Abstract
Most of the substrates degraded by the proteasome are marked with polyubiquitin chains. However, there are a limited number of examples of nonubiquitinated proteins that are degraded by the proteasome. Here, we describe the degradation of the retinoblastoma family of tumor suppressor proteins by the proteasome in the absence of polyubiquitination. The retinoblastoma protein (p105), p107, and p130 are each targeted for degradation by the pp71 protein, which is encoded by the UL82 gene of human cytomegalovirus. It functions to direct their degradation in the absence of other viral proteins. While the pp71-mediated degradation of the retinoblastoma family of proteins requires proteasome function, it occurs without the attachment of ubiquitin to the substrates and in the absence of a functioning ubiquitin-conjugation system.
Protein degradation plays a role in many cellular processes, such as cell cycle regulation, antigen presentation, and the disposal of denatured, unfolded, or oxidized proteins (1–3). Several diseases, including certain types of cancers and neurological disorders, can result from aberrant protein degradation. Most intracellular protein degradation is handled by the ubiquitin–proteasome pathway (3, 4). The proteasome is a large, multisubunit complex that exists in cells in two main forms, a 20S and 26S species. The 20S proteasome is the catalytic core particle. It is shaped like a hollow barrel, with six active protease sites sequestered in its central cavity, so that only proteins entering this chamber are degraded. The openings to this cavity permit only denatured proteins to enter, where they are processively cleaved to small peptides. The addition of a 19S regulator to either or both ends of the 20S proteasome creates the 26S proteasome. The 19S regulator contains ATPases and other proteins and serves numerous functions, including the recognition of the substrate and its translocation to the catalytic core (5).
The majority of proteins degraded by the proteasome are tagged with polyubiquitin, which serves as a signal for their degradation (3). The cellular machinery that adds ubiquitin to substrates consists of three main enzyme classes. The single E1 ubiquitin-activating enzyme primes ubiquitin monomers, allowing their covalent attachment to substrate proteins. Inactivation of the E1 enzyme rapidly depletes the pool of activated ubiquitin, inhibiting the synthesis of polyubiquitin chains and ubiquitin-mediated proteolysis. The E1 enzyme transfers the activated ubiquitin to one of many E2 carrier enzymes, which, in combination with one of many E3 ubiquitin ligases, transfer the ubiquitin monomer to the substrate. The E3 ubiquitin ligase binds to both the substrate and the E2 enzyme, assembling them in the correct geometry for ubiquitin transfer. E3 ubiquitin ligases can either be single proteins or protein complexes.
Ubiquitin is attached to amino groups of either the N-terminal residue (6–8) or an internal lysine residue of the target substrate (3). Additional ubiquitin molecules are added to the lysine 48 residue of the previous ubiquitin in the chain (9). A polyubiquitin chain of at least four residues is required to target a protein to the proteasome for destruction (10). Subunits of the 19S regulator bind to the polyubiquitin chains and catalyze substrate deubiquitination, denaturation, and translocation of the unfolded substrate into the catalytic core for degradation.
The role of polyubiquitination in degradation mediated by the proteasome seems to be 2-fold: target proteins to proteasomes and assist in their unfolding. The actual degradation step requires that ubiquitinated proteins first be denatured and deubiquitinated, to allow them to fit into the narrow channel of the core particle. Thus, the substrates for the actual proteolysis are unfolded and nonubiquitinated. Therefore, if a protein can be delivered to the proteasome in a denatured or partially unfolded state, ubiquitination should not be required for its degradation. In fact, purified 20S and 26S proteasomes can degrade nonubiquitinated, denatured substrates in vitro (11–14). Recently, there have been several reports of proteasome-dependent, ubiquitin-independent degradation of proteins in vivo (15–19). This type of degradation seems to be much less common than the proteasomal degradation of ubiquitinated proteins (20). Here, we provide an example of proteasome-dependent, ubiquitin-independent degradation of cellular proteins mediated by a protein encoded by human cytomegalovirus (HCMV).
HCMV is the prototypical member of the β-herpes virus family (21). The majority of the human population has been infected with HCMV, which establishes a life-long, latent association with its host. Infection is of little clinical consequence in healthy individuals, but primary infection, or reactivation of latent virus, can cause severe disease in patients with immature or compromised immune systems, such as newborns, AIDS patients, or those receiving immunosuppressive therapy during tissue or organ transplants.
HCMV subverts the cell cycle of infected host cells (22) to promote its replication and spread. Infection of quiescent cells induces them to exit the G0 compartment, traverse through G1, and subsequently arrest at the G1/S border. This cell cycle position is presumably favorable for viral DNA replication, as all of the building blocks required for DNA synthesis would be present, but not used for the synthesis of the host cell's genome. HCMV encodes multiple proteins that can stimulate and arrest cell cycle progression. One such protein is encoded by the viral UL82 gene and is termed pp71. Expression of pp71 in quiescent cells stimulates these cells to reenter the cell cycle, progress through the G1 compartment, and enter the S phase (23). pp71 accomplishes this by binding to and subsequently directing the degradation of the hypophosphorylated forms of the retinoblastoma (Rb) family of tumor suppressors, the Rb protein (p105), p107, and p130 (23). The expression of pp71 in asynchronous cells also accelerates them through the G1 phase of the cell cycle, apparently through a mechanism that is independent of its ability to target the Rb family (24).
Here, we show that the HCMV pp71 protein targets the hypophosphorylated forms of the Rb family of proteins for proteasome-dependent degradation through a ubiquitin-independent pathway.
Materials and Methods
Cell Culture.
The human osteosarcoma cell line, U-2 OS (American Type Culture Collection) was maintained in DMEM supplemented with 10% (vol/vol) FBS (HyClone) in an atmosphere of 5% CO2 at 37°C. The mouse ts20 cell line, which contains a temperature-sensitive E1 ubiquitin-activating enzyme, and the H38 control cell line, in which the ts20 mutation was repaired, were gifts from H. Ozer (New Jersey Medical School, Newark, NJ). These cells were maintained at 35°C (the permissive temperature); their restrictive temperature was 39°C.
Degradation Analysis.
For cotransfection assays, U-2 OS cells were transfected with the indicated plasmids by the calcium phosphate method. The DNA precipitate was removed 12 h later, and cells were rinsed and cultured in 10% (vol/vol) FBS-containing medium for an additional 48 h before protein lysates were harvested at 60 h after transfection. The ts20 cells were transfected at 35°C on 10-cm plates with FuGENE 6 (Roche Applied Sciences) for 24 h, after which the cells were split into two dishes. After incubation at the permissive temperature (35°C) for an additional 12 h, some plates were moved to the restrictive temperature (39°C). Lysates were harvested 24 h later. Protein levels were quantified by the Bradford assay, polypeptides in lysates were separated by electrophoresis in an SDS-containing polyacrylamide gel, and Western blots were probed with the indicated antibodies. For the pulse–chase analysis, at 24 h after transfection, U-2 OS cells cultured in 10% (vol/vol) serum-containing medium were incubated in cysteine- and methionine-free medium containing 10% (vol/vol) dialyzed FBS for 1 h before a 30-min pulse with 4 ml of medium containing 0.2 mC/ml 35S-Express (Perkin-Elmer). Cultures were chased in medium containing 10% (vol/vol) FBS, and lysates were immunoprecipitated with the hemagglutinin (HA)-specific antibody. Radioactive bands in blots were quantified by using a PhosphorImager (Molecular Dynamics).
Plasmids.
pSG5-pp71, pSG5-pp71-C219G, pSG5L-HApRb, pCMVp130, and pHAp130 have been described (refs. 23 and 24 and references therein). The pSG5 vector is from Stratagene. pHA-ubq (provided by H. Laman, Imperial Cancer Research Fund, London) encodes HA-tagged ubiquitin. pCW7 and pCW8 (provided by R. Kopito, Stanford University, Stanford, CA) encode yeast wild-type and dominant-negative (K48R mutant) ubiquitins, respectively (25).
Assay for in Vivo Ubiquitination.
Transfected U-2 OS cells were lysed in 100 μl of denaturation buffer (100 mM Tris, pH 7.4/1 mM EDTA/100 mM NaCl/2 mM DTT/3.5% SDS) with mild sonication and boiling (10 min). Debris was removed by centrifugation, and 5-μl aliquots were used for Western blot analysis. The remaining portions of denatured lysates were diluted to 6 ml in HK buffer (50 mM Hepes, pH 7.9/50 mM KCl/1 mM EDTA/0.5% Nonidet P-40/2 mM DTT/1 mM sodium orthovanadate/1 mM PMSF/25 mM NaF/10 μg/ml pepstatin A/10 μg/ml aprotinin/25 μg/ml leupeptin/25 μg/ml trypsin inhibitor) and incubated for 30 min on ice to allow for renaturation. Lysates were incubated with 20 μl of rabbit polyclonal antibody for 2 h. Immune complexes were captured on protein A agarose beads (Amersham Pharmacia), washed four times with lysis buffer and once with PBS. Bound proteins were separated by electrophoresis in an SDS-containing polyacrylamide gel and visualized by Western blot assay.
Antibodies and Inhibitors.
The following antibodies were obtained from commercial sources: tubulin (DM 1A, Sigma), HA (16B12, Babco, Richmond, CA), and p130 (C-20, Santa Cruz Biotechnology). The p53 antibody was 421, and the pp71 antibody was 4F7 (23). Lactacystin and E64 were from Calbiochem.
Results
Degradation of the Rb Family of Proteins by pp71 Occurs Without Detectable Polyubiquitination.
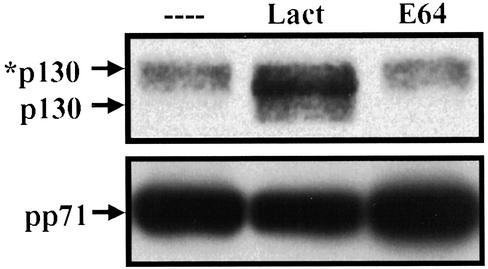
The HCMV pp71 protein binds directly to Rb, p107 and p130 through an Rb-binding LxCxD motif, targeting them for degradation (23). When expression vectors for pp71 and an Rb family member are cotransfected into cells, the Rb family member accumulates to much lower levels than when it is cotransfected with either an empty vector, the adenovirus E1A protein, or HCMV pp65, a protein related in sequence to pp71, but unable to modulate the cell cycle (23). The reduced level to which the Rb family members accumulate in the presence of pp71 results from their increased turnover. We have shown that both Rb and p130 degradation by pp71 is inhibited by multiple proteasome inhibitors (23). Fig. 1 displays an experiment that confirms this earlier result within transfected U-2 OS cells; the pp71-induced degradation of the hypophosphorylated form of p130 was inhibited by the proteasome inhibitor lactacystin but not by E64, an inhibitor of nonproteasomal proteases. Thus, pp71-mediated degradation of p130 requires proteasome function.
Figure 1.
The pp71-mediated degradation of p130 requires proteasome function. U-2 OS cells, cotransfected with vectors expressing HA epitope-tagged p130 and pp71, were cultured for 36 h before the addition of lactacystin (Lact), E64, or diluent lacking drug (– – – –). After an additional 12 h, protein lysates were harvested, and epitope-tagged p130 was analyzed by Western blot with an HA-specific antibody. The asterisk identifies the hyperphosphorylated form of the p130 protein. Expression of pp71 in the transfected cultures was confirmed by Western blot assay.
The majority of proteasomal substrates are targeted for degradation by the addition of polyubiquitin chains (3, 4), a reaction that is catalyzed by a variety of classes of ubiquitin ligase complexes. To determine whether pp71 might act as a component of a ubiquitin ligase, we searched in vivo for pp71-induced polyubiquitinated species of the Rb family members.
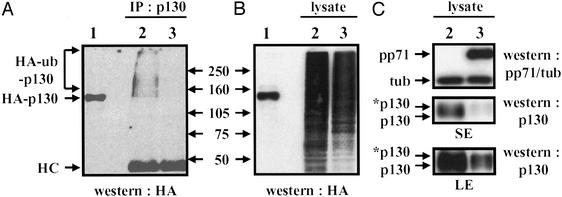
The hyperphosphorylated form of p130 is thought to be degraded by the ubiquitin–proteasome pathway because its level drops dramatically as cells exit G0 and progress through G1, and it can be stabilized by proteasome inhibitors (26). Recently, p130 has been shown to be ubiquitinated in vitro when phosphorylated on serine 672, and mutation of this residue to alanine increased the protein's stability in vivo (27), providing additional evidence for its degradation in vivo by the ubiquitin–proteasome pathway. To probe the role of ubiquitination in the degradation of p130 in vivo, we transfected U-2 OS cells with expression plasmids encoding untagged p130 and HA epitope-tagged ubiquitin, immunoprecipitated p130 from denatured lysates with a polyclonal antibody to p130, and probed with an antibody specific for the HA epitope by Western blot assay to detect polyubiquitinated proteins. We detected polyubiquitinated species (Fig. 2A, lane 2) that migrate in the gels more slowly than p130 (as compared with nonubiquitinated, HA-tagged p130, Fig. 2A, lane 1), providing direct evidence that p130 is ubiquitinated in vivo before degradation by the proteasome. Because pp71 increased the rate of p130 turnover, we expected that, if pp71 acted as a ubiquitin ligase, we would detect an increase in polyubiquitinated p130 in the presence of pp71. However, not only did we not detect an increase, we were unable to detect any polyubiquitinated species of p130 in the presence of pp71 (Fig. 2A, lane 3), in experiments where pp71 was shown to decrease the levels of p130 but not totally eliminate it from the lysate (Fig. 2C). This does not seem to be the result of a global inhibition of ubiquitination by pp71 because roughly similar levels of total ubiquitinated proteins were observed both in the presence and absence of pp71 (Fig. 2B).
Figure 2.
The pp71-mediated degradation of p130 occurs without the accumulation of a detectable polyubiquitinated intermediate. U-2 OS cells were cotransfected with a plasmid encoding p130 that was not tagged with an epitope, a plasmid encoding HA epitope-tagged ubiquitin, and either the expression vector without an inserted coding region (lane 2) or the vector encoding pp71 (lane 3). At 60 h after transfection, lysates were prepared and subjected to immunoprecipitation with a polyclonal p130-specific antibody (A, IP: p130) before Western blot analysis with an HA epitope-specific antibody (Western: HA) to detect p130 containing epitope-tagged ubiquitin (HA-ub-p130). Bands corresponding to antibody heavy chains (HC) are labeled. Aliquots of the same lysates also were analyzed directly with the HA antibody to detect all proteins with ubiquitin conjugates (B, lysate). Lysates expressing HA-tagged p130 (HA-p130) were analyzed to mark the size of nonubiquitinated p130 (lane 1), and the molecular masses (kDa) of marker proteins are indicated. The steady state levels of pp71 plus tubulin as a loading control (Western: pp71/tub) and p130 (Western: p130) were assayed by Western blot in the same lysates (C). Both short (SE) and long (LE) exposures of the p130-specific Western blot are shown, and the asterisk indicates the hyperphosphorylated form of the p130 protein.
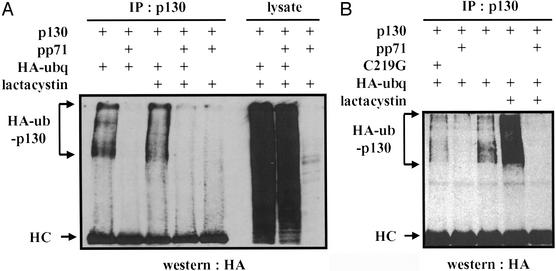
Inhibition of proteasome function can stabilize some polyubiquitinated species and cause them to accumulate, whereas others become deubiquitinated upon prolonged proteasome inhibition. To determine whether proteasome inhibition could stabilize a putative pp71-induced polyubiquitinated p130 intermediate, we looked for such a species in lysates of transfected cells that were treated with lactacystin for either a short or long period. In the absence of pp71, we observed a small increase in polyubiquitinated p130 after 5 h (Fig. 3A) and a substantial increase after 14 h of lactacystin treatment (Fig. 3B). However, we did not detect polyubiquitinated species of p130 in the presence of pp71 in cells treated with lactacystin for either time period (Fig. 3), and, as before, roughly similar levels of total ubiquitinated proteins were observed in the presence and absence of pp71 (Fig. 3A). Thus, we cannot detect polyubiquitination of p130 in the presence of pp71.
Figure 3.
A pp71-induced polyubiquitinated p130 intermediate is not detected in the presence of a proteasome inhibitor. (A) U-2 OS cells were cotransfected with some or all of the plasmids expressing p130, pp71, and HA epitope-tagged ubiquitin (HA-ubq); expression vector without an inserted coding region was added to transfections lacking plasmids encoding pp71 or HA-ubq, and, where indicated, lactacystin was added for 5 h before lysates were harvested. At 60 h after transfection, lysates were prepared and subjected to immunoprecipitation with the p130 antibody (IP: p130) before Western blot analysis with an HA epitope-specific antibody (Western: HA) to detect epitope-tagged ubiquitin conjugated to p130 (HA-ub-p130). Bands corresponding to antibody heavy chains (HC) are labeled. Aliquots of the same lysates also were analyzed directly with the HA antibody to detect all proteins with ubiquitin conjugates (lysate). (B) Lysates of U-2 OS cells, which in some cases included transfection with a plasmid expressing the C219G mutant derivative of pp71, were prepared and analyzed as above, except that, where indicated, lactacystin was added 14 h before the lysates were harvested.
Conceivably, pp71 degrades hypophosphorylated p130 in the absence of polyubiquitination and before it can become phosphorylated when it would be subject to normal cell cycle-mediated polyubiquitination. In support of this hypothesis, we can detect polyubiquitinated p130 in the presence of a pp71 variant, C219G (Fig. 3B), whose mutation resides in the Rb-binding LxCxD motif. This mutant does not stimulate cell cycle progression out of G0, and it fails to degrade Rb and p130 (23). Polyubiquitinated species of p130 are observed in the presence of the C219G pp71 protein, presumably because degradation of the hypophosphorylated form of p130 is prevented. This could allow p130 to be phosphorylated and hence become a substrate for a cellular ubiquitin ligase. We also were unable to detect polyubiquitinated species of Rb or p107 either in the presence or absence of pp71, with or without proteasome inhibition (data not shown). Because Rb and p107 are thought to be regulated only by phosphorylation and not by targeted destruction, and because pp71 does not seem to function as a ubiquitin ligase, this was the expected result.
Rb Family Degradation by pp71 Is Maintained During Conditions That Inhibit the Polyubiquitination of Proteins.
Our analysis of the ubiquitination status of p130 points to a proteasome-dependent, ubiquitin-independent mechanism for the pp71-mediated degradation of the Rb family. An alternative explanation is that pp71-induced ubiquitination of p130 may be undetectable even in this sensitive assay because of technical reasons, such as the low levels of p130 present in the lysates, the short-lived nature of a putative pp71-induced polyubiquitinated p130 intermediate, or the fact that such a species is not stabilized by lactacystin. To distinguish between these two models, we used two independent methods to determine whether polyubiquitination is required for the pp71-mediated degradation of the Rb family.
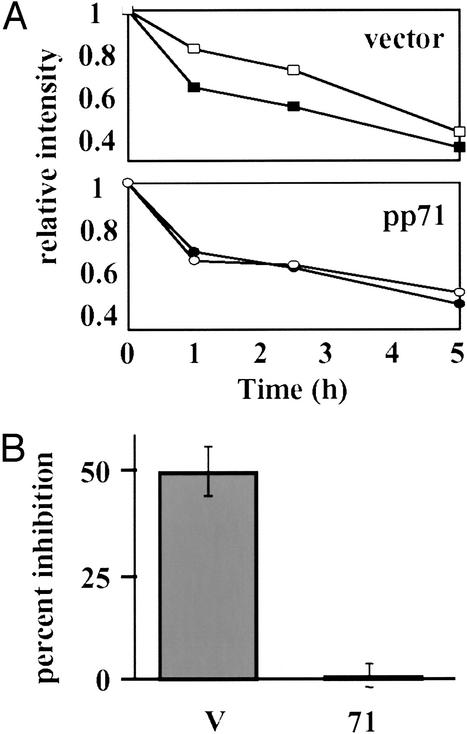
In the first approach, we asked whether a dominant-negative ubiquitin molecule (DN-ubq) could inhibit the pp71-mediated degradation of p130. Ubiquitin chains are extended by the covalent polymerization of ubiquitin molecules through their terminal glycine residue to the lysine at amino acid position 48 of the last ubiquitin in the existing chain. When added to existing chains, a ubiquitin mutant with lysine 48 replaced by arginine, K48R, exerts a dominant-negative, chain-terminating effect and has been shown to inhibit the proteasomal degradation of several proteins (9, 25, 28). We chose to examine p130 because, in the absence of pp71, it is polyubiquitinated and degraded by the proteasome and thus provides an internal positive control in this assay. A pulse–chase protocol was used because competition between exogenous DN-ubq and the endogenous wild-type molecule for incorporation into chains often leads to substantially less than 100% inhibition (25) and thus might not be evident by Western blot analysis. In the absence of pp71, an inhibitory effect on p130 degradation of DN-ubq was observed when compared with cotransfection with an expression plasmid for wild-type ubiquitin (Fig. 4A Upper). However, DN-ubq had no effect on the pp71-mediated degradation of p130 (Fig. 4A Lower), which, as previously illustrated (23), occurred at a much faster rate than in the absence of pp71 (data not shown). In three independent experiments, we observed an ≈50% inhibition of p130 degradation by DN-ubq in the absence of pp71 and less than 2% inhibition in the presence of pp71 (Fig. 4B). As DN-ubq cannot inhibit the pp71-mediated degradation of p130, the simplest conclusion is that this degradation occurs in the absence of polyubiquitination.
Figure 4.
The pp71-mediated degradation of p130 is not inhibited by dominant-negative ubiquitin. (A) U-2 OS cells were cotransfected with a plasmid encoding HA epitope-tagged p130 together with an expression vector lacking a coding region (Upper, vector) or a pp71 expression vector (Lower, pp71). The cultures also received plasmids expressing wild-type ubiquitin (filled symbols), or K48R dominant-negative ubiquitin (open symbols). At 24 h after transfection, the cultures were pulse labeled with 35S-labeled methionine and cysteine for 30 min and then chased for the indicated times in hours (h). The total amount of radioactive protein (hypo- and hyperphosphorylated forms) for each experimental condition relative to the wild-type ubiquitin zero time point was determined by PhosphorImager analysis. (B) The percent inhibition of degradation by the dominant-negative ubiquitin was determined for cultures receiving the expression vector lacking a coding region (V) and for cultures receiving the pp71 expression vector (71) by dividing the percent of p130 degraded in the presence of wild-type ubiquitin by the percent degraded in the presence of the dominant-negative ubiquitin after a 1-h chase period. The mean plus standard deviation for three independent experiments is displayed.
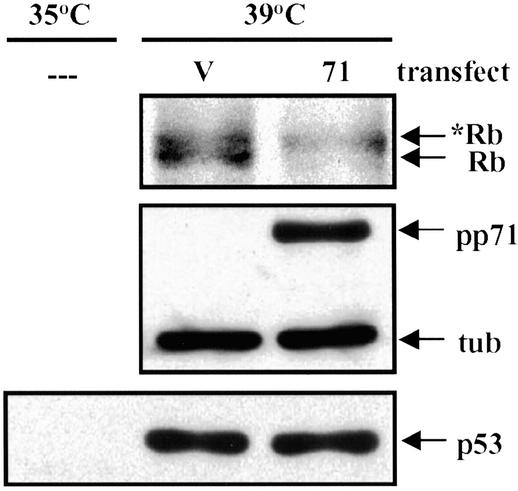
Finally, we performed degradation experiments in the mouse ts20 cell line (29). These cells have a temperature-sensitive E1 ubiquitin-activating enzyme, which is nonfunctional at the restrictive temperature of 39°C. At this temperature, ubiquitin monomers cannot be activated, polyubiquitin chains are not synthesized, and proteins degraded by the ubiquitin–proteasome pathway, such as p53, are stabilized (29). In these cells, we were able to detect the pp71-mediated degradation of the hypophosphorylated form of Rb at the restrictive temperature, under conditions where the p53 protein was stabilized (Fig. 5). Because, under these conditions, polyubiquitination is prevented but pp71-mediated degradation of Rb still occurs, we conclude that this degradation occurs through a ubiquitin-independent mechanism.
Figure 5.
pp71 directs the degradation of the hypophosphorylated form of Rb in the absence of a functioning ubiquitination system. Mouse ts20 cells cultured at the permissive temperature (35°C) were transfected with a plasmid encoding HA epitope-tagged Rb and either an expression vector lacking a coding region (V) or a plasmid expressing pp71 (71). After 24 h at the indicated temperature, lysates were harvested and proteins were analyzed by Western blot with antibody specific for the Rb protein, pp71, tubulin (tub), or the p53 tumor suppressor protein. The asterisk identifies the hyperphosphorylated form of the Rb protein, and – – – designates untransfected cells.
Discussion
The HCMV pp71 protein stimulates quiescent cells to reenter the cell cycle, proceed through G1 and enter the S phase (23). It accomplishes this by degrading the hypophosphorylated forms of the Rb family of tumor suppressors in a proteasome-dependent manner. Here, we show that although inhibitors of the proteasome prevent the pp71-mediated degradation of Rb and p130, this degradation occurs in the absence of detectable polyubiquitination. Thus, we propose that the pp71-mediated degradation of the Rb family occurs through a proteasome-dependent, ubiquitin-independent pathway. This is the first of example of a viral protein that targets cellular proteins for degradation by this mechanism.
In vitro proteasomal degradation of nonubiquitinated model substrates, which have been denatured before addition to the assay, has been well documented (11–14). As ubiquitination is thought only to mediate protein unfolding and targeting to proteasomes and to prevent translocation into the catalytic core, this is not surprising. Nevertheless, a number of recent papers describe the in vitro proteasomal degradation of nonubiquitinated proteins that have not been denatured before addition to the assay (30–34). Although it remains to be determined whether these proteins are degraded by the same mechanism within cells, there is evidence for ubiquitin-independent action of proteasomes in vivo. Indeed, proteasomes are conserved in archaebacteria, where ubiquitin is absent (20). Furthermore, several eukaryotic proteins, namely ornithine decarboxylase (11, 35, 36), the tumor suppressor p53 (15), the cyclin-dependent kinase inhibitor p21 (19, 37), IκBα (16), and minute virus of mice NS2 protein (18), have been shown to be degraded by the proteasome through ubiquitin-independent mechanisms in vivo. Whether these examples are exceptions to the rule or whether proteasome-dependent, ubiquitin-independent degradation is a widespread, physiologically relevant event that occurs in vivo in higher eukaryotes remains a matter of conjecture.
Some proteins seem to undergo proteasomal degradation by both ubiquitin-dependent and ubiquitin-independent mechanisms. For example, IκBα is a protein with a short half-life, and its constitutive degradation has recently been shown to be mediated by the proteasome through a ubiquitin-independent mechanism (16). IκBα binds to the transcription factor NF-κB and sequesters it in the cytoplasm. Upon cellular stimulation, it first becomes phosphorylated and then ubiquitinated, resulting in its proteasomal degradation and the liberation of NF-κB. Conversely, the levels of the short-lived p53 protein are normally kept low by the mdm-2 ubiquitin ligase that stimulates p53 turnover by ubiquitin-mediated proteolysis. However, treatment of cells with dicoumarol, an inhibitor of NAD(P)H quinone oxidoreductase 1, induces p53 degradation by a proteasome-dependent, ubiquitin-independent mechanism (15). For the p21 protein, which is degraded by the proteasome when conjugated to ubiquitin or nonubiquitinated, the signal (if any) that determines which mode of degradation is used is not known. Our results with the Rb family member p130 follow this trend of a substrate being degraded by ubiquitin-dependent and -independent mechanisms. In the absence of pp71, p130 is degraded by a ubiquitin-dependent pathway, and when targeted by pp71, p130 is degraded independently of ubiquitination. It is unclear why cells have chosen to use these two distinct pathways for the degradation of the same protein.
Multiple assays have been used to examine the ubiquitin dependence of proteasomal degradation. One approach employs mutant proteins in which all of the lysine residues have been mutated, theoretically preventing the polyubiquitination of the mutant protein. The continued degradation of such mutants, despite the inability to demonstrate polyubiquitinated species of these proteins, has been put forward as evidence for ubiquitin-independent degradation (15, 16, 19). However, degradation of a lysine-less variant of the α subunit of the T cell receptor was shown to still require the ubiquitin machinery, even though the level of polyubiquitinated intermediate was reduced over 90% compared with the wild-type protein (25). This seems possible because the N-terminal residue of some proteins can serve as a ubiquitin acceptor (6–8). Although this may be an isolated case, and the degradation of most substrates lacking lysines probably results from ubiquitin-independent degradation, results from this type of experiment should be interpreted with caution. The other assays use cells with a temperature-sensitive E1 ubiquitin-activating enzyme (15–18) or dominant-negative ubiquitin mutants (18) to demonstrate that degradation of the target proteins is ubiquitin independent. The application of multiple assays can provide the strongest argument for ubiquitin-independent degradation.
We have used a dominant-negative ubiquitin (Fig. 4) and the ts20 cell line that is E1-deficient at the restrictive temperature (Fig. 5) to demonstrate the in vivo ubiquitin-independent degradation of the Rb family by pp71. How, in the absence of polyubiquitination, does pp71 deliver its substrates to the proteasome? Polyubiquitin chains have been shown to promote the binding of attached proteins to the proteasome (10). Perhaps pp71 or a member of a putative pp71 degradation complex can mediate this interaction, obviating the need for ubiquitin. This is the method by which ornithine decarboxylase is degraded. When bound to antizyme, ornithine decarboxylase is recognized and degraded by the proteasome in the absence of ubiquitination (20). Mutations in either binding partner that disrupt proteasome binding inhibit degradation. p21 binds to the proteasome C8 subunit in vitro (37). The rate of turnover in vivo of mutant p21 proteins directly correlated with their binding affinity for C8 in vitro. Likewise, binding to C8 was required for p21 ubiquitin-independent degradation by purified 20S proteasomes in vitro (37). Thus, proteasome binding may be a common theme for ubiquitin-independent degradation. However, the p21–C8 interaction has not been detected in vivo, which may indicate that these associations are transient or labile, and thus may be difficult to demonstrate. Interestingly, the papillomavirus E7 protein, an LxCxE-containing protein that binds the Rb family and stimulates cell cycle progression like pp71, also binds to the proteasome and directs the degradation of the Rb protein (38). However, proteasome binding has not yet been shown to be required for degradation, and the dependence of the degradation on polyubiquitination has not been examined.
Polyubiquitination may also help to unfold proteins before proteasome degradation (39). If pp71 were to assist in this process, ubiquitin may not be required for pp71-mediated protein degradation. Unfolded or poorly structured substrates such as p21 are degraded by proteasomes in vitro and in vivo in the absence of polyubiquitination. Furthermore, simian virus 40 (SV40) T antigen, which is an LxCxE-containing protein that stimulates cell cycle progression like HCMV pp71 and papillomavirus E7, also degrades the p130 protein and has a DNA J domain that is required for p130 degradation (40). DNA J motifs are found on chaperon-like proteins that can modulate protein folding. The dependence of T antigen-induced degradation of p130 on polyubiquitination has not been investigated, and whether or not pp71 or SV40 T antigen unfolds proteins before inducing their degradation by the proteasome remains to be determined.
Finally, there are many small ubiquitin-like proteins whose functions have not been completely characterized (41). pp71 may induce a covalent modification of the Rb family by one of these ubiquitin-like proteins, which then targets the Rb family for degradation. As many of these proteins have their own E1-activating enzymes, such a process would not be inhibited in the ts20 cells at the restrictive temperature, nor be sensitive to dominant-negative ubiquitin.
In summary, we have identified an additional pathway through which a virus can manipulate the cell cycle: degradation of the hypophosphorylated forms of the Rb family members that is mediated by a virus-coded LxCxD-domain protein, through a proteasome-dependent, ubiquitin-independent mechanism.
Acknowledgments
We thank R. Kopito, H. Laman, and H. Ozer for reagents, and J. Alwine and A. Levine for critical reading of the manuscript. This work was supported by National Institutes of Health Grant CA82396.
Abbreviations
- HCMV
human cytomegalovirus
- HA
hemagglutinin
- Rb
retinoblastoma
References
- 1.Davies K J A. Biochimie. 2001;83:301–310. doi: 10.1016/s0300-9084(01)01250-0. [DOI] [PubMed] [Google Scholar]
- 2.DeMartino G N, Slaughter C A. J Biol Chem. 1999;274:22123–22126. doi: 10.1074/jbc.274.32.22123. [DOI] [PubMed] [Google Scholar]
- 3.Glickman M H, Ciechanover A. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 4.Ciechanover A, Orian A, Schwartz A L. BioEssays. 2000;22:442–451. doi: 10.1002/(SICI)1521-1878(200005)22:5<442::AID-BIES6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 5.Voges D, Zwickl P, Baumeister W. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 6.Aviel S, Winberg G, Massucci M, Ciechanover A. J Biol Chem. 2000;275:23491–23499. doi: 10.1074/jbc.M002052200. [DOI] [PubMed] [Google Scholar]
- 7.Breitschopf K, Bengal E, Ziv T, Admon A, Ciechanover A. EMBO J. 1998;17:5964–5973. doi: 10.1093/emboj/17.20.5964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reinstein E, Scheffner M, Oren M, Ciechanover A, Schwartz A. Oncogene. 2000;19:5944–5950. doi: 10.1038/sj.onc.1203989. [DOI] [PubMed] [Google Scholar]
- 9.Chau V, Tobias J W, Bachmair A, Marriot D, Ecker D J, Gonda D K, Varshavsky A. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 10.Thrower J S, Hoffman L, Rechsteiner M, Pickart C M. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bercovich Z, Rosenberg-Hasson Y, Ciechanover A, Kahana C. J Biol Chem. 1989;264:15949–15952. [PubMed] [Google Scholar]
- 12.Dick L R, Aldrich C, Jameson S C, Moomaw C R, Pramanik B C, Doyle C K, DeMartino G N, Bevan M J, Forman J M, Slaughter C A. J Immunol. 1994;152:3884–3894. [PMC free article] [PubMed] [Google Scholar]
- 13.Driscoll J, Goldberg A L. Proc Natl Acad Sci USA. 1989;86:787–791. doi: 10.1073/pnas.86.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kisselev A F, Akopian T N, Woo K M, Goldberg A L. J Biol Chem. 1999;274:3363–3371. doi: 10.1074/jbc.274.6.3363. [DOI] [PubMed] [Google Scholar]
- 15.Asher G, Lotem J, Sachs L, Kahana C, Shaul Y. Proc Natl Acad Sci USA. 2002;99:13125–13130. doi: 10.1073/pnas.202480499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krappmann D, Wulczyn F G, Scheidereit C. EMBO J. 1996;15:6716–6726. [PMC free article] [PubMed] [Google Scholar]
- 17.Michalek M T, Grant E P, Rock K L. J Immunol. 1996;157:617–624. [PubMed] [Google Scholar]
- 18.Miller C L, Pintel D J. Virology. 2001;285:346–355. doi: 10.1006/viro.2001.0966. [DOI] [PubMed] [Google Scholar]
- 19.Sheaff R J, Singer J D, Swanger J, Smitherman M, Roberts J M, Clurman B E. Mol Cell. 2000;5:403–410. doi: 10.1016/s1097-2765(00)80435-9. [DOI] [PubMed] [Google Scholar]
- 20.Verma R, Deshaies R J. Cell. 2000;101:341–344. doi: 10.1016/s0092-8674(00)80843-0. [DOI] [PubMed] [Google Scholar]
- 21.Mocarski E S, Courcelle C T. In: Fields Virology. Knipe D M, Howley P, editors. Philadelphia: Lippincott; 2001. pp. 2629–2673. [Google Scholar]
- 22.Kalejta R F, Shenk T. Front Biosci. 2002;7:295–306. doi: 10.2741/kalejta. [DOI] [PubMed] [Google Scholar]
- 23. Kalejta, R. F., Bechtel, J. T. & Shenk, T. (2003) Mol. Cell. Biol., in press. [DOI] [PMC free article] [PubMed]
- 24. Kalejta, R. F. & Shenk, T. (2003) J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 25.Yu H, Kopito R R. J Biol Chem. 1999;274:36852–36858. doi: 10.1074/jbc.274.52.36852. [DOI] [PubMed] [Google Scholar]
- 26.Grana X, Garriga J, Mayol X. Oncogene. 1998;17:3365–3383. doi: 10.1038/sj.onc.1202575. [DOI] [PubMed] [Google Scholar]
- 27.Tedesco D, Lukas J, Reed S I. Genes Dev. 2002;16:2946–2957. doi: 10.1101/gad.1011202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ward C L, Omura S, Kopito R R. Cell. 1995;83:121–127. doi: 10.1016/0092-8674(95)90240-6. [DOI] [PubMed] [Google Scholar]
- 29.Chowdary D R, Dermody J J, Jha K K, Ozer H L. Mol Cell Biol. 1994;14:1997–2003. doi: 10.1128/mcb.14.3.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.David D C, Layfield R, Serpell L, Narain Y, Goedert M, Spillantini M G. J Neurochem. 2002;83:176–185. doi: 10.1046/j.1471-4159.2002.01137.x. [DOI] [PubMed] [Google Scholar]
- 31.Jariel-Encontre I, Pariat M, Martin F, Carillo S, Salvat C, Piechaczyk M. J Biol Chem. 1995;270:11623–11627. doi: 10.1074/jbc.270.19.11623. [DOI] [PubMed] [Google Scholar]
- 32.Nunan J, Shearman M S, Checler F, Cappai R, Evin G, Beyreuther K, Masters C L, Small D H. Eur J Biochem. 2001;268:5329–5336. doi: 10.1046/j.0014-2956.2001.02465.x. [DOI] [PubMed] [Google Scholar]
- 33.Tarcsa E, Szymanska G, Lecker S, O'Conner C M, Goldberg A L. J Biol Chem. 2000;275:20295–20301. doi: 10.1074/jbc.M001555200. [DOI] [PubMed] [Google Scholar]
- 34.Tofaris G K, Layfield R, Spillantini M G. FEBS Lett. 2001;509:22–26. doi: 10.1016/s0014-5793(01)03115-5. [DOI] [PubMed] [Google Scholar]
- 35.Glass J R, Gerner E W. J Cell Physiol. 1987;130:133–141. doi: 10.1002/jcp.1041300119. [DOI] [PubMed] [Google Scholar]
- 36.Murakami Y, Matsufuji S, Kameji T, Hayashi S, Igarashi K, Tamura T, Tanaka K, Ichihara A. Nature. 1992;360:597–599. doi: 10.1038/360597a0. [DOI] [PubMed] [Google Scholar]
- 37.Touitou R, Richardson J, Bose S, Nakanishi M, Rivett J, Allday M J. EMBO J. 2001;20:2367–2375. doi: 10.1093/emboj/20.10.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berezutskaya E, Bagchi S. J Biol Chem. 1997;272:30135–30140. doi: 10.1074/jbc.272.48.30135. [DOI] [PubMed] [Google Scholar]
- 39.Ghislain M, Dohmen R J, Levy F, Varshavsky A. EMBO J. 1996;15:4884–4899. [PMC free article] [PubMed] [Google Scholar]
- 40.Stubdal H, Zalvide J, Campbell K S, Schweitzer C, Roberts T M, DeCaprio J A. Mol Cell Biol. 1997;17:4979–4990. doi: 10.1128/mcb.17.9.4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jentsch S, Pyrowolakis G. Trends Cell Biol. 2000;10:335–342. doi: 10.1016/s0962-8924(00)01785-2. [DOI] [PubMed] [Google Scholar]