Abstract
Like other guanine nucleotide-exchange proteins (GEPs) that activate ADP-ribosylation factor (ARF) GTPases, brefeldin A-inhibited GEP2, BIG2, contains an ≈200-aa Sec7 domain that is responsible for this catalytic activity and its inhibition by brefeldin A. The Sec7 domain is located near the center of the molecule and serves to accelerate replacement of GDP bound to ARF with GTP. To explore possible functions of the N-terminal region of BIG2 (1–832), we used three coding-region constructs as bait to screen a human heart cDNA library in a yeast two-hybrid system, retrieving two unique clones that encode a type I protein kinase A (PKA) regulatory subunit, RIα. Coimmunoprecipitation experiments confirmed interaction of in vitro translated BIG2 and RIα, as well as of the endogenous proteins in cytosol of cultured HepG2 cells. Using 28 deletion mutants, we found three regions of BIG2 that interacted with R subunits of PKA. Residues 27–48 (domain A) interacted with RIα and RIβ, 284–301 (domain B) interacted with RIIα and RIIβ, and 517–538 (domain C) interacted with RIα, RIIα, and RIIβ. Sequence analysis and helical wheel projection of amino acids in the three domains revealed potential amphipathic wheel structures characteristic for binding of PKA R subunits. Western blot analysis of subcellular fractions demonstrated translocation of BIG2 (and BIG1) from cytosol to the Golgi and other membrane structures after incubation of cells with 8-Br-cAMP or forskolin. All findings are consistent with a role for BIG2 as an A kinase-anchoring protein (or AKAP) that could coordinate cAMP and ARF regulatory pathways.
The ADP-ribosylation factors (ARFs) are 20-kDa GTPases that have key roles in intracellular vesicular trafficking. They are activated when GTP is bound and associate with membranes to initiate vesicle formation (1, 2). Activation of ARFs requires a guanine nucleotide-exchange protein (GEP) to accelerate release of bound nucleotide from inactive cytosolic ARF-GDP, thereby promoting formation of ARF-GTP, which is membrane bound. ARF GEPs differ in size, functional domain structure, and sensitivity to brefeldin A (BFA), a fungal fatty acid metabolite that blocks protein secretion (3). Mammalian BFA-inhibited GEPs, BIG1 and BIG2, were initially purified as components of an ≈670-kDa cytosolic macromolecular complex (4, 5). Microscopically, endogenous BIG1 and BIG2 in resting HeLa S3 and HepG2 cells were distributed in a punctate pattern throughout the cytoplasm and partially concentrated in the perinuclear Golgi region (6). The presence of BIG2 in cytosol and Golgi is consistent with its dynamic regulation and putative transport functions involving mechanisms that remain to be defined. BIG2, like all ARF GEPs, contains a Sec7 domain that accelerates guanine nucleotide exchange and is responsible for its BFA inhibition, but little is known about functions of the rest of the BIG2 molecule.
Activation of protein kinase A (PKA) was reported to enhance the association of ARF with Golgi membranes (7). Addition of PKA catalytic (C) subunit increased the binding of cytosolic, as well as recombinant, ARF1 to Golgi membranes in vitro, and increasing intracellular cAMP induced translocation of ARF from cytosol to Golgi membranes in cultured cells (7). Thus, a Golgi-localized PKA might play a role in the regulation of coat component recruitment to Golgi membranes. The broad spectrum of signaling events controlled by cAMP (8) requires compartmentalization of PKA and its substrates along with other molecules involved in specific intracellular functions. This is accomplished through association of the regulatory (R) subunits with A kinase-anchoring proteins (AKAPs). AKAPs are a structurally diverse group of proteins with the common function of binding of R subunits and confining PKA activation to discrete locations in the cell (9–11). PKAs are tetrameric serine/threonine kinases with two C and two R subunits, RIα and RIβ in type I and RIIα and RIIβ in type II. Type II PKA is generally associated with specific cellular structures and organelles, whereas type I, which is more sensitive to cAMP, is predominantly cytosolic (8). Classical AKAPs that anchor type II PKA holoenzymes, contain an amphipathic helix of 14–18 amino acids that binds the N termini of homodimers of RIIα or RIIβ (12, 13). So-called dual AKAPs interact with both RI and RII subunits (14, 15).
Here, we report that BIG2 interacted with RIα in a yeast two-hybrid system. Its interaction with RIβ, as well as with RII subunits, was shown later. Three binding domains with different R subunit specificities and sequences consistent with amphipathic helix formation were identified. Incubation of HepG2 cells with 8-Br-cAMP or forskolin increased the amounts of BIG2 that were membrane associated. These data are consistent with a role for BIG2 as an AKAP and with PKA as a component of a macromolecular complex of BIG1 and BIG2.
Materials and Methods
Yeast Two-Hybrid System.
Yeast strains EGY48 [MATα, his3, trp1, ura3, LexAop (x6)-LEU2] and YM4271 (MATa, ura3–52, his3–200, lys2–801, ade2–101, ade5, trp1–901, leu2–3, 112, tyr1–501, gal4-Δ512, gal80-Δ538, ade5∷hisG), MATCHMAKER vectors, and the lacZ reporter plasmid for the LexA system were purchased from CLONTECH and used according to the manufacturer's instructions in published procedures (16).
Antibodies.
Mouse monoclonal antibodies against RIα, RIIα, and C subunits were purchased from BD Signal Transduction (Lexington, KY), chicken polyclonal anti-RIα antibody from Biomol (Plymouth Meeting, PA), horseradish peroxidase-conjugated anti-rabbit IgG, anti-mouse IgG, and anti-chicken IgY from Promega, normal mouse and rabbit IgG from Vector Laboratories, monoclonal LexA and c-myc antibodies from CLONTECH, polyclonal anti-c-myc and anti-hemagglutinin (HA) antibodies from Santa Cruz Biotechnology, and anti-FLAG antibodies from Sigma. Polyclonal antibodies against BIG1 and BIG2 are described in ref. 6.
Cloning of BIG2 Fragments.
Three fragments of BIG2 (see Fig. 1A) were amplified from plasmid pBluescript-BIG2full (provided by Ryoichi Yamaji, see ref. 6) with the following primers (restriction enzyme sites italicized). Forward primer 1 has an EcoRI site (5′-GCGGAATTCATGCAGGAGAGCCAG), reverse primers 2 and 3 have XhoI sites (5′-CGCCTCGAGCTATTGTTGCTTGATG and 5′-CGCCTCGAGCTATGTTTCTTTCATTG), and forward primer 4 has an EcoRI site (5′-GCGGAATTCCAAAAAGAAATCATTGAAC). Primers 1 and 2 were used to prepare constructs encoding BIG2 amino acids 1–643, primers 1 and 3 for 1–832, and primers 4 and 3 for 644–832 (Sec 7 domain). Initiation and termination codons are underlined. PCR products were excised with EcoRI plus XhoI and ligated into pGilda. LexA-BIG2 constructs were sequenced and transformed into the yeast EGY48 to determine that fusion proteins were translocated into the nucleus and had no intrinsic transactivation capability. Stable expression of LexA fusion proteins was verified by Western blotting.
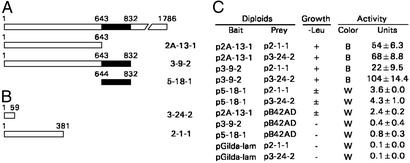
Figure 1.
Interaction between BIG2 and PKA RIα in yeast two-hybrid screen. (A) Three BIG2 fragments encoded by cDNAs used as bait in yeast two-hybrid screen of human heart cDNA library. (B) RIα, full-length (p2-1-1) and amino acids 1–59 (p3-24-2) encoded by cDNAs isolated in the screen. (C) Yeast mating experiments. pGilda (DNA-BD vector only, not shown), pLexA-bait plasmids p2A-13-1, p3-9-2, and p5-18-1, and pGilda-Lam were transformed singly into YM4271 and selected on SD/-His plates. pB42AD (empty vector) and the positive plasmids rescued in KC8 E. coli were transformed separately into EGY48[p8op-lacZ] and selected on SD/-Trp/-Ura plates. β-Galactosidase activity was assayed in solution with o-nitrophenyl-β-D-galactoside as substrate (CLONTECH Yeast Protocol Handbook instructions). Activity = 1,000 × OD420/(t × V × OD600). t = incubation time, V = 0.1 × concentration factor. One unit equals hydrolysis of 1 μmol of substrate per min per OD600 (44). True positives exhibit growth on −Leu plates and β-galactosidase activity (mean ± SD, n = 3). B, blue; W, white.
cDNA Library Screening for BIG2 Interactions.
Library plasmids were extracted, purified (Qiagen, Chatsworth, CA; mega kit), and transformed into yeast EGY48, which had been transformed with the bait plasmids and the lacZ reporter plasmid (p8op-lacZ). Yeast were grown on SD/-His/-Trp/-Ura glucose plates and selected on SD/-His/-Trp/-Ura/-Leu galactose and raffinose plates. Approximately 5 × 107 transformants were screened for interactions evidenced by growth in leucine-free medium and β-galactosidase activity that were dependent on galactose. Specificity of the interaction was confirmed by using as bait cDNAs encoding human lamin C and murine p53. Plasmids were isolated from the positive colonies, inserts amplified by PCR, and PCR products characterized by digestion with AluI, HaeIII, EcoRI, and XhoI to sort 51 positive colonies from p2A-13-1 into seven unique clones; 48 positive colonies from p3-9-2 represented five clones. Positive AD/library plasmids were rescued in KC8 Escherichia coli.
DNA Sequencing and Analysis.
Purified positive AD/library plasmids were initially sequenced by automated sequencing (373 DNA sequenator, Applied Biosystems) with pB42AD sequencing primer 5′-CAGCCTCTTGCTGAGTGGAGATGCC to obtain the 5′ regions of the inserts. Multiple primers located inside of the inserts were used to complete insert sequences. Sequence data were analyzed using GENEWORKS 2.5.1 and compared with nucleotide sequence databases (all GenBank, RefSeq Nucleotides, EMBL, DDBJ, and PDB) by using BLAST (www.ncbi.nlm.nih.gov/BLAST/). p2-1-1 encoded full-length human PKA RIα (NM_002734) and p3-24-2 amino acids 1–59.
Protein Synthesis in Reticulocyte Lysate.
Positive library inserts were excised from pB42AD with EcoRI plus XhoI and inserted into plasmid pGADT7 (CLONTECH) to add an N-terminal HA tag. Bait inserts were excised with EcoRI plus SalI and inserted into the plasmid pGBKT7 (CLONTECH) to add an N-terminal c-myc tag. T7-coupled reticulocyte lysate system (TNT, Promega) was used to synthesize [35S]methionine-labeled bait and library proteins according to the manufacturer's instructions. Samples (5 μl) of in vitro translated bait and prey proteins, combined as indicated, were incubated at 30°C for 1 h before the addition of 470 μl of immunoprecipitation (IP) buffer (20 mM Tris⋅HCl, pH 7.5/150 mM NaCl/1 mM DTT/0.5 mM phenylmethylsulfonyl fluoride/0.1% Tween 20/aprotinin, 5 μg/ml) containing 10 μl (1 μg) of anti-c-myc mAb, anti-HA IgG, or anti-FLAG IgG and 10 μl of protein G-agarose beads. After rocking for 2 h at 4°C, beads were washed three times with 500 μl of TBST buffer (20 mM Tris⋅HCl, pH 7.5/150 mM NaCl/0.1% Tween 20). Proteins eluted in 10 μl of sample buffer were separated by SDS/PAGE (8–16% or 10–20% gel). Dried gels were exposed to x-ray film.
Cloning of PKA Regulatory Subunits.
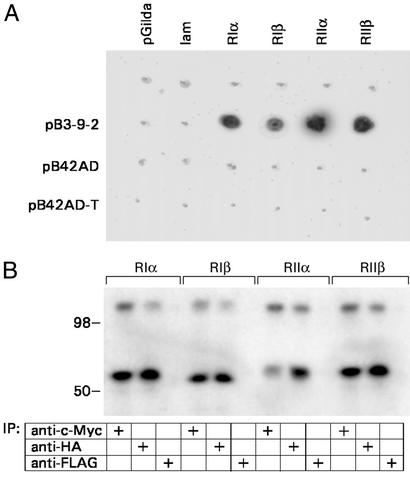
cDNAs for R subunits were amplified by PCR from a human heart cDNA library (CLONTECH) using for RIα (NM_002734), forward primer 5′-GCGGGATCCATGGAGTCTGGCAGTAC with a BamHI site and reverse primer 5′-CGCCTCGAGTCAGACAGACAGTGACAC with a XhoI site; and for RIIβ (M31158), forward primer 5′-GCGGAATTCATGAGCATCGAGATCCCG with an EcoRI site and reverse primer 5′-CGCCCATGGTCATGCAGTGGGTTCAAC with a NcoI site. RIβ (M65066) and RIIα (X14968) were amplified from a human testis cDNA library (CLONTECH) with forward primers (5′-GCGGAATTCATGGCCTCCCCGCC and 5′-GCGGGATCCCCCAACCCGTCTATC) and reverse primers with XhoI sites (5′-CGCCTCGAGTCAGACGGTGAGGGAG and 5′-CGCCTCGAGCACAGCAATGGCAGCAG), respectively. PCR products were excised with appropriate enzymes and inserted into vector pGilda to be transformed into yeast YM4271. BIG2 (1–832), excised from pGilda with EcoRI and XhoI, was ligated to pB42AD to produce pB3-9-2 and transformed into EGY48[p8op-lacZ] for mating experiments (see Fig. 4A).
Figure 4.
BIG2 interacts with PKA RIβ, RIIα, and RIIβ as well as RIα. (A) Yeast diploids formed by mating cells expressing BIG2 (1–832) and those expressing pGilda-RIα, -RIβ, -RIIα, or -RIIβ grew on SD/Gal/Raf/-His/-Leu/-Trp/-Ura plates, and colonies were blue in the presence of X-gal. Data were replicated twice. (B) Samples (5 μl) of [35S]methionine-labeled c-myc-BIG2 (1–832) and the indicated full-length R subunit with an N-terminal HA tag (synthesized in reticulocyte lysate system) were mixed as described for Fig. 2. Proteins immunoprecipitated (2 h, 4°C) with anti-myc, anti-HA, or anti-FLAG antibodies, as indicated, were separated by SDS/PAGE (10–20% gel) before autoradiography. The experiment was replicated twice.
For the experiment in Fig. 4B, PCR with pGilda-RIα, -RIβ, -RIIα, and -RIIβ templates was used to construct pGADT7 plasmids with the corresponding inserts. Forward primers with NdeI sites (5′-GCGCATATGGAGTCTGGCAGTACCGCC, 5′-GCGCATATGGCCTCCCCGCCCGCCTGC, and 5′-GCGCATATGAGCCACATCCAGATCCCG) and reverse primers with XhoI sites (5′-CGCCTCGAGTCAGACAGACAGTGACAC, 5′-CGCCTCGAGTCAGACGGTGAGGGAG, and 5′-CGCCTCGAGCTACTGCCCGAGGTTGC) were used for RIα, RIβ, and RIIα, respectively. For RIIβ, forward primer 5′-GCGGAATTCATGAGCATCGAGATCCCG with an EcoRI site and reverse primer 5′-CGCATCGATTCATGCAGTGGGTTCAAC with a ClaI site were used.
Immunoprecipitation and Western Blot.
To prepare cytosol, confluent HepG2 cells, grown on dishes coated with type I collagen in DMEM (Invitrogen) containing 25 mM Hepes, with 1 mM sodium pyruvate, 10% FBS, penicillin G (100 units/ml), and streptomycin (100 μg/ml) at 37°C in an atmosphere of 5% CO2/95% air, were harvested by scraping in cold PBS. Cells from ≈40 dishes were homogenized in 5 ml of TENDS buffer (20 mM Tris·HCl, pH 8.0/1 mM EDTA/1 mM NaN3/2 mM DTT/0.25 M sucrose), 8 ml per dish, with 0.5 mM 4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF), 0.5 mM benzamidine hydrochloride, and protease inhibitors (soybean and lima bean trypsin inhibitors, leupeptin, and aprotinin, each 1 μg/ml). The homogenate (30 strokes, glass Dounce tissue grinder) was centrifuged (105,000 × g for 1.5 h) to yield cytosol. Samples of cytosol (1.2 mg of protein) were incubated (rocking, 4°C, 4 h) with 5 μg of affinity-purified anti-BIG1, 6 μg of anti-BIG2 IgG, or 5 μg of monoclonal antibodies against RIα, RIIα, or C subunit in TENDS buffer with 2 μM MgCl2, 0.1% ovalbumin, and 0.5 mM AEBSF (total volume 0.5 ml). Controls were normal rabbit or mouse IgG or no antibody. After adding 60 μl of a 50% slurry of protein G-Sepharose CL-4B (Amersham Pharmacia) in PBS and incubation at 4°C overnight, beads were washed three times with 1 ml of ice-cold PBS containing 0.5 mM AEBSF. Proteins bound to beads were eluted in 80 μl of loading buffer at 65°C for 10 min, separated by SDS/PAGE in 4–12% gradient or 8% gels, and transferred to nitrocellulose membrane for incubation with rabbit anti-BIG1 or anti-BIG2 IgG, chicken anti-RIα IgY, or mAb against RIIα or C subunit, followed by appropriate horseradish peroxidase-conjugated second antibodies and detection using Super Signal Chemiluminescent substrate (Pierce).
After determination of the amount of anti-BIG2 IgG or RIα mAb needed for complete IP, samples (50 μg of protein) of HepG2 cytosol were incubated (4°C, 4 h, rocking) with 2 μg of anti-BIG2 IgG or 1.5 μg of anti-RIα mAb in TENDS buffer plus 2 μM MgCl2, 0.5 mM AEBSF, and 0.1% ovalbumin (total volume 250 μl), followed by addition of 40 μl of protein G-Sepharose CL-4B (50% slurry in PBS) and incubation overnight. After centrifugation, the supernatant was subjected to a second IP in the same medium with 2 μg of anti-BIG2 IgG or 1.5 μg of anti-RIα mAb (total volume 0.5 ml), followed by incubation overnight at 4°C with 40 μl of protein G-Sepharose CL-4B (50% slurry in PBS). After washing beads three times with 1 ml of ice-cold PBS containing 0.5 mM AEBSF, bound proteins were eluted with 30 μl of SDS sample buffer and separated by SDS/PAGE (8% gel). Immunoreactive proteins were quantified by using a ChemiImager 5500 (Alpha Innotech, San Leandro, CA).
Results
Interaction of BIG2 with RIα in Vitro and in Vivo.
In a yeast two-hybrid screen of a human heart cDNA library (≈5 × 107 transformants), BIG2 constructs (Fig. 1A) encoding amino acids 1–832 (3-9-2) and 1–643 (2A-13-1) each yielded multiple positive clones, two of which were p2-1-1, encoding full-length PKA RIα, and p3-24-2, encoding the first 59 amino acids of RIα (Fig. 1B). The Sec 7 domain (644–832), which is responsible for ARF GEP activity, yielded no RIα clones. Specificity of the interactions was confirmed by mating experiments (Fig. 1C).
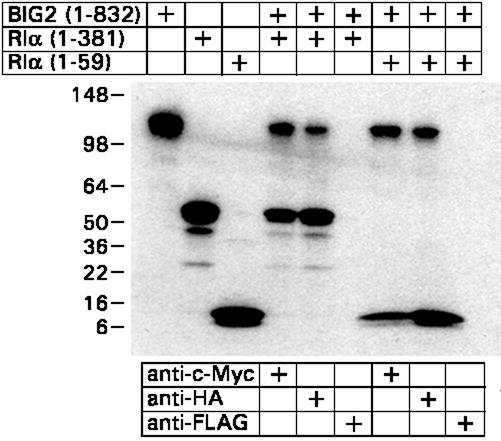
Interaction of BIG2 and RIα was also demonstrated by coimmunoprecipitation of [35S]methionine-labeled epitope-tagged proteins synthesized in a reticulocyte lysate system (Fig. 2). myc-BIG2 (1–832) and HA-RIα (1–381) or HA-RIα (1–59) were individually synthesized and mixed as indicated. Proteins immunoprecipitated with anti-myc, anti-HA, or anti-FLAG (control) antibodies and separated by SDS/PAGE demonstrated an interaction of the N-terminal fragment of recombinant RIα (1–59) and BIG2 (1–832).
Figure 2.
Coimmunoprecipitation of in vitro-translated BIG2 and PKA RIα. Samples (5 μl) of [35S]methionine-labeled bait [c-myc-BIG2 (1–832)] and prey [HA-RIα (1–381), HA-RIα (1–59)] proteins synthesized in TNT T7 reticulocyte lysate system (Promega) were combined as indicated and incubated for 1 h at 30°C before IP, separation of proteins by SDS/PAGE (8–16% gel), and radioautography. The first three lanes are 10-μl samples of the indicated total lysate. Observations were repeated three times.
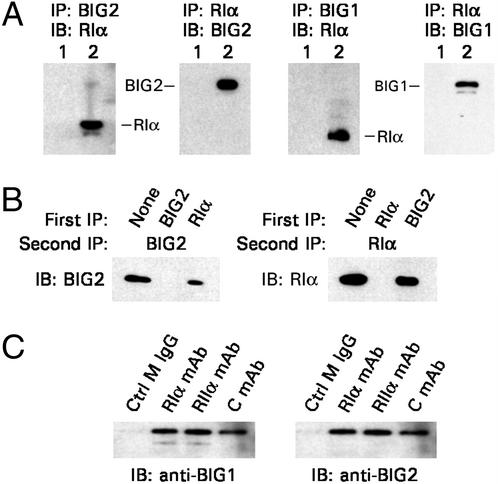
To investigate the association of endogenous BIG2 and RIα in human cells, proteins precipitated from HepG2 cell cytosol with anti-BIG2, anti-BIG1, or anti-RIα antibodies were separated by SDS/PAGE followed by immunoblotting. Antibodies against RIα precipitated BIG1 as well as BIG2, and RIα was precipitated by both anti-BIG1 and anti-BIG2 antibodies (Fig. 3A). Coimmunoprecipitation of significant fractions of BIG1 and BIG2 from HepG2 cells had already been reported (6), but whether or not they interact directly (or whether RIα interacts directly with BIG1) remains to be determined.
Figure 3.
IP of endogenous BIG2, BIG1, and RIα from HepG2 cell cytosol. (A) HepG2 cell cytosol (1.2 mg of protein) was incubated (lane 1) with control antibodies (normal rabbit or mouse) or (lane 2) with anti-BIG2 IgG (6 μg), anti-BIG1 IgG (5 μg), or anti-RIα mAb (5 μg). Precipitated proteins were separated by SDS/PAGE before immunoblotting (IB) with the indicated antibodies. (B) Samples (50 μg) of HepG2 cytosol proteins were incubated (first IP) for 4 h without or with 2 μg of anti-BIG2 IgG or 1.5 μg of anti-RIα mAb (total volume, 500 μl), followed by addition of 40 μl of protein G-Sepharose CL-4B beads and incubation overnight. Beads were collected by centrifugation, and supernatants were incubated (second IP) overnight as described in Materials and Methods. Proteins eluted from washed beads in 30 μl of sample buffer were separated by SDS/PAGE (8% gel) before IB with rabbit anti-BIG2 IgG or chicken anti-RIα IgY antibodies. (C) Samples of proteins precipitated (as in A) from HepG2 cell cytosol (1.2 mg of proteins, 500 μl) with 5 μg of control (Ctrl M) IgG or monoclonal antibodies against RIα, RIIα, or C subunit, as indicated, were separated by SDS/PAGE and immunoblotted with antibodies against BIG1 or BIG2.
To estimate the fractions of total BIG2 and RIα that were associated, HepG2 cytosolic proteins were immunoprecipitated with anti-BIG2 or anti-RIα antibodies followed by a second immunoprecipitation with the indicated antibodies (Fig. 3B) and Western blot analysis of precipitated proteins. Immunoreactive BIG2 was precipitated by anti-RIα, RIα was precipitated by anti-BIG2, and neither BIG2 nor RIα was detected on second immunoprecipitation with the same antibody (Fig. 3B). In three experiments with densitometric quantification, ≈40% RIα was precipitated along with 100% of BIG2, and antibodies against RIα coprecipitated 30–70% of BIG2.
The association of endogenous BIG2 and C subunit of PKA was demonstrated by immunoprecipitation from HepG2 cell cytosol. Antibody against C subunit precipitated both BIG2 and BIG1 (Fig. 3C).
BIG2 Interaction with RIβ, RIIα, and RIIβ.
In additional immunoprecipitation experiments (Fig. 3C), monoclonal antibodies against RIIα subunit, like those against RIα, precipitated BIG1 and BIG2 from HepG2 cell cytosol. To evaluate potential interactions of BIG2 with different R subunits, yeast mating experiments were carried out with transformants expressing BIG2 (1–832) and those expressing RIα, RIβ, RIIα, or RIIβ (Fig. 4A). All diploid colonies were positive for β-galactosidase activity, and controls were negative. Interactions of BIG2 with each of the R subunits was also observed in experiments like that in Fig. 2, in which [35S]methionine-labeled epitope-tagged R subunits and BIG2 (1–832) synthesized in reticulocyte lysates were mixed and immunoprecipitated (Fig. 4B).
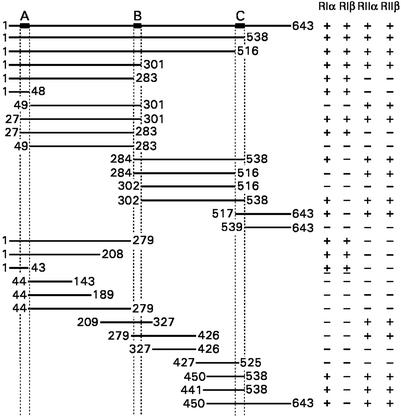
Multiple PKA R Subunit-Binding Sites in BIG2.
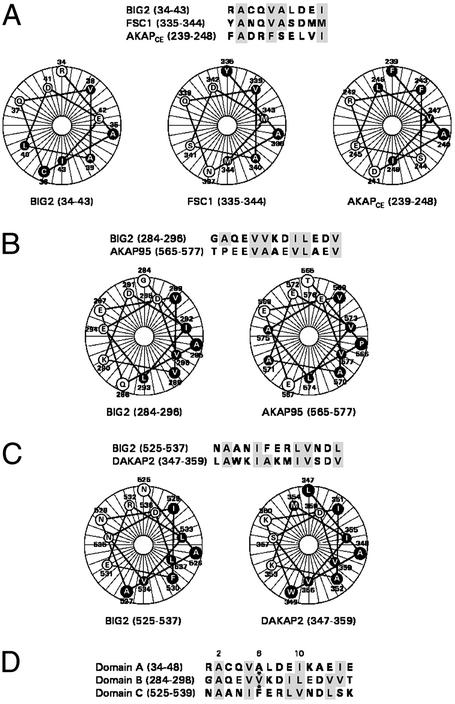
To map the R subunit-binding site(s) within the first 643 residues of BIG2, 28 different fragments were expressed in plasmid pB42AD, and the four R subunits were expressed in the BD fusion plasmid pGilda. They were individually transformed, respectively, into yeast YM4271 and EGY48[p8op-lacZ], and interactions were evaluated in mating experiments (Fig. 5). Three R-binding regions were identified in BIG2. Domain A (amino acids 27–48) interacted with RIα and RIβ, domain B (amino acids 284–301) interacted with RIIα and RIIβ, and domain C (amino acids 517–538) interacted with RIα, RIIα, and RIIβ. An AKAP consensus sequence for RII binding contains conserved hydrophobic residues and forms an amphipathic helix (12, 13). Alignment of BIG2 domains A, B, and C with R-binding domains of known AKAPs shows conserved hydrophobic amino acids and helical structures (Fig. 6).
Figure 5.
R-binding domains in BIG2 (1–643). The indicated fragments of the N-terminal region of BIG2 were expressed in AD fusion plasmid pB42AD and transformed into yeast EGY48[p8op-lacZ]. Four R subunits were expressed in BD fusion plasmid pGilda and transformed into yeast YM4271. Mating experiments were performed to monitor interactions. Data summarized are from one experiment, representative of two, with assays in duplicate.
Figure 6.
Sequence comparison of BIG2 and other putative AKAPs with helical wheel representations. Conserved hydrophobic amino acids are highlighted in the alignments. Filled circles indicate hydrophobic residues, and open circles indicate hydrophilic amino acids. (A) BIG2 domain A, FSC1 (26), and AKAPCE (19). (B) BIG2 domain B and AKAP95 (18). (C) BIG2 domain C and DAKAP2 (28). (D) BIG2 domains A–C.
Translocation of BIG2 to Membranes in HepG2 Cells.
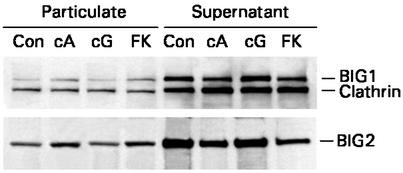
BIG2 and BIG1 had been initially localized by confocal immunofluorescence microscopy in Golgi structures and in a punctate distribution throughout the cytosol in resting conditions (6). Incubation of cells with 8-Br-cAMP or forskolin resulted in movement of BIG1 and BIG2 from cytosolic to membrane fractions, whereas clathrin was unchanged; no effect of 8-Br-cGMP was seen (Fig. 7).
Figure 7.
Effect of cAMP and forskolin on intracellular distribution of BIG1 and BIG2. HepG2 cells (≈4 × 106) were incubated (10 min, 37°C) without or with 1 mM 8-Br-cAMP, 1 mM 8-Br-cGMP, or 50 μM forskolin, as indicated, before homogenization and separation of supernatant and particulate fractions by centrifugation (105,000 × g, 1.5 h). Samples of particulate (60 μg) and supernatant (50 μg) proteins were separated by SDS/PAGE (4–12% gel) and immunoblotted with anti-BIG1 antibody (which reacted also with clathrin) or anti-BIG2 IgG. Observations were replicated three times. Con, control; cA, 8-Br-cAMP; cG, 8-Br-cGMP; FK, forskolin.
Discussion
Although the Sec7 domain is sufficient for ARF activation and its inhibition by BFA, functions of other parts of BIG2 (and BIG1) molecules are poorly understood. We identified three short sequences in the N-terminal 30% of the BIG2 molecule that can bind PKA R subunits, from which we infer that BIG2 may serve as an AKAP and perhaps be regulated by the kinase that it anchors. We demonstrated first the association of RIα with BIG2 (1–643) in a yeast two-hybrid screen and then with endogenous proteins in HepG2 cells where elevation of cAMP caused translocation of BIG2 from cytosol to Golgi and/or other membranes. RIα (1–59) was responsible for this interaction, consistent with a model for association between an AKAP amphipathic helix and the four-helix bundle formed by RIIα N termini in a homodimer (17).
PKA is a tetramer of pairs of identical C and AKAP-binding R subunits. Mammalian type I and type II R subunits each exist in α and β isoforms. Most analyses of PKA anchoring have focused on AKAPs that interact with RII, especially RIIα (9, 18). So-called dual AKAPs, such as D-AKAP1 and D-AKAP2, can interact with both RI and RII (14, 15), and there are several AKAPs that associate specifically with RI or RI-like subunits, such as AKAPCE (19, 20). RIβ has been implicated in the presynaptic regulation of hippocampal LTP/LTD and in vivo with synaptic plasticity in the visual cortex (21–23), but no AKAP for RIβ was identified in those systems. Three positions that are identical in RIα (Val-22, Ile-27, and Cys-39) and RICE (Val-27, Ile-32, and Cys-44) govern their interaction with DAKAP1 and AKAPCE (17, 20), respectively. Presence of the same residues in RIβ (Val-22, Ile-27, and Cys-39) presumably provides a structural basis for AKAP interactions. The reported existence of RIα–RIβ heterodimers in human neoplastic B and peripheral blood T cells (24) invites speculation that BIG2 might interact also with such heterodimers.
Although most AKAPs seem to have single R-binding sites, there are multiple R-binding sites in DAKAP550 (25), FSC1/AKAP82 (26), and BIG2. In BIG2, domain A (amino acids 27–48) interacted with RIα and RIβ. Alignment of amino acid sequence of domain A with those of FSC1 (amino acids 335–344) and AKAPCE (amino acids 239–248), all three of which interact preferentially with RI or RI-like subunits, revealed the conserved amino acids (Fig. 6A). BIG2 domain B (amino acids 284–301) interacted with RIIα and RIIβ. Sequence alignment and helical wheel comparison with AKAP95 (amino acids 565–577) revealed a perfect match (Fig. 6B) with the RII-binding motif, X-φ-X-X-X-(A or S)-X-X-φ-φ-X-X-φ-φ-X-X-(A or S)-φ, described by Carr and colleagues (27), where A is alanine, S is serine, X is any amino acid, and φ is leucine, isoleucine, or valine. Sequence within BIG2 domain C (amino acids 525–537) that interacted with RIα, RIIα, and RIIβ was aligned with DAKAP2, which also interacts with RIα, RIIα, and RIIβ. Amino acids of DAKAP2 critical for R binding, as suggested by Hamuro et al. (28), are highlighted in Fig. 6C.
After study of RI/RII binding and sequence specificity by Ala- and Val-scanning mutagenesis, Miki and Eddy (29) had proposed a “three amino acid” hypothesis, suggesting critical determinants at positions 2, 6, and 10 within the binding sites (Fig. 6D). Position 2 (a small side chain, Ala) and position 10 (a large hydrophobic chain) were required for amphipathic helix formation with the amino acid at position 6 determining R subunit specificity. Ser at position 6 enabled binding of RI or RII with similar affinity. Ala, with a small side chain amino acid, favored RI binding, whereas a large hydrophobic moiety favored RII (29). In the three R-binding domains of BIG2 (Fig. 6D), positions 2 and 10 are consistent with that structure, with Ala in position 2 and Ile, Leu, or Val in position 10. Domain A (34–48), with Ala in position 6, interacted with RIα and RIβ, but not RIIα or RIIβ, whereas domain B, BIG2 (284–298), with Val in position 6, interacted with RII rather than RI subunits, which seems to fit a “three amino acid” rule (29). AKAPCE (239–248), with Ser at position 6, did not, however, have similar affinities for RI and RII as expected by the rule, but interacted only with RI or RI-like subunits (20). BIG2 domain C (525–539), which interacted with both RIα and RII, has Phe, a hydrophobic amino acid with an aromatic side chain at position 6, adjacent to Glu-531 (position 7), which may or may not serve as “gatekeeper” to form a binding pocket accommodating a portion of the R docking domain in a manner similar to that of Phe-243 and Ser-244 in AKAPCE (20). Further mutagenesis and structural experiments are needed to resolve the question. Investigation of interactions of R subunits and BIG1, with domain C sequence identical to that in BIG2 and only conservative differences in domain A sequence, is also imperative.
BIG2 is an AKAP that can interact with all four mammalian R subunits. From the yeast two-hybrid screen, however, we retrieved only RIα, indicating perhaps that its association with BIG2 differs from that of the other R subunits. In our experiments, endogenous C, as well as RI (and other R subunits), was immunoprecipitated with antibodies against BIG2, consistent with AKAP localization of the PKA holotetramer. AKAPs, although they differ widely in overall structure, are related functionally by their ability to immobilize PKA at specific subcellular locations (8, 9, 30). Compartmentalization of cAMP action by maintaining PKA in close proximity to substrates and regulatory or catalytic molecules is critical for specificity and efficiency of regulatory systems (8, 9). A recent report of overexpression of RIIα in an RIIα-deficient B lymphoid cell line (Reh) indicated a regulatory role for Golgi-associated PKA in transport of ricin, a plant toxin, from endosomes to Golgi (31). Two other Golgi-localized AKAPs, myeloid translocation gene 16b (32) and AKAP350 (33), have also been described.
Phosphorylation of yeast Sec7 had been demonstrated in early studies by Franzusoff et al. (34). Two consensus sites for PKA phosphorylation are present in the sequence of human BIG2, 611RRCS614 and 1675RRDS1678, but we are, thus far, unsuccessful in demonstrating an effect of PKA-catalyzed phosphorylation on its GEP activity. BIG2 moved from cytosol to membrane fractions in HepG2 cells incubated with 8-Br-cAMP. Martin et al. (7) had reported that PKA promoted the binding of ARF1 to Golgi membranes in vitro and in intact cells. Our data are consistent with a localization of PKA at the Golgi via BIG2. With three AKAP domains that seem to differ in R specificity, BIG2 could have scaffolding functions in more than one type of cAMP signaling complex, only some of which may include BIG1. Because the antibodies that we used for both immunoblotting and immunoprecipitation were produced with a peptide representing amino acids 232–241 in BIG2, it is obvious that some forms of BIG2 proteins might have been missed in these experiments. Dong et al. (35), for example, described six isoforms of AKAP-KL generated by alternative mRNA splicing. This could be an important mechanism for increasing diversity of BIG2 targeting and functional specificity.
Although AKAPs are defined by their ability to bind to PKA, there is accumulating evidence that they interact simultaneously with multiple regulatory enzymes, such as other kinases and phosphatases, to integrate different signal pathways (9, 30). AKAP79 associated with calcineurin protein phosphatase 2B (36) and PKC (37), yotiao and AKAP220 with phosphatase 1 (38, 39), gravin with PKC (40), and mAKAP with PDE4D3 phosphodiesterase (41). BIG2, in its GEP function, associates, of course, with the ARFs that it activates, through which it is involved in recruitment of the AP-1 γ-adaptin complex to form clathrin-coated vesicles from TGN membranes (42, 43). Other functional domains and molecular interactions of the 190-kDa BIG2 molecule remain to be identified. Among the goals of current work is a complete description of the BIG2 AKAP complexes that may serve to integrate cAMP and small G protein signaling pathways.
Acknowledgments
We thank Carol Kosh for expert secretarial assistance.
Abbreviations
- ARF
ADP-ribosylation factor
- GEP
guanine nucleotide-exchange protein
- BFA
brefeldin A
- PKA
protein kinase A or cAMP-dependent protein kinase
- AKAP
A kinase-anchoring protein
- AEBSF
4-(2-aminoethyl)benzenesulfonyl fluoride
- IP
immunoprecipitation
- HA
hemagglutinin
References
- 1.Moss J, Vaughan M. J Biol Chem. 1995;270:12327–12330. doi: 10.1074/jbc.270.21.12327. [DOI] [PubMed] [Google Scholar]
- 2.Moss J, Vaughan M. J Biol Chem. 1998;273:21431–21434. doi: 10.1074/jbc.273.34.21431. [DOI] [PubMed] [Google Scholar]
- 3.Misumi Y, Misumi Y, Miki K, Takatsuki A, Tamura G, Ikehara Y. J Biol Chem. 1986;261:11398–11403. [PubMed] [Google Scholar]
- 4.Morinaga N, Tsai S-C, Moss J, Vaughan M. Proc Natl Acad Sci USA. 1996;93:12856–12860. doi: 10.1073/pnas.93.23.12856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Togawa A, Morinaga N, Ogasawara M, Moss J, Vaughan M. J Biol Chem. 1999;274:12308–12315. doi: 10.1074/jbc.274.18.12308. [DOI] [PubMed] [Google Scholar]
- 6.Yamaji R, Adamik R, Takeda K, Togawa A, Pacheco-Rodriguez G, Ferrans V J, Moss J, Vaughan M. Proc Natl Acad Sci USA. 2000;97:2567–2572. doi: 10.1073/pnas.97.6.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin M E, Hidalgo J, Rosa J L, Crottet P, Velasco A. J Biol Chem. 2000;275:19050–19059. doi: 10.1074/jbc.275.25.19050. [DOI] [PubMed] [Google Scholar]
- 8.Taylor S S, Buechler J A, Yonemoto W. Annu Rev Biochem. 1990;59:971–1005. doi: 10.1146/annurev.bi.59.070190.004543. [DOI] [PubMed] [Google Scholar]
- 9.Michel J J, Scott J D. Annu Rev Pharmacol Toxicol. 2002;42:235–257. doi: 10.1146/annurev.pharmtox.42.083101.135801. [DOI] [PubMed] [Google Scholar]
- 10.Theurkauf W E, Vallee R B. J Biol Chem. 1982;257:3284–3290. [PubMed] [Google Scholar]
- 11.Sarkar D, Erlichman J, Rubin C S. J Biol Chem. 1984;259:9840–9846. [PubMed] [Google Scholar]
- 12.Newlon M G, Roy M, Morikis D, Carr D W, Westphal R, Scott J D, Jennings P A. EMBO J. 2001;20:1651–1662. doi: 10.1093/emboj/20.7.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feliciello A, Gottesman M E, Avvedimento E V. J Mol Biol. 2001;308:99–114. doi: 10.1006/jmbi.2001.4585. [DOI] [PubMed] [Google Scholar]
- 14.Huang L J, Durick K, Weiner J A, Chun J, Taylor S S. J Biol Chem. 1997;272:8057–8064. doi: 10.1074/jbc.272.12.8057. [DOI] [PubMed] [Google Scholar]
- 15.Wang L, Sunahara R K, Krumins A, Perkins G, Crochiere M L, Mackey M, Bell S, Ellismanm M H, Taylor S S. Proc Natl Acad Sci USA. 2001;98:3220–3225. doi: 10.1073/pnas.051633398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Golemis E, Gyuris J, Brent R. Current Protocols in Molecular Biology. New York: Wiley; 1996. pp. 20.1.1–20.1.35. [Google Scholar]
- 17.Banky P, Newlon M G, Roy M, Garrod S, Taylor S S, Jennings P A. J Biol Chem. 2000;275:35146–35152. doi: 10.1074/jbc.M003961200. [DOI] [PubMed] [Google Scholar]
- 18.Eide T, Coghlan V, Orstavik S, Holsve C, Solberg R, Skalhegg B S, Lamb N J, Langeberg L, Fernandez A, Scott J D, et al. Exp Cell Res. 1998;238:305–316. doi: 10.1006/excr.1997.3855. [DOI] [PubMed] [Google Scholar]
- 19.Angelo R G, Rubin C S. J Biol Chem. 1998;273:14633–14643. doi: 10.1074/jbc.273.23.14633. [DOI] [PubMed] [Google Scholar]
- 20.Angelo R G, Rubin C S. J Biol Chem. 2000;275:4351–4362. doi: 10.1074/jbc.275.6.4351. [DOI] [PubMed] [Google Scholar]
- 21.Hensch T K, Gordon J A, Brandon E P, McKnight G S, Idzerda R L, Stryker M P. J Neurosci. 1998;18:2108–2117. doi: 10.1523/JNEUROSCI.18-06-02108.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brandon E P, Zhuo M, Huang Y Y, Qi M, Gerhold K A, Burton K A, Kandel E R, McKnight G S, Idzerda R L. Proc Natl Acad Sci USA. 1995;92:8851–8855. doi: 10.1073/pnas.92.19.8851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herberg F W, Maleszka A, Eide T, Vossebein L, Tasken K. J Mol Biol. 2000;298:329–339. doi: 10.1006/jmbi.2000.3662. [DOI] [PubMed] [Google Scholar]
- 24.Tasken K, Skalhegg B S, Solberg R, Andersson K B, Taylor S S, Lea T, Blomhoff H K, Jahnsen T, Hansson V. J Biol Chem. 1993;268:21276–21283. [PubMed] [Google Scholar]
- 25.Han J D, Baker N E, Rubin C S. J Biol Chem. 1997;272:26611–26619. doi: 10.1074/jbc.272.42.26611. [DOI] [PubMed] [Google Scholar]
- 26.Miki K, Eddy E M. J Biol Chem. 1998;273:34384–34390. doi: 10.1074/jbc.273.51.34384. [DOI] [PubMed] [Google Scholar]
- 27.Vijayaraghavan S, Liberty G A, Mohan J, Winfrey V P, Olson G E, Carr D W. Mol Endocrinol. 1999;13:705–717. doi: 10.1210/mend.13.5.0278. [DOI] [PubMed] [Google Scholar]
- 28.Hamuro Y, Burns L, Canaves J, Hoffman R, Taylor S, Woods V. J Mol Biol. 2002;321:703–714. doi: 10.1016/s0022-2836(02)00419-9. [DOI] [PubMed] [Google Scholar]
- 29.Miki K, Eddy E M. J Biol Chem. 1999;274:29057–29062. doi: 10.1074/jbc.274.41.29057. [DOI] [PubMed] [Google Scholar]
- 30.Edwards A S, Scott J D. Curr Opin Cell Biol. 2000;12:217–221. doi: 10.1016/s0955-0674(99)00085-x. [DOI] [PubMed] [Google Scholar]
- 31. Birkeli, K. A., Llorente, A., Torgersen, M. L., Keryer, G., Tasken, K. & Sandvig, K. (2003) J. Biol. Chem., in press. [DOI] [PubMed]
- 32.Schillace R V, Andrews S F, Liberty G A, Davey M P, Carr D W. J Immunol. 2002;168:1590–1599. doi: 10.4049/jimmunol.168.4.1590. [DOI] [PubMed] [Google Scholar]
- 33.Shanks R A, Steadman B T, Schmidt P H, Goldenring J R. J Biol Chem. 2002;277:40967–40972. doi: 10.1074/jbc.M203307200. [DOI] [PubMed] [Google Scholar]
- 34.Franzusoff A, Redding K, Crosby J, Fuller R S, Schekman R. J Cell Biol. 1991;112:27–37. doi: 10.1083/jcb.112.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong F, Feldmesser M, Casadevall A, Rubin C S. J Biol Chem. 1998;273:6533–6541. doi: 10.1074/jbc.273.11.6533. [DOI] [PubMed] [Google Scholar]
- 36.Coghlan V M, Perrino B A, Howard M, Langeberg L K, Hicks J B, Gallatin W M, Scott J D. Science. 1995;267:108–111. doi: 10.1126/science.7528941. [DOI] [PubMed] [Google Scholar]
- 37.Faux M C, Rollins E N, Edwards A S, Langeberg L K, Newton A C, Scott J D. Biochem J. 1999;343:443–452. [PMC free article] [PubMed] [Google Scholar]
- 38.Marx S O, Kurokawa J, Reiken S, Motoike H, D'Armiento J, Marks A R, Kass R S. Science. 2002;295:496–499. doi: 10.1126/science.1066843. [DOI] [PubMed] [Google Scholar]
- 39.Schillace R V, Scott J D. Curr Biol. 1999;9:321–324. doi: 10.1016/s0960-9822(99)80141-9. [DOI] [PubMed] [Google Scholar]
- 40.Nauert J B, Klauck T M, Langeberg L K, Scott J D. Curr Biol. 1997;7:52–62. doi: 10.1016/s0960-9822(06)00027-3. [DOI] [PubMed] [Google Scholar]
- 41.Dodge K L, Khouangsathiene S, Kapiloff M S, Mouton R, Hill E V, Houslay M D, Langeberg L K, Scott J D. EMBO J. 2001;20:1921–1930. doi: 10.1093/emboj/20.8.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shinotsuka C, Yoshida Y, Kawamoto K, Takatsu H, Nakayama K. J Biol Chem. 2002;277:9468–9472. doi: 10.1074/jbc.M112427200. [DOI] [PubMed] [Google Scholar]
- 43.Shinotsuka C, Waguri S, Wakasugi M, Uchiyama Y, Nakayama K. Biochem Biophys Res Commun. 2002;294:254–260. doi: 10.1016/S0006-291X(02)00456-4. [DOI] [PubMed] [Google Scholar]
- 44.Miller J H. A Short Course in Bacterial Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. p. 74. [Google Scholar]