1097-0061(199904)15:6<507::AID-YEA369>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Graham LA, Hill KJ, Stevens TH. Assembly of the yeast vacuolar H+-ATPase occurs in the endoplasmic reticulum and requires a Vma12p/Vma22p assembly complex. J Cell Biol. 1998;142:39–49. doi: 10.1083/jcb.142.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulli MP, Jaquenoud M, Shimada Y, Niederhauser G, Wiget P, Peter M. Phosphorylation of the Cdc42 exchange factor Cdc24 by the PAK-like kinase Cla4 may regulate polarized growth in yeast. Mol Cell. 2000;6:1155–1167. doi: 10.1016/s1097-2765(00)00113-1. [DOI] [PubMed] [Google Scholar]
- Heil-Chapdelaine RA, Oberle JR, Cooper JA. The cortical protein Num1p is essential for dynein-dependent interactions of microtubules with the cortex. J Cell Biol. 2000;151:1337–1344. doi: 10.1083/jcb.151.6.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges PE, McKee AH, Davis BP, Payne WE, Garrels JI. The Yeast Proteome Database (YPD): a model for the organization and presentation of genome-wide functional data. Nucleic Acids Res. 1999;27:69–73. doi: 10.1093/nar/27.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holly SP, Blumer KJ. PAK-family kinases regulate cell and actin polarization throughout the cell cycle of Saccharomyces cerevisiae. J Cell Biol. 1999;147:845–856. doi: 10.1083/jcb.147.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DI, Pringle JR. Molecular characterization of CDC42, a Saccharomyces cerevisiae gene involved in the development of cell polarity. J Cell Biol. 1990;111:143–152. doi: 10.1083/jcb.111.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei T, Tanaka K, Hihara T, Umikawa M, Imamura H, Kikyo M, Ozaki K, Takai Y. Interaction of Bnr1p with a novel Src homology 3 domain-containing Hof1p. Implication in cytokinesis in Saccharomyces cerevisiae. J Biol Chem. 1998;273:28341–28345. doi: 10.1074/jbc.273.43.28341. [DOI] [PubMed] [Google Scholar]
- Kranz JE, Holm C. Cloning by function: an alternative approach for identifying yeast homologs of genes from other organisms. Proc Natl Acad Sci USA. 1990;87:6629–6633. doi: 10.1073/pnas.87.17.6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberer E, Wu C, Leeuw T, Fourest-Lieuvin A, Segall JE, Thomas DY. Functional characterization of the Cdc42p binding domain of yeast Ste20p protein kinase. EMBO J. 1997;16:83–97. doi: 10.1093/emboj/16.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechler T, Shevchenko A, Li R. Direct involvement of yeast type I myosins in Cdc42-dependent actin polymerization. J Cell Biol. 2000;148:363–373. doi: 10.1083/jcb.148.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L, Klee SK, Evangelista M, Boone C, Pellman D. Control of mitotic spindle position by the Saccharomyces cerevisiae formin Bni1p. J Cell Biol. 1999;144:947–961. doi: 10.1083/jcb.144.5.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew DJ, Reed SI. A cell cycle checkpoint monitors cell morphogenesis in budding yeast. J Cell Biol. 1995a;129:739–749. doi: 10.1083/jcb.129.3.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew DJ, Reed SI. Cell cycle control of morphogenesis in budding yeast. Curr Opin Genet Dev. 1995b;5:17–23. doi: 10.1016/s0959-437x(95)90048-9. [DOI] [PubMed] [Google Scholar]
- Li R, Zheng Y, Drubin DG. Regulation of cortical actin cytoskeleton assembly during polarized cell growth in budding yeast. J Cell Biol. 1995;128:599–615. doi: 10.1083/jcb.128.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Longtine MS, Theesfeld CL, McMillan JN, Weaver E, Pringle JR, Lew DJ. Septin-dependent assembly of a cell cycle-regulatory module in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:4049–4061. doi: 10.1128/mcb.20.11.4049-4061.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan JN, Sia RA, Lew DJ. A morphogenesis checkpoint monitors the actin cytoskeleton in yeast. J Cell Biol. 1998;142:1487–1499. doi: 10.1083/jcb.142.6.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RK, Matheos D, Rose MD. The cortical localization of the microtubule orientation protein, Kar9p, is dependent upon actin and proteins required for polarization. J Cell Biol. 1999;144:963–975. doi: 10.1083/jcb.144.5.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DA, Sprague GF. The phosphotyrosyl phosphatase activator, Ncs1p (Rrd1p), functions with Cla4p to regulate the G(2)/M transition in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:488–500. doi: 10.1128/MCB.21.2.488-500.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosch HU, Kohler T, Braus GH. Different domains of the essential GTPase Cdc42p required for growth and development of Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:235–248. doi: 10.1128/MCB.21.1.235-248.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulholland J, Preuss D, Moon A, Wong A, Drubin D, Botstein D. Ultrastructure of the yeast actin cytoskeleton and its association with the plasma membrane. J Cell Biol. 1994;125:381–391. doi: 10.1083/jcb.125.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P, Botstein D. Phenotypic analysis of temperature-sensitive yeast actin mutants. Cell. 1985;40:405–416. doi: 10.1016/0092-8674(85)90154-0. [DOI] [PubMed] [Google Scholar]
- Ozaki-Kuroda K, Yamamoto Y, Nohara H, Kinoshita M, Fujiwara T, Irie K, Takai Y. Dynamic localization and function of Bni1p at the sites of directed growth in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:827–839. doi: 10.1128/MCB.21.3.827-839.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter M, Neiman AM, Park HO, van Lohuizen M, Herskowitz I. Functional analysis of the interaction between the small GTP binding protein Cdc42 and the Ste20 protein kinase in yeast. EMBO J. 1996;15:7046–7059. [PMC free article] [PubMed] [Google Scholar]
- Pringle JR, Preston RA, Adams AE, Stearns T, Drubin DG, Haarer BK, Jones EW. Fluorescence microscopy methods for yeast. Methods Cell Biol. 1989;31:357–435. doi: 10.1016/s0091-679x(08)61620-9. [DOI] [PubMed] [Google Scholar]
- Pruyne D, Bretscher A. Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J Cell Sci. 2000;113:365–375. doi: 10.1242/jcs.113.3.365. [DOI] [PubMed] [Google Scholar]
- Pruyne DW, Schott DH, Bretscher A. Tropomyosin-containing actin cables direct the Myo2p-dependent polarized delivery of secretory vesicles in budding yeast. J Cell Biol. 1998;143:1931–1945. doi: 10.1083/jcb.143.7.1931. [DOI] [PubMed] [Google Scholar]
- Ramer SW, Davis RW. A dominant truncation allele identifies a gene, STE20, that encodes a putative protein kinase necessary for mating in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1993;90:452–456. doi: 10.1073/pnas.90.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman TJ, Johnson DI. Saccharomyces cerevisiae Cdc42p GTPase is involved in preventing the recurrence of bud emergence during the cell cycle. Mol Cell Biol. 2000;20:8548–8559. doi: 10.1128/mcb.20.22.8548-8559.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richman TJ, Sawyer MM, Johnson DI. The Cdc42p GTPase is involved in a G2/M morphogenetic checkpoint regulating the apical-isotropic switch and nuclear division in yeast. J Biol Chem. 1999;274:16861–16870. doi: 10.1074/jbc.274.24.16861. [DOI] [PubMed] [Google Scholar]
- Roberts CJ, Raymond CK, Yamashiro CT, Stevens TH. Methods for studying the yeast vacuole. Methods Enzymol. 1991;194:644–661. doi: 10.1016/0076-6879(91)94047-g. [DOI] [PubMed] [Google Scholar]
- Roberts RL, Fink GR. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- Rose, M.D., Winston, F., Hieter, P., (ed.). (1990). Methods in Yeast Genetics, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.
- Rubin GM. Preparation of RNA and ribosomes from yeast. Methods Cell Biol. 1975;12:45–64. doi: 10.1016/s0091-679x(08)60951-6. [DOI] [PubMed] [Google Scholar]
- Sagot I, Klee SK, Pellman D. Yeast formins regulate cell polarity by controlling the assembly of actin cables. Nat Cell Biol. 2002;4:42–50. doi: 10.1038/ncb719. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sheu YJ, Barral Y, Snyder M. Polarized growth controls cell shape and bipolar bud site selection in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:5235–5247. doi: 10.1128/mcb.20.14.5235-5247.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu YJ, Santos B, Fortin N, Costigan C, Snyder M. Spa2p interacts with cell polarity proteins and signaling components involved in yeast cell morphogenesis. Mol Cell Biol. 1998;18:4053–4069. doi: 10.1128/mcb.18.7.4053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M. The Spa2 protein of yeast localizes to sites of cell growth. J Cell Biol. 1989;108:1419–1429. doi: 10.1083/jcb.108.4.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague GF. Assay of yeast mating reaction. Methods Enzymol. 1991;194:77–93. doi: 10.1016/0076-6879(91)94008-z. [DOI] [PubMed] [Google Scholar]
- Tjandra H, Compton J, Kellogg D. Control of mitotic events by the Cdc42 GTPase, the Clb2 cyclin and a member of the PAK kinase family. Curr Biol. 1998;8:991–1000. doi: 10.1016/s0960-9822(07)00419-8. [DOI] [PubMed] [Google Scholar]
- Toenjes KA, Sawyer MM, Johnson DI. The guanine-nucleotide-exchange factor Cdc24p is targeted to the nucleus and polarized growth sites. Curr Biol. 1999;9:1183–1186. doi: 10.1016/S0960-9822(00)80022-6. [DOI] [PubMed] [Google Scholar]
- Tong AH, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Vallen EA, Caviston J, Bi E. Roles of Hof1p, Bni1p, Bnr1p, and Myo1p in cytokinesis in Saccharomyces cerevisiae. Mol Biol Cell. 2000;11:593–611. doi: 10.1091/mbc.11.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valtz N, Herskowitz I. Pea2 protein of yeast is localized to sites of polarized growth and is required for efficient mating and bipolar budding. J Cell Biol. 1996;135:725–739. doi: 10.1083/jcb.135.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss EL, Bishop AC, Shokat KM, Drubin DG. Chemical genetic analysis of the budding-yeast p21-activated kinase Cla4p. Nat Cell Biol. 2000;2:677–685. doi: 10.1038/35036300. [DOI] [PubMed] [Google Scholar]
- Wu C, Lytvyn V, Thomas DY, Leberer E. The phosphorylation site for Ste20p-like protein kinases is essential for the function of myosin-I in yeast. J Biol Chem. 1997;272:30623–30626. doi: 10.1074/jbc.272.49.30623. [DOI] [PubMed] [Google Scholar]
- Zahner JE, Harkins HA, Pringle JR. Genetic analysis of the bipolar pattern of bud site selection in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:1857–1870. doi: 10.1128/mcb.16.4.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziman M, Preuss D, Mulholland J, O'Brien JM, Botstein D, Johnson DI. Subcellular localization of Cdc42p, a Saccharomyces cerevisiae GTP-binding protein involved in the control of cell polarity. Mol Biol Cell. 1993;4:1307–1316. doi: 10.1091/mbc.4.12.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.