Abstract
Rice was chosen as a model organism for genome sequencing because of its economic importance, small genome size, and syntenic relationship with other cereal species. We have constructed a bacterial artificial chromosome fingerprint–based physical map of the rice genome to facilitate the whole-genome sequencing of rice. Most of the rice genome (∼90.6%) was anchored genetically by overgo hybridization, DNA gel blot hybridization, and in silico anchoring. Genome sequencing data also were integrated into the rice physical map. Comparison of the genetic and physical maps reveals that recombination is suppressed severely in centromeric regions as well as on the short arms of chromosomes 4 and 10. This integrated high-resolution physical map of the rice genome will greatly facilitate whole-genome sequencing by helping to identify a minimum tiling path of clones to sequence. Furthermore, the physical map will aid map-based cloning of agronomically important genes and will provide an important tool for the comparative analysis of grass genomes.
INTRODUCTION
Rice is the principal food crop of half of the world's population and also serves as a crop research system to understand yield, hybrid vigor, and disease resistance. Rice has emerged as a model system for studying cereal genomics because of its small genome size (430 Mb) (Arumuganathan and Earle, 1991), syntenic relationship with other agronomically important cereal species (Bennetzen et al., 1998; Gale and Devos, 1998), and the availability of genome resources such as well-defined genetic maps (Causse et al., 1994; Harushima et al., 1998), an extensive collection of expressed sequence tags (ESTs) (Kurata et al., 1994; Yamamoto and Sasaki, 1997; http://rgp.dna.affrc.go.jp/), the TIGR Rice Gene Index (Quackenbush et al., 2000; http://www.tigr.org/tdb/tgi.html), and a yeast artificial chromosome (YAC) map (Saji et al., 2001; http://rgp.dna.affrc.go.jp/publicdata/physicalmap99/yacall.html).
Determination of the complete genomic sequence of rice is the objective of the International Rice Genome Sequencing Project (IRGSP) (Sasaki and Burr, 2000; http://rgp.dna.affrc.go.jp/cgi-bin/statusdb/seqcollab.pl), which is led by Japan and involves Brazil, China, Great Britain, France, India, Korea, Taiwan, Thailand, and the United States. The IRGSP is using a clone-by-clone strategy to sequence the rice genome. This approach, which has proven effective for the human genome (International Human Genome Mapping Consortium, 2001; International Human Genome Sequencing Consortium, 2001), the Caenorhabditis elegans genome (Coulson et al., 1986), and the Arabidopsis genome (Marra et al., 1999; Mozo et al., 1999), relies on the identification of a tiling path composed of large-insert clones that spans a given genomic region with minimal overlaps. Therefore, a comprehensive physical map is essential for this efficient and thorough approach to genome sequencing. Furthermore, an established correlation between the physical and genetic maps also is essential for performing efficient map-based gene cloning and associating candidate genes with important biological or agronomic traits.
A YAC physical map covering 63% of the rice genome was constructed recently (Saji et al., 2001). However, instability, high chimera frequency, and difficulties in manipulation and purification make YAC clones less than ideal substrates for genome sequencing. Instead, large-insert, low-copy-number bacterial clones, namely bacterial artificial chromosomes (BACs) and P1-derived artificial chromosomes (PACs), are the substrates of choice. A recent article has reported on the fingerprinting of 21,087 BAC clones from indica rice cv Teqing (Tao et al., 2001). However, very limited genetic anchoring information is available. We have constructed two deep-coverage BAC libraries from japonica rice cv Nipponbare that were fingerprinted with HindIII and assembled into a physical map of the rice genome. Our rice physical map consists of 65,287 fingerprinted BAC clones (including 2778 singletons) thought to represent 20-fold redundant coverage of the genome. The 62,509 BAC clones have been organized into 458 ordered sets of overlapping clones (contigs), and 284 of these BAC clone contigs, estimated to contain 362.9 Mb of the rice genome, have been correlated to the genetic map. Therefore, an estimated 90.6% of the rice genome is represented by genetically anchored BAC contigs, based on an estimated genome size of ∼400 Mb. The integrated physical and genetic map can be accessed with WebFPC (Soderlund et al., 2002) at http://www.genome.clemson.edu/projects/rice/fpc/integration.
RESULTS
BAC Library Construction and Fingerprinting
Two deep-coverage BAC libraries were constructed from high-molecular-weight DNA from rice embedded in agarose plugs. The DNA was partially digested with HindIII or EcoRI, double size selected, and ligated into pBeloBAC11 or pBACIndigo, respectively. The ligation reactions were transformed into Escherichia coli, plated on selective media, and arrayed. The HindIII library consists of 36,864 clones with an average insert size of 129 kb, whereas the EcoRI library consists of 55,296 clones with an average insert size of 121 kb. Approximately 5% of the clones from each library are considered contaminants, either containing organelle DNA or no inserts altogether. The coverage for the HindIII and EcoRI BAC libraries is estimated at 10.6 and 15.0 haploid genome equivalents, respectively, thus providing 25-fold redundant coverage when combined.
A total of 73,728 BAC clones were fingerprinted using the method described by Marra et al. (1997). Briefly, purified BAC clone DNA was digested to completion with HindIII, the fragments were run on high-resolution agarose gels, and the fingerprint of each clone was formulated with IMAGE software (Sulston et al., 1989) based on the mi-gration distances of restriction fragments with extensive manual editing. A total of 65,287 BAC clones were fingerprinted successfully by these methods. The average fragment number per clone was 28. The HindIII fingerprint data were subjected to overlap analysis using the FingerPrinted Contig software package FPC version 4.7 (Soderlund et al., 2000). Automated assembly of the fingerprint data using the Sulston score cutoff of 1e-12 (scientific notation) and a fixed tolerance of 7 resulted in 1019 BAC contigs, whereas 2778 clones (4.2%) remained as singletons. This contig collection was estimated to represent 453 Mb of rice genomic DNA (based on 92,866 nonredundant bands with an average band size of 4878 bp), whereas the average contig contained 60 BAC clones representing 445 kb. This is an overestimate because of the unrecognized overlaps between the contigs. Further refinement of the rice physical map was accomplished by simultaneously anchoring BAC contigs to the rice genetic map while editing contigs manually to consolidate smaller contigs to improve the overall contiguity of the physical map resource.
Manual Editing of Contigs
Manual editing improves the physical map in two ways. First, identifying potential joins between contigs, and performing merges, increases the overall contiguity of the resource. Second, the manual editing phase identifies potential chimeric contigs by revealing incorrectly overlapped fingerprint data or by highlighting conflicting marker data. Problematic contigs were resolved by breaking them at those sites recognized by marker or fingerprint conflicts. Potential contig merges were recognized by searching the entire FPC fingerprint database for matches to fingerprints from clones representing contig termini above the Sulston score cutoff of 1e-10. Those contig pairs whose overall fingerprint patterns supported joins were merged, and the total clone order was recalculated. One round of such analysis reduced the total number of BAC contigs to 581 from the original 1069. A second round of merging using a Sulston score cutoff of 1e-08 further reduced the number of contigs to 458. Only genetically anchored contigs, whose map positions supported such actions, were considered during the second round of merging to ensure data integrity.
Anchoring of Rice BAC Contigs to the Rice Genetic Map
Four approaches to correlate the genetic and physical maps were used. First, we generated DNA probes from a subset of the genetic markers serving as landmarks on the Japanese RGP (Rice Genome Program) rice genetic map (Harushima et al., 1998; http://rgp.dna.affrc.go.jp/). DNA probes for either DNA gel blot hybridization or overgo hybridization (http://genome.wustl.edu/gsc/overgo/overgo.html) were constructed from markers selected at 3- to 5-centimorgan (cM) intervals along each of the 12 rice chromosomes (Table 1) to cover the entire genome. Additional markers were selected between the intervals to anchor additional contigs.
Table 1.
Genetic Markers, Probes, Chromosome Size, and Coverage Data
| Chromosome | Genetic Markers |
Probes (Markers Included) |
Contig No. | Previously Estimated Chromosome Size (Mb)a |
Predicted Chromosome Size (Mb)b |
Size of Anchored Contigs (Mb) |
Coverage (%) |
|---|---|---|---|---|---|---|---|
| 1 | 231 | 413 | 32 | 51.5 | 44 | 42.7 | 97 |
| 2 | 184 | 316 | 26 | 43.4 | 39.8 | 35.8 | 90 |
| 3 | 224 | 364 | 26 | 47.5 | 40.8 | 35.7 | 87.5 |
| 4 | 119 | 273 | 24 | 36.8 | 39 | 34.5 | 88.5 |
| 5 | 139 | 239 | 27 | 33.6 | 33.2 | 30.9 | 93.1 |
| 6 | 129 | 229 | 22 | 35.1 | 31.8 | 28.2 | 88.7 |
| 7 | 158 | 292 | 25 | 33.1 | 35 | 30.3 | 86.6 |
| 8 | 88 | 181 | 18 | 33.6 | 27.6 | 25.8 | 93.5 |
| 9 | 80 | 139 | 16 | 27 | 21.6 | 20.3 | 94 |
| 10 | 136 | 337 | 20 | 23.7 | 26.8 | 24.6 | 91.8 |
| 11 | 118 | 245 | 28 | 33.7 | 30.3 | 28.6 | 94.4 |
| 12 | 98 | 171 | 20 | 30.9 | 30.6 | 25.5 | 83.3 |
| Total | 1704 | 3199 | 284 | 430 | 400.5 | 362.9 | 90.6 |
Estimated chromosome size is based on the YAC map (Saji et al., 2001).
Predicted chromosome size is based on this physical map.
Second, in silico hybridization was used to anchor physical contigs genetically. We sequenced both ends of every BAC clone insert in the HindIII and EcoRI libraries and generated 110,438 sequence-tagged connectors (STCs) (Mao et al., 2000; http://www.genome.clemson.edu/projects/rice/rice_bac_end). All 110,438 STCs were used to tentatively anchor contigs based on sequence homology with sequenced restriction fragment length polymorphism (RFLP) markers detected in silico (Yuan et al., 2000). By this method, 418 rice genetic markers were associated with BAC end sequences with high confidence. Comparison of the in silico anchored data with the overgo and DNA gel blot hybridization data served to rectify conflicts and anchor additional contigs. Remaining conflicts were resolved by DNA gel blot hybridization with probes constructed from appropriate genetic markers.
Third, contig end walking was performed with overgo primer pairs (http://genome.wustl.edu/gsc/overgo/overgo.html) designed from the end sequences of clones at the termini of anchored contigs. These probes were hybridized to high-density filter sets of the HindIII and EcoRI BAC libraries to identify potentially overlapping and extending clones. Fingerprints were consulted to confirm all potential clone extensions, or contig merges, identified with end-walking probes. The end-walking effort was focused on the short arms of chromosomes 3 (0 to 55.8 cM) and 10 (0 to 30.2 cM) in support of the CCW (Clemson University, Cold Spring Harbor Laboratory, Washington University School of Medicine Genome Sequencing Center) Rice Genome Sequencing Consortium (http://www.genome.clemson.edu/projects/rice/ccw/) to sequence these regions of the rice genome.
Fourth, we integrated portions of the Monsanto draft rice genome sequence data (Barry, 2001). Associations between the sequenced Monsanto BAC clones and our rice physical map (Clemson University Genome Institute [CUGI]) were identified through in silico searches for high-quality CUGI STC matches to the Monsanto BAC clone sequence. Aggressive filtering was performed to identify and remove questionable matches attributable to known repetitive sequences; this involved finding Monsanto clones that match CUGI STCs from multiple unrelated contigs or clones or CUGI STCs that match Monsanto clones thought to be unrelated in the genome (see Methods). A total of 38,287 individual Monsanto-to-CUGI clone associations were established, representing 2146 unique Monsanto clones and 19,113 unique CUGI clones.
Of the 2146 Monsanto clones, 1636 had high-confidence associations with 1442 unique RGP genetic markers. The positions of the CUGI clones with STCs that matched Monsanto clone sequences integrate the corresponding Monsanto BAC clones into the CUGI physical map. This integration process resulted in placing one or more Monsanto clones into 174 previously anchored contigs and into an additional 43 previously unanchored contigs. Conflicts arising from the integration process were resolved manually by extensive review of the in silico anchored data and performing targeted overgo hybridization with RGP genetic markers. Integration resulted in map position conflicts for 44 contigs, of which 30 conflicts arose from errors in Monsanto clone positions. Eleven of these conflicts were simple shifts in genetic map position that arose from inaccuracies in correlating genetic marker data from two different publicly available genetic maps (RGP [http://rgp.dna.affrc.go.jp/publicdata/geneticmap2000/] and Gramene [ftp://brie.cshl.org/pub/gramene/maps/]). These conflicts were resolved to reflect the RGP genetic map position. The remaining 19 conflicts resulted from gross errors in map position assignments for 23 Monsanto clones. The accuracy of the Monsanto clone position assignments is 98% based on the conflict analysis (2056 clones accurately mapped in anchored contigs of 2093 clones mapped into anchored contigs). The integrated physical and genetic map is shown in Figure 1.
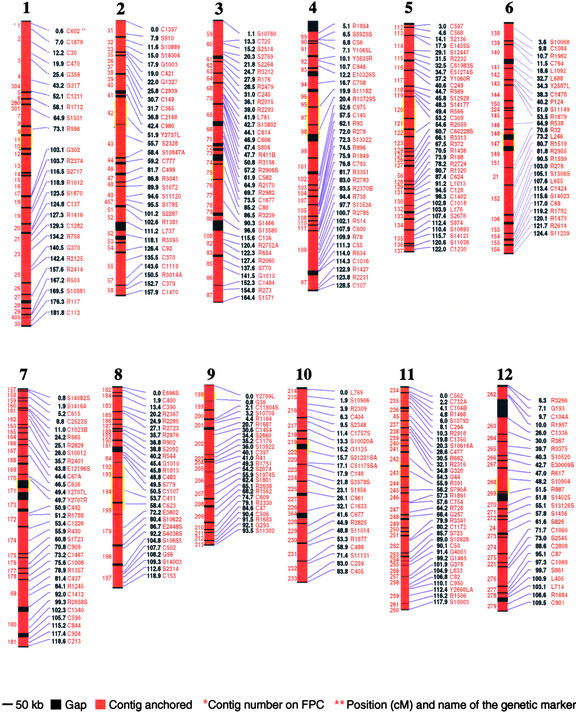
Figure 1.
BAC-Based Physical Map of the Rice Genome.
The RGP genetic markers used for anchoring are listed at right in red. Next to the RGP genetic markers are the genetic positions in centimorgans. In the middle is the physical map on scale. The anchored portions are shown in red and the gaps are shown in black. At left are the contigs in order. The positions of the centromeres are shown in yellow; they are based on the RGP genetic map (Harushima et al., 1998; Saji et al., 2001). The centromeres of chromosomes 1, 6, 7, and 9 are mapped to single genetic positions at 73.4, 65.8, 49.7, and 0.8 cM, respectively. The centromeres of chromosomes 3, 4, 5, 8, 11, and 12 are mapped in genetic intervals of 0.8 cM (85.2 to 86 cM), 3.7 cM (19.6 to 23.3 cM), 1.4 cM (53.2 to 54.6 cM), 3.5 cM (50.8 to 54.3 cM), 1.1 cM (54.8 to 55.9 cM), and 3.3 cM (48.2 to 51.5 cM), respectively. The centromere of chromosome 10 is mapped to 15.4 to 15.9 cM by FISH using a centromere-specific repeat as a probe (Cheng et al., 2001). The centromere of chromosome 2 was mapped originally to 50 to 50.3 cM (Harushima et al., 1998). However, this region (∼900 kb) is located within a single contig with a physical-to-genetic distance ratio of 369 kb/cM, whereas the neighboring contig has severely suppressed recombination (>1 Mb/cM). Therefore, we have included 50 to 54.6 cM as the centromeric region for chromosome 2.
Physical Map Accuracy
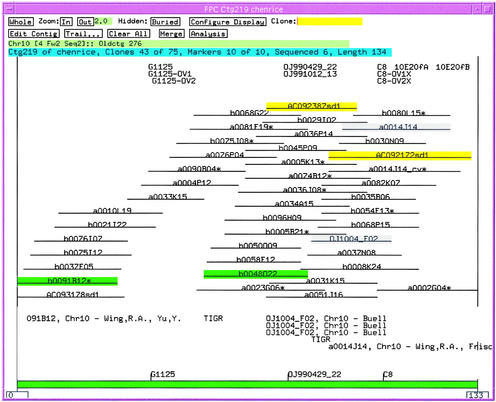
For rice chromosome 1, 346 BAC or PAC clones have been sequenced and mapped by the RGP and deposited into GenBank. In an attempt to independently assess the fidelity of the integrated physical map, these clones were digested in silico, converted to migration rates, and incorporated into our fingerprinted BAC clone physical map of rice at a Sulston score cutoff of 1e-12 (Soderlund et al., 2002). The map locations of the integrated in silico digests were in agreement with the chromosome anchoring and marker orders determined during physical map construction of their contig targets for 305 of these clones, leaving 41 singletons. Using a Sulston score cutoff of 1e-10, 23 of the remaining 41 singletons integrated into the physical map at locations consistent with the clone order and anchoring information, 1 mapped to the wrong location, and 17 remained as singletons. Among the 17 singletons, 12 could be assigned to contig termini (possible low-coverage regions) based on the RGP finished sequence information. Four of the remaining five clones, located in the middle of contigs, were between 40 and 75 kb in size and thus could be integrated only at a very low Sulston score. The final clone (OJ1316_H05) appears to be misassembled, thereby producing an aberrant in silico fingerprint. An example of this integration is shown in Figure 2. The current physical map with all integrated public clones is available at http://www.genome.clemson.edu/projects/rice/fpc.
Figure 2.
FPC Display of Contig 219.
Clones highlighted in green are in shotgun sequencing. Clones highlighted in yellow are redigested finished clones from GenBank, and the corresponding BAC clones are shown in gray.
In addition, our efforts at anchoring the physical map to chromosome 10 of the rice genetic map are in agreement with the chromosome 10 fluorescence in situ hybridization (FISH) studies by Cheng et al. (2001).
Genome Coverage
A local estimate of the degree of rice genome coverage actually represented in the physical map was obtained by examining the almost completely sequenced rice chromosome 1 (http://rgp.dna.affrc.go.jp/). The length of the pseudomolecule of chromosome 1 (nonoverlapping sequences) is ∼43 Mb, with 12 gaps (not including telomeric ends). Three gaps are located in the middle of the FPC contigs; therefore, they do not represent physical gaps in our map. The other nine gaps correspond to the same physical gaps that are found in our physical map. Six contiguous regions of the pseudomolecule cover more than one contig. STC analysis of clones located on contig ends against each corresponding ungapped region determined that only three gaps remain and the other contigs overlap, although they were separate initially, based on poor fingerprint (and clone) coverage at their junctions. The length of the three remaining gaps is ∼320 kb. Assuming that the 43-Mb nonoverlapped pseudomolecule of chromosome 1 is representative of the euchromatic portion of the rice genome, our physical map covers 99.3% of the euchromatic portion of the rice genome.
Wet bench and in silico marker anchoring analysis anchored 284 of the 458 BAC contigs representing the CUGI physical map of rice. On the basis of the number of bands derived from FPC contigs covering the 43-Mb nonoverlapping sequences of chromosome 1, we determined that the average HindIII band size in FPC is 4878 bp. Based on the length of each contig, which is the approximate number of nonredundant fragments, and the metric of 4878 bp per fragment, we calculated that 362.9 Mb of rice genomic DNA was represented in our anchored contigs. The sizes of gaps between contigs were estimated on the basis of a local ratio of physical distance to genetic distance. For regions with reduced recombination frequency, the estimation of gap sizes was based on the YAC physical map (Saji et al., 2001). On the basis of the estimated sizes of the anchored contigs and the remaining gaps in our physical map, we estimate the size of the euchromatic portion of the rice genome to be ∼400 Mb. Therefore, based on these estimates, ∼90.6% of the rice genome is represented in fingerprinted BAC clone contigs anchored to the genetic map.
Genetic Recombination
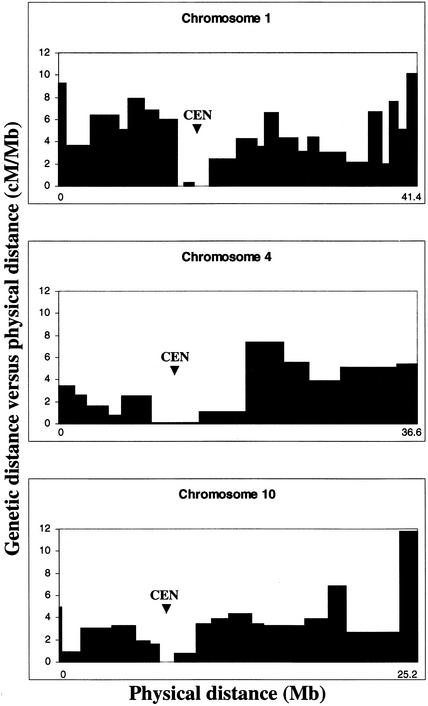
The comparison of the physical map and the genetic map has enabled us to reveal the relationship between physical distance and genetic distance for the entire rice genome. The average physical distance per centimorgan is observed to be 244 kb for the rice genome, which varies with position along the chromosome, with centromere regions exhibiting >1 Mb/cM (Figure 3). The severe reduction of recombination frequency within centromeric regions suggests that these represent sites of suppressed genetic recombination. This phenomenon also is observed on the short arms of chromosomes 4 and 10 (Figure 3). Contig 124 (1395 kb, 10.1 to 12.2 cM on chromosome 4) and contig 272 (1431 kb, 1.1 to 2.2 cM on chromosome 10) have a physical-to-genetic distance ratio of >1 Mb/cM and likely represent extensive heterochromatic regions, which may prove difficult to sequence.
Figure 3.
Genetic Recombination in Chromosomes 1, 4, and 10.
The x axis represents the physical distance in megabases along the chromosome. The y axis represents the ratio of genetic distance to physical distance (cM/Mb). The approximate centromere positions are marked with arrowheads (CEN). The physical distance intervals for which cM/Mb was estimated are based on the size of a single contig if two or more genetic markers were used in contig anchoring or the sizes of two or more contigs if a single genetic marker was used in contig anchoring. Estimated cM/Mb was extended to neighboring gaps in the physical map.
DISCUSSION
We have built a robust rice physical map. More than 65,000 BAC clones representing 20-fold coverage have been fingerprinted successfully and assembled into physical contigs. The integrity of the contig assembly and clone order has been confirmed independently by FPC Simulated Digest using sequenced BAC and PAC clones from GenBank. Approximately 90% of the rice genome has been anchored genetically. Among the genetically anchored contigs, ∼80% are anchored by two or more genetic markers and therefore are oriented properly, whereas >80% are anchored by multiple methods (i.e., marker hybridization, in silico hybridization, FISH, and sequenced clones).
On the basis of the physical map, we estimated the euchromatic portion of the rice genome to be ∼400 Mb, whereas earlier studies estimated the rice genome to be 430 Mb, based on DNA content (Arumuganathan and Earle, 1991; Saji et al., 2001). In contrast to the previous estimate of 51.5 Mb for chromosome 1 (Table 1) (Saji et al., 2001), our size estimate of 44 Mb is in agreement with that derived from the nearly completed sequence of chromosome 1 (43 Mb). The previously estimated chromosome size appears to be based on genetic distance rather than physical distance. Based on the total number of bands covered by all of the physical contigs (82,580) and the metric of 4878 bp per fragment, our physical map covers ∼403 Mb of genomic DNA, which is consistent with our estimation of 400 Mb, based on estimated contig and gap sizes. However, our estimation of the rice genome size may be an underestimate because the nuclear organizer region on the tip of chromosome 9 (Shishido et al., 2000) appears to have been excluded from our physical map (our unpublished data). Also, centromeric and other highly repetitive genomic regions tend to be compressed in fingerprint-based physical maps because of either identical HindIII fingerprints from highly repetitive regions or the absence of the HindIII restriction site from large genomic regions.
Our BAC fingerprint–based rice physical map serves as a foundation on which to organize and assist the sequencing of the rice genome and is being used for this purpose by IRGSP members. Minimal tiling paths can be selected rapidly from all unsequenced portions of the rice genome, and clones that bridge gaps can be identified from the current set of sequenced clones. Simultaneously, genomic DNA sequence data generated by the IRGSP are retrieved daily, digested in silico, converted into migration rates, and assembled into our physical map (Soderlund et al., 2002). This process has been used to check the integrity of clone order and contig assembly. Sequenced clones from other sources also can be anchored to our physical map using FPC Simulated Digest (Soderlund et al., 2002). This process also is facilitating the identification of misnamed and misassembled clones.
This physical map will influence our understanding of rice genome organization profoundly. For example, preliminary analysis has identified complete chloroplast and mitochondrial genome insertions into the rice nuclear genome (our unpublished data). Furthermore, a complete physical-genetic map of rice is necessary for comparative genomics studies with other grass genomes. We have used the Japanese RGP high-density genetic map (Harushima et al., 1998) to integrate with the physical map. More than 2000 well-mapped genetic markers are available. Many of these markers are conserved among the grass genomes because they represent expressed genes (cDNAs or ESTs). These markers can be used to integrate the physical and genetic maps of rice, sorghum, maize, and other grasses. This rice physical map can be used to build comparative physical maps of sorghum, maize, and other cereal species. High-resolution comparative physical maps will reveal regions of colinearity and rearrangement and will have important implications for the use of rice as a model system to study other important cereal species. This will facilitate map-based cloning of agronomically important genes in species with large genome sizes, such as maize, wheat, and barley, using rice as a surrogate.
We have surveyed the whole-genome genetic recombination based on the integrated physical and genetic map. Genetic recombination has been suppressed at centromeric regions as well as on the short arms of chromosomes 4 and 10. However, the degree of suppression is more similar to that observed in the genome of Arabidopsis (Schmidt et al., 1995) than to that observed in the genomes of wheat, barley, or maize. Recombinationally inactive regions in rice are limited to a few megabases. However, in the wheat genome, most genes are clustered in the distal regions of chromosomes, although the large centromeric regions (as large as 100 Mb) are gene poor and recombinationally inactive (Gill et al., 1996). Translocation studies have suggested that this phenomenon occurs in the maize genome as well (Coe et al., 1988).
We will continue to update our physical map as sequencing progresses and more anchoring information becomes available. Further refinements of the physical map can be accessed with WebFPC at http://www.genome.clemson.edu/projects/rice/fpc. One potential improvement is through additional contig anchoring using simple sequence repeats. Through data mining of the rice STC database, >3000 simple sequence repeats have been identified, of which ∼90% are located on genetically anchored contigs (our unpublished data). The remaining simple sequence repeats (10%) are located on contigs not yet anchored genetically; thus, they can be used as markers to place these contigs onto the genetic map.
In summary, this paper represents a publicly funded account of a whole-genome BAC physical map of the rice genome integrated extensively with the genetic map. This resource, estimated to cover 90.6% of the rice genome in genetically anchored overlapping BAC fingerprint contigs, will be invaluable for the ongoing genome sequencing project and will serve as a framework in support of the rice genome sequencing project. The extensive correlation of this resource to the genetic map will aid the map-based cloning of agronomically important genes in rice, revealing themes in genome organization and accelerating the functional analysis of genes in other cereal species.
METHODS
Bacterial Artificial Chromosome Library Construction, Fingerprinting, and Contig Assembly
The bacterial artificial chromosome (BAC) vectors pBeloBAC11 and pBACIndigo were used to construct the HindIII and EcoRI libraries, respectively, and were prepared as described previously (Woo et al., 1994). Megabase plant DNA embedded in agarose plugs was obtained from 4- to 5-week-old greenhouse-grown rice seedlings (Oryza sativa ssp japonica cv Nipponbare) as described by Peterson et al. (2000) using option Y for plant tissues containing low levels of secondary compounds. Partial digestion of megabase DNA (using HindIII or EcoRI), size selection, and ligation were performed as described in detail by Peterson et al. (2000). Recombinant colonies were chosen using the Genetix Q-bot and stored individually in 384-well microtiter plates (Genetix, Hampshire, UK). BAC libraries, filters, and clones are available upon request from the Clemson University Genome Institute BAC/Expressed Sequence Tag (CUGI BAC/EST) Resource Center (http://www.genome.clemson.edu).
Fingerprinting of the BAC clones was performed according to Marra et al. (1997). Restriction fragment identification was performed using IMAGE software (Sulston et al., 1989) with extensive manual editing. Automatic assembly of the fingerprinted clones was performed as described by Soderlund et al. (2000).
Marker Hybridization
Hybridizations were performed on high-density filters containing the clones of the fingerprinted libraries. To locate the Rice Genome Program (RGP) rice genetic markers (http://rgp.dna.affrc.go.jp/publicdata/geneticmap2000) on the contigs, the cDNA clones were digested with restriction endonucleases to excise the insert, which was separated electrophoretically and gel isolated with the QIAEX II kit (Qiagen, Valencia, CA). The probes were labeled radioactively using the Ambion random priming kit (Austin, TX). In cases in which a restriction fragment length polymorphism (RFLP) clone was not available but its sequence was, two overgo primers were designed using the script Overgo maker (http://genome.wustl.edu/gsc/overgo/overgo.html). Radioactive labeling was performed as described (Ross et al., 1999). To close gaps between anchored contigs, the overgo strategy was used, but the primers were designed based on the sequence-tagged connector (STC) from the most terminal clones in the target contigs. For the cDNA probes, hybridization was performed at 65°C, and the filters were washed at the same temperature twice (15 min each) in 1 × SSC (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate) and 0.1% SDS and once (15 to 20 min) in 0.1 × SSC and 0.1% SDS. For overgo probes, hybridization was performed at 60°C, and the filters were washed at 60°C for 15 min on the rotary oven with 4 × SSC and 0.1% SDS and once for 15 to 20 min on a shaker at 60°C with 1.5 × SSC and 0.1% SDS. The labeled membranes were exposed to x-ray films or to phosphorimager screens overnight.
In Silico Anchoring
The STC search against RFLP markers was performed as described previously (Yuan et al., 2000). National Center for Biotechnology Information BLASTN (http://ncbi.nlm.nih.gov) was used to align CUGI BAC end sequences to the Monsanto rice BAC sequence resource (Barry, 2001). All matches were screened to remove those alignments that failed to meet a minimum match identity of 95% over a minimum of 100 bp. Map information associated with both the Monsanto BAC resource and the CUGI BAC end sequence resource was consulted in an attempt to further screen for erroneous matches. Any CUGI STC sequence that aligned to more than two nonoverlapping Monsanto BACs (overlap based on their relationship within the Monsanto physical map fingerprinted contigs, or sequence overlap) was discarded. Likewise, alignments between a Monsanto BAC sequence and STCs from more than two unrelated CUGI contigs also were discarded. Furthermore, as a result of the high redundancy associated with the CUGI contigs, we required a given Monsanto BAC sequence to hit multiple STCs from overlapping CUGI clones before we considered integrating it into the target contig. Therefore, integration of a Monsanto BAC into a CUGI contig was considered only if it exhibited sequence overlap with a minimum of 12 overlapping or neighboring clones from within the same contig. This requirement was reduced to nine if the clones that were hit clustered at the end of a contig.
Acknowledgments
We thank Dr. Takuji Sasaki and the Japanese Ministry of Agriculture, Forestry, and Fisheries DNA Bank for the rice RFLP markers used in this study. This work was supported by grants from the U.S. Department of Agriculture, the National Science Foundation, the Department of Energy Rice Genome Sequencing Project (U.S. Department of Agriculture, Cooperative State Research, Education, and Extension Service Grant No. 99-3517-8505), and the Rockefeller Foundation. Any opinions, findings, and conclusions or recommendations expressed herein are those of the authors and do not necessarily reflect the views of the U.S. Department of Agriculture, the National Science Foundation, or the Rockefeller Foundation. We thank Novartis and Steve Goff for financial support to generate the BAC fingerprint data and preliminary contig analysis. Work at the John Innes Centre was funded by the Biotechnology and Biological Sciences Research Council through Investigating Gene Function Initiative Grant No. 208/IGF12449. The Indian Initiative on Rice Genome Sequencing is supported by a grant from the Department of Biotechnology, Government of India.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.010485.
References
- Arumuganathan, K., and Earle, E.D. (1991). Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9, 208–218. [Google Scholar]
- Barry, G. (2001). The use of the Monsanto draft rice genome sequence in research. Plant Physiol. 125, 1164–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen, J.L., SanMiguel, P., Chen, M.S., Tikhonov, A., Francki, M., and Avramova, Z. (1998). Grass genomes. Proc. Natl. Acad. Sci. USA 95, 1975–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causse, M.A., et al. (1994). Saturated molecular map of the rice genome based on an interspecific backcross population. Genetics 138, 1251–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Z., Presting, G.G., Buell, C.R., Wing, R.A., and Jiang, J. (2001). High-resolution pachytene chromosome mapping of bacterial artificial chromosomes anchored by genetic markers reveals the centromere location and the distribution of genetic recombination along chromosome 10 of rice. Genetics 157, 1749–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coe, E.H., Neuffer, M.G., and Hoishington, D.A. (1988). The genetics of corn. In Corn and Corn Improvement, 3rd ed, G.F. Sprague and J.W. Dudley, eds (Madison, WI: American Society of Agronomy/Crop Science Society of America/Soil Science Society of America), pp. 81–236.
- Coulson, A., Sulston, J., Brenner, S., and Karn, J. (1986). Toward a physical map of the genome of the nematode Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 83, 7821–7825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale, M.D., and Devos, K.M. (1998). Plant comparative genetics after 10 years. Science 282, 656–659. [DOI] [PubMed] [Google Scholar]
- Gill, K.S., Gill, B.S., Endo, T.R., and Taylor, T. (1996). Identification and high-density mapping of gene-rich regions in chromosome group 1 of wheat. Genetics 144, 1883–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harushima, Y., et al. (1998). A high-density rice genetic linkage map with 2275 markers using a single F2 population. Genetics 148, 479–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata, N., et al. (1994). A 300 kilobase interval genetic-map of rice including 883 expressed sequences. Nat. Genet. 8, 365–372. [DOI] [PubMed] [Google Scholar]
- International Human Genome Mapping Consortium (2001). A physical map of the human genome. Nature 409, 934–941. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium (2001). Initial sequencing and analysis of the human genome. Nature 409, 860–920.11237011 [Google Scholar]
- Mao, L., Wood, T.C., Yu, Y.S., Budiman, M.A., Tomkins, J., Woo, S.S., Sasinowski, M., Presting, G., Frisch, D., Goff, S., Dean, R.A., and Wing, R.A. (2000). Rice transposable elements: A survey of 73,000 sequence-tagged-connectors. Genome Res. 10, 982–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra, M., Dewar, K., Dunn, P., Ecker, J.R., Fischer, S., Kloska, S., Lehrach, H., Marra, M., Martienssen, R., Meier-Ewert, S., and Altmann, T. (1997). High throughput fingerprint analysis of large-insert clones: Contig construction and selection of clones for DNA-sequencing. Genome Res. 7, 1072–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marra, M., et al. (1999). A map for sequence analysis of the Arabidopsis thaliana genome. Nat. Genet. 22, 265–270. [DOI] [PubMed] [Google Scholar]
- Mozo, T., Dewar, K., Dunn, P., Ecker, J.R., Fischer, S., Kloska, S., Lehrach, H., Marra, M., Martienssen, R., Meier-Ewert, S., and Altmann, T. (1999). A complete BAC-based physical map of the Arabidopsis thaliana genome. Nat. Genet. 22, 271–275. [DOI] [PubMed] [Google Scholar]
- Peterson, D.G., Tomkins, J.P., Frisch, D.A., Wing, R.A., and Paterson, A.H. (2000). Construction of plant bacterial artificial chromosome (BAC) libraries: An illustrated guide. J. Agric. Genomics 5, (www.ncgr.org/research/jag).
- Quackenbush, J., Liang, F., Holt, I., Pertea, G., and Upton, J. (2000). The TIGR gene indices: Analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res. 28, 141–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, M., LaBrie, T., McPherson, S., and Stanton, V.P. (1999). Screening large-insert libraries by hybridization. In Current Protocols in Human Genetics, A. Boyl, ed (New York: Wiley), pp 5.6.1–5.6.32.
- Saji, S., Umehara, Y., Antonio, B., Yamane, H., Tanoue, H., Baba, T., Aoki, H., Ishige, N., Wu, J.Z., Koike, K., Matsumoto, T., and Sasaki, T. (2001). A physical map with yeast artificial chromosome (YAC) clones covering 63% of the 12 rice chromosomes. Genome 44, 32–37. [DOI] [PubMed] [Google Scholar]
- Sasaki, T., and Burr, B. (2000). International Rice Genome Sequencing Project: The effort to completely sequence the rice genome. Curr. Opin. Plant Biol. 3, 138–141. [DOI] [PubMed] [Google Scholar]
- Schmidt, R., West, J., Love, K., Lenehan, Z., Lister, C., Thompson, H., Bouchez, D., and Dean, C. (1995). Physical map and organization of Arabidopsis thaliana chromosome-4. Science 270, 480–483. [DOI] [PubMed] [Google Scholar]
- Shishido, R., Sano, Y., and Fukui, F. (2000). Ribosomal DNAs: An exception to the conservation of gene order in rice genomes. Mol. Gen. Genet. 263, 586–591. [DOI] [PubMed] [Google Scholar]
- Soderlund, C., Humphray, S., Dunham, A., and French, L. (2000). Contigs built with fingerprints, markers and FPC V4.7. Genome Res. 10, 1772–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderlund, C., Engler, F., Hatfield, J., Blundy, S., Chen, M., Yu, Y., and Wing, R. (2002). Mapping sequence to rice FPC. In Computational Biology and Genome Informatics, P. Wang, J. Wang, and C. Wu, eds (World Scientific Publishing), in press.
- Sulston, J., Mallett, F., Durbin, R., and Horsnell, T. (1989). Image analysis of restriction enzyme fingerprint autoradiograms. Comput. Appl. Biosci. 13, 101–106. [DOI] [PubMed] [Google Scholar]
- Tao, Q., Chang, Y.L., Wang, J.Z., Chen, H.M., Islam-Faridi, M.N., Scheuring, C., Wang, B., Stelly, D.M., and Zhang, H.B. (2001). Bacterial artificial chromosome-based physical map of the rice genome constructed by restriction fingerprint analysis. Genetics 158, 1711–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo, S.S., Jiang, J., Gill, B.S., Patterson, A.H., and Wing, R.A. (1994). Construction and characterization of a bacterial artificial chromosome library for Sorghum bicolor. Nucleic Acids Res. 22, 4922–4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, K., and Sasaki, T. (1997). Large-scale EST sequencing in rice. Plant Mol. Biol. 35, 135–144. [PubMed] [Google Scholar]
- Yuan, Q., Liang, F., Hsiao, J., Zismann, V., Benito, M.I., Quackenbush, J., Wing, R., and Buell, R. (2000). Anchoring of rice BAC clones to the rice genetic map in silico. Nucleic Acids Res. 28, 3636–3641. [DOI] [PMC free article] [PubMed] [Google Scholar]