Abstract
The tobacco N gene confers resistance to Tobacco mosaic virus (TMV) and encodes a toll–interleukin-1 receptor/nucleotide binding/Leu-rich repeat class protein. Recent evidence indicates that the Nicotiana benthamiana Rar1 gene (NbRar1), which encodes a protein with a zinc finger motif called CHORD (Cys- and His-rich domain), is required for the function of N. To investigate the role of NbRar1 in plant defense, we identified its interaction partners. We show that the NbRar1 protein interacts with NbSGT1, a highly conserved component of the SCF (Skp1/Cullin/F-box protein)-type E3 ubiquitin ligase complex involved in protein degradation. In addition, we show that NbSGT1 interacts with NbSKP1. Suppression of NbSGT1 and NbSKP1 shows that these genes play an important role in the N-mediated resistance response to TMV. Both NbRar1 and NbSGT1 associate with the COP9 signalosome, another multiprotein complex involved in protein degradation via the ubiquitin-proteasome pathway. Silencing of the NbCOP9 signalosome also compromises N-mediated resistance to TMV. Our results reveal new roles for SCF and the COP9 signalosome in plant defense signaling.
INTRODUCTION
Plants mount various defense responses to survive the challenge of pathogen attack. One such response involves recognition of pathogen-encoded ligands by plant disease resistance (R) gene products (Gabriel and Rolfe, 1990). This recognition event initiates signaling pathways that lead to hypersensitive response (localized cell death) at the site of pathogen ingress, rapid oxidative burst, cell wall strengthening, protein phosphorylation, and activation of various defense response genes (McDowell and Dangl, 2000). These events are followed by a nonspecific general defense response throughout the plant called systemic acquired resistance (Ryals et al., 1996). During systemic acquired resistance, salicylic acid levels increase throughout the plant, defense genes such as pathogenesis-related genes are expressed, and the plant becomes increasingly resistant to further pathogen attack. Evidence suggests that in addition to salicylic acid, jasmonic acid and ethylene play roles in plant defense (Thomma et al., 2001).
In the last decade, the cloning of a number of plant R genes that confer resistance to various pathogens has brought significant progress in our understanding of host–pathogen interactions (Dangl and Jones, 2001). Different classes of cloned R proteins contain similar structural features. Members of the most prominent class of R genes, the NB-LRRs, contain a nucleotide binding domain (NB) and C-terminal Leu-rich repeats (LRRs) of various lengths. The NB-LRR class of R genes can be further classified into toll–interleukin-1 receptor (TIR)–NB-LRR or coiled-coil (CC)–NB-LRR based on their N-terminal sequences. Members of the TIR-NB-LRR class of proteins contain an N-terminal domain that is similar to the cytoplasmic domains of Toll, interleukin-1 receptor (TIR domain), and Myd88 (Rock et al., 1998). The CC-NB-LRR proteins contain a CC domain at the N terminus. Other classes of R genes contain a kinase and/or an LRR domain.
The conserved domains of R proteins from different plant species suggest that activation of common signaling pathways occurs upon pathogen perception (Baker et al., 1997). For example, the functional TIR-NB-LRR class of R genes requires EDS1, and a subset of the CC-NB-LRR class of R genes requires NDR1, for resistance to pathogens (Aarts et al., 1998). Another example of a converging point in disease resistance is the requirement for the Rar1 gene in resistance to powdery mildew by several unlinked CC-NB-LRR genes (Schulze-Lefert and Vogel, 2000). Recent evidence suggests that Nicotiana benthamiana Rar1 (NbRar1) also is an essential component of the N gene–mediated resistance response to Tobacco mosaic virus (TMV) (Liu et al., 2002). This is interesting because the N gene belongs to the TIR-NBS-LRR class of R genes (Whitham et al., 1994). Thus, Rar1 may represent an example of a signaling component involved in pathways shared by both the CC-NB-LRR and the TIR-NB-LRR resistance gene classes.
Genetic studies in barley suggest that Rar1 functions downstream of pathogen perception and upstream of H2O2 accumulation and host cell death (Shirasu et al., 1999). Rar1 encodes a protein with a novel zinc finger motif called CHORD (Cys- and His-rich domain) that is present in all eukaryotes except yeast. Rar1 lacks a CS domain that is found in metazoan CHORD proteins. The CS domain shares homology with yeast SGT1 and human SIP, both of which are novel subunits of a multiprotein ubiquitin ligase (E3) complex (the SCF [Skp/Cullin/F-box] complex) (Kitagawa et al., 1999; Matsuzawa and Reed, 2001). This complex mediates the degradation of multiple proteins involved in diverse signaling pathways through a ubiquitin-proteasome pathway (Deshaies, 1999). Different components of the SCF also have been shown to interact with the COP9 signalosome (Lyapina et al., 2001), a multiprotein complex involved in protein degradation through the 26S proteasome (Wei and Deng, 1999).
Shirasu et al. (1999) hypothesized that SGT1-like proteins in plants that contain a CS domain might provide a link between Rar1 and the SCF complex and to the protein degradation pathway. To understand the role and regulation of NbRar1 in protein degradation and plant defense mechanisms, we used a yeast two-hybrid screen to identify its interactors. We report here that NbRar1 interacts directly with NbSGT1 of the SCF ubiquitin ligase complex. In turn, NbSGT1 interacts with NbSKP1. Both NbRar1 and NbSGT1 associate with the COP9 signalosome. The N gene plants that are knocked out for the expression of NbSGT1, NbSKP1, and the COP9 signalosome are compromised in their resistance response to TMV. These findings are consistent with the most recent work published by Azevedo et al. (2002) and Austin et al. (2002).
RESULTS
Identification of NbRar1-Interacting Proteins
Previous studies in our laboratory suggested that NbRar1 is an essential component of N-mediated signaling (Liu et al., 2002). To identify proteins that interact with NbRar1, we conducted a yeast two-hybrid screen (Kolonin et al., 2000). The NbRar1 sequence was fused to a nuclear localization signal sequence (NLS) and a LexA DNA binding domain (NLS-LexA-BD) under the control of a Gal-inducible promoter (GAL10). This construct was used as the bait to screen a tomato cDNA library that was fused to the B42 activation domain (AD) under the control of a Gal-inducible promoter (GAL1).
We identified 24 Leu prototrophs that showed activation of the LacZ reporter gene. The cDNA inserts of these 24 positive clones were sequenced. A database search for homology using the BLAST program suggested three classes of NbRar1 interactors: SGT1-like (7 clones), hsp90 (13 clones), and OTU-like Cys protease (4 clones).
We focused specifically on the NbRar1 interactors that showed significant homology with the SGT1 proteins found in yeast, human, Arabidopsis, and barley. SGT1 in yeast is a novel subunit of the SCF complex that is involved in kinetochore assembly (Kitagawa et al., 1999). In human, an SGT1 homolog called SIP (Siah-interacting protein) complements defects in yeast strains that contain SGT1 mutant alleles (Kitagawa et al., 1999; Matsuzawa and Reed, 2001). SIP interacts with the SCF complex and controls β-catenin levels, which affect the activity of β-catenin–dependent Tcf/LEF transcription factors (Matsuzawa and Reed, 2001). The longest SGT1 cDNA clone we obtained from the yeast two-hybrid screen contained only 237 amino acids corresponding to the C-terminal region.
We cloned the full-length cDNA of SGT1 from N. benthamiana using reverse transcriptase–mediated (RT) PCR. The predicted N. benthamiana SGT1 protein (NbSGT1) encodes an open reading frame of 370 amino acids and shows substantial homology with SGT1 proteins from different organisms (Figure 1). The NbSGT1 protein shares 90% identity and 95% similarity with tomato SGT1. In addition, it shares homology with barley SGT1 (61% identity and 75% similarity) (Azevedo et al., 2002) and Arabidopsis SGT1a and SGT1b (60% identity and 70% similarity) (Austin et al., 2002). It also shares homology with human SGT1 (44% identity and 63% similarity), human SIP (20% identity and 31% similarity), and yeast SGT1 (27% identity and 44% similarity) (Kitagawa et al., 1999; Matsuzawa and Reed, 2001).
Figure 1.
Comparison of the N. benthamiana SGT1 Amino Acid Sequence with Those of Its Homologs from Other Organisms.
Alignment of the predicted N. benthamiana SGT1 (NbSGT1) protein with its homologs from tomato (tomSGT1), barley (HvSGT1), Arabidopsis (AtSGT1a and AtSGT1b), yeast (ScSGT1), and human (hSGT1 and hSIP1). Numbers at left indicate the positions of the amino acid residues. Identical residues are shaded in black, and similar residues are shaded in gray.
NbSGT1 Interacts Directly with NbRar1
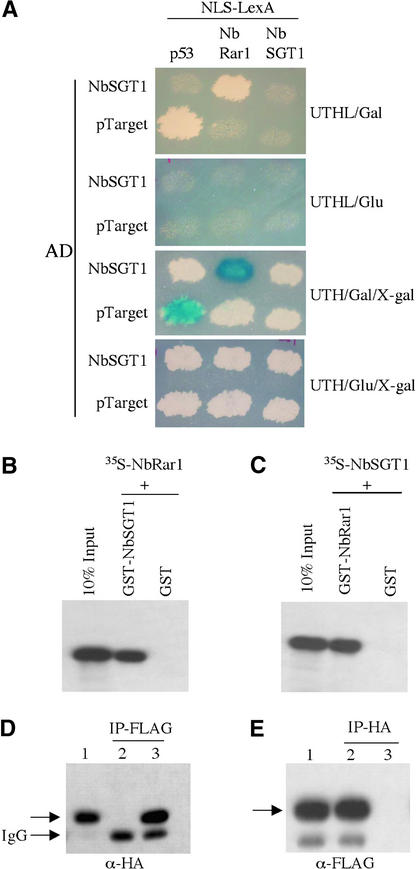
Specific interaction between full-length NbRar1 and NbSGT1 proteins was confirmed using yeast two-hybrid, in vitro pulldown, and in planta interaction assays. Yeast colonies containing both LexA-BD-NbRar1 and AD-NbSGT1 grew on Leu− plates containing Gal (Figure 2A, top) and turned dark blue on 5-bromo-4-chloro-3-indolyl β-d-galactoside plates (Figure 2A, third from top). Yeast colonies containing both LexA-BD-NbRar1 and AD-NbSGT1 did not grow on Leu− plates containing Glc (Figure 2A, second from top) and were white on Leu+ plates containing Glc and 5-bromo-4-chloro-3-indolyl β-d-galactoside (Figure 2A, bottom). Furthermore, no interaction was observed between LexA-BD-NbRar1 and control bait pLexA-p53 (Figure 2A). These results suggest that the expression of both reporter genes (Leu and LacZ) is dependent on the interaction and the Gal-inducible expression of NbRar1 and NbSGT1.
Figure 2.
Analysis of NbRar1 Interaction with NbSGT1.
(A) Growth of yeast strains containing NLS-LexA-p53 (control) or NLS-LexA-NbRar1 or NLS-LexA-NbSGT1 baits transformed with AD-NbSGT1 or AD-pTarget (control) on Leu-deficient medium containing Gal or Glc after 3 days at 30°C (top and second from top). Induction of LacZ expression was monitored on X-Gal–containing medium with Gal or Glc after 3 days at 30°C (third from top and bottom).
(B) One microgram of GST-NbSGT1 or GST was incubated with 35S-labeled NbRar1. Beads were washed and analyzed by SDS-PAGE and fluorography. As a control, 10% of input 35S-labeled NbRar1 was loaded directly on the same gel.
(C) One microgram of GST-NbRar1 or GST was incubated with 35S-labeled NbSGT1 and analyzed as in (B). As a control, 10% of input 35S-labeled NbSGT1 was loaded directly on the same gel.
(D) N. benthamiana plants were transfected with TMV expression vectors, producing FLAG-tagged NbRar1 alone (lane 2) or together with HA-tagged NbSGT1 (lane 3). Total cell extracts were normalized for protein quantity, and immunoprecipitation performed using anti-FLAG antibody–conjugated Sepharose beads. After washing, immune complexes were analyzed by SDS-PAGE followed by immunoblot analysis using an anti-HA antibody. As a control, input total cell extract containing HA-tagged NbSGT1 was loaded directly on the same gel (lane 1). IgG corresponds to the light IgG chain of the anti-FLAG antibody.
(E) N. benthamiana plants were transfected with TMV expression vectors producing HA-tagged NbSGT1 alone (lane 3) or together with FLAG-tagged NbRar1 (lane 2). The immunoprecipitation of total cell extracts using anti-HA antibody–conjugated Sepharose beads followed by immunoblot analysis using an anti-FLAG antibody was performed as described for (D). As a control, input total cell extract containing FLAG-tagged NbRar1 was loaded directly on the same gel (lane 1).
NbRar1 interaction with NbSGT1 was further confirmed by in vitro binding assays. NbRar1 and NbSGT1 were expressed in Escherichia coli as glutathione S-transferase (GST) fusion proteins and purified using glutathione-Sepharose beads. The identity of these fusion proteins was confirmed using a GST antibody (data not shown). Purified GST-NbSGT1 and GST-NbRar1 fusion proteins were mixed with 35S-radiolabeled NbRar1 or NbSGT1 translated in vitro. Analysis of the bound fraction by SDS-PAGE and autoradiography showed that GST-NbSGT1 bound to NbRar1 (Figure 2B) and GST-NbRar1 bound to NbSGT1 (Figure 2C). Neither NbRar1 nor NbSGT1 protein translated in vitro interacted with the GST control alone (Figures 2B and 2C).
We performed in planta interaction assays using a TMV-based viral expression system to further confirm the interaction between NbRar1 and NbSGT1. Hemagglutinin (HA)-tagged NbSGT1 and FLAG-tagged NbRar1 were expressed alone or together by transfecting N. benthamiana leaves with TMV-based viral vectors. Immunoblot analysis of total protein extracts from the leaves transfected with HA-tagged NbSGT1 or FLAG-tagged NbRar1 detected single protein bands of the expected size for NbSGT1 (Figure 2D, lane 1) and NbRar1 (Figure 2E, lane 1), respectively.
Immunoprecipitation of protein extracts derived from cotransfected leaves using FLAG monoclonal antibody followed by immunoblot analysis with anti-HA antibody detected NbSGT1 (Figure 2D, lane 3). This finding demonstrates that NbSGT1 and NbRar1 proteins interact in plant cells. In a reciprocal experiment, the HA antibody immunoprecipitated NbRar1 (Figure 2E, lane 2). This interaction is specific because control immunoprecipitation experiments performed using tissue transfected with TMV vector carrying FLAG-tagged NbRar1 alone (Figure 2D, lane 2) and TMV vector carrying HA-tagged NbSGT1 alone (Figure 2E, lane 3) failed to detect any protein bands.
NbSTG1 Interacts Directly with the SCF Component NbSKP1
In yeast and human, SGT1 interacts with SKP1, a component of the SCF-type E3 ubiquitin ligase complex (Kitagawa et al., 1999; Matsuzawa and Reed, 2001). Therefore, we performed yeast two-hybrid library screening to search for NbSGT1-interacting partners. Our two-hybrid screen yielded 20 NbSGT1-interacting partners: 14 were homologs of Rar1, and 6 clones encoded an unknown protein. Because our screen failed to identify SKP1, we tested direct interaction between SKP1 and NbSGT1.
We cloned NbSKP1, which encodes a protein of 153 amino acids and shares substantial homology with Arabidopsis SKP1/ASK1 (74% similarity and 83% identity) and ASK2 (67% identity and 79% similarity) (Figure 3). Homology with other Arabidopsis SKP1-like proteins ranges from 35 to 57% identity and 52 to 71% similarity. NbSKP1 bears homology with human SKP1 (58% identity and 71% similarity) and yeast SKP1 (46% identity and 67% similarity).
Figure 3.
Comparison of the N. benthamiana SKP1 Amino Acid Sequence with Those of Its Homologs from Other Organisms.
Alignment of the predicted N. benthamiana SKP1 (NbSKP1) protein with its homologs from Arabidopsis (AtSKP1 and AtSKP2), yeast (ScSKP1), and human (hSKP1). Numbers at left indicate the positions of the amino acid residues. Identical residues are shaded in black, and similar residues are shaded in gray.
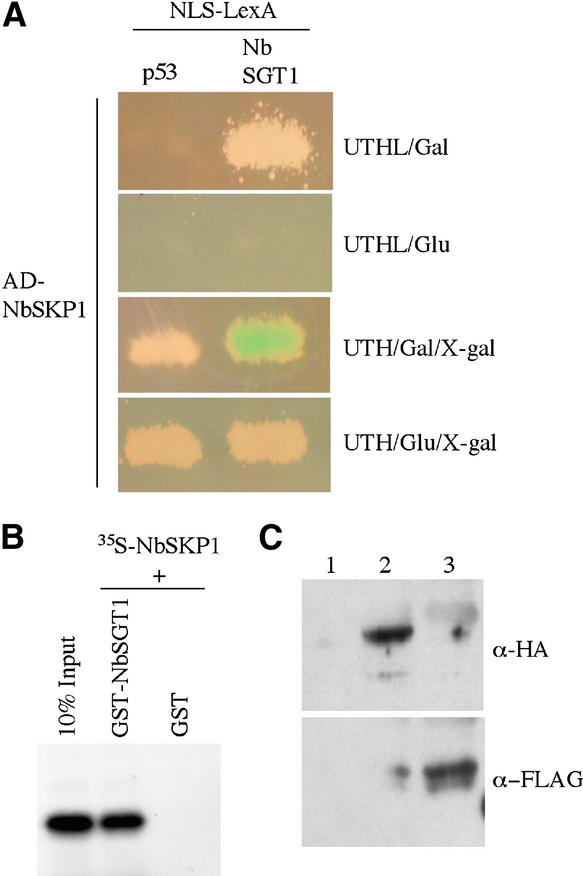
NbSKP1 interacted specifically with NbSGT1 in a yeast two-hybrid assay (Figure 4A). This interaction was confirmed by in vitro binding assays using 35S-labeled NbSKP1 and purified GST-NbSGT1 fusion protein (Figure 4B). These results show that NbSGT1 interacts directly with the SCF protein complex component NbSKP1, as its counterparts do in yeast and human.
Figure 4.
Analysis of NbSGT1 Interaction with NbSKP1.
(A) Yeast strains containing NLS-LexA-NbRar1 or NLS-LexA-p53 (control) bait were transformed with AD-NbSKP1. Growth on Leu-deficient medium containing Gal or Glc was examined for 3 days at room temperature (top and second from top). Induction of LacZ expression was monitored on X-Gal–containing medium with Gal or Glc for 3 days at room temperature (third from top and bottom).
(B) One microgram of GST-NbSGT1 or GST was incubated with 35S-labeled NbSKP1. Beads were washed and analyzed by SDS-PAGE and fluorography. As a control, 10% of input 35S-labeled NbSKP1 was loaded directly on the same gel.
(C) N. benthamiana plants were transfected with TMV expression vector carrying Myc tag alone (lane1), TMV expression vector producing Myc-tagged NbSKP1 and HA-tagged NbSGT1 (lane 2), or TMV expression vector producing Myc-tagged NbSKP1 and FLAG-tagged NbRar1 (lane 3). The immunoprecipitation of total cell extracts using anti-Myc antibody–conjugated Sepharose beads followed by immunoblot analysis using anti-HA (top) and anti-FLAG (bottom) antibodies was performed as described for Figure 2D.
We performed in planta interaction assays using a TMV-based viral expression system to further confirm the interaction between NbSKP1 and NbSGT1 or NbRar1. Myc-tagged NbSKP1 and HA-tagged NbSGT1 or FLAG-tagged NbRar1 were coexpressed by transfecting N. benthamiana leaves. Immunoprecipitation of protein extracts derived from cotransfected leaves using anti-Myc antibody followed by immunoblot analysis with anti-HA or anti-FLAG antibodies detected NbSGT1 (Figure 4C, top, lane 2) and NbRar1 (Figure 4C, bottom, lane 3), respectively. Control immunoprecipitation experiments performed using tissue transfected with TMV vector carrying Myc tag alone failed to detect any protein bands (Figure 4C, top and bottom, lane 1). These results suggest that NbSKP1 exists in a complex with NbSGT1 and NbRar1 in planta.
NbRar1 and NbSGT1 Interact with the Components of SCF in Vivo
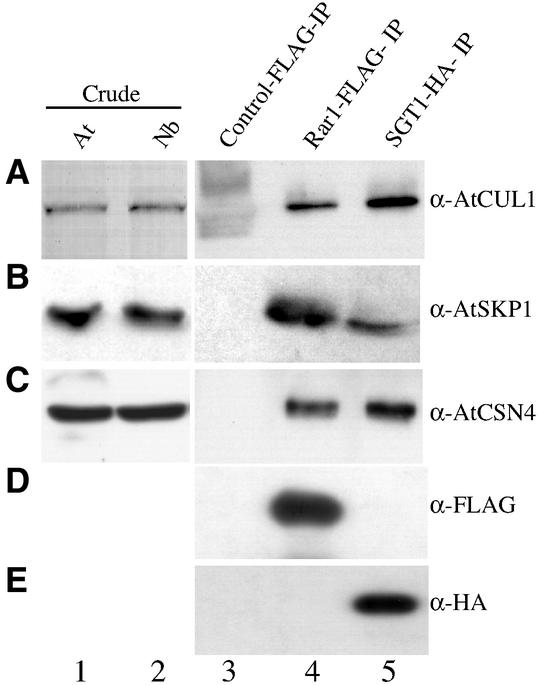
SGT1 associates with the SCF-type E3 ubiquitin ligase complex proteins SKP1 and Cdc53p (a homolog of Cullin1) in yeast (Kitagawa et al., 1999). This prompted us to investigate whether NbRar1 and NbSGT1 associate with components of the SCF ubiquitin ligase complex in planta. We explored the hypothesis that Cullin1 (CUL1) and NbSKP1 coimmunoprecipitate with NbRar1 and NbSGT1. For this purpose, we used Arabidopsis Cullin1 (AtCUL1) and AtSKP1 antibodies because they immunoreact with N. benthamiana Cullin1 (NbCUL1) (Figure 5A, lane 2) and NbSKP1 (Figure 5B, lane 2), respectively.
Figure 5.
NbRar1 and NbSGT1 Association with SCF and the COP9 Signalosome.
N. benthamiana plants were transfected with empty TMV expression vector carrying FLAG tag alone (lane 3), TMV expression vector producing FLAG-tagged NbRar1 (lane 4), or TMV expression vector producing HA-tagged NbSGT1 (lane 5). Total cell extracts were normalized for protein quantity, and immunoprecipitation was performed using anti-FLAG or anti-HA antibody–conjugated Sepharose beads. After washing, immune complexes were eluted using FLAG or HA peptides. The resulting complex was analyzed by SDS-PAGE followed by immunoblot analysis using anti-AtCUL1 (A), anti-AtSKP1 (B), anti-AtCSN4 (C), anti-FLAG (D), and anti-HA (E) antibodies. As a control, whole cell extracts from Arabidopsis (At; lane 1) and N. benthamiana (Nb; lane 2) plants were analyzed by immunoblotting.
First, we transfected FLAG-tagged Rar1 and HA-tagged SGT1 into N. benthamiana leaves using a TMV-based viral expression vector. Total protein extracts derived from these transfected leaves were immunoprecipitated using anti-FLAG or anti-HA antibodies linked covalently to agarose beads and probed with anti-AtCUL1 or anti-AtSKP1 antibodies. Our results suggest that NbRar1 and NbSGT1 exist in a complex with NbCUL1 (Figure 5A, lanes 4 and 5) and NbSKP1 (Figure 5B, lanes 4 and 5). Control immunoprecipitations with TMV vector carrying FLAG tag alone did not detect NbCUL1 (Figure 5A, lane 3) or NbSKP1 (Figure 5B, lane 3).
NbRar1 and NbSGT1 Interact with the COP9 Signalosome in Vivo
Recently, the Cullin1 subunit of the SCF complex has been shown to interact with the COP9 signalosome (CSN) (Lyapina et al., 2001). The COP9 signalosome was discovered originally in Arabidopsis as a suppressor of photomorphogenesis (Wei and Deng, 1999). The eukaryotic COP9 signalosome is a multiprotein complex with eight distinct subunits (CSN1 to CSN8) that is involved in protein degradation via the ubiquitin proteasome pathway (Wei and Deng, 1999). We tested NbRar1 and NbSGT1 interaction with all eight subunits of the COP9 signalosome using the yeast two-hybrid system. None of the individual subunits interacted with NbRar1 or NbSGT1 (data not shown).
We performed immunoprecipitation assays to investigate whether NbRar1 and NbSGT1 form a complex with the COP9 signalosome in vivo. Immunoblot analysis of total protein extract derived from N. benthamiana leaves detected NbCSN4 using AtCSN4 antibodies (Figure 5C, lanes 1 and 2). We probed the immunocomplex of NbRar1 and NbSGT1 with anti-AtCSN4 antibodies and observed the association of NbCSN4 with NbRar1 and NbSGT1 (Figure 5C, lanes 4 and 5). Control immunoprecipitation with TMV vector carrying the FLAG tag alone did not detect NbCSN4 (Figure 5C, lane 3). These results suggest that NbRar1 and NbSGT1 associate with the COP9 signalosome because the CSN4 subunit is always present in the COP9 multiprotein complex.
Suppression of NbSGT1 and NbSKP1 Leads to the Loss of N-Mediated Resistance to TMV
We used a Tobacco rattle virus (TRV)–based virus-induced gene silencing (VIGS) technique as a gene knockout system to examine the biological role of NbSGT1 and NbSKP1 in N-mediated resistance to TMV (Ratcliff et al., 2001; Liu et al., 2002). VIGS is initiated when a plant is infected with recombinant TRV carrying an insert homologous with a host gene. Endogenous gene transcripts homologous with the VIGS vector insert are degraded by a post-transcriptional gene silencing mechanism or by cosuppression (Baulcombe, 1999). VIGS is a rapid, transient assay with the phenotype of the silenced plant mimicking a loss-of-function phenotype.
We performed VIGS to silence NbSGT1 and NbSKP1 genes in transgenic N. benthamiana lines containing the tobacco N gene (referred to as NN). Transgenic NN plants confer resistance to TMV, as well as to recombinant TMV carrying the green fluorescent protein (TMV-GFP), by inducing the hypersensitive response at the site of infection and containing virus at the infection site (Liu et al., 2002). To suppress NbSGT1 and NbSKP1 using TRV-VIGS, we cloned fragments of these genes into pTRV2 (RNA2 cDNA of TRV) as described in Methods. Mixtures of Agrobacterium tumefaciens cultures containing pTRV1 (RNA1 cDNA of TRV) with pTRV2, pTRV2-N, pTRV2-NbSGT1, or pTRV2-NbSKP1 T-DNAs were infiltrated onto NN plants at the four-leaf stage.
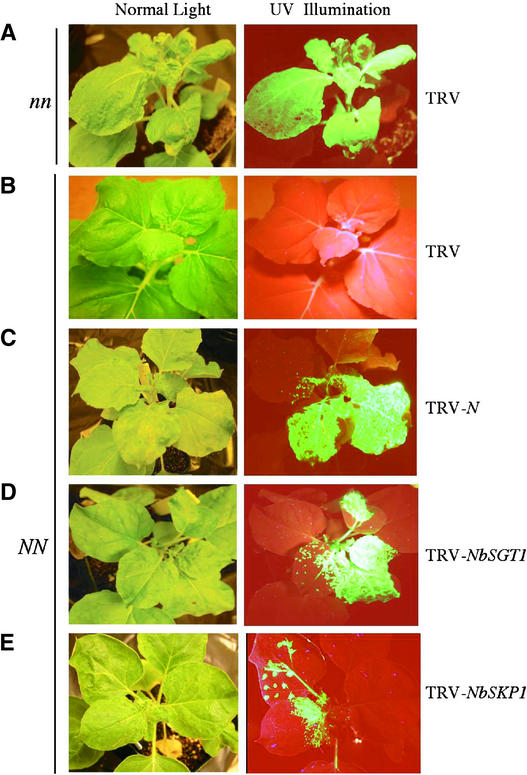
Ten days after infiltration, the upper leaves of these plants were infected with TMV-GFP. These plants were visualized under UV light for at least 20 days to observe whether TMV-GFP spread from the infection site. In wild-type N. benthamiana plants (referred to as nn) infected with TRV alone, TMV-GFP spread to the upper leaves (Figure 6A). In NN plants, TMV-GFP was restricted to the infection site (Figure 6B). Interestingly, in NbSGT1- and NbSKP1-suppressed NN plants, TMV-GFP spread into the upper leaves (Figures 6D and 6E). Loss of resistance to TMV-GFP in these plants was similar to that observed in NN plants suppressed for the N transgene (Figure 6C). Therefore, the phenotypic data of the VIGS assays suggest that the NbSGT1 and NbSKP1 genes are required for N gene–mediated TMV resistance.
Figure 6.
Analysis of the Effect of NbSGT1 and NbSKP1 Suppression on N Gene–Mediated Resistance to TMV.
Wild-type nn (A) and transgenic NN ([B] to [E]) N. benthamiana plants were first silenced for N (C), NbSGT1 (D), and NbSKP1 (E) at the four-leaf stage. (A) and (B) show nonsilenced controls. The upper leaves of these plants then were infected with TMV-GFP virus to monitor resistance or susceptibility responses. The spread of TMV-GFP from the inoculated leaf into the upper uninoculated leaves indicates loss of resistance to TMV.
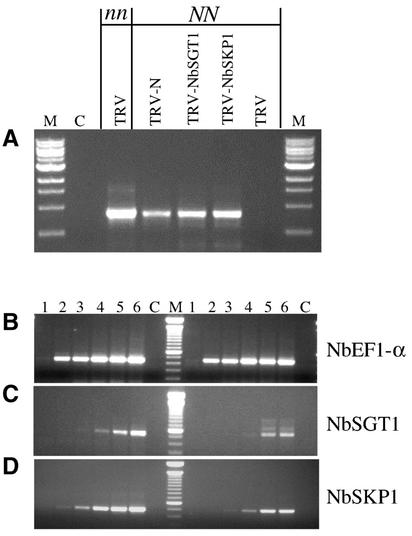
We confirmed the presence of TMV-GFP RNA in the upper uninfected leaves of N-, NbSGT1-, and NbSKP1-silenced plants by RT-PCR analysis using primers that anneal to the TMV movement protein gene (Figure 7A). No TMV-RNA was detected in NN control plants infected with TRV alone (Figure 7A).
Figure 7.
Molecular Analysis of NbSGT1 and NbSKP1 Suppression.
(A) RT-PCR results for the presence of TMV-GFP RNA in NbSGT1- and NbSKP1-suppressed plants. First-strand cDNA was synthesized using total RNA from N-, NbSGT1-, and NbSKP1-silenced NN plants and nonsilenced (nn and NN) plants. This cDNA was used in PCR to amplify the TMV movement protein (M) gene. For the control reaction (C), the RT mix without reverse transcriptase was used as a template.
(B) to (D) RT-PCR to confirm the silencing of NbSGT1 and NbSKP1 expression. First-strand cDNA was synthesized using total RNA isolated from NbSGT1- and NbSKP1-silenced NN plants. This cDNA was used in PCR with gene-specific primers.
(B) Typical PCR products for EF1α derived from NbSGT1-silenced (right) and nonsilenced (left) plants.
(C) PCR products of NbSGT1 derived from NbSGT1-silenced (right) and nonsilenced (left) plants.
(D) PCR products of NbSKP1 derived from NbSKP1-silenced (right) and nonsilenced (left) plants.
Lanes 1 to 6 correspond to products from PCR cycles 15, 18, 21, 24, 27, and 30. Lane C represents the control reaction, in which the RT mix without reverse transcriptase was used as a template.
We performed semiquantitative RT-PCR to examine the effects of VIGS-induced suppression on the endogenous amounts of mRNA for the NbSGT1 and NbSKP1 genes. In NbSGT1- and NbSKP1-silenced plants, the SGT1 and SKP1 messages were reduced by >82 and 71%, respectively, compared with the TRV-infected control (Figures 7C and 7D). However, EF1α RNA levels were similar (Figure 7B, TRV-NbSGT1) and served as an internal control for RNA quality and RT-PCR amplification.
Suppression of the COP9 Signalosome Leads to the Loss of N-Mediated Resistance to TMV
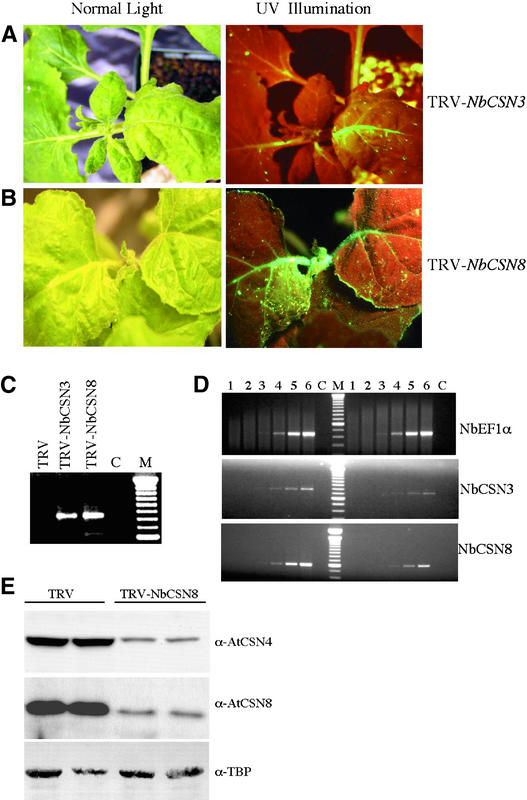
We used the VIGS assay described above to examine the biological role of the COP9 signalosome in N-mediated resistance to TMV. Suppression of the NbCSN3 and NbCSN8 genes in NN plants resulted in the spread of TMV-GFP to the upper leaves (Figures 8A and 8B). We confirmed the presence of TMV-GFP RNA in these upper leaves (Figure 8C). RT-PCR analysis found 79% reduction of NbCSN3 and 72% reduction of NbCSN8 endogenous mRNA levels (Figure 8D).
Figure 8.
Analysis of the Effect of COP9 Signalosome Suppression on the N-Mediated Resistance to TMV.
(A) and (B) Transgenic NN plants were first silenced for NbCSN3 (A) or NbCSN8 (B) at the four-leaf stage. The upper leaves of these plants were infected with TMV-GFP virus to monitor resistance or susceptibility responses as described for Figure 6.
(C) RT-PCR to detect TMV-GFP RNA in NbCSN3- and NbCSN8-suppressed plants, as described for Figure 7A.
(D) RT-PCR to confirm the silencing of NbCSN3 and NbCSN8 expression, as described for Figures 7B to 7D. The top shows typical PCR products for EF1α derived from NbCSN3-silenced (right) and nonsilenced (left) plants. The middle shows PCR products of NbCSN3 derived from NbCSN3-silenced (right) and nonsilenced (left) plants. The bottom shows typical PCR products of NbCSN8 derived from NbCSN8-silenced (right) and nonsilenced (left) plants. Lanes 1 to 6 correspond to products from PCR cycles 15, 18, 21, 24, 27, and 30. Lane C represents the control reaction, in which the RT mix without reverse transcriptase was used as a template.
(E) NbCSN4 and NbCSN8 silencing results at the protein level. Total protein extracted from nonsilenced and NbCSN8-silenced plants was normalized for protein quantity and analyzed by SDS-PAGE followed by immunoblot analysis using anti-AtCSN4 and anti-AtCSN8 antibodies. As a loading control, an anti-TATA box binding protein (TBP) antibody was used.
Studies from COP9 signalosome mutants suggest that the loss of one subunit of the CSN results in the loss of the entire protein complex (Schwechheimer et al., 2001). We tested the level of the COP9 signalosome subunits CSN8 and CSN4 in NbCSN8-silenced plants. Total protein was extracted from NbCSN8-silenced and nonsilenced NN plants, and immunoblot analyses using anti-AtCSN4 and anti-AtCSN8 antibodies were performed. We observed ∼75 and 70% reduction of NbCSN8 and NbCSN4 subunit levels, respectively, in NbCSN8-silenced plants compared with nonsilenced NN plants (Figure 8E). Together, our phenotypic and molecular data suggest that suppression of the COP9 signalosome compromises the N-mediated resistance response to TMV.
Arabidopsis SGT1b Rescues the NbSGT1 Suppression Phenotype
SGT1 is a highly conserved component of the SCF-type E3 ubiquitin ligase complex in eukaryotes (Kitagawa et al., 1999; Lyapina et al., 2001; Matsuzawa and Reed, 2001). In fact, human SGT1 and SIP complement yeast sgt1 mutant defects (Kitagawa et al., 1999; Matsuzawa and Reed, 2001). Because NbSGT1 has similarity to two Arabidopsis SGT1-like genes (AtSGT1a and AtSGT1b), we tested whether these Arabidopsis genes can complement the NbSGT1 suppression phenotype in NN plants. We overexpressed AtSGT1a and AtSGT1b using a Potato virus X (PVX) expression system. NN plants were coinfiltrated with a mixture of Arabidopsis containing pTRV-NbSGT1 (silencing construct) and pPVX-AtSGT1a-2xHA or pPVX-AtSGT1b-2xHA (overexpression constructs).
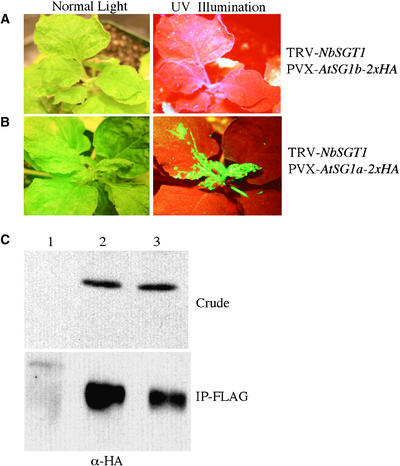
Ten days after infiltration, these plants were challenged with TMV-GFP. AtSGT1b fully rescued the loss-of-resistance phenotype induced by the suppression of NbSGT1 (cf. Figures 9A and 6D). However, AtSGT1a failed to complement the suppression effect of NbSGT1, because we observed TMV-GFP on the upper, uninfected leaves of the plants containing pPVX-AtSGT1a-2xHA (Figure 9B). Infection of pPVX-AtSGT1a-2xHA and pPVX-AtSGT1b-2xHA alone in NN and wild-type nn plants had no effect on TMV-GFP resistance or susceptibility (data not shown).
Figure 9.
Conserved SGT1 Function in Plant Defense.
(A) and (B) Transgenic NN plants were first silenced for NbSGT1. In these plants, AtSGT1b (A) and AtSGT1a (B) were overexpressed using a PVX-based expression vector. The upper leaves of these plants were infected with TMV-GFP virus to monitor resistance or susceptibility responses, as described for Figure 6.
(C) N. benthamiana plants were transfected with the TMV expression vector carrying FLAG tag alone (lane 1) or the TMV expression vector producing HA-tagged AtSGT1a (lane 2) and HA-tagged AtSGT1b (lane 3). Whole cell extracts from transfected plants were analyzed by immunoblotting using HA antibodies (top). Total cell extracts were normalized for protein quantity and immunoprecipitation using anti-FLAG antibody–conjugated Sepharose beads. After washing, immune complexes were eluted using FLAG peptide. The resulting complex was analyzed by SDS-PAGE followed by immunoblot analysis with anti-HA antibodies (bottom).
To exclude the possibility that rescue of the suppression phenotype by AtSGT1b is not attributable simply to the lack of suppression of endogenous NbSGT1, we performed semiquantitative RT-PCR. We found that NbSGT1 is suppressed in these plants (data not shown). In addition, we reasoned that the region targeted for NbSGT1 silencing shows only 71% homology with AtSGT1a and AtSGT1b at the nucleotide level; therefore, TRV-NbSGT1 does not suppress the PVX constructs. We confirmed the expression of AtSGT1a and AtSGT1b proteins using HA antibodies (Figure 9C, top). In addition, our immunoprecipitation experiments suggest that AtSGT1a and AtSGT1b associate with NbRar1 (Figure 9C, bottom). Together, these results suggests that the function of SGT1b is highly conserved in plants and plays a role in plant defense. In addition, AtSGT1a may function in different defense pathways or have some other function.
DISCUSSION
SCF Function in TMV Resistance
We report here that a CHORD-containing NbRar1 protein that is required for the N-mediated resistance to TMV interacts with NbSGT1 in yeast two-hybrid and in vitro coimmunoprecipitation assays. These results are consistent with the most recent findings by Azevedo et al. (2002) in that HvRar1 has been shown to interact directly with HvSGT1. SGT1 is a novel, highly conserved component of the SCF-type E3 ubiquitin ligase complex (Kitagawa et al., 1999; Lyapina et al., 2001; Matsuzawa and Reed, 2001). SGT1, which plays an important role in kinetochore function in yeast, forms complexes with the other SCF components SKP1 and Cdc53p, a Cullin1 homolog (Kitagawa et al., 1999). In human, an SGT1 homolog, SIP, which regulates β-catenin levels in the Wnt signaling pathway, also forms a complex with SKP1 (Matsuzawa and Reed, 2001).
We show that NbSGT1 associates physically with NbSKP1 in vitro and in vivo. Suppression of NbSGT1 and NbSKP1 in NN transgenic plants by VIGS resulted in the loss of resistance to TMV. This finding indicates that NbSGT1 and NbSKP1 are required for N function. VIGS- or RNA interference-like gene knockout systems are known to silence highly homologous genes. This makes the interpretation of our results difficult. To overcome this shortcoming, we overexpressed SGT1 homologs from heterologous Arabidopsis in NbSGT1-silenced NN plants.
Arabidopsis has two SGT1-like genes, AtSGT1a and AtSGT1b (Austin et al., 2002; Azevedo et al., 2002). Our N. benthamiana genomic DNA gel blot analysis suggested that NbSGT1 is a single-copy gene (data not shown). Consistent with this observation, we recovered only a single sequence from our RT-PCR cloning effort of NbSGT1. In addition, all seven clones derived from our tomato two-hybrid library also belong to a single sequence. Of the two Arabidopsis SGT1-like genes, only AtSGT1b was able to rescue the NbSGT1 suppression phenotype.
It is not clear why AtSGT1a failed to rescue the suppression phenotype even though NbSGT1 shares 60% identity with AtSGT1b and AtSGT1a at the amino acid level. Interestingly, Austin et al. (2002) recently reported that AtSGT1b is required for the function of another TIR-NB-LRR–containing RPP5 resistance gene in Arabidopsis. Even though AtSGT1a and AtSGT1b are highly similar, the presence of a functional copy of AtSGT1a in a AtSGT1b mutant background failed to provide RPP5-mediated resistance to Peronospora parasitica.
Together, our results from protein–protein interactions, the suppression phenotype, and its rescue indicate that NbSGT1 is required for N gene function. We did not attempt to rescue the NbSKP1 suppression phenotype because there are 19 SKP1-like genes in Arabidopsis. In our VIGS of NbSKP1, we cannot exclude the possibility of the silencing of other SKP1-like genes. Therefore, the data presented here indicate that the NbSKP1-like gene is required for the function of the N gene. Thus, SCF seems to play an important role in plant defense, in addition to the auxin response in plants.
COP9 Signalosome Function in TMV Resistance
The role of the COP9 signalosome in plant photomorphogenesis is well established (Wei and Deng, 1999). Because mutations in the CSN subunits of the COP9 signalosome result in lethality, it is difficult to investigate the role of the COP9 signalosome in other signal transduction processes. However, recently, Schwechheimer et al. (2001) made a nonlethal phenotype of CSN5 in an antisense transgenic plant, showing that the COP9 signalosome plays an important role in the degradation of the SCFTIR substrate PSIAA6 in the auxin response. VIGS, used as a reverse genetics approach as described here, provides a new way to study the function of a gene whose knockout leads to a lethal phenotype.
Recent evidence indicates that the subunits of the COP9 signalosome interact directly with components of the SCF-type E3 ubiquitin ligase complex (Lyapina et al., 2001). Because NbSKP1 is a component of the SCF complex and interacts with NbSGT1, we tested whether NbSGT1 interacts with subunits of the COP9 signalosome using a yeast two-hybrid assay. We found no interaction between NbSGT1 and any of the eight subunits of the COP9 signalosome. In human, Cullin1 (also a subunit of the SCF) has been shown to interact strongly with the CSN2 subunit and weakly with the CSN6 subunit in yeast two-hybrid assays, but the SKP1 component of SCF fails to interact with any subunit of the COP9 signalosome.
However, in vivo, the SCF components Cullin1, SKP1, HRT1/Rbx1, and SKP2 (F-box protein) associate with the COP9 signalosome (Lyapina et al., 2001). In addition, SKP2 also associates with SGT1 and hsp90β. These results prompted us to test the association of NbSGT1 with the COP9 signalosome in vivo. Our studies using immune complexes from plant cells suggest that NbSGT1 associates with the COP9 signalosome. Interestingly, a recent report from Azevedo et al. (2002) indicates that HvSGT1 also associates with the COP9 signalosome. In addition, suppression of the CSN3 and CSN8 subunits of the COP9 signalosome resulted in loss of resistance to TMV. Our results provide evidence for the new role of the COP9 signalosome as an integral part of plant defense.
What Is the Role of SCF Ubiquitin Ligase and the COP9 Signalosome In Plant Defense?
The SCF-type E3 ubiquitin protein ligases act with E2 enzymes to ubiquitinate short-lived cytoplasmic and nuclear proteins and target them for degradation (Hochstrasser, 2000). Recent evidence indicates that the COP9 signalosome plays an important role in mediating SCF-type E3 ubiquitin ligase function. In the SCF model, Cullin and Rbx1/Hrt/Roc (RING finger proteins) first dock SKP1 with an E2 enzyme, then SKP1 binds to an F-box protein, which then binds the substrate that is targeted for degradation. Therefore, in SCF, a variable F-box protein component serves as a substrate receptor for ubiquitination and subsequent degradation (Deshaies, 1999).
There are >300 F-box proteins in plants (Arabidopsis Genome Initiative, 2000). Only a few plant F-box proteins, such as TIR1 (Gray et al., 1999), COI1 (Xie et al., 1998), UFO (Samach et al., 1999), ZTL (Somers et al., 2000), and FKF1 (Nelson et al., 2000), have been studied. COI1 is involved in defense, and others are involved in developmental processes. Moreover, only TIR1 has been shown to assemble into SCF complexes in vivo.
Based on the model discussed above, R proteins may relay signals through Rar1 and SGT1 into a plant SCF complex for ubiquitin-mediated protein degradation. Our results suggest that Rar1 interacts directly with SGT1 and exists in a functional SCF complex. However, it is not clear whether Rar1 is an additional component of the SCF that participates in SCF ubiquitination activity or is simply an interaction partner of SGT1. SGT1 is found in the SKP2 (F-box) complex from human (Lyapina et al., 2001). However, it was not detected in the Grr1 (F-box) complex in yeast and does not participate in SCF ubiquitination activity in vitro (Kitagawa et al., 1999). Therefore, it is unclear at present whether SGT1 is a core component of the SCF ligase complex or participates independently. As shown here and in other systems, SGT1 seems to interact with potentially functional forms of SCF complexes.
Unlike yeast SGT1, plant and human SGT1 proteins contain three TPR repeats at the N terminus. TPR domains play an important role in protein–protein interactions that mediate binding to other proteins (Lamb et al., 1995). Interestingly, in our yeast two-hybrid assays, NbRar1 interacts with the C-terminal domain (amino acid residues 134 to 370) of NbSGT1 but not with the N-terminal domain (residues 1 to 133) that contains TPR (Y. Liu and S.P. Dinesh-Kumar, unpublished observation). It is possible that the TPR domain of NbSGT1 interacts with proteins that target Rar1 and SGT1 to the functional SCF complex. In fact, in the anaphase-promoting E3 ligase complex, many TPR-containing proteins (Apc3/Cdc27, Apc6/Cdc16, and Apc8/Cdc23) form an independent complex and are predicted to serve as a scaffold for the anaphase-promoting E3 ligase complex (Jackson et al., 2000).
What are the targets of protein degradation in plant defense? In the SCF-mediated protein degradation pathways, SKP1 binds to F-box proteins, which in turn bind to the specific substrate that is targeted for degradation (Hochstrasser, 2000). Recent evidence suggests that Skp1 or Skp1 homologs also can bind to non-F-box–containing proteins such as Rav1 (Seol et al., 2001) and adenovirus E4orf6 (Querido et al., 2001). Further studies will be required to determine whether F-box proteins or other candidate proteins involved in the Rar1-SGT1-SKP1 complex function in plant defense.
It is possible that this complex may recruit target substrates, presumably negative regulators of defense response, and subject them to degradation through the COP9 signalosome via the 26S proteasome. These negative regulators may be inhibitors of cell death, transcription factors that are required for the induction of defense response genes, or other regulatory proteins.
In the future, it will be interesting to determine to what extent the role of protein degradation in plant defense is general. It is necessary to determine the roles of SGT1, SCF, and the COP9 signalosome in other R gene–mediated resistance signaling pathways. Identification of the SCF targets for plant defense using interaction cloning, affinity purification of protein complexes followed by mass spectrometry, and VIGS knockout approaches will increase our understanding of the role of the SCF-type E3 ligase and the COP9 signalosome in plant disease resistance signaling.
METHODS
Plasmid Construction
Yeast Two-Hybrid Vectors
The LexA DNA binding domain containing bait vectors was prepared by cloning full-length NbRar1 and NbSGT1 PCR products into pTBS1, in which the nuclear localization signal sequence LexA DNA binding domain is under the control of a GAL10 promoter (T. Burch-Smith and S.P. Dinesh-Kumar, unpublished data). The B42 DNA activation domain (AD) containing prey vectors was prepared by cloning full-length NbSKP1 and NbSGT1 into pJG4-5 (Kolonin et al., 2000). Plasmids pLexA-p53 and pAD-Target were purchased from Origene Technologies (Rockville, MD).
Bacterial Expression Vectors
NbSGT1 and NbRar1 were amplified by PCR and recombined into pDEST15 using the GATEWAY system (Invitrogen, Carlsbad, CA) to express glutathione S-transferase (GST)–tagged fusion proteins in Escherichia coli.
T7 Promoter–Based Vectors for in Vitro Translation
NbSGT1, NbRar1, and NbSKP1 were amplified by PCR and recombined into pDEST14 using the GATEWAY system.
Tobacco mosaic virus–Based Expression Vectors
pYL257, a Tobacco mosaic virus (TMV)–based expression vector, was created by cloning the attR1-Cmr-ccdB-attR2 PCR product from GATEWAY conversion system cassette B into TMV binary vector pSPDK661 (Liu et al., 2002). pTMV-NbSGT1-2xHA, pTMV-NbRar1-2xFLAG, and pTMV-NbSKP1-2xMyc were created by recombining full-length PCR products of NbSGT1 with 2 × hemagglutinin (HA) tag, NbRar1 with 2 × FLAG tag, and NbSKP1 with 2 × Myc tag, respectively, into pYL257 using the GATEWAY system.
Potato virus X–Based Expression Vectors
pSPDK658, a Potato virus X (PVX) T-DNA vector containing the full-length PVX cDNA from pCambia2300 (from David Baulcombe, Sainsbury Lab, Norwich, UK), was cloned into pYL41, a pBin19 derivative carrying the duplicated 35S promoter of Cauliflower mosaic virus and nopaline synthase terminator to give pSPDK658. attR1-Cmr-ccdB-attR2 was amplified by PCR from GATEWAY conversion system cassette B and cloned into pSPDK658 to give pYL254, a PVX-based expression vector. pPVX-AtSGT1a-2xHA and pPVX-AtSGT1b-2xHA were generated by recombining full-length Arabidopsis thaliana SGT1a and SGT1b, respectively, with 2 × HA tag into pYL254 using the GATEWAY system.
Tobacco rattle virus 2 Derivatives
cDNA fragments of putative Nicotiana benthamiana SGT1 (nucleotides 1 to 479), SKP1 (nucleotides 1 to 411), CSN3 (nucleotides 43 to 582), and CSN8 (nucleotides 1 to 298) were amplified from cDNA clones and cloned into pTRV2 (Liu et al., 2002). pTRV2-N has been described (Liu et al., 2002). The identities of the vectors described above were confirmed by sequencing. Primer sequences used for this purpose will be available upon request.
Yeast Two-Hybrid Screen and Interaction Assays
The yeast two-hybrid prey library was constructed in pJG4-5. A cDNA library construction kit (Stratagene) was used to generate cDNAs using poly(A)+ RNA isolated from tissues taken from TMV-resistant (VF36::NN) (Whitham et al., 1996) and TMV-susceptible (VF36) plants 1, 4, and 8 h after TMV infection. The cDNA library was fused to the B42 AD under the control of a Gal-inducible promoter (GAL1). The yeast two-hybrid screen and interaction assays were performed as described (Kolonin et al., 2000).
Isolation of cDNAs from N. benthamiana
Total RNA was extracted from N. benthamiana leaves using RNAwiz solution (Ambion, Austin, TX), and single-stranded cDNA was prepared from 2 μg of total RNA with Superscript II reverse transcriptase (Invitrogen) and oligo(dT) primer. Full-length cDNAs of putative NbCullin1, NbCSN3, NbCSN4, and NbCSN8 were amplified by PCR with primer combinations based on putative tomato Cullin1 (TC84853), putative tomato CSN3 (TC94454), putative tomato CSN4 (BG134323 and TC91324), and CSN8 (AW030374 and AW624151). The 5′ and 3′ ends of NbSGT1 were cloned using the SMRT rapid amplification of cDNA ends kit (Clontech, Palo Alto, CA). PCR products were cloned into pCR-TOPO vector (Invitrogen), and their identities were verified by DNA sequencing. Primer sequences used for this purpose will be available upon request.
GST Pulldown Assays
GST-NbSGT1 and GST-NbRar1 fusion proteins were produced in BL21 codon plus cells (Stratagene) and affinity purified using glutathione-Sepharose beads. Approximately 1 μg of purified GST fusion proteins were incubated for 3 h with 35S-Met–labeled in vitro–translated proteins (TNT; Promega) at 4°C in 0.2 mL of buffer A (100 mM NaCl, 50 mM Tris-Cl, pH 7.5, 0.1 mM EDTA, 0.1 mM EGTA, 50 mM NaF, 0.2% Triton X-100, 0.1% β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride, and complete protease inhibitor). The beads were washed four times with ice-cold buffer B (100 mM NaCl, 50 mM Hepes, pH 7.5, 0.1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, and complete protease inhibitor) at 4°C. The washed beads were boiled in SDS sample buffer, and proteins were separated by SDS-PAGE. Gels were fixed, fluorographed using Amplify solution (Amersham Pharmacia), dried, and exposed to x-ray film.
Immunoprecipitation
Total protein from nontransfected and transfected leaves was extracted using the protocol described in Serino et al. (1999). Immunoprecipitations were performed using EZview Red anti-FLAG M2 agarose (Sigma) and anti-HA antibody affinity matrix (CRP, Berkeley, CA) at 4°C overnight. After washing four times with PBS solution, proteins were eluted using 3 × FLAG or HA peptide (Sigma). Immune complexes were analyzed by SDS-PAGE and immunoblotting using various antibodies and were detected using Westpico SuperSignal enhanced chemiluminescence (Pierce).
Virus-Induced Gene Silencing Assay, Green Fluorescent Protein Imaging, and Detection of TMV by Reverse Transcriptase–Mediated PCR
Wild-type nn and transgenic NN plants were grown in pots at 25°C in a growth chamber under a 16-h-light/8-h-dark cycle. For the virus-induced gene silencing assay, pTRV1 or pTRV2 and its derivatives were introduced into Agrobacterium tumefaciens strain GV2260. Agrobacterium cultures (OD600 = 0.8) containing pTRV1 or pTRV2 derivative plasmids were mixed at a 1:1 ratio and infiltrated into the lower leaves of four-leaf-stage plants using a 1-mL needleless syringe. Ten days after TRV infiltration, the upper leaves of the plants were infected with TMV–green fluorescent protein virus. In the overexpression experiments, pPVX derivatives and pTRV derivatives were coinfiltrated. Each silencing experiment was repeated at least five times, and each experiment included at least four independent plants. Green fluorescent protein imaging was performed using UV illumination, and photographs were taken using an Olympus Camedia E10 digital camera (Tokyo, Japan).
To detect the presence of TMV, total RNA extracted from upper uninoculated leaves was used to generate the first-strand cDNA using a primer that anneals to the 3′ end of the movement protein gene of TMV. This first strand was used in PCR reactions to amplify the movement protein gene, and the resulting PCR products were analyzed by agarose gel electrophoresis.
Analysis of Transcript Levels by Reverse Transcriptase–Mediated PCR
Total RNA was extracted from N. benthamiana leaves and treated using the Message Clean Kit (Genhunter, Houston, TX) to remove DNA contamination. This RNA (1 μg) was used to synthesize the first-strand cDNA. Samples from each reaction (1.5 μL) were used in a 30-μL PCR mixture containing the cDNA template and Taq DNA polymerase. Primers that anneal outside the region of the virus-induced gene silencing target sequence were used. The sequences of primers used in PCR will be available upon request. PCR was performed for 15, 18, 21, 24, 27, and 30 cycles. Control PCR (using RNA without reverse transcription) was performed for 30 cycles. The intensities of the PCR bands were analyzed and quantified using Gel Doc 2000 and Quantity One Version 4.2.1 (Bio-Rad).
Accession Numbers
The accession numbers for Arabidopsis SGT1a and SGT1b are A439975 and AF43976, respectively.
Acknowledgments
We thank Janet Stewart for editing the manuscript. We thank the members of our laboratory for comments and critical reading of the manuscript. We thank Barbara Baker for VF36::NN transgenic tomato seeds, David Baulcombe for vector pPC2S (T7-PVX), Tessa Burch-Smith for vector pTBS1, and William Crosby for AtSKP1 antibodies. The COP9 signalosome work in X.-W.D.'s laboratory was supported by National Science Foundation Grant MCD-0077217 and U.S.–Israel Binational Agricultural Research and Development Fund Grant IS-3123-99. This work in S.P.D.-K.'s laboratory was supported by National Institutes of Health Grant RO1 GM62625-01 and National Science Foundation Grant DBI-0077510.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.002493.
References
- Aarts, N., Metz, M., Holub, E., Staskawicz, B.J., Daniels, M.J., and Parker, J.E. (1998). Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc. Natl. Acad. Sci. USA 95, 10306–10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative. (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815. [DOI] [PubMed] [Google Scholar]
- Austin, M.J., Muskett, P., Kahn, K., Feys, B.J., Jones, J.D.G., and Parker, J.E. (2002). Regulatory role of SGT1 in early R gene-mediated plant defense. Science 295, 2077–2080. [DOI] [PubMed] [Google Scholar]
- Azevedo, C., Sadanandom, A., Kitagawa, K., Freialdenhoven, A., Shirasu, K., and Schulze-Lefert, P. (2002). The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295, 2073–2076. [DOI] [PubMed] [Google Scholar]
- Baker, B., Zambryski, P., Staskawicz, B., and Dinesh-Kumar, S.P. (1997). Signaling in plant-microbe interactions. Science 276, 726–733. [DOI] [PubMed] [Google Scholar]
- Baulcombe, D.C. (1999). Fast forward genetics based on virus-induced gene silencing. Curr. Opin. Plant Biol. 2, 109–113. [DOI] [PubMed] [Google Scholar]
- Dangl, J.L., and Jones, J.D.G. (2001). Plant pathogens and integrated defense responses to infection. Nature 411, 826–833. [DOI] [PubMed] [Google Scholar]
- Deshaies, R.J. (1999). SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol. 15, 435–467. [DOI] [PubMed] [Google Scholar]
- Gabriel, D., and Rolfe, B. (1990). Working models of specific recognition in plant-microbe interactions. Annu. Rev. Phytopathol. 28, 365–391. [Google Scholar]
- Gray, W.M., del Pozo, J.C., Walker, L., Hobbie, L., Risseeuw, E., Banks, T., Crosby, W.L., Yang, M., Ma, H., and Estelle, M. (1999). Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev. 13, 1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser, M. (2000). Evolution and function of ubiquitin-like protein-conjugation systems. Nat. Cell Biol. 2, E153–E157. [DOI] [PubMed] [Google Scholar]
- Jackson, P.K., Eldridge, A.G., Freed, E., Furstenthal, L., Hsu, J.Y., Kaiser, B.K., and Reimann, J.D.R. (2000). The role of the RINGs: Substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol. 10, 429–439. [DOI] [PubMed] [Google Scholar]
- Kitagawa, K., Skowyra, D., Elledge, S.J., Harper, J.W., and Hieter, P. (1999). SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex. Mol. Cell 4, 21–33. [DOI] [PubMed] [Google Scholar]
- Kolonin, M.G., Zhong, J., and Finley, R.L. (2000). Interaction mating methods in two-hybrid systems. Methods Enzymol. 328, 26–46. [DOI] [PubMed] [Google Scholar]
- Lamb, J.R., Tugendreich, S., and Hieter, P. (1995). Tetratrico peptide repeat interactions: To TPR or not to TPR? Trends Biochem. Sci. 20, 257–259. [DOI] [PubMed] [Google Scholar]
- Liu, Y., Schiff, M., Marathe, R., and Dinesh-Kumar, S.P. (2002). Tobacco Rar1, EDS1 and NPR1/NIM1 like genes are required for N-mediated resistance to tobacco mosaic virus. Plant J. 30, 415–429. [DOI] [PubMed] [Google Scholar]
- Lyapina, S., Cope, G., Shevchenko, A., Serino, G., Tsude, T., Zhou, C., Wolf, D.A., Wei, N., Shevchenko, A., and Deshaies, R.J. (2001). Promotion of NEDD8-CUL1 conjugate cleavage by COP9 signalosome. Science 292, 1382–1385. [DOI] [PubMed] [Google Scholar]
- Matsuzawa, S.-I., and Reed, J.C. (2001). Siah-1, SIP, and Ebi collaborate in a novel pathway for β-catenin degradation linked to p53 responses. Mol. Cell 7, 915–926. [DOI] [PubMed] [Google Scholar]
- McDowell, J.M., and Dangl, J.L. (2000). Signal transduction in the plant immune response. Trends Biochem. Sci. 25, 79–82. [DOI] [PubMed] [Google Scholar]
- Nelson, D.C., Lasswell, J., Rogg, L.E., Cohen, M.A., and Bartel, B. (2000). FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 101, 331–340. [DOI] [PubMed] [Google Scholar]
- Querido, E., Blanchette, P., Yan, Q., Kamura, T., Morrison, M., Boivin, D., Kaelin, W.G., Conaway, R.C., Conaway, J.W., and Branton, P.E. (2001). Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 15, 3104–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff, F., Martin-Hernandez, A.M., and Baulcombe, D.C. (2001). Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 25, 237–245. [DOI] [PubMed] [Google Scholar]
- Rock, F.L., Hardiman, G., Timans, J.C., Kastelein, R.A., and Bazan, J.F. (1998). A family of human receptors structurally related to Drosophila Toll. Proc. Natl. Acad. Sci. USA 95, 588–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryals, J.A., Neuenschwander, U.H., Willits, M.G., Molina, A., Steiner, H.-Y., and Hunt, M.D. (1996). Systemic acquired resistance. Plant Cell 8, 1809–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach, A., Klenz, J.E., Kohalmi, S.E., Risseeuw, E., Haughn, G.W., and Crosby, W.L. (1999). The UNUSUAL FLORAL ORGANS gene of Arabidopsis thaliana is an F-box protein required for normal patterning and growth in the floral meristem. Plant J. 20, 433–445. [DOI] [PubMed] [Google Scholar]
- Schulze-Lefert, P., and Vogel, J. (2000). Closing the ranks to attack by powdery mildew. Trends Plant Sci. 5, 343–348. [DOI] [PubMed] [Google Scholar]
- Schwechheimer, C., Serino, G., Callis, J., Crosby, W.L., Lyapina, S., Deshaies, R.J., Gray, W.M., Estelle, M., and Deng, X.-W. (2001). Interaction of the COP9 signalosome with the E3 ubiquitin ligase SCFTIR1 in mediating auxin response. Science 292, 1379–1382. [DOI] [PubMed] [Google Scholar]
- Seol, J.H., Shevchenko, A., Shevchenko, A., and Deshaies, R.J. (2001). Skp1 forms multiple protein complexes, including RAVE, a regulator of V-ATPase assembly. Nat. Cell Biol. 3, 384–391. [DOI] [PubMed] [Google Scholar]
- Serino, G., Tsuge, T., Kwok, S., Matsui, M., Wei, N., and Deng, X.-W. (1999). Arabidopsis cop8 and fus4 mutations define the same gene that encodes subunit 4 of the COP9 signalosome. Plant Cell 11, 1967–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasu, K., Lahaye, T., Tan, M.-W., and Schulze-Lefert, P. (1999). A novel class of eukaryotic zinc-binding proteins is required for disease resistance signaling in barley and development in C. elegans. Cell 99, 355–366. [DOI] [PubMed] [Google Scholar]
- Somers, D.E., Schultz, T.F., Milnamow, M., and Kay, S. (2000). ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101, 319–329. [DOI] [PubMed] [Google Scholar]
- Thomma, B.P.H.J., Penninckx, I.A.M.A., Broekaert, W.F., and Cammue, B.P.A. (2001). The complexity of disease signaling in Arabidopsis. Curr. Opin. Immunol. 13, 63–68. [DOI] [PubMed] [Google Scholar]
- Wei, N., and Deng, X.-W. (1999). Making sense of the COP9 signalosome, a conserved regulatory protein complex from Arabidopsis to human. Trends Genet. 15, 98–103. [DOI] [PubMed] [Google Scholar]
- Whitham, S., Dinesh-Kumar, S.P., Choi, D., Hehl, R., Corr, C., and Baker, B. (1994). The product of the tobacco mosaic virus resistance gene N: Similarity to toll and the interleukin-1 receptor. Cell 78, 1101–1115. [DOI] [PubMed] [Google Scholar]
- Whitham, S., McCormick, S., and Baker, B. (1996). The N gene of tobacco confers resistance to tobacco mosaic virus in transgenic tomato. Proc. Natl. Acad. Sci. USA 93, 8776–8781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, D., Feys, B.F., James, S., Nieto-Rostro, M., and Turner, J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094. [DOI] [PubMed] [Google Scholar]