Abstract
Hansenula polymorpha ass3 mutants are characterized by the accumulation of inactive alcohol oxidase (AO) monomers in the cytosol, whereas other peroxisomal matrix proteins are normally activated and sorted to peroxisomes. These mutants also have a glutamate or aspartate requirement on minimal media. Cloning of the corresponding gene resulted in the isolation of the H. polymorpha PYC gene that encodes pyruvate carboxylase (HpPyc1p). HpPyc1p is a cytosolic, anapleurotic enzyme that replenishes the tricarboxylic acid cycle with oxaloacetate. The absence of this enzyme can be compensated by addition of aspartate or glutamate to the growth media. We show that HpPyc1p protein but not the enzyme activity is essential for import and assembly of AO. Similar results were obtained in the related yeast Pichia pastoris. In vitro studies revealed that HpPyc1p has affinity for FAD and is capable to physically interact with AO protein. These data suggest that in methylotrophic yeast pyruvate carboxylase plays a dual role in that, besides its well-characterized metabolic function as anapleurotic enzyme, the protein fulfils a specific role in the AO sorting and assembly process, possibly by mediating FAD-binding to AO monomers.
INTRODUCTION
The yeast Hansenula polymorpha is able to use methanol as sole carbon and energy source. Growth on this compound is accompanied by the induction of peroxisomes that contain the key enzymes of methanol metabolism. Alcohol oxidase (AO) is a major constituent of these organelles and catalyzes the oxidation of methanol into formaldehyde and hydrogen peroxide. Inactive AO monomers are synthesized in the cytosol and posttranslationally imported into the target organelle, where the protein is activated. The active enzyme is an octamer, containing eight identical subunits, which each contains a FAD molecule as cofactor (reviewed by van der Klei et al., 1991a).
Both in vivo and in vitro experiments suggested that assembly of AO into active octamers is most likely not a spontaneous process (Distel et al., 1987; van der Klei et al., 1989b). Several independent experiments suggested that specific helper proteins (tentatively called assembly factors) are required to mediate AO assembly. Studies on an H. polymorpha riboflavin (Rf) auxotrophic mutant revealed that Rf limitation interfered with the assembly and the import of AO (Evers et al., 1994) and suggested that cofactor binding, oligomerization, and translocation of AO are tightly coupled processes. However, in all H. polymorpha peroxisome-deficient (pex) mutants analyzed so far AO is normally assembled and active in the cytosol. This suggests that AO assembly does not require the specific (acidic) microenvironment of the peroxisomal matrix (van der Klei et al., 1991c).
Previous biochemical approaches to identify AO assembly factors failed so far. We therefore sought to isolate these components by a genetic approach. To this end we have isolated a collection of mutants that displayed reduced AO activities (van Dijk et al., 2002).
Here, we report the functional complementation of one of these mutants. We show that the protein product of the complementing gene, pyruvate carboxylase, has a dual function in that the protein, but not the enzyme activity, is crucial for sorting and subsequent assembly of AO protein in peroxisomes of H. polymorpha.
MATERIALS AND METHODS
Organisms and Growth Conditions
Escherichia coli strains DH5α and C600 were cultivated as described (Sambrook et al., 1989). The H. polymorpha strains used in this study are NCYC 495 (leu1.1), NCYC 495 (leu1.1 ura3; Gleeson and Sudbery, 1988) and mutants derived from these strains: ass3-110.leu 1.1 (van Dijk et al., 2002), Δpex3 ura3 (Baerends et al., 1996), Δpyc1 leu1.1, Δpex3 Δpyc1, and Δpyc1.PAMOPYC.
H. polymorpha cells were grown on minimal media containing 0.67% (wt/vol) Yeast Nitrogen Base without amino acids (Difco, Sparks, MD) containing 1% glucose (YND) or 0.5% methanol (YNM); on YPD containing 1% yeast extract, 1% peptone, and 1% glucose or mineral medium (van Dijken et al., 1976) supplemented with 0.5% carbon source and 0.25% nitrogen source. For the induction of peroxisomes mutant strains were precultivated in YPD medium and shifted to methanol-containing mineral medium for 16 h. To accumulate monomeric AO in the cytosol of Δpyc1::PAMOPYC, cells were grown for 6 h on media containing 0.1% glycerol/0.5% methanol/0.25% ammonium sulfate. Subsequently, the cells were incubated for 30 min in media without carbon or nitrogen sources (Waterham et al., 1993) followed by transfer to mineral media containing 0.5% glucose and 0.25% ethylamine.
Pichia pastoris wild-type MP 36 (his3)and MP 36-Δpyc(pyc::his3) (Menendez et al., 1998) were cultivated as described by Faber et al. (1998).
When needed uracil (20 mg/l), leucine (20 mg/l), histidine (40 mg/l), aspartate (60 mg/l), or glutamate (60 mg/l) were added to the media.
Isolation and Characterization of the Pyruvate Carboxylase (PYC1) Gene
Genetic manipulations of H. polymorpha were performed as described previously (Gleeson and Sudbery, 1988; Faber et al., 1992; Titorenko et al., 1993; Faber et al., 1994) Standard recombinant DNA techniques were carried out essentially as described (Sambrook et. al, 1989). Endonuclease restriction enzymes and biochemicals were obtained from Roche (Almere, the Netherlands) and used as detailed by the manufacturer. To clone the complementing genomic fragment, mutant ass3-110 was transformed with an H. polymorpha genomic library (Tan et al., 1995). Leucine prototrophs were tested on YNM plates for the ability to grow on methanol (Mut+). Mut+ transformants were selected. Their plasmid content was isolated and reintroduced in mutant ass3-110. Four plasmids that complemented ass3-110 again were selected for further analysis. These plasmids contained overlapping genomic fragments ranging in size from 6.5 to 9.0 kb. A 4.2-kb complementing DNA fragment was subcloned as an EcoRI-XbaI fragment into pBluescript II KS+ (pBSII KS+; Stratagene Inc., San Diego, CA). Sequencing of both strands was carried out on a LiCor automated DNA-sequencer using dye-primer chemistry (LI-COR Inc., Lincoln, NE). For DNA and amino acid sequence analysis, the PC-GENETM program (Release 6.70; IntelliGenetics, Mountain View, CA) was used. The TBLASTN algorithm (Altschul et al., 1997) was used to search the databases at the National Center for Biotechnology Information (Bethesda, MD). The nucleotide sequence of H. polymorpha PYC1 (HpPYC1) was deposited at GenBank and was assigned Accession No. AF 221670.
Construction of Mutants
A PYC1 disruption strain was constructed as follows: the H. polymorpha URA3 gene (Merckelbach et al., 1993) was isolated as a BglII (blunt ended)–PstI fragment and ligated into pBKS-PYC (StyI [blunt ended]–PstI fragment). From this plasmid (pBKS-ΔPYC::URA3) a 3.1-kb BglII fragment was isolated and used to transform H. polymorpha NCYC 495 (leu1.1 ura3) or pex3::LEU2 (ura3 Δpex3). Transformants were selected for uracil prototrophy and inability to grow on minimal methanol media. Correct integration was confirmed by Southern blotting.
A mutated HpPYC1 gene (R316Q), in which the codon encoding residue 316 (arginine) was substituted by glutamine, was constructed by overlap extension PCR (Horton et al., 1990) using primer PYC-1 (5′GACATTATTTCATCGAAATTAATCCTCAGATCCAGGTC GAGCACACC3′) and both pBKS-40 universal (MF) and -50 reverse primers (MR). From the PCR product a 0.3-kb PstI/NarI fragment was exchanged with the same fragment in pBKS-PYC, resulting in pBKS-PYCR316Q. For introduction into H. polymorpha NCYC 495 (leu1.1 Δpyc1) plasmid pYT3-PYCR316Q was constructed by ligation of the full-length mutated HpPYC1 gene (XbaI/EcoRI blunt ended) from the pBKS plasmid into pYT3 (XbaI/BamHI blunt ended; Tan et al., 1995).
Plasmid pHIPX5 carrying HpPYC1 under control of AMO promoter (PAMO) was constructed as follows: PYC1, amplified by PCR using primers “PYC-ATG” (CTTCCATGGCCCAGGTCG) and “PYC-STOP” (CCGCATGCGCAGAGCGAGACGC), was digested by NcoI and SphI and cloned into pHIPX5 digested with the same restriction enzymes. The resulting plasmid was linearized with BsiWI and transformed into H. polymorpha NCYC 495 Δpyc1(leu1.1). A strain in which a single copy of the expression construct was integrated was selected, based on Southern blot analysis.
Isolation of His6-tagged HpPyc1p
For isolation of C-terminal His6-tagged HpPyc1p, PYC1 was amplified by PCR, using primers “PYC-ATG” and “PYC-STOP,” digested with NcoI/PvuII and inserted into NcoI/BglII (blunt ended)–digested pQE-60 (Qiagen GmbH, Hilden, Germany), resulting in plasmid pQE60-PYC. E. coli Sq13009[pREP4] containing pQE60-PYC was grown as detailed in The QIAexpressionist. Cell pellets were suspended in a 50 mM potassium phosphate buffer, pH 7.4, containing 300 mM NaCl; 0.2 mM b-ME, 10% glycerol, 0.2 mM EDTA, and Complete (Roche). Cells were disrupted using a French press. Cell debris and other insoluble material were removed by centrifugation. The cell extract was loaded onto a 1 ml Ni2+ containing HiTrap Chelating column (Amersham Pharmacia Biotech AB, Uppsala, Sweden), washed with 10 ml buffer containing 75 mM imidazole and 150 mM NaCl in 50 mM potassium phosphate, pH 7.4, and eluted with the same buffer containing 500 mM imidazole. The peak fractions contained highly purified HpPyc1p as determined by SDS-PAGE and Coomassie brilliant blue staining (unpublished data).
Biochemical Methods
Crude extracts were prepared according to van der Klei et al.(1991b). AO activity was measured as described by Verduyn et al. (1984). AO monomers and octamers were separated by sucrose density centrifugation (Goodman et al., 1984). Protein concentrations were determined using the Bio-Rad protein assay kit (Bio-Rad Gmbh, Munich, Germany) using bovine serum albumin (BSA) as standard. The FAD content of AO was determined in immunoprecipitates by fluorescence spectroscopy as detailed previously (van der Klei et al., 1989a). SDS-PAGE (Laemmli, 1970) and Western blotting (Kyhse-Andersen, 1984) was performed as described. Blots were decorated using antisera against various H. polymorpha proteins or Saccharomyces cerevisiae pyruvate carboxylase (Rohde et al., 1991) and the chromogenic or chemiluminescent Western blotting kit (Boehringer Mannheim BV, Almere, the Netherlands). Cell fractionation was performed as described by van der Klei et al. (1998). A postnuclear supernatant (10 ml in total) was loaded onto a discontinuous sucrose gradient (25 ml). After centrifugation 1.5-ml fractions were taken from the bottom.
AO/HpPyc1p Binding Studies
AO and BSA columns were prepared as described by Evers et al. (1993). On binding the proteins were denatured by incubation for 16 h at 4°C in a buffer containing 8 M urea in 25 mM Tris-HCl, pH 7.0, followed by extensive washing with buffer A (25 mM Tris-HCl, 50 mM KCl, 1 mM DTT, pH 7.0) to remove the urea. For binding studies 100 μl purified HpPyc1p (50 μg/ml) was loaded onto 50-μl columns, followed by washing with 20 column volumes buffer A and elution with 20 column volumes of a solution containing 8 M urea. Proteins were precipitated with TCA and analyzed by Western blotting.
Fluorescence Correlation Spectroscopy
The fluorescence correlation spectroscopy (FCS) setup was basically as described by Hink and Visser (1998). For excitation of FAD, argon ion laser lines of 488 nm were used. The light intensity was adjusted by using various neutral density filters. Measurements were made in a 96-well chamber in 50 mM potassium phosphate buffer, pH 7.0, at room temperature. For the analysis of FAD binding to HpPyc1p, purified HpPyc1p (200 or 400 nM) was incubated with FAD (100 nM) for 1 h at 37°C before the measurements. The concentrations of FAD and pyruvate carboxylase were calculated based on their extinction coefficients (FAD ε450 = 11.3 mM−1 cm−1, HpPyc1p monomer ε280 = 77 mM−1 cm−1). Diffusion constants of individual fluorescent molecules were calculated from the time-dependent fluctuation of the fluorescent signal. Experimental autocorrelation curves were then fitted by theoretical autocorrelation functions using the FCS Data Processor 1.3 software. In all series of experiments the alignment and focusing of the setup was frequently checked by measuring the autocorrelation function of 7.6 nM rhodamin 110. The dimensions of the excitation volume were determined by the known diffusion coefficient of rhodamin 110.
Electron Microscopy
Whole cells were fixed and prepared for electron microscopy as described (Waterham et al., 1994). Immunolabeling was performed on ultrathin sections of Unicryl-embedded cells, using specific polyclonal antibodies against various H. polymorpha and S. cerevisiae proteins and gold-conjugated goat anti-rabbit (GAR) antibodies (Waterham et al., 1994). Cytochemical staining experiments for the detection and localization of AO activity were performed by the CeCl3-based method (Veenhuis et al., 1976).
RESULTS
The H. polymorpha Pyruvate Carboxylase Gene Functionally Complements a Mutant Defective in AO Assembly
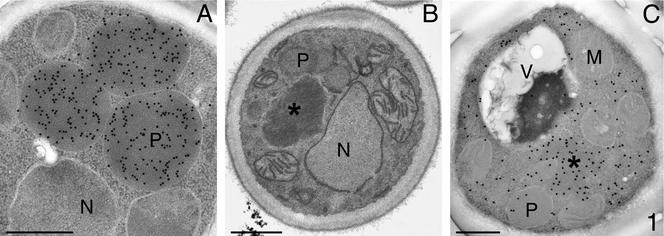
In a genetic approach to identify proteins involved AO assembly/activation, we have isolated a collection of H. polymorpha mutants that are impaired to utilize methanol as sole carbon source (Mut− phenotype) because of strongly reduced or absent AO activities (Van Dijk et al., 2002). Four mutants were characterized by normal AO protein levels but strongly reduced AO enzyme activities. Localization studies revealed that the import of AO into peroxisomes was specifically blocked in these mutants (Van Dijk et al., 2002). The overall morphology of methanol-induced cells of a representative strain of these mutants, ass3-110, compared with WT cells, is shown in Figure 1.
Figure 1.
Morphology and immunocytochemistry of methanol-induced cells of WT H. polymorpha (Figure 1A) and ass3-110 cells (B and C). WT cells (A) are characterized by the presence of several large peroxisomes that harbor AO protein (α-AO antibodies). Only small peroxisomes are found in cells of the mutant strain (B; KMnO4-fixation). Electron micrographs are taken of glutaraldehyde-fixed cells, poststained with uranylacetate unless otherwise indicated. M, mitochondrion; N, nucleus; P, peroxisome, V, vacuole, *AO protein aggregate. Bar, 0.5 μm.
Growth experiments revealed that these four mutants also displayed, in addition to the Mut− phenotype, a severe growth defect on minimal glucose-ammonium sulfate media, which could be restored by addition of aspartate or glutamate. Addition of these amino acids, however, did not result in the complementation of the Mut− phenotype. The amino acid requirement (Asp−/Glu−) could not be separated from the Mut− phenotype through backcrosses with parental strains, indicating that both phenotypes were closely linked. Complementation analysis revealed that all four mutants with this phenotype (Mut−, Asp−/Glu−) fell in one complementation group, designated ass3.
To isolate the defective gene, strain ass3-110 was transformed with a genomic H. polymorpha library. Transformants capable to grow on mineral media containing methanol (Mut+, Asp+/Glu+) were selected. Subcloning and reintroduction of the complementing fragments into ass3-110 resulted in a 4.2-kb genomic fragment that contained the complementing activity. This fragment was sequenced and the sequence was deposited at GenBank (Accession No. AF 221670).
Sequence analysis revealed that the complementing fragment contained a potential open reading frame (ORF) encoding a protein of 1175 amino acids with a predicted MW of 130 kDa. A database search revealed that this protein was highly similar throughout the entire protein to pyruvate carboxylases (Pyc) from various organisms ranging from bacteria (e.g., Bacillus subtilis 50% identity) and yeast (P. pastoris, 81% identity; S. cerevisiae Pyc1p and Pyc2p, both 77% identity, to man (53% identity). On the basis of the results of this search, we designated the gene PYC1 and its translation product HpPyc1p.
An H. polymorpha PYC1 disruption mutant (Δpyc1) was constructed in which approximately half of the H. polymorpha PYC1 gene was deleted (the region encoding amino acids 298–905). Growth experiments indicated that cells of Δpyc1 showed the same phenotype as the original mutant ass3-110: no growth on minimal media containing glucose and ammonium sulfate unless aspartate or glutamate were added and a defect in growth on methanol independent of the presence of aspartate or glutamate.
Mating of the Δpyc1 strain with the original ass3-110 mutant resulted in diploids that were all Mut−. After sporulation, no Mut+ cells were observed, demonstrating that ass3-110 and Δpyc1 are closely linked and most likely are alleles of the same gene.
Pyc is an anapleurotic enzyme that replenishes the tricarboxylic acid (TCA) cycle with oxaloacetate from pyruvate. For S. cerevisiae it has been shown that the absence of Pyc results in the inability of cells to grow on minimal media containing glucose and ammonium sulfate, although they do grow on glucose-aspartate containing media (Stucka et al., 1991). Also P. pastoris mutants lacking Pyc were reported to be unable to grow on glucose-ammonium sulfate media, whereas growth is possible with aspartate or glutamate as nitrogen source.
These data indicate that the Asp−/Glu− phenotype of H. polymorpha Δpyc1 is due to the absence of Pyc enzyme activity. However, it does not explain why H. polymorpha Δpyc1 cells are unable to grow on methanol media that contain Asp or Glu.
Properties of AO in H. polymorpha Δpyc1 Cells
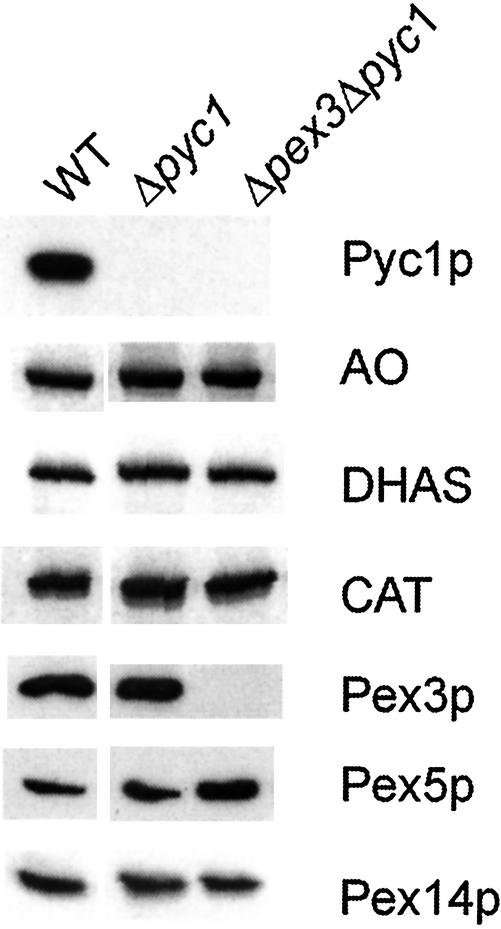
AO enzyme activity measurements in crude extracts prepared from methanol-induced Δpyc1 and WT control cells, revealed that the AO activity of Δpyc1 cells was <2% of the activity found in WT cells (0.07 and 4.1 U/mg protein, respectively). Western blot analysis of these extracts revealed, however, that the AO protein levels were normal in Δpyc1 cells (Figure 2). Also, the amounts of other peroxisomal matrix enzymes (dihydroxyacetone synthase [DHAS] and catalase [CAT]) and the peroxins Pex3p, Pex5p, and Pex14p were virtually identical in WT and Δpyc1 cells (Figure 2).
Figure 2.
Western blot analysis of crude extracts prepared from methanol-induced H. polymorpha WT, Δpyc1, and Δpex3Δpyc1 cells. The blots were decorated with antibodies against various H. polymorpha proteins. HpPyc1p was detected using antibodies against S. cerevisiae Pycp that cross-react with the H. polymorpha protein. The proteins are present in virtually equal amounts except that Pex5p levels are slightly enhanced in Δpex3Δpyc1cells. Equal amounts of protein were loaded per lane.
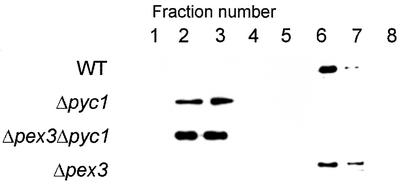
To analyze whether the absence of AO activity was due to a defect in AO oligomerization, crude extracts were subjected to sucrose density gradient centrifugation in order to separate AO monomers from octamers (Goodman et al., 1984). Western blot analysis revealed that in gradients prepared from WT control cells almost all AO protein was found in the bottom fractions (Figure 3, fraction 6) where octameric AO sediments. However, in gradients prepared from methanol-induced Δpyc1 cells AO protein was found in the top fractions, indicative for a monomeric state (Figure 3, fractions 2 and 3). In the fractions where octameric AO sediments no AO protein was detected. However, because the cells still display some enzyme activity (<2% of the enzyme activity in WT cells) and only octameric AO is active, the octameric AO apparently is below the level of detection in the experiment shown in Figure 3.
Figure 3.
Analysis of the oligomerization state of AO protein in H. polymorpha WT, Δpyc1, Δpex3Δpyc1, and Δpex3 cells by sucrose density gradient centrifugation of crude extracts prepared from methanol-induced cells. All fractions of the gradient were analyzed for the presence of AO protein by Western blotting. Fraction 1 represents the top fraction, fraction 8 the bottom fraction. Monomeric AO sediments to fractions 2–3, octameric AO to fractions 6–7. Equal portions of the fractions were loaded per lane.
Fluorescence analysis of the FAD content of AO protein, immunoprecipitated from crude extracts of WT and Δpyc1 cells, revealed that in precipitates of equal amounts of AO protein from crude extracts prepared from Δpyc1 or WT cells the concentration of FAD was ∼25-fold lower in precipitates from Δpyc1 cells compared with WT controls.
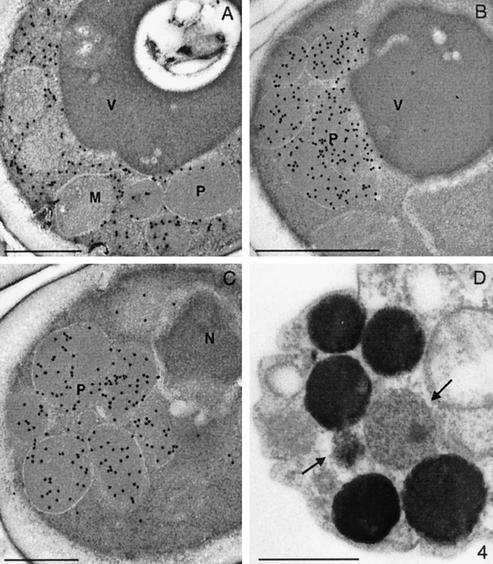
The overall morphology of methanol-induced H. polymorpha Δpyc1 cells was highly comparable to that of the original ass3-110 cells. Immunocytochemically, anti-AO–specific labeling was predominantly localized in the cytosol, with very little labeling on peroxisomes (Figure 4A). However, the other major PTS1 proteins DHAS (Figure 4B) and CAT (Figure 4C) showed a normal peroxisomal location. Cytochemical staining experiments revealed that in Δpyc1 AO enzyme activity was, like in WT cells, confined to peroxisomes (Figure 4D), although the amount of activity fluctuated between individual organelles, reflected by variations in staining intensity. Although the level of AO activity is very low in Δpyc1 cells, this can easily be detected by cytochemical staining, because this technique is extremely sensitive.
Figure 4.
Immunocytochemical demonstration of peroxisomal matrix proteins in methanol-induced H. polymorpha Δpyc1 cells. Using anti-AO antisera labeling was predominantly observed in the cytosol, whereas only a minor portion of the protein was localized at peroxisomes (A). Characteristically, the labeling intensities of the peroxisomal profiles strongly varied. Specific anti-DHAS (B) or anticatalase (C) dependent labeling was confined to peroxisomes. Cytochemical staining for the detection of AO enzyme activity (D) revealed that the enzyme activity was invariably confined to peroxisomes. Like AO protein, also the staining intensity varied (indicated by arrows) among the peroxisomal population, present in one cells, indicative for variations in the levels of active AO protein.
Taken together, these data indicate that in H. polymorpha Δpyc1 cells bulk of the AO protein is in the cytosol in an inactive, FAD-lacking, monomeric form while a minor fraction is present as enzymatically active, FAD-containing octamers inside peroxisomes.
The AO Assembly Failure in Δpyc1 Cells Is Not Indirect and Due to an Import Defect
The failure of AO assembly in Δpyc1 cells may be related to a spatial separation of the AO monomers (in the cytosol) and putative peroxisomal assembly factor(s). In H. polymorpha pex mutants these peroxisomal factor(s) most likely are also mislocalized to the cytosol, thus explaining why in these cells AO assembly/activation normally occurs in this compartment (van der Klei et al., 1991c). In Δpyc1 cells, however, normal peroxisomes are still present that may contain the putative AO assembly factor(s), as a peroxisomal protein import defect other than for AO protein was not observed. To test this possibility, we constructed a H. polymorpha Δpex3Δpyc1 double mutant, in which all peroxisomal matrix proteins are predicted to be mislocalized to the cytosol (Baerends et al., 1996). Biochemical analysis of crude extracts of methanol-induced Δpex3Δpyc1 cells showed that the level of various peroxisomal enzymes and peroxins was normal (Figure 2). However, AO assembly was not restored, because very low specific AO activities were detected (unpublished data). Also, sucrose density gradient analysis of crude extracts prepared from methanol-induced Δpex3Δpyc1 cells revealed that, like in Δpyc1, AO was predominantly present in a monomeric state (Figure 3). Controls, prepared from crude extracts of Δpex3 cells, confirmed that the absence of peroxisomes in these cells did not influence AO oligomerization, because bulk of the AO protein was found in fractions where octameric AO sediments (Figure 3, fraction 6 and 7). These results suggest that the AO assembly defect in Δpyc1 cells is not an indirect effect, due to a specific AO protein import block.
AO Assembly Does not Require HpPyc1p Enzyme Activity
To test whether AO assembly is dependent on HpPyc1p enzyme activity, we introduced a point mutation in H. polymorpha PYC1 that replaces the active site residue arginine 316 by glutamine (R316Q). Indeed, this mutant was unable to grow on minimal medium containing glucose unless supplemented with aspartate or glutamate (unpublished data). However, in the presence of these amino acids, cells of the mutant strain could grow on methanol, indicative for the restoration of the AO assembly defect (Figure 5A). Western blotting experiments using antibodies against S. cerevisiae Pyc protein, which cross-react with HpPyc1p (see Figure 2), revealed that HpPyc1pR316Q was synthesized in methanol-induced cells to levels comparable to WT cells (Figure 5B). Immunocytochemical analysis confirmed that AO protein was exclusively present in peroxisomes of these cells, indistinguishable from WT cells (Figure 5C). Hence, not the enzyme activity but another function of the protein is required for AO assembly.
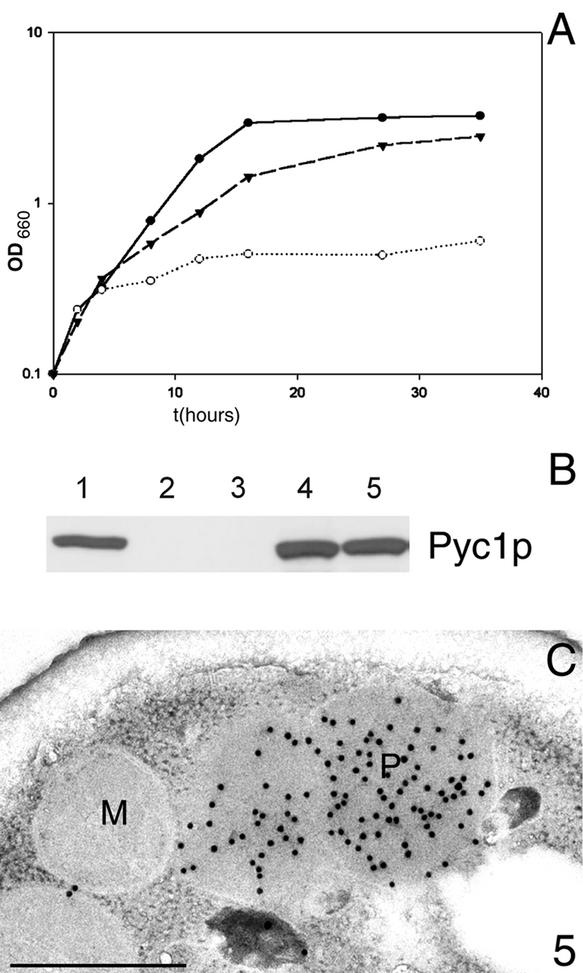
Figure 5.
Enzymatically active HpPyc1p is not required for AO assembly. (A) Growth of H. polymorpha WT (●), H. polymorpha Δpyc1 producing the mutant protein HpPyc1pR316Q (▾) and H. polymorpha Δpyc1 containing an empty expression plasmid (○) on mineral media containing methanol supplemented Asp and Glu. H. polymorpha Δpyc1 cells are severely hampered in growth (OD660 = 0.6 after 36 h). The initial increase in cell density is due to the presence of small amounts of yeast extract in the media. In contrast, cells of H. polymorpha WT and the Δpyc1 strain producing HpPyc1pR316Q are able to grow on methanol and reached a comparable final yield (OD660 3.2 and 2.5 after 36 h of growth, respectively; the cell density is expressed as optical density at 660 nm; OD660). (B) Comparison of the levels of HpPyc1p in crude extracts prepared from methanol-induced H. polymorpha strains using Western blotting and antibodies against S. cerevisiae Pycp. Lane 1, WT; lanes 2–5, Δpyc1 cells containing no plasmid (2), an empty expression plasmid (3), a plasmid containing WT HpPYC1 (4) or a plasmid containing mutant PYC1-R316Q (5). Equal amounts of protein were loaded per lane. (C) Immunocytochemical demonstration of AO protein in methanol-induced Δpyc1 cells producing HpPyc1p containing the mutation R316Q. Anti-AO specific labeling was confined to peroxisomes.
HpPyc1p Is a Cytosolic Enzyme
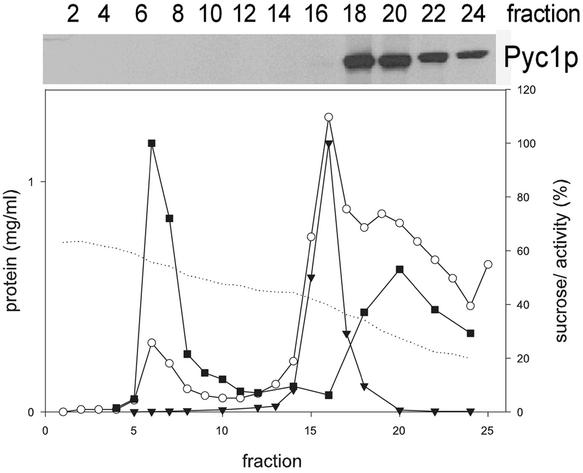
To determine the subcellular location of HpPyc1p, homogenized protoplasts prepared from methanol-grown WT cells were subjected to sucrose density centrifugation followed by Western blot analysis of the various fractions obtained. As shown in Figure 6, HpPyc1p was only detected in the low-density fractions at the top of the gradient (Figure 6, fractions 18–24), indicative for a cytosolic location. Analysis of the peroxisomal peak fractions did not reveal any HpPyc1p, indicating that the protein does not partially cosediment with peroxisomes. A cytosolic location is in line with the reported location of Pyc protein in S. cerevisiae (Walker et al., 1991).
Figure 6.
Sucrose gradient, prepared from a postnuclear supernatant obtained from methanol-grown H. polymorpha WT cells. The graph shows the distribution of the peroxisomal marker enzyme AO (+), the mitochondrial marker enzyme cytochrome c oxidase ([itrig]), the protein (0) and sucrose concentrations (dotted line). The Western blot shows the distribution of HpPyc1p in the even fractions of the gradient. The protein was only detected in the upper part of the gradient (fractions 18–24), which corresponds to the cytosol. Sucrose concentrations are expressed as % (wt/wt), the protein concentrations as mg/ml and the specific activities of AO and cytochrome c oxidase as percentages of the value in the peak fractions that were arbitrarily set at 100.
Newly Induced HpPyc1p Can Mediate Assembly of Cytosolically Accumulated AO Monomers
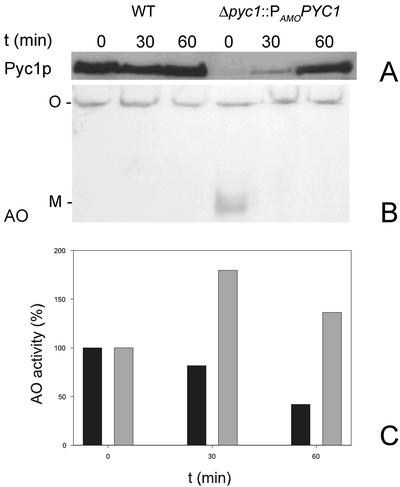
The intriguing question concerns the function of HpPyc1p in AO assembly: does it act like a chaperone or does it serve other functions, e.g., in cofactor binding? This question was addressed in a Δpyc1 strain that contained a copy of the PYC1 gene under control of the inducible amine oxidase promoter (PAMO). In this strain, Δpyc1::PAMOPYC1, the synthesis of AO monomers and the HpPyc1p protein can be separated in time. First, cells of this strain were induced on methanol/ammonium sulfate, conditions that induce the synthesis of AO monomers, but largely represses HpPyc1p synthesis due to the presence of ammonium sulfate. Subsequently, the cells were shifted to glucose/ethylamine, conditions that strongly repress AO synthesis (due to the presence of glucose) but induce HpPyc1p synthesis due to the induction of the PAMO by ethylamine. This allowed addressing the question whether existing, monomeric AO molecules that had accumulated in the cytosol, were still accessible for the HpPyc1p function in AO assembly. The results depicted in Figure 7 show that HpPyc1p was below the limit of detection in Δpyc1::PAMOPYC1 cells before the shift (Figure 7A, lane 4, T = 0 min). After the shift to ethylamine-containing media, PYC1 was rapidly induced and HpPyc1p levels comparable to WT were detected within 60 min. (compare lanes 1–3 with lane 6). Western blot analysis of a native gel confirmed that significant amounts of monomeric AO had accumulated before the shift to ethylamine as nitrogen source. However, within 30 min after the induction of HpPyc1p synthesis, the monomeric AO band had disappeared. At the same time the level of AO enzyme activity (Figure 7C) had significantly increased. In a control experiment using WT cells, the level of AO activity and octameric AO decreased as a result of glucose-induced degradation of peroxisomes. This process most likely also occurs in Δpyc1::PAMOPYC1 cells, as indicated by the subsequent reduction in AO protein and activity levels that follow the initial strong increase (Figure 7C).
Figure 7.
Biochemical demonstration of the assembly of cytosolically accumulated monomeric AO upon subsequent artificial induction of PYC1. Cells of the Δpyc1::PAMOPYC1 strain were pregrown on methanol/ammonium sulfate to induce AO synthesis under conditions that PYC1 expression is strongly repressed by ammonium sulfate. Subsequently the cells were shifted (at T = 0 min) to media containing glucose and ethylamine to induce PAMO, and thus HpPyc1p production and to repress AO synthesis. (A) A Western blot of an SDS-PAA gel prepared from crude extracts of WT and Δpyc1::PAMOPYC1 cells taken before (T = 0 min) or 30 and 60 min after the shift. The blot, decorated with anti-Pyc antibodies, shows the induction profile of HpPyc1p in Δpyc1::PAMOPYC1 cells, compared with WT levels. (B) A Western blot of a native gel of the same samples. This blot, decorated with anti-AO antibodies, which allows to detect both octameric AO (O) or monomeric AO (M), shows that monomeric AO present at T = 0 min is undetectable at T = 30 min. (C) The enzyme activities of AO in the same samples (black bar: WT cells; gray bar: Δpyc1::PAMOPYC1 cells). The reduction in AO activities in WT cells is due to selective peroxisome degradation, induced by glucose. The values in both strains at T = 0 min were set to 100% (absolute values at T = 0 h for WT ,2.0 U/mg protein and Δpyc1::PAMOPYC1 cells 0.5 U/mg protein).
HpPyc1p Physically Interacts with AO and FAD
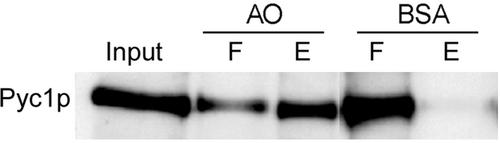
To study whether HpPyc1p has affinity for AO protein, in vitro binding studies were performed. To this purpose a Sepharose column containing AO protein was prepared (Evers et al., 1993). As a control, immobilized BSA was used. Purified HpPyc1p was loaded onto these columns. On extensive washing, the bound protein was eluted by a buffer containing 8 M urea. As shown in Figure 8, a significant portion of the loaded HpPyc1p protein had bound to the AO column, whereas in the control experiment using BSA all HpPyc1p was found in the flow-through fraction. These findings indicate that HpPyc1p is capable of binding to AO protein.
Figure 8.
In vitro interaction of AO and HpPyc1p. Purified HpPyc1p was loaded onto columns containing AO or BSA protein. The columns were subsequently extensively washed. The flow through and wash fractions were pooled (F). Bound HpPyc1p was eluted using a buffer containing 8 M urea (E). Equal portions of F and E were subjected to SDS-PAGE and blotted. The blots were decorated using anti-Pyc antibodies. Most HpPyc1p was found in the elution fraction (E) when AO columns were used, in contrast all HpPyc1p was found in the flow through and wash fraction (F) when a control column containing BSA was used.
To study whether HpPyc1p could play a role in the association of FAD to AO, we tested whether HpPyc1p, which contains an ATP-binding motif, is capable of binding FAD. FAD binding was measured using fluorescence correlation spectroscopy (FCS), a technique that allows to measure diffusion constants of fluorophores. Analysis of normalized fluorescence autocorrelation curves of FAD in the absence of HpPyc1p revealed that the average diffusion time of FAD, obtained by a one-component fit analysis, was 41.53 μs. This value is in agreement with the molecular weight of FAD molecules. After addition of purified HpPyc1p, the fluorescence autocorrelation curve changed and could be best fitted with a two-component fit. Fixing the average diffusion time of FAD, the diffusion time of the second component was 191–213 μs. Using the equation MWPYC = (τPYC/τFAD)3 × MWFAD the molecular weight of the second component was calculated to be 105–130 kDa. Assuming the HpPyc1p molecules to be spherical in shape, this is in agreement with the apparent molecular weight of monomeric HpPyc1p calculated from its amino acid sequence (130 kDa). Hence, these data indicate that part of the FAD had bound to the added HpPyc1p. In control experiments using lysozyme instead of HpPyc1p no change of fluorescence autocorrelation curve and diffusion time were observed.
Also in P. pastoris Pyc1p Is Essential for AO Import and Assembly
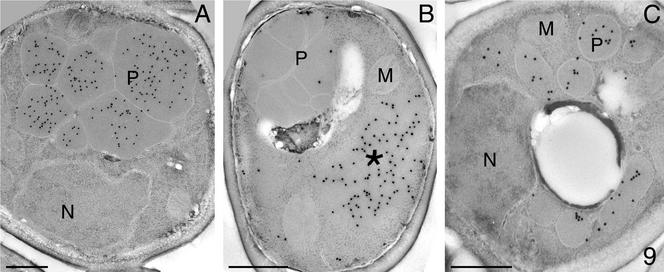
To determine whether the Pyc1p-dependent AO assembly defect is limited to H. polymorpha or represents a common feature of methylotrophic yeast, we analyzed a PYC deletion strain of P. pastoris (Menendez et al., 1998). The P. pastoris Δpyc strain, like its H. polymorpha counterpart, was unable to grow on glucose unless aspartate or glutamate was added. Growth on methanol was fully prevented, irrespective of the presence of these amino acids in the media. Immunocytochemical experiments revealed that, like in H. polymorpha, P. pastoris Δpyc cells did not import AO in peroxisomes (Figure 9B) as in WT cells (Figure 9A).
Figure 9.
Immunocytochemical localization of AO (A and B) and acyl-CoA oxidase (C) in P. pastoris WT (A) and Δpyc cells (B and C). In methanol-grown WT cells anti-AO-dependent labeling is confined to peroxisomes (A), whereas in Δpyc cells (B) the protein was predominantly detected in a cytosolic aggregate (*) but not in peroxisomes. In oleic acid-grown Δpyc cells the antiacyl-CoA oxidase–dependent labeling is confined to peroxisomal profiles (C).
Moreover, a P. pastoris pyc1 suppressor mutant (Menendez et al., 1998), in which the aspartate requirement was restored, still failed to assemble AO and thus, to grow on methanol (unpublished data), indicating that also in P. pastoris Pyc protein, but not the enzyme activity, is required for AO assembly.
Remarkably, P. pastoris Δpyc cells grew normally on oleic acid, at rates similar to WT controls. Hence, the absence of Pyc1p has no general deteriorating effect on peroxisome biogenesis or function. Also, one of the key enzymes of oleate metabolism is a peroxisomal flavoprotein, namely acyl-CoA oxidase. This protein displays normally activities (unpublished data) and is located in peroxisomes (Figure 9C). Therefore, the PYC1 deletion seems to interfere specifically with AO assembly and not with that of other peroxisomal flavin proteins.
DISCUSSION
We have identified pyruvate carboxylase (Pyc) as the first protein that has an essential function in assembly of peroxisomal AO in methylotrophic yeast. Pyc is an anapleurotic enzyme that replenishes the tricarboxylic acid cycle with oxaloacetate from pyruvate. As a consequence, yeasts lacking Pyc enzyme activity cannot grow on minimal glucose media unless aspartate or glutamate is added, amino acids that can be converted into oxaloacetate. Unexpectedly, H. polymorpha and P. pastoris strains lacking Pyc are also unable to grow on methanol, independent of the presence of aspartate or glutamate in the medium. We demonstrated that this growth defect is due to a severe block in the assembly of AO, a key enzyme in methanol metabolism. Our data convincingly show that Pyc protein but not its enzyme activity is necessary for AO assembly. The import and activation of another peroxisomal flavin enzyme, acyl-CoA oxidase, was not affected in the absence of Pyc. Hence, Pyc seems to function specifically in the AO assembly pathway in methylotrophic yeast.
The current model of AO assembly hypothesizes that AO monomers but not octamers are imported into peroxisomes. This is based on the finding that octameric AO protein is not incorporated into peroxisomes in vivo (Douma et al., 1990; Waterham et al., 1993) and on the results of elegant pulse chase experiments that provided evidence that assembly into the active octamer takes place inside the organelle (Goodman et al., 1984; Stewart et al., 2001).
FAD most likely binds to monomeric AO, as is indicated by the finding that FAD cannot reassociate in vivo to AO octamers, from which FAD has been chemically removed (van der Klei et al., 1989a). Whether in H. polymorpha FAD binds to AO monomers before or upon translocation across the peroxisomal membrane was so far still speculative.
We show here that in H. polymorpha Δpyc1 cells bulk of the AO protein accumulated as inactive, FAD-lacking monomers in the cytosol. Only a minor portion had assembled into FAD-containing, enzymatically active octamers, which—based on cytochemical experiments—were localized in the peroxisomal matrix. This suggests that the presence of HpPyc1p is important for FAD-binding to AO monomers, the subsequent translocation into the peroxisomal matrix and finally the assembly into octamers.
Interestingly, the phenotype of H. polymorpha Δpyc1 cells is highly comparable to that of the H. polymorpha riboflavin auxotrophic mutant, rif1 (Evers et al., 1996), in which also bulk of the AO protein accumulates as FAD-lacking monomers in the cytosol. A likely way to explain both observations is that FAD binding to AO monomers in the cytosol is a prerequisite to allow efficient translocation into peroxisomes. Our current findings suggest that cytosolically located HpPyc1p is required to bind FAD to AO monomers.
We found that in HpPyc1p-deficient cells soluble, monomeric AO accumulated that could be activated upon subsequent artificial induction of PYC1 expression. Together with the observation that HpPyc1p physically interacts with AO protein and is capable of binding FAD, our current data strongly suggest that HpPyc1p functions as a FAD-binding protein in the cytosol.
Relatively little is known on proteins that play a role in cofactor binding. It is generally assumed that cofactor binding occurs spontaneously upon formation of the correct binding site in a protein molecule. So far no proteins have been described that play a role in noncovalent binding of FAD; also, only one is yet identified that is essential for covalent FAD binding (Kim et al., 1995). Several examples are known of proteins involved in binding of heme, for instance, mitochondrial heme lyases that are necessary for the attachment of heme to cytochrome c or cytochrome c1 (Page et al., 1998). However, despite extensive research molecular details on their mode of action are still lacking.
An alternative explanation for the phenotype of H. polymorpha Δpyc1 is that HpPyc1p is essential to mediate binding of AO to the receptor Pex5p and that FAD-binding occurs after import mediated by a peroxisomal factor. However, this explanation is less likely in view of the fact that in cells of the Δpex3Δpyc1 double mutant AO remains monomeric, whereas it is normally active in single pex mutants, e.g., Δpex3 (compare Figure 3) and Δpex5 (van der Klei et al., 1995).
We showed before, that FAD-containing AO monomers can assemble spontaneously into octamers in vitro (Evers et al., 1995). Hence, it can be envisaged that this also can take place in intact cells in vivo. However, when FAD-binding indeed occurs in the cytosol the cell has to deal with the problem how to prevent premature spontaneous assembly of the FAD-containing monomers. There is a strong metabolic need for the cell to postpone this event until import has occurred, because only minor amounts of active AO in the cytosol give rise to severe energetical disadvantages due to a cytosolic H2O2 metabolism that would retard or even prevent growth on methanol (van der Klei et al., 1991b). One possibility is that HpPyc1p remains bound to AO after cofactor binding. A second option is that, upon FAD binding, the protein is immediately donated to Pex5p, which prevents octamer formation. Because other PTS1 proteins (DHAS, CAT) are normally imported in Δpyc1 cells and also their Pex5p levels are similar to WT cells, it is indeed unlikely that Pex5p molecules are bound to the large pool of FAD-lacking AO monomers in the cytosol of Δpyc1 cells.
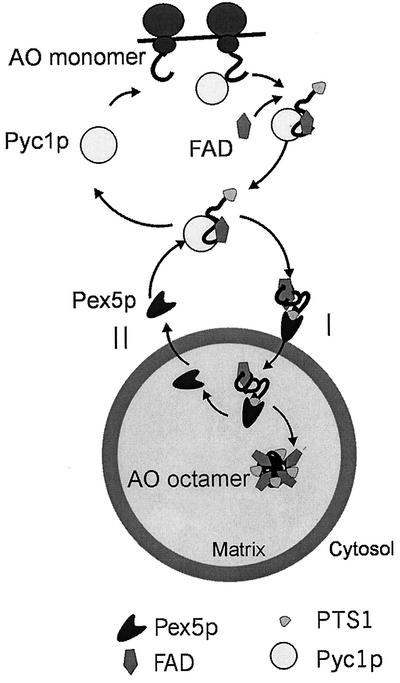
Based on the above reasoning, our adapted hypothetical model of AO assembly predicts that the first step in AO import is HpPyc1p-mediated FAD binding to newly synthesized AO protein in the cytosol (Figure 10). Subsequently, the FAD-containing monomers bind to the PTS1 receptor Pex5p, followed by the translocation of the Pex5p-AO cargo complex into peroxisomes. After dissociation of the cargo from Pex5p in the organellar matrix, the FAD-containing AO monomers may spontaneously oligomerize into enzymatically active octamers followed by shuttling of Pex5p back to the cytosol to mediate another round of PTS1 protein import. This model is in line with the previously proposed “extended shuttle model” for Pex5p (van der Klei et al., 1995; van der Klei and Veenhuis, 1996), which was recently experimentally proven by Dammai and Subramani (2001) for human cells.
Figure 10.
Hypothetical model of AO import and assembly in H. polymorpha. AO monomers are synthesized on free ribosomes in the cytosol. In the cytosol HpPyc1p assists in binding of the cofactor FAD to newly synthesized AO monomers. Subsequently, the FAD-containing monomers are bound to the PTS1 receptor, Pex5p. Both the receptor and its cargo are translocated across the peroxisomal membrane (I), followed by dissociation of the AO-Pex5p complex in the organellar matrix. The FAD-containing AO monomers then assemble into octamers. Pex5p recycles to the cytosol (II) to mediate another round of PTS1 protein import.
A further implication of the present study is that in methylotrophic yeasts Pyc has multiple functions. The relevance of this is obvious in view of the data on the human genome that have become available recently (Lander et al., 2001; Venter et al., 2001). The 30,000 genes that have been identified cannot cope for the multitude of functions that are predicted to be essential in men unless specific genes encode multiple proteins or proteins with multiple functions. Proteins that fulfill two different functions are the housekeeping enzyme lactate dehydrogenase B in duck, which is also the lens structural protein epsilon-crystallin (Hendriks et al., 1988); glyceraldehyde 3-phosphate dehydrogenase, which has been shown to play a role in endocytosis in CHO cells (Robbins et al., 1995); and the α-subunit of phosphofructokinase (but not enzyme activity), which is required for the onset of glucose-mediated selective pexophagy (microautophagy; Yuan et al., 1997). The switch in function may even be temperature dependent, as demonstrated for heat shock protein DegP, which normally functions as a chaperone but displays proteolytic activity at elevated temperatures (Spiess et al., 1999).
Our collection of Ass mutants that comprises 10 different complementation groups (Van Dijk et al., 2002), suggests that additional proteins may be involved in AO biosynthesis (e.g., targeting or translocation). This view is also based on the finding that in P. pastoris the carboxyterminal PTS1 signal is not the only targeting information essential for import (Waterham et al., 1997). Therefore, specific additional proteins may exist that exclusively function in AO import and assembly. We are currently trying to identify these proteins by cloning the encoding genes by functional complementation of the other ass mutants.
ACKNOWLEDGMENTS
We thank Nina Visser for her help in the fluorescence analysis and Ineke Keizer-Gunnink and Anita Kram for skilful technical assistance in different parts of this work. S. cerevisiae Pycp antibodies were kindly supplied by J. C. Wallace, University of Adelaide, Australia. R.v.D. is supported by a grant of the Netherlands Technology Foundation (STW) and I.J.v.d.K. by a PIONIER-fellowship from ALW/NWO. D.Y.W. is supported by an ALW grant.
Abbreviations used:
- AO
alcohol oxidase
- BSA
bovine serum albumin, CAT, catalase
- DHAS
dihydroxyacetone synthase
- FCS
fluorescence correlation spectroscopy
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–07–0417. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–07–0417.
REFERENCES
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerends RJS, et al. The Hansenula polymorpha PER9 gene encodes a peroxisomal membrane protein essential for peroxisome assembly and integrity. J Biol Chem. 1996;271:8887–8894. doi: 10.1074/jbc.271.15.8887. [DOI] [PubMed] [Google Scholar]
- Dammai V, Subramani S. The human peroxisomal targeting signal receptor, Pex5p, is translocated into the peroxisomal matrix and recycled to the cytosol. Cell. 2001;105:187–196. doi: 10.1016/s0092-8674(01)00310-5. [DOI] [PubMed] [Google Scholar]
- Distel B, Veenhuis M, Tabak HF. Import of alcohol oxidase into peroxisomes of Saccharomyces cerevisiae. EMBO J. 1987;6:3111–3116. doi: 10.1002/j.1460-2075.1987.tb02620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douma AC, Veenhuis M, Driessen AJM, Harder W. Liposome mediated introduction of proteins into protoplasts of the yeast Hansenula polymorpha as a possible tool to study peroxisome biogenesis. Yeast. 1990;6:99–105. [Google Scholar]
- Evers ME, Huhse B, Titorenko VI, Kunau WH, Hartl FU, Harder W, Veenhuis M. Affinity purification of molecular chaperones of the yeast Hansenula polymorpha using immobilized denatured alcohol oxidase. FEBS Lett. 1993;321:32–36. doi: 10.1016/0014-5793(93)80615-2. [DOI] [PubMed] [Google Scholar]
- Evers ME, Titorenko VI, van der Klei IJ, Harder W, Veenhuis M. Assembly of alcohol oxidase in peroxisomes of the yeast Hansenula polymorpha requires the cofactor flavin adenine dinucleotide. Mol Biol Cell. 1994;5:829–837. doi: 10.1091/mbc.5.8.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers ME, Harder W, Veenhuis M. In vitro dissociation and re-assembly of peroxisomal alcohol oxidases of Hansenula polymorpha and Pichia pastoris. FEBS Lett. 1995;368:293–296. doi: 10.1016/0014-5793(95)00653-q. [DOI] [PubMed] [Google Scholar]
- Evers ME, Titorenko VI, Harder W, van der Klei IJ, Veenhuis M. Flavin adenine dinucleotide binding is the crucial step in alcohol oxidase assembly in the yeast Hansenula polymorpha. Yeast. 1996;12:917–923. doi: 10.1002/(SICI)1097-0061(199608)12:10%3C917::AID-YEA984%3E3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Faber KN, Haima P, Harder W, Veenhuis M, AB G. Highly-efficient electrotransformation of the yeast Hansenula polymorpha. Curr Genet. 1994;25:305–310. doi: 10.1007/BF00351482. [DOI] [PubMed] [Google Scholar]
- Faber KN, Swaving GJ, Faber F, AB G, Harder W, Veenhuis M, Haima P. Chromosomal targeting of replicating plasmids in the yeast Hansenula polymorpha. J Gen Microbiol. 1992;138:2405–2416. doi: 10.1099/00221287-138-11-2405. [DOI] [PubMed] [Google Scholar]
- Faber KN, Heyman JA, Subramani S. Two AAA family peroxins, PpPex1p and PpPex6p, interact with each other in an ATP-dependent manner and are associated with different subcellular membranous structures distinct from peroxisomes. Mol Cell Biol. 1998;18:936–943. doi: 10.1128/mcb.18.2.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleeson MAG, Sudbery PE. Genetic analysis in the methylotrophic yeast Hansenula polymorpha. Yeast. 1988;4:293–303. [Google Scholar]
- Goodman JM, Scott CW, Donahue PN, Atherton JP. Alcohol oxidase assembles post-translationally into the peroxisome of Candida boidinii. J Biol Chem. 1984;259:8485–8493. [PubMed] [Google Scholar]
- Hendriks W, Mulders JW, Bibby MA, Slingsby C, Bloemendal H, de Jong WW. Duck lens epsilon-crystallin and lactate dehydrogenase B4 are identical: a single-copy gene product with two distinct functions. Proc Natl Acad Sci USA. 1988;85:7114–7118. doi: 10.1073/pnas.85.19.7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hink M, Visser AJWG. Fluorescence correlation spectroscopy of flavins and flavoenzymes: photochemical and photophysical aspects. In: Rettig W, Struhmel B, Schrader S, editors. Applied Fluorescence in Chemistry, Biology, and Medicine. Berlin: Springer Verlag; 1998. , pp 101–118. [DOI] [PubMed] [Google Scholar]
- Horton RM, Cai ZL, Ho SN, Pease LR. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques. 1990;8:528–535. [PubMed] [Google Scholar]
- Kim J, Fuller JH, Kuusk V, Cunane L, Chen ZW, Mathews FS, McIntire WS. The cytochrome subunit is necessary for covalent FAD attachment to the flavoprotein subunit of p-cresol methylhydroxylase J. Biol Chem. 1995;270:31202–31209. doi: 10.1074/jbc.270.52.31202. [DOI] [PubMed] [Google Scholar]
- Kyhse-Andersen J. Electroblotting of multiple gels: a simple apparatus without buffer tank for rapid transfer of proteins from polyacrylamide to nitrocellulose. J Biochem Biophys Methods. 1984;10:203–209. doi: 10.1016/0165-022x(84)90040-x. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Menendez J, Delgado J, Gancedo C. Isolation of the Pichia pastoris PYC1 gene encoding pyruvate carboxylase and identification of a suppressor of the pyc phenotype. Yeast. 1998;14:647–654. doi: 10.1002/(SICI)1097-0061(199805)14:7<647::AID-YEA269>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Merckelbach A, Godecke S, Janowicz ZA, Hollenberg CP. Cloning and sequencing of the ura3 locus of the methylotrophic yeast Hansenula polymorpha and its use for the generation of a deletion by gene replacement. Appl Microbiol Biotechnol. 1993;40:361–364. doi: 10.1007/BF00170393. [DOI] [PubMed] [Google Scholar]
- Page MD, Sambongi Y, Ferguson SJ. Contrasting routes of c-type cytochrome asembly in mitochondria, chloroplasts and bacteria. Trends Biochem Sci. 1998;23:103–108. doi: 10.1016/s0968-0004(98)01173-6. [DOI] [PubMed] [Google Scholar]
- Robbins AR, Ward RD, Oliver C. A mutation in glyceraldehyde 3-phosphate dehydrogenase alters endocytosis in CHO cells. J Cell Biol. 1995;130:1093–1104. doi: 10.1083/jcb.130.5.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde M, Lim F, Wallace JC. Electron microscopic localization of pyruvate carboxylase in rat liver and Saccharomyces cerevisiae by immunogold procedures. Arch Biochem Biophys. 1991;290:197–201. doi: 10.1016/0003-9861(91)90608-l. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Stewart MQ, Esposito RD, Gowani J, Goodman JM. Alcohol oxidase and dihydroxyacetone synthase, the abundant peroxisomal poteins of methylotrophic yeasts, assemble in different cellular compartments. J Cell Sci. 2001;114:2863–2868. doi: 10.1242/jcs.114.15.2863. [DOI] [PubMed] [Google Scholar]
- Stucka R, Dequin S, Salmon JM, Gancedo C. DNA sequences in chromosomes II and VII code for pyruvate carboxylase isoenzymes in Saccharomyces cerevisiae: analysis of pyruvate carboxylase-deficient strains. Mol Gen Genet. 1991;229:307–315. doi: 10.1007/BF00272171. [DOI] [PubMed] [Google Scholar]
- Spiess C, Beil A, Ehrmann M. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell. 1999;97:339–347. doi: 10.1016/s0092-8674(00)80743-6. [DOI] [PubMed] [Google Scholar]
- Tan X, Waterham HR, Veenhuis M, Cregg JM. The Hansenula polymorpha PER8 gene encodes a novel peroxisomal integral membrane protein involved in proliferation. J Cell Biol. 1995;128:307–319. doi: 10.1083/jcb.128.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titorenko VI, Waterham HR, Cregg JM, Harder W, Veenhuis M. Peroxisome biogenesis in the yeast Hansenula polymorpha is controlled by a complex set of interacting gene products. Proc Natl Acad Sci USA. 1993;90:7470–7474. doi: 10.1073/pnas.90.16.7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Klei IJ, Veenhuis M, Nicolay K, Harder W. In vivo inactivation of peroxisomal alcohol oxidase in Hansenula polymorpha by KCN is an irreversible process. Arch Microbiol. 1989a;151:26–33. doi: 10.1007/BF00444664. [DOI] [PubMed] [Google Scholar]
- van der Klei IJ, Veenhuis M, van der Ley I, Harder W. Heterologous expression of alcohol oxidase in Saccharomyces cerevisiae: properties of the enzyme and implications for microbody development. FEMS Microbiol Lett. 1989b;48:133–137. doi: 10.1111/j.1574-6968.1989.tb03287.x. [DOI] [PubMed] [Google Scholar]
- van der Klei IJ, Harder W, Veenhuis M. Biosynthesis and assembly of alcohol oxidase, a peroxisomal matrix protein in methylotrophic yeasts: a review. Yeast. 1991a;7:195–209. doi: 10.1002/yea.320070302. [DOI] [PubMed] [Google Scholar]
- van der Klei IJ, Harder W, Veenhuis M. Methanol metabolism in a peroxisome-deficient mutant of Hansenula polymorpha: a physiological study. Arch Microbiol. 1991b;156:15–23. doi: 10.1007/BF00418181. [DOI] [PubMed] [Google Scholar]
- van der Klei IJ, Sulter GJ, Harder W, Veenhuis M. Assembly of alcohol oxidase in the cytosol of a peroxisome-deficient mutant of Hansenula polymorpha—properties of the protein and architecture of the crystals. Yeast. 1991c;7:195–209. [Google Scholar]
- van der Klei IJ, Hilbrands RE, Swaving GJ, Waterham HR, Vrieling EG, Titorenko VI, Cregg JM, Harder W, Veenhuis M. The Hansenula polymorpha PER3 gene is essential for the import of PTS1 proteins into the peroxisomal matrix. J Biol Chem. 1995;270:17229–17236. doi: 10.1074/jbc.270.29.17229. [DOI] [PubMed] [Google Scholar]
- van der Klei IJ, Veenhuis M. Peroxisome biogenesis in the yeast Hansenula polymorpha: a structural and functional analysis. Ann NY Acad Sci. 1996;804:47–59. doi: 10.1111/j.1749-6632.1996.tb18607.x. [DOI] [PubMed] [Google Scholar]
- van der Klei IJ, van der Heide M, Baerends RJS, Rechinger KB, Nicolay K, Kiel JAKW, Veenhuis M. The Hansenula polymorpha per6 mutant is affected in two adjacent genes which encode dihydroxyacetone kinase and a novel protein, Pak1p, involved in peroxisome integrity. Curr Genet. 1998;34:1–11. doi: 10.1007/s002940050360. [DOI] [PubMed] [Google Scholar]
- van Dijk R, Lahchev KL, Kram AM, van der Klei IJ, Veenhuis M. Isolation of mutants defective in the assembly of octameric alcohol oxidase of Hansenula polymorpha. FEMS Yeast Res. 2002;1:257–263. doi: 10.1111/j.1567-1364.2002.tb00043.x. [DOI] [PubMed] [Google Scholar]
- van Dijken JP, Otto R, Harder W. Growth of Hansenula polymorpha in a methanol-limited chemostat. Physiological responses due to the involvement of methanol oxidase as a key enzyme in methanol metabolism. Arch Microbiol. 1976;111:137–144. doi: 10.1007/BF00446560. [DOI] [PubMed] [Google Scholar]
- Veenhuis M, van Dijken JP, Harder W. Cytochemical studies on the localization of methanol oxidase and other oxidases in peroxisomes of methanol-grown Hansenula polymorpha. Arch Microbiol. 1976;111:123–135. doi: 10.1007/BF00446559. [DOI] [PubMed] [Google Scholar]
- Venter JC, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Verduyn C, van Dijken JP, Scheffers WA. Colorometric alcohol assays with alcohol oxidase. J Microbiol Methods. 1984;2:15–25. [Google Scholar]
- Walker ME, Val DL, Rohde M, Devenish RJ, Wallace JC. Yeast pyruvate carboxylase: identification of two genes encoding isoenzymes. Biochem Biophys Res Commun. 1991;176:1210–1217. doi: 10.1016/0006-291x(91)90414-3. [DOI] [PubMed] [Google Scholar]
- Waterham HR, Titorenko VI, Swaving GJ, Harder W, Veenhuis M. Peroxisomes in the methylotrophic yeast Hansenula polymorpha do not necessarily derive from pre-existing organelles. EMBO J. 1993;12:4785–4794. doi: 10.1002/j.1460-2075.1993.tb06167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterham HR, Titorenko VI, Haima P, Cregg JM, Harder W, Veenhuis M. The Hansenula polymorpha PER1 gene is essential for peroxisome biogenesis and encodes a peroxisomal matrix protein with both carboxy- and amino-terminal targeting signals. J Cell Biol. 1994;127:737–749. doi: 10.1083/jcb.127.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterham HR, Russell KA, de Vries Y, Cregg JM. Peroxisomal targeting, import and assembly of alcohol oxidase in Pichia pastoris. J Cell Biol. 1997;139:1419–1431. doi: 10.1083/jcb.139.6.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Tuttle DL, Shi YJ, Ralph GS, Dunn WA, Jr. Glucose-induced microautophagy in Pichia pastoris requires the alpha-subunit of phosphofructokinase. J Cell Sci. 1997;110:1935–1945. doi: 10.1242/jcs.110.16.1935. [DOI] [PubMed] [Google Scholar]