Abstract
Plant cells contain several types of vacuoles with specialized functions. Although the biogenesis of these organelles is well understood at the morphological level, the machinery involved in plant vacuole formation is largely unknown. We have recently identified an Arabidopsis mutant, vcl1, that is deficient in vacuolar formation. VCL1 is homologous to a protein that regulates membrane fusion at the tonoplast in yeast. On the basis of these observations, VCL1 is predicted to play a direct role in vacuolar biogenesis and vesicular trafficking to the vacuole in plants. In this work, we show that VCL1 forms a complex with AtVPS11 and AtVPS33 in vivo. These two proteins are homologues of proteins that have a well-characterized role in membrane fusion at the tonoplast in yeast. VCL1, AtVPS11, and AtVPS33 are membrane-associated and cofractionate with tonoplast and denser endomembrane markers in subcellular fractionation experiments. Consistent with this, VCL1, AtVPS11, and AtVPS33 are found on the tonoplast and the prevacuolar compartment (PVC) by immunoelectron microscopy. We also show that a VCL1-containing complex includes SYP2-type syntaxins and is most likely involved in membrane fusion on both the PVC and tonoplast in vivo. VCL1, AtVPS11, and AtVPS33 are the first components of the vacuolar biogenesis machinery to be identified in plants.
INTRODUCTION
The efficient and accurate fusion of membranes is a vital requirement for cells to maintain their essential compartmentalization. It is estimated that several hundred genes are involved in trafficking and membrane fusion in plants, humans, insects, or fungi (Bock et al., 2001). In vitro, purified soluble N-ethyl maleimide–sensitive factor adaptor protein receptor (SNARE) proteins alone are sufficient to drive the fusion of artificial membranes and to recapitulate the specificity of fusion reactions that are observed in vivo. SNAREs are also essential for membrane fusion in vivo, but additional proteins play an important role both in giving specificity to the reaction and in driving the fusion of the membranes. Factors such as NSF, Rab GTPases, members of the Sec1 family of proteins, and large multiprotein complexes (Sacher et al., 1998; Guo et al., 1999; Sato et al., 2000) contribute to the specificity and kinetics of vesicular transport. Moreover, it has been shown that the homotypic fusion of vacuoles in yeast requires SNAREs for docking membranes, but the fusion itself may be driven by proteolipid channels formed by V0 trans-complexes in the opposing membranes (Peters et al., 2001). It is clear that SNAREs are necessary but not sufficient to account for the complexity of vesicular trafficking within the cell.
We recently reported the identification of the vacuoleless1 (vcl1) mutant of Arabidopsis (Rojo et al., 2001). This mutant exhibits an extremely aberrant development that leads to embryo lethality at late torpedo stage. At the subcellular level, the most striking phenotype is the absence of vacuoles in the embryo and the accumulation of small vesicles and autophagosomes. In addition, a vacuolar soluble protein, AtALEU, is abnormally secreted in the mutant. These phenotypes are consistent with a role of VCL1 in positively regulating membrane fusion at the tonoplast. The VCL1 protein shares some similarity with Vps16p from yeast. Mutations in Vps16p also block vacuole biogenesis and affect all known vacuolar protein transport pathways in yeast; however, in contrast to VCL1, VPS16 gene product is not essential (Horazdovsky and Emr, 1993). The similarity of the proteins (24% identity through the entire protein) (Rojo et al., 2001) and the phenotypes of the two mutants indicates that VCL1 and Vps16p are homologues. Vps16p forms part of the C–Vps protein complex in yeast that also includes Vps11p, Vps18p, Vps33p, Vps39p, and Vps41p. The C–Vps complex is required for the docking stage during homotypic fusion of vacuoles and heterotypic fusion of vesicles at the tonoplast (Rieder and Emr, 1997; Sato et al., 2000; Peterson and Emr, 2001). In the initial priming stage of vacuolar fusion, Vam3p syntaxin is disassembled from a cis-SNARE complex by the action of Sec18p and most likely is maintained in an unpaired active state by associating with C–Vps complex (Price et al., 2000; Sato et al., 2000). It is not yet known how the C–Vps complex is bound to the vacuole before the priming phase. After the priming, the C–Vps complex functions as an activator of Ypt7p, a small GTP-binding protein of the Rab family, and as an effector of the activated GTP-bound form of Ypt7p. Activated Ypt7p is required for the formation of trans-SNARE pairs during the docking stage of vacuole fusion (Sato et al., 2000; Wickner, 2002). In Drosophila, the eye pigment mutants light, deep orange, and carnation have defective pigment granules and carry mutations in their VPS41, VPS18, and VPS33 homologues, respectively (Warner et al., 1998; Sevrioukov et al., 1999). Deep orange and carnation are part of a multiprotein complex that regulates membrane fusion events at endosomal compartments. In mammals, the human homologues of the C-Vps proteins were shown to interact with syntaxin-7, a SNARE showing the highest level of homology to yeast Vam3p (Kim et al., 2001). Thus, the presence of large protein complexes that interact with syntaxins and regulate vesicle fusion at late endosomes or lysosomes seems to be conserved in eukaryotic organisms.
We now provide evidence that VCL1 forms a complex with both AtVPS11 and AtVPS33, the Arabidopsis homologues of Vps11p and Vps33p, respectively. VCL1, AtVPS11, and AtVPS33 are peripherally associated with membranes and cofractionate with vacuolar markers on sucrose density gradients and in preparations containing purified vacuoles. The yeast homologues of these proteins and the other members of the C–Vps complex also cofractionate with vacuolar markers, but no direct evidence of their tonoplast association has been reported. We show here by immunoelectron microscopy that VCL1, AtVPS11, and AtVPS33 localize to the tonoplast, supporting a role for the AtC–VPS complex in regulating membrane fusion at the plant vacuole. These findings are consistent with the absence of vacuoles and accumulation of small vesicles observed in the vcl1 mutant. In addition, we also show localization of the AtC–VPS complex to the prevacuolar compartment (PVC), suggesting that AtVPS33 plays the role of a still-unidentified protein from the Sec1p family on this organelle. We also present data showing the coimmunoprecipitation of VCL1 with either SYP21 or SYP22 syntaxins.
MATERIALS AND METHODS
Sequence Analyses
Protein sequences of Vps11p and Vps33p and their closest homologues were acquired from GenBank (AtVPS11, AF436824; MmVPS11, BC016258; hVPS11, AB027508; NcVPS11, AL355933; SpVPS11, AL355013; Vps11p, X54466; CeVPS11, Z46794; DmVPS11, AY094791; AtVPS33, AF357527; MmVPS33, AK019463; hVPS33, BC016617; SpVPS33, AL136536; Vps33p, U19729; CeVPS33, U29488; car, AF133260). Full-length sequences were aligned using the ClustalW algorithm (Thompson et al., 1994), and a phylogenetic tree was prepared using the Drawgram algorithm (Felsenstein, 1989). The percentage of identity toward the Arabidopsis protein, indicated in parentheses, was calculated using the Align algorithm (Myers and Miller, 1988). All sequence analyses were performed in the Biology Workbench web-based environment (http://workbench.sdsc.edu) under default conditions.
Antibody Production
AtVPS11 (amino acids 151–932) and AtVPS33 (amino acids 59–336) were cloned into pET28b (Novagen, Madison, WI). These plasmids resulted in a translational fusion of a fragment of either AtVPS11 or AtVPS33 with six histidine residues. Overexpressed fusion proteins were purified under denaturing conditions by nickel affinity chromatography as described by the manufacturer (Novagen) and used to immunize rabbits at Cocalico Biologicals (Reamstown, PA). The antibodies were affinity-purified as described by Bassham and Raikhel (1998). Briefly, the purified recombinant proteins were separated by SDS-PAGE and transferred to nitrocellulose, and the strip containing the fusion protein was cut out. After blocking in milk, crude serum was incubated with the strip. After extensive washing, the specific antibodies were eluted with 100 mM glycine, pH 2.5. The eluate was neutralized and used in all further experiments.
Membrane Association Studies
Differential centrifugation and extraction of VCL1, AtVPS11, and AtVPS33 from microsomal membranes were performed as described in Bassham and Raikhel (1998). Sucrose density gradients were done essentially as described previously by Sanderfoot et al. (1998). Protein samples were analyzed by immunoblotting with antibodies to VCL1 (Rojo et al., 2001), AtALEU (Ahmed et al., 2000), γ-TIP (Avila-Teeguarden and Raikhel, unpublished data), AtVPS45 (Bassham and Raikhel, 1998), SYP41 (Bassham et al., 2000), SYP21/22 (Sanderfoot et al., 1999), AtSEC12 (Bar-Peled and Raikhel, 1997), AtELP (Ahmed et al., 1997), and SYP111 (Lauber et al., 1997).
Expression of Green Fluorescent Protein–tagged VCL1 in Arabidopsis
The PCR-modified VCL1 cDNA was inserted as a XhoI-BamHI fragment behind the constitutive cauliflower mosaic virus 35S promoter in pEZT–NL binary vector (Cutler and Ehrhardt, Carnegie Institution of Washington, Stanford, CA). The construct was transformed into Agrobacterium tumefaciens and subsequently vacuum-infiltrated into Arabidopsis ecotype Columbia as described previously (Bent et al., 1994).
Electron Microscopy Procedures
Cryosections of Arabidopsis root tips were prepared as described by Sanderfoot et al. (1998) and used for all immunogold labeling experiments as in Sanderfoot et al. (1998). A monoclonal antibody to green fluorescent protein (GFP) was acquired from Clontech Laboratories (Palo Alto, CA). For double-labeling experiments, after incubation of the grids with the first antibody, a second fixation step followed by a second blocking step was used to prevent cross-reactivity of the antibodies at later stages of the protocol. Many sections from independent plants were observed for each combination of antibodies. Controls were performed with the use of the corresponding preimmune serum substituted for one or both of the antisera. In all cases, these controls demonstrated high specificity of the labeling.
Immunoprecipitation Procedures
Detergent extracts of microsomes from Arabidopsis tissues and subsequent immunoprecipitations were performed as described in Bassham and Raikhel (1998) and Sanderfoot et al. (2001a). Briefly, detergent extracts were incubated with specific antibodies or preimmune sera at 4°C, followed by Protein A–Sepharose beads (Amersham Pharmacia Biotech, Piscataway, NJ). Beads were pelleted and extensively washed, and bound proteins were eluted in SDS sample buffer. Each immunoprecipitation experiment was repeated at least twice from independent plants. A monoclonal antibody to T7 tag was acquired from Novagen.
RESULTS
Identification and Expression Studies of AtVPS11 and AtVPS33
In yeast, Vps16p forms the C–Vps complex, including Vps33p, a protein from the Sec1p family. On the basis of the homology between Vps16p and VCL1, an Arabidopsis Sec1p homologue is probably found in a similar complex with VCL1. Six genes are annotated as members of the Sec1p family of proteins in the Arabidopsis genome. On the basis of sequence analysis, these Arabidopsis genes can be classified into four groups. The Arabidopsis genes belonging to each group are more closely related to a yeast homologue from this family (Sec1p, Vps33p, Sly1p, or Vps45p) than to other Arabidopsis paralogues (Sanderfoot et al., 2000). By this criterion, a single gene homologue of Vps33p (At3g54860) can be found in Arabidopsis. Because of sequence information and functional homology, we renamed this gene AtVPS33. We have obtained the complete cDNA corresponding to the AtVPS33 gene by RT-PCR on root tissue. The cDNA sequence was deposited in the GenBank database (accession number AF357527). The AtVPS33 cDNA differs substantially from the cDNA predicted from the genomic sequence in the Arabidopsis database and contains a 592–amino acid open reading frame.
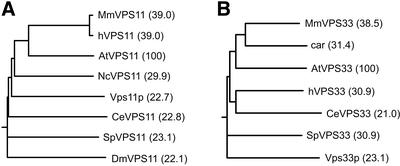
We have also identified a single-gene homologue of Vps11p in the Arabidopsis genome (At2g05170). The full-length cDNA was acquired from an EST clone (AV529217). The gene is named AtVPS11 and contains a 932–amino acid open reading frame. Both AtVPS11 and AtVPS33 show higher sequence identity to the homologues from Mus musculus and Homo sapiens than to the yeast Vps11p and Vps33p proteins, respectively (Figure 1).
Figure 1.
Phylogenetic analysis of (A) selected Vps11p-like proteins and (B) Vps33p-like proteins from Arabidopsis thaliana (At), Caenorhabditis elegans (Ce), Drosophila melanogaster (Dm), Homo sapiens (h), Mus musculus (Mm), Neurospora crassa (Nc), and Schizosaccharomyces pombe (Sp) (names of the species are listed in the alphabetical order). Full-length sequences were aligned using ClustalW algorithm, and rooted phylogenetic tree was prepared using Drawgram algorithm. Percentage of identity toward Arabidopsis homologues, shown in parentheses, was calculated using Align algorithm. All algorithms were applied under default conditions in the Biology Workbench web-based environment (http://workbench.sdsc.edu). See MATERIALS AND METHODS for details and protein accession numbers.
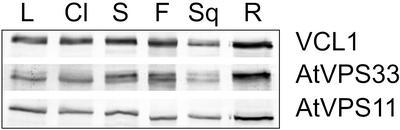
To study AtVPS11 and AtVPS33 in more detail, we raised antibodies in rabbits against protein fragments expressed as His fusions in Escherichia coli. The crude sera were affinity-purified and used for all subsequent experiments. We investigated the expression pattern of VCL1, AtVPS11, and AtVPS33 in different tissues by Western blot analysis (Figure 2). The proteins were ubiquitously expressed in all tissues analyzed, although a higher expression was observed in roots, a pattern similar to that of other Arabidopsis genes involved in vesicular trafficking, such as AtVPS45 (Bassham and Raikhel, 1998), SYP21 (Bassham et al., 1995), and SYP51, SYP61, and SYP71 (Sanderfoot et al., 2001a).
Figure 2.
VCL1, AtVPS11, and AtVPS33 have similar tissue distribution patterns. Equal amounts of total protein extracted from leaves (L), cauline leaves (Cl), stems (S), flowers (F), siliques (Sq), and roots (R) were separated by SDS-PAGE and analyzed by immunoblotting with antisera to VCL1, AtVPS11, and AtVPS33.
VCL1, AtVPS11, and AtVPS33 are Peripheral Membrane-associated Proteins
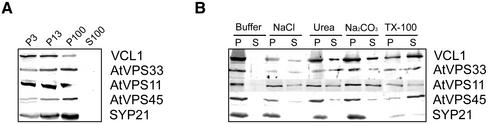
VCL1, AtVPS11, and AtVPS33 proteins contain no hydrophobic stretches that may function as signal peptides or transmembrane domains, or sequences that resemble transit peptides for transport into plastids or nuclear localization signals. Therefore, they are predicted to be cytosolic proteins. Many cytosolic proteins involved in vesicle trafficking associate with membranes to perform their function. We analyzed the presence of these proteins in soluble and membrane fractions from Arabidopsis cell cultures by differential centrifugation experiments. As shown in Figure 3A, VCL1, AtVPS11, and AtVPS33 were present entirely in the microsomal fractions, similar to known microsomal proteins like SYP21 (PVC) and AtVPS45 (trans-Golgi network, TGN).
Figure 3.
Association of VCL1, AtVPS11, and AtVPS33 with membranes. (A) AtC–VPS complex associates with microsomal membranes. An Arabidopsis cell suspension extract was separated into a 3000-g membrane fraction (P3), a 13,000-g membrane fraction (P13), a 100,000-g membrane fraction (P100), and a soluble fraction (S100). Equal amounts of protein from each fraction were analyzed by immunoblotting with the antisera to AtC-VPS proteins, AtVPS45, and SYP21. (B) VCL1, AtVPS11, and AtVPS33 are peripheral membrane proteins. A 100,000-g membrane pellet from an Arabidopsis suspension cell extract was resuspended in extraction buffer alone, 1 M NaCl, 0.1 M Na2CO3, 2 M urea, or 1% Triton X-100. Insoluble material was repelleted, and soluble and pellet fractions were analyzed by SDS-PAGE and with the indicated antibodies.
VCL1, AtVPS11, and AtVPS33 were solubilized from the membranes by urea, NaCl, and under alkaline conditions but to varying extents. The extraction efficiency resembles the solubilization of the previously described peripheral membrane protein AtVPS45 (Bassham and Raikhel, 1998), unlike the integral membrane protein SYP21, which is solubilized only with detergent (Figure 3B). Thus, VCL1, AtVPS11, and AtVPS33 are unlikely to be integral membrane proteins. Lacking detectable transmembrane domains, they appear to be peripherally associated with membranes, probably by interacting with other membrane-anchored proteins.
VCL1, AtVPS11, and AtVPS33 Are Associated with the Tonoplast and the PVC
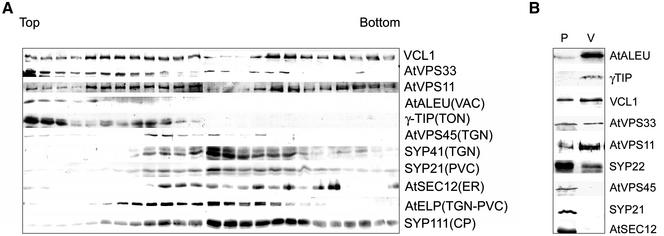
To discern the subcellular localization of VCL1, AtVPS11, and AtVPS33, we fractionated microsomes from Arabidopsis in linear sucrose density gradients, and their distribution in the gradient was compared with previously characterized markers of different compartments of the Arabidopsis endomembrane system (Figure 4A). The vacuolar markers γ-TIP and AtALEU fractionate at the top of the gradient, whereas markers from other compartments, such as AtVPS45 and SYP41 (TGN) (Bassham et al., 2000), AtSEC12 (endoplasmic reticulum, ER) (Bar-Peled and Raikhel, 1997), SYP21 (PVC) (Sanderfoot et al., 1998) and KNOLLE/SYP111 (cell plate) (Lukowitz et al., 1996), are present in denser fractions. VCL1, AtVPS11, and AtVPS33 were present at the top of the gradient together with AtALEU and γ-TIP. In addition, they were also present in denser fractions together with AtVPS45, SYP21, and SYP41. These results indicate that VCL1, AtVPS11, and AtVPS33 associate with the tonoplast membrane as well as with some denser compartments.
Figure 4.
(A) Sucrose-density fractionation of VCL1, AtVPS11, and AtVPS33. A 100,000-g membrane fraction from an Arabidopsis cell suspension extract was analyzed by fractionation on a sucrose density gradient. Fractions were analyzed by SDS-PAGE and immunoblotting with the antisera to VCL1, AtVPS11, AtVPS33, and indicated subcellular markers. (B) VCL1, AtVPS11, and AtVPS33 are associated with the tonoplast membrane and denser endomembrane compartments. The protein samples from protoplasts (P) and vacuoles (V) from an Arabidopsis cell suspension were volume-normalized to reflect the organelle redistribution during the vacuole isolation and analyzed by SDS-PAGE and immunoblotting with the indicated antisera.
To confirm the observed cofractionation of VCL1, AtVPS11, and AtVPS33 with vacuolar markers, we isolated vacuoles from Arabidopsis cell cultures as previously described (Ahmed et al., 2000). The fractions enriched in vacuoles were selected under the microscope by staining with neutral red (data not shown). We examined the presence of different markers of the endomembrane system in the vacuolar fractions. As expected, the vacuolar markers AtALEU and γ-TIP were enriched in the vacuolar fractions (Figure 4B), but neither AtVPS45 (TGN), SYP21 (PVC), nor AtSEC12 (ER) were present at detectable levels in the vacuole-enriched fractions, indicating that they were not contaminated with those endomembranes. The vacuole-enriched fractions contained a significant amount of VCL1, AtVPS11, AtVPS33, and SYP22 (Figure 4B), indicating that these proteins were associated with the tonoplast membrane. SYP22 was previously localized to the PVC (Sanderfoot et al., 1999), but it may also be found on the tonoplast in the apical meristem (Sato et al., 1997). However, the enrichment factor was not as high as in the case of γ-TIP or AtALEU. The previously described localization of SYP22 strongly suggests that SYP22 may be associated with two membrane populations: the PVC and probably the tonoplast.
To further determine the localization of AtVPS33, AtVPS11, and VCL1, we performed immunogold electron microscopy on Arabidopsis roots. Double-immunolabeling with AtVPS11 and AtVPS33 antisera showed that both colocalized extensively. The colocalization was found consistently over the tonoplast and a dense membrane structure, probably the PVC (Figure 5A). However, not all vacuoles were equally labeled (Figure 5A, top left corner, and 5B). The anti-VCL1 antibodies proved to be unsuitable for immunoelectron microscopy (data not shown). Thus, we decided to use transgenic Arabidopsis plants that express a VCL1-GFP fusion protein. Using the GFP monoclonal antibodies, we were able to show that VCL1 labeling was associated with the tonoplast and the same dense compartments (Figure 5C), consistent with the localization of AtVPS11 and AtVPS33. Tonoplast localization of VCL1 in purified vacuoles and in plants expressing a VCL1-GFP fusion protein allowed us to conclude that VCL1 localization was not altered in transgenic Arabidopsis plants. To decide unambiguously whether the dense subcellular structure labeled with VCL1, AtVPS11, and AtVPS33 proteins is the PVC, we performed colocalization of AtVPS33 and T7-tagged SYP22 in transgenic Arabidopsis expressing T7-SYP22 (Sanderfoot et al., 1999). It was previously shown that localization of T7-tagged SYP2-type syntaxins to the PVC is not affected in the transgenic Arabidopsis plants (Sanderfoot et al., 1999). Using T7 monoclonal antibodies and affinity-purified AtVPS33 antibodies, we confirmed that the AtC–VPS complex was localized to the PVC (Figure 5D). Interestingly, although SYP22 was previously localized to the tonoplast in the apical meristem by immunoelectron microscopy (Sato et al., 1997), no labeling of the tonoplast was detected by use of either SYP22 or T7 antibodies in plants expressing T7-SYP22 (Sanderfoot et al., 1999).
Figure 5.
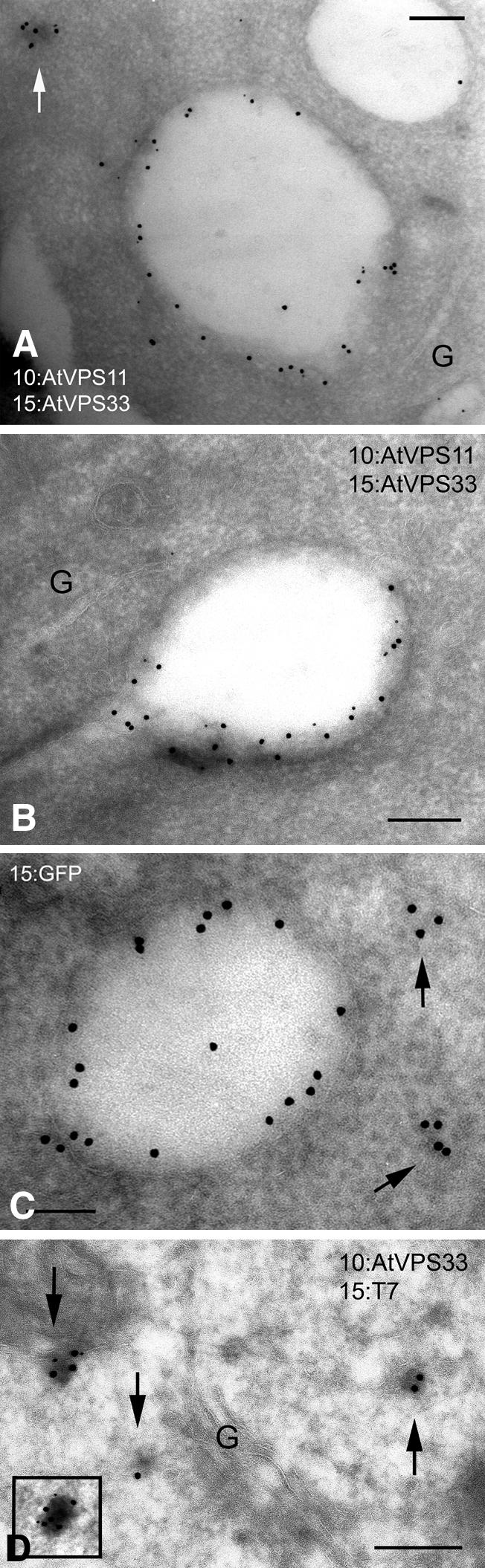
VCL1, AtVPS11, and AtVPS33 localize to the tonoplast and the PVC. (A and B) Cryosections of Arabidopsis root cells were double-immunolabeled with affinity-purified AtVPS11 and AtVPS33 antibodies. AtVPS11 was detected with 10 nm of gold; AtVPS33 was detected with 15 nm of gold. G, Golgi. An arrow highlights the PVC. (C) A cryosection of transgenic Arabidopsis root cell expressing VCL1-GFP fusion was immunolabeled with monoclonal GFP antibodies. VCL1 was detected with 15 nm of gold. Arrows indicate the PVC. (D) A cryosection of transgenic Arabidopsis root cell expressing T7-SYP22 was double-immunolabeled with monoclonal T7 and affinity-purified AtVPS33 antibodies. AtVPS33 was detected with 10 nm of gold; T7-SYP22 was detected with 15 nm of gold. Control sections showed no detectable labeling (data not shown). G, Golgi. Arrows and inset show the PVCs. Bars, 200 nm.
VCL1, AtVPS11, and AtVPS33 Form Complexes in Arabidopsis, Coimmunoprecipitating SYP2-Type Syntaxins
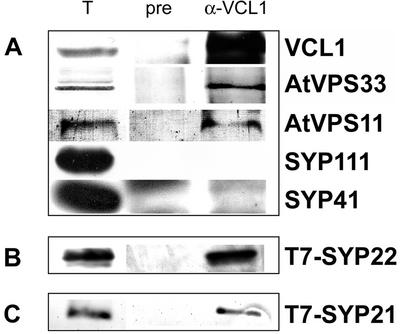
In yeast, Vps11p, Vps16p, and Vps33p form part of a multiprotein complex called the C–Vps complex (Rieder and Emr, 1997; Sato et al., 2000; Peterson and Emr, 2001). We investigated whether the VCL1, AtVPS11, and AtVPS33 proteins assemble into a complex in Arabidopsis. We used the VCL1 antibodies to immunoprecipitate VCL1-interacting proteins from detergent extracts of Arabidopsis microsomes. We studied the presence of candidate proteins in the VCL1-immunoprecipitated fraction by immunoblotting. As shown in Figure 6A, VCL1, AtVPS11, and AtVPS33 were coimmunoprecipitated with the VCL1 antibody, indicating that they interact in Arabidopsis, forming the AtC–VPS complex.
Figure 6.
(A) VCL1, AtVPS11, and AtVPS33 coimmunoprecipitate with the use of VCL1 antibodies. Detergent-solubilized membrane preparations from Arabidopsis suspension cultures were subjected to immunoprecipitation using VCL1 antibodies. Aliquots of total extract (T), a control, using the preimmune serum (pre), and the eluate (α-VCL1) were analyzed by immunoblotting with indicated antibodies. (B and C) AtC–VPS complex interacts with SYP2 syntaxins. VCL1 protein was immunoprecipitated from total membrane proteins prepared from transgenic Arabidopsis plants expressing either T7-SYP22 (B) or T7-SYP21 (C). Total protein (T), a preimmune control (pre), and the eluate (α-VCL1) were analyzed with T7-monoclonal antibodies.
The C–Vps complex in yeast interacts with the Vam3p syntaxin, facilitating its assembly into a trans-SNARE complex, and promotes membrane fusion. The closest homologues of Vam3p in Arabidopsis are SYP2-type syntaxins (Sanderfoot et al., 1999). We analyzed whether the AtC–VPS complex interacts with SYP2-type syntaxins and/or other syntaxins from Arabidopsis (SYPs). No interaction of SYP21 (PVC, Sanderfoot et al., 1998), SYP22 (vacuole, Sato et al., 1997; PVC, Sanderfoot et al., 1998), SYP41 (TGN, Bassham et al., 2000), SYP51 and SYP61 (TGN/PVC, Sanderfoot et al., 2001a), or KNOLLE/SYP111 (cell plate, Lukowitz et al., 1996) with AtC-VPS proteins was observed when VCL1 antibodies or AtVPS33 antibodies were used for the immunoprecipitation (Figure 6A or data not shown). To avoid the possibility that our assay is not sensitive enough to detect the interactions, we decided to analyze the complex formation in transgenic Arabidopsis plants that overexpress epitope-tagged T7-SYP21 and T7-SYP22 (Sanderfoot et al., 1999), the most likely candidate SNAREs. The localization of both T7-tagged SYP2-type syntaxins is not altered in these transgenic plants, as was shown previously (Sanderfoot et al., 1999). The use of highly sensitive T7 antibodies further facilitated the detection of SYP2-type syntaxins in eluates. Using VCL1 antiserum, we were able to precipitate both T7-SYP21 and T7-SYP22 from detergent extracts isolated from Arabidopsis microsomes (Figure 6, B and C).
DISCUSSION
A number of proteins that may be involved in vesicular trafficking in plants have been identified. These include a putative sorting receptor for vacuolar proteins (Ahmed et al., 2000), as well as proteins involved in vesicle formation (Pimpl et al., 2000, Kang et al., 2001) and in vesicle targeting and fusion (Bassham et al. 1995, 2000; Lukowitz et al., 1996; Sato et al., 1997; Assaad et al., 2001; Heese et al., 2001; Sanderfoot et al., 2001a). These studies revealed that the protein families that mediate vesicle trafficking in plants are similar to those in other eukaryotic organisms. However, it has also become apparent that it is not possible to predict the precise function of plant proteins on the basis of the function of their closest sequence homologues from yeast or animals (Bassham and Raikhel, 2000). Moreover, the secretory pathway has many unique features in plants, including plant-specific processes such as cell plate formation during cytokinesis and secretion of plant cell-wall polysaccharides, as well as plant cell-specific compartments. In particular, certain cell types contain specialized vacuoles called protein storage vacuoles (PSVs) that appear to be compound organelles (Jiang et al., 2001). Several types of vacuoles, such as PSVs and lytic vacuoles (LVs), may coexist within an individual plant cell (for review, see Vitale and Raikhel, 1999). Microscopy observations indicate that proteins are targeted to this PSV from the TGN or directly from the ER in dense vesicles that are distinct from the clathrin-coated vesicles that transport other proteins to the LV (Hoh et al., 1995; Robinson et al., 1998; Ahmed et al., 2000; Hillmer et al., 2001). In addition, these pathways have been dissected pharmacologically (Matsuoka et al., 1995). However, very few genetic data on the components involved in vesicular trafficking to vacuoles or in the biogenesis of these organelles in plants is available. Several mutants in putative components of the vesicular trafficking machinery in plants have been identified (Shevell et al., 1994; Lukowitz et al., 1996; Assaad et al. 2001; Heese et al., 2001; Rojo et al., 2001; Sanderfoot et al., 2001a; Kato et al., 2002). Vcl1 and zig1/vti11 mutants have defects in vacuole formation, and their gene products are predicted to have a direct function in membrane docking/fusion at the vacuole and the PVC (Rojo et al., 2001; Kato et al., 2002). Null mutations in SYP41, SYP42, SYP21, and SYP22, which may be involved in a vacuolar transport pathway, are lethal during early pollen development (Sanderfoot et al., 2001b), and homozygous mutant plants cannot be recovered.
To identify other proteins involved in Arabidopsis vacuolar formation, we searched for VCL1-interacting proteins on the basis of the hypothesis that VCL1 forms a complex similar to that of the C–Vps complex. VCL1 is homologous to yeast Vps16p, a component of the 38S C–Vps complex that includes Vps11p, Vps18p, Vps33p, Vps39p, and Vps41p and regulates the docking stage of membrane fusion at the tonoplast (Rieder and Emr, 1997; Sato et al., 2000; Peterson and Emr, 2001). We found AtVPS33, a member of the Sec1p family of proteins homologous to yeast Vps33p, to be membrane-associated in Arabidopsis cell cultures, whereas most of Vps33p/Carnation is soluble in yeast and Drosophila. Similarly, both VCL1 and AtVPS11 were identified to be associated with membranes, but the human homologues hVPS16 and hVPS11, respectively, were detected in both cytosolic and membrane fractions (Kim et al., 2001). The predominance of the membrane-associated form of AtC-VPS proteins in Arabidopsis cell cultures may indicate that the vacuolar trafficking is very active in those cells. It will be of interest to test whether the membrane association of AtC–VPS complex changes in different tissues and whether this can be correlated with vacuolar membrane fusion activity.
A GFP-tagged Vps33p was recently shown to label vacuoles in yeast (Wang et al., 2002), but the localization of the endogenous Vps33p protein or the other C–Vps–complex proteins was not directly assayed. Now we have provided compelling immunoelectron microscopy data showing that the AtC–VPS complex is associated with the tonoplast, consistent with its proposed role in vacuole membrane fusion in fungi. In addition, we provide data that the AtC–VPS complex is also localized to the PVC. In yeast, Vps45p from the Sec1p family was identified to regulate vesicle fusion on both the TGN and PVC. The AtVPS45, an Arabidopsis homologue of Vps45p, is localized exclusively to the TGN (Bassham et al., 2000). Our data indicate that the Sec1p homologue AtVPS33 may have functions on both the PVC and the tonoplast in Arabidopsis.
Members of the Sec1p family of proteins are thought to regulate membrane fusion through their direct interaction with syntaxins. In Arabidopsis, two members of this family have been shown to interact with syntaxins: AtVPS45 interacts with SYP41 and SYP42 in the TGN, and KEULE interacts with KNOLLE/SYP111, presumably in the cell plate. Similarly, the AtC–VPS complex may interact via AtVPS33 with the tonoplast/PVC syntaxins and promote SNARE-complex assembly during docking/fusion. We tested the association of several syntaxins with the AtC–VPS complex and found no evidence of any interaction. On the basis of their localization and of analogy to their yeast and animal counterparts, the syntaxins analyzed were the most likely candidates to interact with the AtC–VPS complex. In this respect, it is important to note that no true homologue of Vam3p, the tonoplast syntaxin that interacts with Vps33p in yeast, has been found in Arabidopsis (Sanderfoot et al., 1999). SYP21 and SYP22 are the closest homologues of Vam3p in Arabidopsis, but they are more closely related to the yeast PVC syntaxin Pep12p. Their localization, primarily in the PVC, also indicates that they are more closely related, with regard to function, to Pep12p than to Vam3p. However, SYP22, which complements the yeast vam3 mutation, has also been localized to the tonoplast in apical meristematic cells (Sato et al., 1997). In this work, we clearly demonstrate the presence of SYP22 in highly purified vacuolar preparations. Both localization and shared homology strongly support the hypothesis that SYP21/22 may interact with AtC–VPS complex. Although the AtC–VPS complex can be associated with the tonoplast/PVC syntaxins only at certain stages of the membrane fusion process, our immunoprecipitation assay may not be sensitive enough to detect the association of a small SYP21/22 pool with the AtC–VPS complex. Therefore, we investigated the possible interactions in transgenic Arabidopsis overexpressing either T7-SYP21 or T7-SYP22. In these plants, a localization of T7-tagged SYP2-type syntaxins remains unaltered (Sanderfoot et al., 1999). In separate experiments, we found that T7-SYP21 and T7-SYP22 coimmunoprecipitated VCL1. These data indicate that SYP2-type syntaxins are t-SNAREs interacting with the AtC–VPS complex. On the basis of the localization topology, we propose that the AtC–VPS complex and SYP2-type syntaxins are involved in heterotypic membrane fusion on the PVC and most likely also in heterotypic and homotypic membrane fusion on the tonoplast.
In summary, we have found novel components of the vesicular trafficking machinery in Arabidopsis, AtVPS11 and AtVPS33. On the basis of sequence, localization, and interaction with the previously characterized protein VCL1, both AtVPS11 and AtVPS33 are probably involved in regulating membrane fusion at the tonoplast and the PVC. Additional biochemical assays to further characterize the complex formation are currently in progress. The identification of these three proteins constitutes a handle to investigate the unique properties of vacuolar biogenesis and vacuolar trafficking in plants. VCL1, AtVPS11, and AtVPS33 are single-copy genes in Arabidopsis, suggesting that there is no redundancy for their function. As in the case of VCL1, null mutations in AtVPS33 and AtVPS11 are expected to be lethal. Therefore, the identification and analysis of leaky VCL1, AtVPS11, and AtVPS33 mutants will be of a special interest. Tilling, a method for the identification of single base changes in a gene of interest, was recently developed for Arabidopsis (Colbert et al., 2001). In this way, it is possible to obtain allelic series of mutants with single amino acid substitutions, which may result in leaky or temperature-sensitive mutations. These mutants may facilitate the investigation of transport pathways with respect to the targeting of vacuolar cargo proteins and the morphology of the endomembrane compartments. In addition, the analysis of the Arabidopsis genome revealed putative single-gene homologues of yeast VPS18, VPS39, and VPS41 genes, At1g12470, At4g36630, and At1g08190, respectively. The role of the corresponding proteins in the formation of the AtC–VPS complex is currently under investigation.
ACKNOWLEDGMENTS
The authors thank Kazusa DNA Research Institute for providing the cloned cDNA of AtVPS11. We also acknowledge Dr. Diane C. Bassham and Dr. Anton A. Sanderfoot for helpful comments on the manuscript and Jocelyn Brimo for helping with the artwork and formatting the manuscript. This work was supported by funds from the Department of Energy (DE-FG03–02ER15295/A000), the United States–Israel Binational Science (1999355), and the National Science Foundation (MCB-0296080) to N.V.R.
Abbreviations used:
- PVC
prevacuolar compartment
- SNARE
soluble N-ethyl maleimide sensitive factor adaptor protein receptor
- SYP
syntaxin of plants
- TGN
trans-Golgi network
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–08–0509. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02–08–0509.
REFERENCES
- Ahmed SU, Bar-Peled M, Raikhel NV. Cloning and subcellular location of an Arabidopsis receptor-like protein that shares common features with protein-sorting receptors of eukaryotic cells. Plant Physiol. 1997;114:325–336. doi: 10.1104/pp.114.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed SU, Rojo E, Kovaleva V, Venkataraman S, Dombrowski JE, Matsuoka K, Raikhel NV. The plant vacuolar sorting receptor AtELP is involved in transport of NH2-terminal propeptide-containing vacuolar proteins in Arabidopsis thaliana. J Cell Biol. 2000;149:1335–1344. doi: 10.1083/jcb.149.7.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaad FF, Huet Y, Mayer U, Jürgens G. The cytokinesis gene KEULE encodes a Sec1 protein that binds the syntaxin KNOLLE. J Cell Biol. 2001;152:531–543. doi: 10.1083/jcb.152.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled M, Raikhel NV. Characterization of AtSEC12 and AtSAR1: proteins likely involved in endoplasmic reticulum and Golgi transport. Plant Physiol. 1997;114:315–324. doi: 10.1104/pp.114.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham DC, Gal S, da Silva Conceição A, Raikhel NV. An Arabidopsis syntaxin homologue isolated by functional complementation of a yeast pep12 mutant. Proc Natl Acad Sci USA. 1995;92:7262–7266. doi: 10.1073/pnas.92.16.7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham DC, Raikhel NV. An Arabidopsis VPS45p homolog implicated in protein transport to the vacuole. Plant Physiol. 1998;117:407–415. doi: 10.1104/pp.117.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham DC, Raikhel NV. Plant cells are not just green yeast. Plant Physiol. 2000;122:999–1001. doi: 10.1104/pp.122.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassham DC, Sanderfoot AA, Kovaleva V, Zheng HY, Raikhel NV. AtVPS45 complex formation at the trans-Golgi network. Mol Biol Cell. 2000;11:2251–2265. doi: 10.1091/mbc.11.7.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent AF, Kunkel BN, Dahlbeck D, Brown KL, Schmidt R, Giraudat J, Leung J, Staskawicz BJ. RPS2 of Arabidopsis thaliana: a leucine-rich repeat class of plant disease resistance genes. Science. 1994;265:1856–1860. doi: 10.1126/science.8091210. [DOI] [PubMed] [Google Scholar]
- Bock JB, Matern HT, Peden AA, Scheller RH. A genomic perspective on membrane compartment organization. Nature. 2001;409:839–841. doi: 10.1038/35057024. [DOI] [PubMed] [Google Scholar]
- Colbert T, Till BJ, Tompa R, Reynolds S, Steine MN, Yeung AT, McCallum CM, Comai L, Henikoff S. High-throughput screening for induced point mutations. Plant Physiol. 2001;126:480–484. doi: 10.1104/pp.126.2.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP: phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Guo W, Roth D, Walch-Solimena C, Novick P. The exocyst is an effector for Sec4p, targeting secretory vesicles to sites of exocytosis. EMBO J. 1999;18:1071–1080. doi: 10.1093/emboj/18.4.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese M, Gansel X, Sticher L, Wick P, Grebe M, Granier F, Jürgens G. Functional characterization of the KNOLLE-interacting t-SNARE AtSNAP33, and its role in plant cytokinesis. J Cell Biol. 2001;155:239–249. doi: 10.1083/jcb.200107126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillmer S, Movafeghi A, Robinson DG, Hinz G. Vacuolar storage proteins are sorted in the cis-cisternae of the pea cotyledon Golgi apparatus. J Cell Biol. 2001;152:41–50. doi: 10.1083/jcb.152.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoh B, Hinz G, Jeong BK, Robinson DG. Protein storage vacuoles form de novo during pea cotyledon development. J Cell Sci. 1995;108:299–310. doi: 10.1242/jcs.108.1.299. [DOI] [PubMed] [Google Scholar]
- Horazdovsky BF, Emr SD. The VPS16 gene product associates with a sedimentable protein complex and is essential for vacuolar protein sorting in yeast. J Biol Chem. 1993;268:4953–4962. [PubMed] [Google Scholar]
- Jiang L, Phillips TE, Hamm CA, Drozdowicz YM, Rea PA, Maeshima M, Rogers SW, Rogers JC. The protein storage vacuole: a unique compound organelle. J Cell Biol. 2001;155:991–1002. doi: 10.1083/jcb.200107012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang BH, Busse JS, Dickey C, Rancour DM, Bednarek SY. The Arabidopsis cell plate-associated dynamin-like protein, ADL1Ap, is required for multiple stages of plant growth and development. Plant Physiol. 2001;126:47–68. doi: 10.1104/pp.126.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Morita MT, Fukaki H, Yamauchi Y, Uehara M, Niihama M, Tasaka M. SGR2, a phospholipase-like protein, and ZIG/SGR4, a SNARE, are involved in the shoot gravitropism of Arabidopsis. Plant Cell. 2002;14:33–46. doi: 10.1105/tpc.010215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BY, Kramer H, Yamamoto A, Kominami E, Kohsaka S, Akazawa C. Molecular characterization of mammalian homologues of class C Vps proteins that interact with syntaxin-7. J Biol Chem. 2001;276:29393–29402. doi: 10.1074/jbc.M101778200. [DOI] [PubMed] [Google Scholar]
- Lauber MH, Waizenegger I, Steinmann T, Schwarz H, Mayer U, Hwang I, Lukowitz W, Jürgens G. The Arabidopsis KNOLLE protein is a cytokinesis-specific syntaxin. J Cell Biol. 1997;139:1485–1493. doi: 10.1083/jcb.139.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W, Mayer U, Jürgens G. Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell. 1996;12:61–71. doi: 10.1016/s0092-8674(00)80993-9. [DOI] [PubMed] [Google Scholar]
- Matsuoka K, Bassham DC, Raikhel NV, Nakamura K. Different sensitivity to wortmannin of two vacuolar sorting signals indicates the presence of distinct sorting machineries in tobacco cells. J Cell Biol. 1995;130:1307–1318. doi: 10.1083/jcb.130.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers EW, Miller W. Optimal alignments in linear space. Comput Appl Biosci. 1988;4:11–17. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- Peters C, Bayer MJ, Buhler S, Andersen JS, Mann M, Mayer A. Trans-complex formation by proteolipid channels in the terminal phase of membrane fusion. Nature. 2001;409:581–587. doi: 10.1038/35054500. [DOI] [PubMed] [Google Scholar]
- Peterson MR, Emr SD. The class C Vps complex functions at multiple stages of the vacuolar transport pathway. Traffic. 2001;2:476–486. doi: 10.1034/j.1600-0854.2001.20705.x. [DOI] [PubMed] [Google Scholar]
- Pimpl P, Movafeghi A, Coughlan S, Denecke J, Hillmer S, Robinson DG. In situ localization and in vitro induction of plant COPI-coated vesicles. Plant Cell. 2000;12:2219–2236. doi: 10.1105/tpc.12.11.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A, Seals D, Wickner W, Ungermann C. The docking stage of yeast vacuole fusion requires the transfer of proteins from a cis-SNARE complex to a Rab/Ypt protein. J Cell Biol. 2000;148:1231–1238. doi: 10.1083/jcb.148.6.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder SE, Emr SD. A novel RING finger protein complex essential for a late step in protein transport to the yeast vacuole. Mol Biol Cell. 1997;8:2307–2327. doi: 10.1091/mbc.8.11.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DG, Baumer M, Hinz G, Hohl I. Vesicle transfer of storage protein to the vacuole: the role of the Golgi apparatus and multivesicular bodies. J Plant Physiol. 1998;152:659–667. [Google Scholar]
- Rojo E, Gillmor CS, Kovaleva V, Somerville CR, Raikhel NV. VACUOLELESS1 is an essential gene required for vacuole formation and morphogenesis in Arabidopsis. Dev Cell. 2001;1:303–310. doi: 10.1016/s1534-5807(01)00024-7. [DOI] [PubMed] [Google Scholar]
- Sacher M, Jiang Y, Barrowman J, Scarpa A, Burston J, Zhang L, Schieltz D, Yates JR, III, Abeliovich H, Ferro-Novick S. TRAPP, a highly conserved novel complex on the cis-Golgi that mediates vesicle docking and fusion. EMBO J. 1998;17:2494–2503. doi: 10.1093/emboj/17.9.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot AA, Ahmed SU, Marty-Mazars D, Rapoport I, Kirchhausen T, Marty F, Raikhel NV. A putative vacuolar cargo receptor partially colocalizes with AtPEP12p on a prevacuolar compartment in Arabidopsis roots. Proc Natl Acad Sci USA. 1998;95:9920–9925. doi: 10.1073/pnas.95.17.9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot AA, Assaad FF, Raikhel NV. The Arabidopsis genome: an abundance of soluble N-ethylmaleimide-sensitive factor adaptor protein receptors. Plant Physiol. 2000;124:1558–1569. doi: 10.1104/pp.124.4.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot AA, Kovaleva V, Bassham DC, Raikhel NV. Interactions between syntaxins identify at least five SNARE complexes within the Golgi/prevacuolar system of the Arabidopsis cell. Mol Biol Cell. 2001a;12:3733–3743. doi: 10.1091/mbc.12.12.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot AA, Kovaleva V, Zheng H, Raikhel NV. The t-SNARE AtVAM3p resides on the prevacuolar compartment in Arabidopsis root cells. Plant Physiol. 1999;121:929–938. doi: 10.1104/pp.121.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot AA, Pilgrim M, Adam L, Raikhel NV. Disruption of individual members of Arabidopsis syntaxin gene families indicates each have essential functions. Plant Cell. 2001b;13:659–666. doi: 10.1105/tpc.13.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Nakamura N, Ohsumi Y, Kouchi H, Kondo M, Hara-Nishimura I, Nishimura M, Wada Y. The AtVAM3 encodes a syntaxin-related molecule implicated in the vacuolar assembly in Arabidopsis thaliana. J Biol Chem. 1997;272:24530–24535. doi: 10.1074/jbc.272.39.24530. [DOI] [PubMed] [Google Scholar]
- Sato TK, Rehling P, Peterson MR, Emr SD. Class C Vps protein complex regulates vacuolar SNARE pairing and is required for vesicle docking/fusion. Mol Cell. 2000;6:661–671. doi: 10.1016/s1097-2765(00)00064-2. [DOI] [PubMed] [Google Scholar]
- Sevrioukov EA, He JP, Moghrabi N, Sunio A, Kramer H. A role for the deep orange and carnation eye color genes in lysosomal delivery in Drosophila. Mol Cell. 1999;4:479–486. doi: 10.1016/s1097-2765(00)80199-9. [DOI] [PubMed] [Google Scholar]
- Shevell DE, Leu WM, Gillmor CS, Xia G, Feldmann KA, Chua NH. EMB30 is essential for normal cell division, cell expansion, and cell adhesion in Arabidopsis and encodes a protein that has similarity to Sec7. Cell. 1994;77:1051–1062. doi: 10.1016/0092-8674(94)90444-8. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale A, Raikhel NV. What do proteins need to reach different vacuoles? Trends Plant Sci. 1999;4:149–155. doi: 10.1016/s1360-1385(99)01389-8. [DOI] [PubMed] [Google Scholar]
- Wang L, Seeley ES, Wickner W, Merz AJ. Vacuole fusion at a ring of vertex docking sites leaves membrane fragments within the organelle. Cell. 2002;108:357–369. doi: 10.1016/s0092-8674(02)00632-3. [DOI] [PubMed] [Google Scholar]
- Warner TS, Sinclair DA, Fitzpatrick KA, Singh M, Devlin RH, Honda BM. The light gene of Drosophila melanogaster encodes a homologue of VPS41, a yeast gene involved in cellular-protein trafficking. Genome. 1998;41:236–243. [PubMed] [Google Scholar]
- Wickner W. Yeast vacuoles and membrane fusion pathways. EMBO J. 2002;21:1241–1247. doi: 10.1093/emboj/21.6.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]