Abstract
L1 and Alu elements are long and short interspersed retrotransposable elements (LINEs and SINEs) in humans, respectively. Proteins encoded in the autonomous L1 mediate retrotransposition of the nonautonomous Alu and cellular mRNAs. Alu is the only active SINE in the human genome and is derived from 7SL RNA of signal recognition particle. In the other eukaryotic genomes, various tRNA- and 5S rRNA-derived SINEs are found. Some of the tRNA- and 5S rRNA-derived SINEs have partner LINEs of which 3′ sequences are similar to those of the SINEs. One of the tRNA-derived SINEs is shown to be mobilized by its partner LINE. Many copies of tRNA and 5S rRNA pseudogenes are present in the human genome. These pseudogenes may have been generated via the retrotransposition process using L1 proteins. Although there are no sequence similarities between L1 and Alu, L1 functionally links with Alu and even cellular genes, impacting on our genome shaping.
INTRODUCTION
In the human genome, coding sequences are less than 5% while repeat sequences are more than 50% [1]. Most of these repeat sequences are derived from retrotransposons, which transpose through RNA intermediates. L1 and Alu elements are the most successful families of non-LTR elements representing approximately 30% of the human genome [1]. L1 is about 6 kb long, has an internal promoter for RNA polymerase II, and encodes two essential polypeptides (ORF1 and ORF2) for retrotransposition (see, eg, [2, 3, 4]). The product of ORF1 is an RNA-binding protein, and ORF2 encodes a protein with endonuclease and reverse transcriptase activities [5, 6, 7, 8, 9]. While L1 moves autonomously, Alu is a nonautonomous element. Alu elements are short (about 300 bp), and have internal promoters for RNA polymerase III [10]. Since Alu elements encode no proteins, it had been presumed that Alu borrows the enzymes like reverse transcriptases from other sources for retrotransposition.
HUMAN L1 CAN MOBILIZE ALU AND PROCESSED PSEUDOGENES
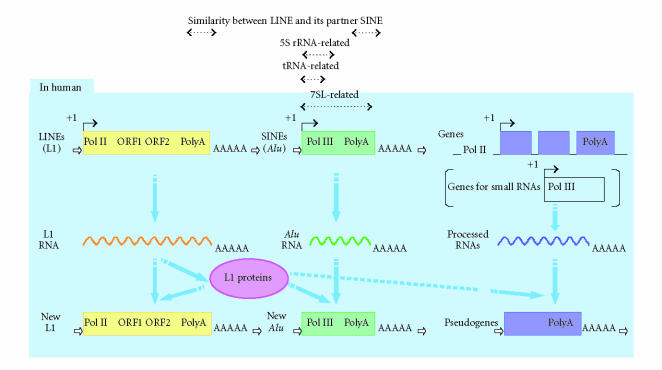
The idea that SINE transposition can be mediated by L1 element was described by Feng et al [8], Jurka [11], and Martin et al [12]. Recently, Dewannieux et al [13] showed that L1 can mobilize Alu in the human cells: neomycin-marked Alu sequences transposed in the Hela cells transiently expressing the L1 ORF2 proteins; and the transposition process included splicing out of the autocatalytic intron, target site duplication, and integrations into consensus A-rich sequences. Reverse transcriptase primes on the 3′ terminal poly A stretch of the L1 mRNA [7]. Also in the experiments using neomycin-marked Alu sequences, the 3′ terminal polyA tracts of the Alus were required for retrotransposition. Moreover, L1 can mediate retrotransposition of a cellular mRNA that is not associated with retrotransposon, although the rate of retrotransposition is 100–1000 fold lower than that in the case of Alu [13, 14, 15]. Very recently, U6 snRNA was reported to be mobilized by L1 [16]. L1 mobilizes Alu and different kinds of cellular RNAs, and plays important roles in human genome shaping. Figure 1 is a schematic representation of the retrotransposition of the human L1 elements and their dependents. Retrotransposition of L1 and Alu (and processed genes) results in insertion mutations, and crossing-over between the homologous elements is one of the sources of genetic variations (see, eg, [17]). Insertions of Alu elements introduce alternative 3′ splice sites into existing genes, possibly resulting in defective splicing [18]. Alu and L1 elements can alter the distribution of methylation in the genome, and possibly transcription of genes [19, 20]. These rearrangements have a great impact on the genome evolution. Most mutations may be harmless, because coding and control sequences comprise less than 5% of the human genome DNA [1]. However, for example, it is reported that Alu insertions cause neurofibromatosis, haemophilia, familial hyperchoresterolaemia, breast cancer, insulin-resistant diabetes type II and Ewing sarcoma [19].
Figure 1.
Retrotransposition of the human LINE-1 (L1) elements and their dependents. L1 contains two open reading frames ORF1 and ORF2. Products of these ORFs associate with the transcripts of L1, Alu, and cellular genes. The RNA-protein complexes bind to another part of the genome, and new elements of L1 and Alu, and pseudogenes which lack introns are generated. Genes for small RNAs like tRNA and 5S rRNA may also retrotranspose. In the other eukaryotic genomes, there are also tRNA- and 5S rRNA-derived SINEs. tRNA- and 5S rRNA-derived SINEs have tRNA- and 5S rRNA-related regions at the 5′ ends, respectively. The 3′ end regions of some of the tRNA- and 5S rRNA-derived SINEs show similarities to the 3′ end regions of their partner LINEs at the nucleotide sequence level.
LINKS BETWEEN LINES AND SINES IN THE OTHER EUKARYOTIC GENOMES
The Alu element which is the only active SINE in the human genome, is thought to be derived from the 7 SL RNA that is a component of signal recognition particle [21]. In the other eukaryotic genomes, there are SINEs which are derived from tRNA genes [22]. Nucleotide sequences of the 5′ regions of tRNA-derived SINEs are similar to those of tRNA genes. Some tRNA-derived SINEs have sequence similarity to LINEs in their 3′ end regions [22]: for example, HE1 SINE and HER1 LINE in higher elasmobranchs (sharks, skates, and rays), tortoise SINE and CR1-like LINE of turtle, salmonid Hpa1 SINE and RSg-1 LINE of rainbow trout, and P.s.1/SINE and Lucy-1 CR1-like LINE in Podarcis sicula. This fact leads us to think that these SINEs are mobilized by the partner LINEs. Indeed, it is demonstrated that in Hela cells, the 3′ end of a fish (eel) SINE is recognized by the reverse transcriptase of its partner LINE, and that the fish SINE can be mobilized by the partner's machinery [23]. In the zebrafish genome, 5S rRNA-derived SINEs have been found [24]: their 5′ end regions are similar to the 5S rRNA at the nucleotide sequence level, and their 3′ regions resemble the 3′ parts of their partner LINEs. One may imagine that LINEs had ever existed (or still exists) with ability to provide their 3′ parts for generation of partner SINEs. All of SINEs, tRNA genes, and 5S rRNA genes are transcribed by RNA polymerase III. Alu elements, tRNA-derived SINEs, and tRNA genes have type 2 internal promoters, while 5S rRNA-derived SINEs and 5S rRNA genes have type 1 internal promoters. Different from the type 1 promoters, the type 2 promoters in 5S rRNA genes synthesize the RNAs with the DNA signals upstream of the transcribed region, which perhaps results in the restricted distribution of 5S rRNA-derived SINEs in eukaryotic species [24].
CONTRIBUTION OF L1 TO OUR GENOME EVOLUTION
Interestingly, there are many copies of pseudogenes, fragments and paralogues of tRNA and 5S rRNA genes in the human genome [1]: 497 copies of the true tRNA genes compared with 324 copies of their related genes; and 4 copies of the true 5S rRNA gene compared with 520 copies of its related genes. As described above, in the human genome, Alu is the only active SINE, and the active tRNA-derived SINE and the 5S rRNA-derived SINE have not been found. However, it is possible that human tRNA and 5S rRNA genes retrotranspose with the L1-encoded proteins, if the transcripts accidentally contain A-rich sequences at the 3′ ends [25]. The human genome is reported to contain many types of chimeric retrogenes that were formed using the L1 integration machinery [26]. It should be noted that a new insertion of Alu to the germline is computationally estimated to occur in about 1 of every 100 births [27]. Directly and indirectly, L1 greatly contributes to the evolution of our genome.
References
- 1.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Athanikar JN, Badge RM, Moran JV. A YY1-binding site is required for accurate human LINE-1 transcription initiation. Nucleic Acids Research. 2004;32(13):3846–3855. doi: 10.1093/nar/gkh698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87(5):917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 4.Swergold GD. Identification, characterization, and cell specificity of a human LINE-1 promoter. Molecular and Cellular Biology. 1990;10(12):6718–6729. doi: 10.1128/mcb.10.12.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hohjoh H, Singer MF. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. The EMBO Journal. 1996;15(3):630–639. [PMC free article] [PubMed] [Google Scholar]
- 6.Martin SL, Cruceanu M, Branciforte D, et al. LINE-1 retrotransposition requires the nucleic acid chaperone activity of the ORF1 protein. Journal of Molecular Biology. 2005;348(3):549–561. doi: 10.1016/j.jmb.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 7.Cost GJ, Feng Q, Jacquier A, Boeke JD. Human L1 element target-primed reverse transcription in vitro. The EMBO Journal. 2002;21(21):5899–5910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87(5):905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 9.Mathias SL, Scott AF, Kazazian HH, Jr, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254(5039):1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- 10.Fuhrman SA, Deininger PL, LaPorte P, Friedmann T, Geiduschek EP. Analysis of transcription of the human Alu family ubiquitous repeating element by eukaryotic RNA polymerase III. Nucleic Acids Research. 1981;9(23):6439–6456. doi: 10.1093/nar/9.23.6439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jurka J. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(5):1872–1877. doi: 10.1073/pnas.94.5.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin F, Olivares M, Lopez MC, Alonso C. Do non-long terminal repeat retrotransposons have nuclease activity? Trends in Biochemical Sciences. 1996;21(8):283–285. [PubMed] [Google Scholar]
- 13.Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nature Genetics. 2003;35(1):41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 14.Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nature Genetics. 2000;24(4):363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 15.Wei W, Gilbert N, Ooi SL, et al. Human L1 retrotransposition: cis preference versus trans complementation. Molecular and Cellular Biology. 2001;21(4):1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilbert N, Lutz S, Morrish TA, Moran JV. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Molecular and Cellular Biology. 2005;25(17):7780–7795. doi: 10.1128/MCB.25.17.7780-7795.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kazazian HH, Jr, Goodier JL. LINE drive. retrotransposition and genome instability. Cell. 2002;110(3):277–280. doi: 10.1016/s0092-8674(02)00868-1. [DOI] [PubMed] [Google Scholar]
- 18.Sorek R, Ast G, Graur D. Alu-containing exons are alternatively spliced. Genome Research. 2002;12(7):1060–1067. doi: 10.1101/gr.229302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nature Reviews Genetics. 2002;3(5):370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- 20.Han JS, Szak ST, Boeke JD. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature. 2004;429(6989):268–274. doi: 10.1038/nature02536. [DOI] [PubMed] [Google Scholar]
- 21.Weiner AM. An abundant cytoplasmic 7S RNA is complementary to the dominant interspersed middle repetitive DNA sequence family in the human genome. Cell. 1980;22(1 pt 1):209–218. doi: 10.1016/0092-8674(80)90169-5. [DOI] [PubMed] [Google Scholar]
- 22.Ohshima K, Okada N. SINEs and LINEs: symbionts of eukaryotic genomes with a common tail. Cytogenetic and Genome Research. 2005;110(1–4):475–490. doi: 10.1159/000084981. [DOI] [PubMed] [Google Scholar]
- 23.Kajikawa M, Okada N. LINEs mobilize SINEs in the eel through a shared 3′ sequence. Cell. 2002;111(3):433–444. doi: 10.1016/s0092-8674(02)01041-3. [DOI] [PubMed] [Google Scholar]
- 24.Kapitonov VV, Jurka J. A novel class of SINE elements derived from 5S rRNA. Molecular Biology and Evolution. 2003;20(5):694–702. doi: 10.1093/molbev/msg075. [DOI] [PubMed] [Google Scholar]
- 25.Szafranski K, Dingermann T, Glockner G, Winckler T. Template jumping by a LINE reverse transcriptase has created a SINE-like 5S rRNA retropseudogene in Dictyostelium. Molecular Genetics and Genomics. 2004;271(1):98–102. doi: 10.1007/s00438-003-0961-9. [DOI] [PubMed] [Google Scholar]
- 26.Buzdin A, Gogvadze E, Kovalskaya E, et al. The human genome contains many types of chimeric retrogenes generated through in vivo RNA recombination. Nucleic Acids Research. 2003;31(15):4385–4390. doi: 10.1093/nar/gkg496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kazazian HH., Jr An estimated frequency of endogenous insertional mutations in humans. Nature Genetics. 1999;22(2):130–130. doi: 10.1038/9638. [DOI] [PubMed] [Google Scholar]