Abstract
Human herpesvirus 8 (HHV-8) or Kaposi's sarcoma-associated herpesvirus, implicated in the pathogenesis of Kaposi's sarcoma, utilizes heparan sulfate-like molecules to bind the target cells via its envelope-associated glycoproteins gB and gpK8.1A. HHV-8-gB possesses the Arg-Gly-Asp (RGD) motif, the minimal peptide region of many proteins known to interact with subsets of host cell surface integrins. HHV-8 utilizes α3β1 integrin as one of the receptors for its entry into the target cells via its gB interaction and induces the activation of focal adhesion kinase (FAK) (S. M. Akula, N. P. Pramod, F.-Z. Wang, and B. Chandran, Cell 108:407-419, 2002). Since FAK activation is the first step in the outside-in signaling necessary for integrin-mediated cytoskeletal rearrangements, cell adhesions, motility, and proliferation, the ability of HHV-8-gB to mediate the target cell adhesion was examined. A truncated form of gB without the transmembrane and carboxyl domains (gBΔTM) and a gBΔTM mutant (gBΔTM-RGA) with a single amino acid mutation (RGD to RGA) were expressed in a baculovirus system and purified. Radiolabeled HHV-8-gBΔTM, gBΔTM-RGA, and ΔTMgpK8.1A proteins bound to the human foreskin fibroblasts (HFFs), human dermal microvascular endothelial (HMVEC-d) cells, human B (BJAB) cells, and Chinese hamster ovary (CHO-K1) cells with equal efficiency, which was blocked by preincubation of proteins with soluble heparin. Maxisorp plate-bound gBΔTM protein induced the adhesion of HFFs and HMVEC-d and monkey kidney epithelial (CV-1) cells in a dose-dependent manner. In contrast, the gBΔTM-RGA and ΔTMgpK8.1A proteins did not mediate adhesion. Adhesion mediated by gBΔTM was blocked by the preincubation of target cells with RGD-containing peptides or by the preincubation of plate-bound gBΔTM protein with rabbit antibodies against gB peptide containing the RGD sequence. In contrast, adhesion was not blocked by the preincubation of plate-bound gBΔTM protein with heparin, suggesting that the adhesion is mediated by the RGD amino acids of gB, which is independent of the heparin-binding domain of gB. Integrin-ligand interaction is dependent on divalent cations. Adhesion induced by the gBΔTM was blocked by EDTA, thus suggesting the role of integrins in the observed adhesions. Focal adhesion components such as FAK and paxillin were activated by the binding of gBΔTM protein to the target cells but not by gBΔTM-RGA protein binding. Inhibition of FAK phosphorylation by genistein blocked gBΔTM-induced FAK activation and cell adhesion. These findings suggest that HHV-8-gB could mediate cell adhesion via its RGD motif interaction with the cell surface integrin molecules and indicate the induction of cellular signaling pathways, which may play roles in the infection of target cells and in Kaposi's sarcoma pathogenesis.
Kaposi's sarcoma (KS) is a common vascular tumor associated with human immunodeficiency virus type 1 (HIV-1) infection (6). In the absence of HIV-1 infection, KS occurs in three distinct epidemiological forms: classic KS (CKS), endemic aggressive KS, and transplantation-associated KS (6). Various models have been proposed for the origin of KS, and none of these factors has been demonstrated to be etiologically associated with KS (44). KS was hypothesized to be mediated by HIV-Tat, since Tat binding to the heparan sulfate (HS) in the extracellular matrix (ECM) was believed to act by the displacement of basic fibroblast growth factor (BFGF) from the matrix (10, 22, 23). In addition, HIV-Tat is also believed to stimulate cell adhesion and growth through its RGD motif interaction with the endothelial cell surface α5β1 and αvβ3 integrin molecules, thus inducing cytokines and basic fibroblast growth factor necessary for KS development (10, 20, 22, 23). Even though HIV infection accelerates KS development, this may not be the sole inciting event in KS etiology, since CKS, endemic aggressive KS, and transplantation-associated KS occur in the absence of HIV-1 infection (6).
Chang et al. (15) reported the identification of novel herpesvirus DNA sequences (human herpersvirus 8 [HHV-8]/KS-associated herpesvirus [KSHV]) in AIDS-associated KS. An explosion of studies following this finding showed that HHV-8/KSHV is etiologically associated with KS (6, 26, 46). HHV-8 DNA has been detected in all epidemiological forms of KS, suggesting that HHV-8 could be a potential common etiological factor for KS (6, 26, 46). Cell lines with B-cell characteristics established from the body cavity-based B-cell lymphomas (BCBL) carry HHV-8 in a latent form, and a lytic cycle can be induced by 12-O-tetradecanoylphorbol-13-acetate (TPA) (26, 46). HHV-8 has a broad in vivo and in vitro cellular tropism. HHV-8 DNA and transcripts have been identified in vivo in human B cells, endothelial cells, epithelial cells, keratinocytes, and macrophages (6, 26, 46). HHV-8 has been shown to infect a variety of human and other mammalian cells, such as human B cells, epithelial cells, human endothelial cells, human foreskin fibroblasts (HFFs), human keratinocytes, CV-1, owl monkey kidney cells and baby hamster kidney (BHK-21) cells (14, 42, 49). If in vitro permissiveness of a cell type is judged by a productive lytic replication of HHV-8 after entry into cells, there is as yet no suitable cell culture system to support a lytic replication of input HHV-8. Only a latent infection is observed in the infected cells (14, 42, 49). However, if in vitro permissiveness is judged by the establishment of HHV-8 latency and the ability to support HHV-8 lytic replication after activation by agents, cells such as HFFs, human carcinoma cells, and endothelial cells are permissive, as evident by the presence of circular latent HHV-8 DNA, by the expression of HHV-8 latency-associated nuclear antigen encoded by the open reading frame (ORF) 73, and by the ability to support lytic replication upon activation by TPA (14, 42, 49).
The broad in vitro cellular tropism of HHV-8 may be in part due to its interaction with the ubiquitous host cell surface HS-like molecules (3). HHV-8 encodes several glycoproteins (43), and researchers have demonstrated the interaction of virion envelope-associated HHV-8 glycoprotein gB (ORF 8) and gpK8.1A with HS molecules (2, 11, 51). Among the alpha-, beta-, and gammaherpesvirus gBs sequenced to date, only HHV-8-gB possesses the RGD amino acids (aa) 27 to 29 at the extracellular amino-terminal coil region after the putative signal sequence (43). Integrins are a large family of heterodimeric receptors containing noncovalently associated transmembrane α and β glycoprotein subunits (28, 40). There are 17 α and 9 β subunits generating more than 22 known combinations of αβ cell surface receptors. Each cell expresses several combinations of αβ integrins (28, 40). Each αβ combination has its own binding specificity and signaling properties (28, 40). The RGD motif is the minimal peptide region of many proteins known to interact with subsets of host cell surface integrins critical for a variety of cell functions, such as the regulation of gene expression, activation of focal adhesion kinases (FAKs), activation of cytoskeleton elements, endocytosis, attachment, motility, cell survival, cell cycle progression, cell growth, apoptosis, and differentiation (28). HHV-8 infectivity was inhibited by RGD peptides, antibodies against RGD-gB peptide, antibodies against RGD-dependent α3β1 integrin, and soluble α3β1 integrin (4). Expression of human α3 integrin increased the infectivity of virus for Chinese hamster ovary cells (4). Anti-HHV-8-gB antibodies immunoprecipitated the virus-α3 and -β1 complexes, and virus binding studies suggest a role for α3β1 integrin in HHV-8's entry (4). Further, HHV-8 infection induced the integrin-mediated activation of FAK. These findings implicated a role for α3β1 integrin and the associated signaling pathways in HHV-8 entry into the target cells (4).
The importance of the HHV-8-gB-RGD motif in viral infection raised the possibility that gB-RGD could mediate cell adhesion. To examine the ability of gB to induce cell adhesion, HHV-8-gB without the transmembrane and cytoplasmic domain (gBΔTM) and a gBΔTM mutant (gBΔTM-RGA) with a single amino acid mutation (RGD to RGA) were expressed in the baculovirus system and purified. Radiolabeled gBΔTM, gBΔTM-RGA, and ΔTMgpK8.1A proteins bound to the target cells with equal efficiency, which was blocked by soluble heparin. However, only gBΔTM mediated the adhesion of target cells. Cell adhesion mediated by gBΔTM was blocked by RGD-containing peptides, rabbit antibodies against gB peptide containing the RGD sequence, and EDTA but not by heparin. HHV-8-gBΔTM activated the focal adhesion (FA) components. Inhibition of FAK phosphorylation by genistein also blocked gBΔTM-induced FAK activation and cell adhesion. These results suggest that HHV-8-gB could induce cellular signaling pathways required for cell adhesion.
MATERIALS AND METHODS
Cells.
HFFs (Clonetics, Walkersville, Md.), HMVEC-d cells (CC-2543; Clonetics), CV-1 cells (ATCC CCL-70), BJAB (HHV-8- and Epstein-Barr virus-negative human B cells) cells, Chinese hamster ovary cells (CHO-K1; ATCC CCL-61), Spodoptera frugiperda ovarian cells (Sf 9) (PharMingen, San Diego, Calif.), and Trichoplusia ni egg cells (High-5) (Invitrogen, Carlsbad, Calif.) were used in this study. HFFs and CV-1 cells were grown in Dulbecco modified Eagle medium (DMEM; Gibco BRL, Grand Island, N.Y.) with 2 mM glutamine, 10% fetal bovine serum (FBS), and antibiotics. HMVEC-d cells were grown in EBM2-MV medium (Clonetics). Monolayers of CHO-K1 were grown in Ham's F12K medium (Gibco BRL). BJAB cells were grown in RPMI 1640 (Gibco BRL). Sf 9 cells were grown in TNM-FH insect medium (PharMingen). High-5 cells were grown in Ultimate Insect Serum Free Medium (Invitrogen).
Ligands, peptides, and antibodies.
Collagen type I and vitronectin were obtained from Chemicon International Inc., Temecula, Calif. Fibronectin was from Life Technologies, Rockville, Md. Gelatin was from Difco Laboratories, Detroit, Mich. Biotin-labeled heparin was from Carbomer, Inc., Westborough, Mass. Rabbit anti-p125FAK antibodies, monoclonal antibody (MAb) to actin (clone AC-40), GRGDSPL and GRADSPL peptides, lysophosphatidic acid (LPA), genistein, heparin, and chondroitin sulfate A (CS-A) were from Sigma, St. Louis, Mo. Mouse anti-phospho-FAK antibody (Y397, BD) and mouse MAb to paxillin (clone 177) were from Transduction Laboratories, Los Angeles, Calif. 125I-labeled epidermal growth factor (EGF) was from ICN, Irvine, Calif.
Construction of recombinant HHV-8-gBΔTM and gBΔTM-RGA mutant.
The HHV-8-gB (ORF 8) gene was amplified from the BCBL-1 cells by PCR with primers gB(F) (5′-AGT GAG GAT CCA CAA TGA CTC CCA GG-3′) with a BamHI site and gB(R) (5′-AGA ATG AAT TCT CAC TCC CCC GTT TCC G-3′) with an EcoRI site. The amplified fragment was cloned into the BamHI-EcoRI sites of pCDNA3.1(+), an eukaryotic expression vector (Invitrogen) containing the human cytomegalovirus immediate-early promoter, to create the gB-pCDNA3.1(+) clone. Clones were verified by sequencing (2). The full-length HHV-8-gB gene was subcloned into the BamHI and EcoRI sites of the pAcGHLT-A baculovirus vector (PharMingen) and was cotransfected with BaculoGold DNA into Sf 9 cells.
The 2,106-bp gBΔTM gene region encoding aa 1 to 702 lacking the transmembrane and cytoplasmic domains was amplified from the gB-pCDNA3.1(+) construct (2) by PCR with primers gB-HIS forward (5′-AG TGA GGA TCC ACA ATG ACT CCC AGG-3′ with a BamHI site) and gB-HIS reverse (5′-TCC GAA TTC TCA ATG ATG ATG ATG ATG ATG GCC ACC CAG GTC CGC CAC TAT CTC-3′ with an EcoRI site). A His6 tag was introduced at the carboxyl terminus of gBΔTM. The amplified fragment (2,124 bp) was cloned into the BamHI-EcoRI sites of pAcGP67-B (gBΔTM/pAcGP67-B; PharMingen) and verified by sequencing. The gBΔTM-RGA mutant was generated by mutating the RGD amino acids in the gBΔTM/pAcGP67-B plasmid to RGA by using primers RGA forward (5′-CAC TCG AGG GGT GCC ACC TTT CAG ACG-3′) and RGA reverse (5′-CGT CTG AAA GGT GGC ACC CCT CGA GTG-3′) and the QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) as per the manufacturer's recommendations to yield the gBΔTM-RGA/pAcGP67-B plasmid. To generate the recombinant baculovirus, gBΔTM/pAcGP67-B and gBTM-RGA/pAcGP67-B plasmids were cotransfected with BaculoGold DNA (PharMingen) into the Sf 9 insect cells (51).
Expression of recombinant HHV-8-gBΔTM, gBΔTM-RGA, and ΔTMgpK8.1A proteins.
Expression and purification of the HHV-8-gBΔTM, gBTΔM-RGA, ΔTMgpK8.1A, and full-length glutathione S-transferase (GST)-gB (51, 54) proteins from the infected High-5 cells by using nickel columns (PharMingen) were done as per procedures described before (51). For obtaining radiolabeled proteins, cells infected with baculovirus for 2 days were labeled with [35S]methionine for 3 days. The purity of labeled and unlabeled proteins was analyzed by Coomassie staining of sodium dodecyl sulfate (SDS)-10% polyacrylamide gel electrophoresis (PAGE) gels, by Western blotting with rabbit anti-gB peptide, rabbit anti-gB antibodies, and anti-gpK8.1A MAbs and by autoradiography (2, 51, 54). All reagents used in the protein purification procedures were prepared in endotoxin-free water. The presence of endotoxin was tested by end-point chromogenic Limulus amebocyte lysate assay as per protocols recommended by the manufacturer (Charles River Laboratories, Charleston, S.C.).
Rabbit anti-full-length gB and anti-gB peptide polyclonal antibodies.
The expression and purification of GST-full-length gB fusion protein were done using procedures described previously (54). HHV-8-gB peptides, gB-N1 (aa 27 to 47; RGDTFQTSSSPTPPGSSSK), gB-N2 (aa 167 to 191; GVENTFTDRDDVNTTVFLQPVEGLT), gB-N3 (aa 573 to 593; ARNEIILTNNQVETCKDTCEH), and gB-C (aa 828 to 845; RGYKPLTQSLDISPETGE) were synthesized and purified by Syn Pep Corp., Dublin, Calif. New Zealand White male rabbits were immunized with the purified GST-gB fusion protein and gB peptides (RGD gB-N1, gB-N2, gB-N3, and gB-C). Immunoglobulin G (IgG) fractions were purified by protein A-Sepharose 4B columns (Amersham Pharmacia Biotech, Piscataway, N.J.). Nonspecific antibodies were removed by columns of cyanogen bromide-activated Sepharose 4B covalently coupled with purified GST protein and BJAB cell lysate.
Western blot assays.
Samples were boiled in sample buffer with (reduced) or without (nonreduced) 2-mercaptoethanol (2-ME), subjected to SDS-10% PAGE, and electrophoretically transferred onto nitrocellulose membranes. Standard prestained and unstained molecular weight markers (Gibco BRL) were included in the parallel lanes. The membranes were soaked in blocking solution (10 mM Tris-HCl, pH 7.2, 150 mM NaCl, 5% skim milk, 0.02% NaN3) at 4°C overnight and then reacted with rabbit antibodies for 3 h at room temperature. The membranes were washed five times with washing buffer (10 mM Tris-HCl, pH 7.2, 150 mM NaCl, 0.3% Tween 20) and incubated for 1 h at room temperature with alkaline phosphatase-conjugated goat anti-rabbit IgG (KPL, Gaithersburg, Md.). The bound enzyme-labeled antibodies were detected by nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate substrate (Sigma). The reactions were stopped by washing the membranes in distilled water.
SIFA.
BJAB and HFFs were used for surface immunofluorescence assay (SIFA) as per procedures described previously (2, 51) with minor modifications. Briefly, 106 BJAB cells were fixed with 0.1% paraformaldehyde, washed, and incubated with different concentrations of purified recombinant gBΔTM or gBΔTM-RGA proteins for 90 min at 4°C. HFFs in eight-well chamber slides (Nalge Nunc International, Naperville, Ill.) were incubated with different concentrations of proteins in serum-free DMEM with glutamine for 90 min at 4°C, washed with PBS, and fixed with 4% paraformaldehyde for 10 min at room temperature. BJAB and HFFs were washed, incubated with rabbit anti-HHV-8-gB antibodies for 60 min at 4°C, washed, and incubated for 60 min at 4°C with prestandardized fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit antibodies. These cells were washed, mounted with SlowFade (Molecular Probes, Eugene, Oreg.), and examined under a fluorescence microscope with the Nikon Magna Firewire digital imaging system (4). Preimmune rabbit antibodies and rabbit anti-gpK8.1A antibodies were used as negative controls.
[35S]methionine-labeled recombinant HHV-8 protein-binding assay.
The recombinant protein-binding assay was performed according to the method described previously (51). Briefly, confluent HFF monolayers in 24-well plates were washed and blocked for 30 min at 4°C with phosphate-buffered saline (PBS) containing 1% FBS, 5 mM bovine serum albumin (BSA), and 0.1 mM CaCl2. Cells were incubated with different concentrations of purified recombinant [35S]methionine-labeled proteins (2,452 cpm of gBΔTM, 1,629 cpm of gBΔTM-RGA, and 2,755 cpm of ΔTMgpK8.1A per μg) in DMEM with 10% FBS and 0.01% NaN3 for 90 min at 4°C. After incubation, cells were washed five times with DMEM, lysed with 1% SDS, and 1% Triton X-100 in distilled water and the cell-bound radioactivity was counted. All experiments were done in triplicate and repeated three times.
To test the ability of heparin or CS-A to inhibit the binding of gBΔTM, gBΔTM-RGA, and ΔTMgpK8.1A proteins, a constant quantity of purified labeled protein (6 μg) was mixed with medium alone or with medium containing different concentrations of heparin and incubated at 4°C for 60 min. These were then added to the target cells and incubated at 4°C for 90 min, and the cells were washed five times with DMEM and lysed with 1% SDS and 1% Triton X-100 in distilled water. The cell-bound protein counts per minute in the presence or absence of heparin and the percentage of inhibition of binding were calculated. All reactions were done in triplicate and repeated three times.
125I-labeled EGF binding assay.
Confluent HFFs in 24-well plates were washed twice with serum-free DMEM and blocked with 0.1% BSA-DMEM for 1 h at 4°C. Different concentrations of 125I-labeled EGF (1 mCi/ml, 200 mCi/mg; ICN, Irvine, Calif.) diluted in 0.1% DMEM were added to the cells (160 μl/well). After incubation at 4°C for 1 h, cells were washed five times with DMEM and harvested with 1% SDS and 1% Triton X-100 in distilled water, and the cell-bound radioactivity was measured. For the heparin-blocking assay, a fixed amount of 125I-labeled EGF (259,272 cpm/ng) was mixed with different concentrations of heparin in 0.1% BSA-DMEM and incubated for 1 h on ice before adding to the cells. All experiments were performed in triplicates and were repeated three times.
Cell adhesion assay.
Cell adhesion assays were performed as per methods described previously with minor modifications (34). Maxisorp enzyme-linked immunosorbent assay (ELISA) plates (Nunc, Roskilde, Denmark) were coated with 100 μl of different concentrations of proteins overnight at 4°C in PBS. After three washes with sterile PBS, plates were blocked with 1% BSA in PBS for 2 h at 4°C and washed three times. Target cells were treated with 0.05% trypsin-0.2% EDTA (34) for 2 min at room temperature, washed, and collected in DMEM with 0.25% BSA and 0.5 mg of trypsin inhibitor (Sigma)/ml. These cells were washed twice with DMEM, resuspended in 0.1% BSA-serum-free DMEM, and plated in the protein-coated plates at a concentration of 2 × 104 cells/well in a 100-μl volume (34). The plate contents were incubated at 37°C in a 5% CO2 atmosphere with 100% humidity for 1 h for CV-1 cells or for 45 min for HFF and HMVEC-d cells and washed four times with serum-free DMEM, and the adherent cells were fixed with 4% paraformaldehyde in PBS for 30 min at room temperature. Adherent cells were washed, stained with 0.5% crystal violet in water with 20% (vol/vol) methanol for 15 min at room temperature, and washed four or five times. Dye was extracted with 0.1 M sodium citrate and quantified by the measurement of absorbance at 595 nm (34) in an ELISA reader (Bio Kinetics Reader EL340; Bio-Tek Instruments, Winooski, Vt.).
To determine the effect of heparin or anti-RGD antibodies, protein-coated plates were incubated with different concentrations of rabbit IgG antibodies or heparin for 1 h at 4°C in 50 μl of 0.1% BSA-serum-free DMEM before addition of the target cells for adhesion assays. For peptide-blocking assays or for analyzing the effect of EDTA, cells were resuspended in DMEM with 0.1% BSA with different concentrations of peptides or EDTA and were incubated for 30 min at 4°C with gentle mixing every 10 min before plating. HFFs were also incubated with noncytotoxic doses of genistein in 4 ml of serum-free DMEM at 37°C for 1 h. These cells were harvested by trypsin treatment, suspended in serum-free DMEM-0.1% BSA with different concentrations of genistein, and seeded into the protein-coated plates for adhesion assays.
ELISA.
To determine the ability of recombinant proteins to bind the Maxisorp plates, ELISA was performed as per procedures described previously (54). Briefly, Maxisorp plates were coated with different concentrations of gBΔTM or gBΔTM RGA or ΔTMgpK8.1A proteins in PBS overnight at 4°C. Plates were washed five times with 0.085% NaCl-0.05% Tween 20, blocked with 10% nonfat milk for 1 h at 37°C, washed, and incubated with rabbit anti-gB or anti-gpK8.1A IgG antibodies (54) for 1 h at 37°C, followed by goat anti-rabbit antibodies coupled to horseradish peroxidase. After reaction with 5-bromo-4-chloro-3-indolylphosphate-nitroblue tetrazolium substrate, the reaction was stopped by 2 N H2SO4 and read at 450 nm.
To determine the ability of Maxisorp plate-bound proteins to bind heparin, plates were coated with fibronectin, gBΔTM, gBΔTM-RGA, ΔTMgpK8.1A, and BSA (4 μg/ml in PBS, 100 μl/well) overnight at 4°C. Plates were washed five times with 0.05% Tween 20-PBS, blocked with 3% BSA-PBS for 2 h at 4°C, washed, and incubated with different concentrations of biotin-labeled heparin for 1 h at 4°C. Plates were washed and incubated with ImmunoPure streptavidin horseradish peroxidase (0.1 μg/ml, 100 μl per well; Pierce, Rockford, Ill.) for 1 h at room temperature, and washed and bound horseradish peroxidase was quantified by reaction with 5-bromo-4-chloro-3-indolylphosphate-nitroblue tetrazolium substrate.
Immunofluorescence examination of FAK activation.
Cells in chamber slides were serum starved for 24 h at 37°C and were incubated with 200 μl of gBΔTM and gBΔTM RGA proteins (50 ng/ml) or LPA (20 ng/ml) in serum-free DMEM for 5 min at 37°C. These cells were washed in PBS, fixed in 3.7% formaldehyde (in PBS) for 10 min at room temperature, permeabilized with 0.1% Triton X-100-PBS for 4 min, and blocked with 1% BSA-PBS for 10 min at room temperature. Cells were then incubated with a prestandardized dilution of mouse antipaxillin antibody for 1 h at room temperature, washed, and incubated with rabbit anti-p125FAK antibodies for an additional 1 h at room temperature. These cells were washed and incubated with goat anti-rabbit tetramethyl rhodamine isothiocyanate (Sigma) and goat anti-mouse FITC antibodies for 30 min at room temperature. Stained cells were washed, counterstained with Evan's blue (1:10,000) for 5 min at room temperature, washed, and viewed with appropriate filters under a fluorescence microscope with the Nikon Magna Firewire digital imaging system.
Immunoblot examination of FAK activation.
Serum-starved HFFs were either untreated or pretreated with 25, 50, and 100 μM concentrations of genistein for 1 h at 37°C. These cells were incubated with DMEM or DMEM containing gBΔTM and gBΔTM RGA proteins (1 μg/ml) for 15 min at 37°C, washed in PBS, lysed on ice for 20 min by 0.5 ml of radioimmunoprecipitation assay lysis buffer, and centrifuged at 13,000 × g for 20 min at 4°C, and the supernatants were collected. Equal quantities (8 μg) of protein samples were resolved by SDS-10% PAGE and transferred to nitrocellulose membranes. The membranes were blocked in blocking buffer overnight at 4°C, reacted with mouse anti-phospho-FAK antibody for 3 h at room temperature followed by incubating with horseradish peroxidase-conjugated goat anti-mouse antibody for 90 min at room temperature, and developed with enhanced chemiluminescence reagent (NEN Life Science, Boston, Mass.). Membranes were stripped and reprobed with MAb to actin. The bands were scanned and the band intensities were assessed with the ImageQuaNT software program (Molecular Dynamics).
Observation of target cell morphology.
Confluent target cells in chamber slides were washed twice with serum-free DMEM and were incubated with different concentrations of recombinant gBΔTM, gBΔTM-RGA, or ΔTMgpK8.1A protein (150 μl/well) diluted in serum-free DMEM for 30 min at room temperature or at 37°C. Slides were observed under a Nikon inverted microscope (ECLIPSE TE 200).
Cytotoxicity assay.
Different concentrations of recombinant proteins were incubated with target cells at 37°C, and at different time points, supernatants were collected and assessed for cellular toxicity with the cytotoxicity assay kit (Promega).
RESULTS
Expression and purification of HHV-8-gBΔTM protein without transmembrane and cytoplasmic domains.
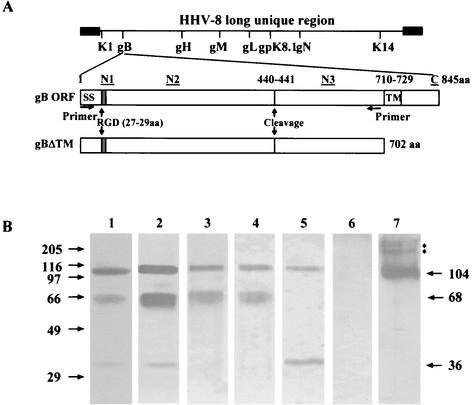
The HHV-8-gB ORF is 845 aa long with a signal sequence (aa 1 to 23) and a transmembrane domain (aa 710 to 729) and with 13 N glycosylation sites (Fig. 1A). At aa 440 to 441, there is a potential proteolytic cleavage site (RKRR/S), and cleavage at this site would result in two proteins with the predicted molecular masses of about 48 and 45 kDa (2, 7). Among the alpha-, beta-, and gammaherpesvirus gBs sequenced to date, only HHV-8-gB possesses the RGD (Arg-Gly-Asp) aa 27 to 29 at the extracellular amino-terminal coil region after the putative signal sequence (Fig. 1A). In HHV-8-carrying BCBL-1 cells, gB is synthesized as a 110-kDa precursor protein and undergoes cleavage and processing, and the envelope-associated gB is made up of 75- and 54-kDa polypeptides forming disulfide-linked heterodimers and multimers (2, 7). Since only about 20% of TPA-induced BCBL-1 cells expressed HHV-8 lytic cycle proteins (2, 46), the yield of purified gB by affinity chromatography was insufficient for functional studies. Hence, a 2,106-bp gBΔTM gene region encoding aa 1 to 702 lacking the transmembrane and the carboxyl domains was amplified by PCR (Fig. 1A), cloned, and expressed in the baculovirus system. The gBΔTM mutant (gBΔTM-RGA) with a single amino acid mutation (RGD to RGA) was also expressed in the baculovirus system. High-5 cells infected with gBΔTM and gBΔTM-RGA-baculovirus were labeled with [35S]methionine, and radiolabeled and unlabeled His-tagged proteins from the cell lysate were purified by nickel columns (51). The purity of the protein was analyzed by Coomassie staining of SDS-PAGE, Western blot reactions with rabbit anti-gB antibodies, and autoradiography. Fractions containing the purified protein were pooled, dialyzed, concentrated, and reanalyzed.
FIG. 1.
(A) Construction of HHV-8-gBΔTM ORF without the transmembrane and carboxyl domains. The top line shows the schematic diagram of HHV-8 genome and the locations of encoded glycoprotein ORFs. HHV-8-gB ORF (ORF 8) is 845 aa long with a signal sequence (SS, aa 1 to 23) and a transmembrane domain (TM, aa 710 to 729). The locations of RGD motif, putative cleavage site, and the four synthetic gB peptides (N1, aa 27 to 47; N2, aa 167 to 191; N3, aa 573 to 593; and C, aa 828 to 845) used for raising rabbit antibodies are indicated. gBΔTM was constructed by using primers amplifying aa 1 to 702 with the signal sequence but lacking the transmembrane and the carboxyl domains and with a His tag at the carboxyl terminus. The gBΔTM/pAcGP67-B plasmid was generated by cloning the amplified fragment (2,124 bp) into the BamHI-EcoRI sites of the pAcGP67-B vector. The gBΔTM-RGA mutant was generated by mutating the RGD amino acids in the gBΔTM/pAcGP67-B plasmid to RGA by site-directed mutagenesis. (B) Expression and purification of gBΔTM protein in the baculovirus system. High-5 cells were infected with baculovirus-gBΔTM for 5 to 7 days, and His-tagged gBΔTM protein from cell pellets was purified by use of a nickel column. Protein purity was analyzed by SDS-10% PAGE gels and Western blotting with anti-gB and anti-gB-peptide rabbit IgG antibodies. Samples were boiled in sample buffer with 2-ME (lanes 1 to 6) or without 2-ME (lane 7). Lane 1, Coomassie stain of purified gBΔTM; lane 2, purified gBΔTM in Western blot reactions with rabbit anti-gB antibodies; lane 3, purified gBΔTM in Western blot reactions with rabbit anti-gB-N1 antibodies; lane 4, purified gBΔTM in Western blot reactions with rabbit anti-gB-N2 antibodies; lane 5, purified gBΔTM in Western blot reactions with rabbit anti-gB-N3 antibodies; lane 6, purified gBΔTM in Western blot reactions with rabbit anti-gB-C antibodies; and lane 7, purified gBΔTM run under nonreducing conditions in Western blot reactions with rabbit anti-gB antibodies. The numbers on the left indicate the molecular masses (in kilodaltons) of the standard protein markers run in parallel lanes. The numbers on the right indicate the approximate molecular masses (in kilodaltons) of the purified gBΔTM protein. The asterisks represent the multimer forms of gBΔTM protein with approximate molecular masses of >180 kDa.
When purified gBΔTM protein treated with sample buffer with 2-ME (reducing conditions) was analyzed by Coomassie staining, three bands of about 36, 68, and 104 kDa were detected (Fig. 1B, lane 1). Contaminating proteins were not detected. Rabbit anti-gB ORF antibodies recognized all three bands of gBΔTM protein (Fig. 1B, lane 2). The predicted molecular mass of gBΔTM protein is about 79 kDa, and cleavage at aa 440 to 441 in gBΔTM would result in two unglycosylated proteins with the predicted molecular masses of about 48 and 30 kDa, with eight and five N glycosylation sites, respectively. To determine whether gBΔTM proteins expressed in the insect cells also undergo proteolytic cleavage and processing like the full-length gB in HHV-8 infected cells, rabbit sera were raised against synthetic peptides corresponding to the amino and carboxyl termini of gB (N1, aa 27 to 47; N2, aa 167 to 191; N3, aa 573 to 593; and C, aa 828 to 845) (Fig. 1A).
In Western blot reactions, rabbit anti-gB-N1 and anti-gB-N2 antibodies against the amino terminus of gBΔTM protein recognized the 68- and 104-kDa proteins but not the 36-kDa protein (Fig. 1B, lanes 3 and 4). In contrast, rabbit anti-N3 antibodies recognizing the gBΔTM protein after the cleavage site recognized only the 36- and 104-kDa proteins but not the 68-kDa protein (Fig. 1B, lane 5). Rabbit anti-gB-C antibodies against the carboxyl terminus of full-length gB did not react with gBΔTM protein (Fig. 1B, lane 6). The specificity of anti-N1, -N2, and -N3 antibodies was also demonstrated by the absence of reactivities by normal rabbit sera and anti-gpK8.1A antibodies (data not shown). When nonreduced samples of gBΔTM protein were analyzed, the 68- and 36-kDa bands were replaced by the broadly migrating 104- to 116-kDa band, as well as multiple polypeptides of more than 180 kDa (Fig. 1B, lanes 7). These results suggest that the 104-kDa protein probably represents the uncleaved gB precursor and that the 68- and 36-kDa proteins probably represent the cleaved and processed amino and carboxyl domains of gBΔTM protein. These data demonstrated that the gBΔTM protein expressed in the insect cells also undergoes proteolytic cleavage and forms disulfide-linked heterodimers and multimers like the full-length gB and thus suggest that the conformation of the gBΔTM protein probably resembles the conformation of the extracellular domain of full-length gB. Identical processing and heterodimer and multimer formation were also seen with the gBΔTM-RGA protein (data not shown), suggesting that the single amino acid change (D to A) did not change the general conformation and characteristics of the gBΔTM-RGA protein. Nonreduced purified gBΔTM, gBΔTM-RGA, and ΔTMgpK8.1A proteins were used throughout the following studies.
HHV-8-gBΔTM and gBΔTM-RGA proteins bind to the target cells.
Our previous studies show that HHV-8 binds and enters a variety of target cells, which include human cells (BJAB, Raji, 293, HFFs, HeLa, and endothelial cells), monkey cells (Vero and CV-1 cells), hamster cells (BHK-21 and CHO), and mouse cells (L) as shown by the detection of DNA, limited HHV-8 gene expression, and green fluorescent protein expression (3). Our studies demonstrated that the broad cellular tropism of HHV-8 may be in part due to its interaction with the ubiquitous host cell surface HS-like molecule (3). This conclusion was based on the following findings: (i) HHV-8 infection of HFF was inhibited in a dose-dependent manner by soluble heparin, a glycosaminoglycan closely related to HS; (ii) enzymatic removal of HFF cell surface HS with heparinases I and III reduced HHV-8 infection; (iii) soluble heparin inhibited the binding of radiolabeled HHV-8 to human B-cell lines, embryonic kidney epithelial (293) cells, and HFFs, suggesting interference at the virus attachment stage; (iv) cell surface- adsorbed HHV-8 was displaced by soluble heparin; and (v) radiolabeled HHV-8 also bound to wild-type HS-expressing CHO-K1 cells. In contrast, binding of virus to mutant CHO cells deficient in HS was significantly reduced. These data suggested that γ2-HHV-8 is adsorbed to cells by binding to cell surface HS-like moieties. In this respect, HHV-8 resembles some members of α (herpes simplex virus type 1, herpes simplex virus type 2, pseudorabies virus, and bovine herpesvirus type 1), β (human cytomegalovirus; HHV-7), and γ2 (bovine herpesvirus type 4) herpesviruses, where the initial virus-cell interaction also involves the binding to the cell surface HS (3).
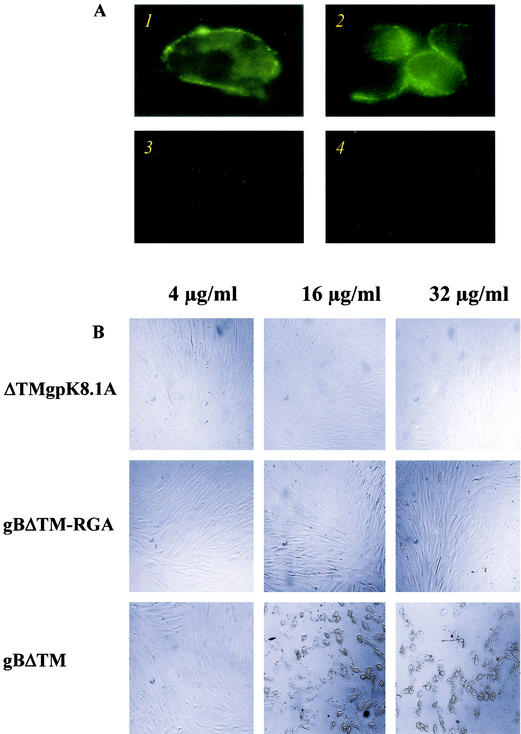
Our studies have also demonstrated the interaction of virion envelope-associated HHV-8 glycoproteins gB(ORF8) and gpK8.1A with HS molecules (2, 51). Using SIFAs and radiolabeled protein-binding assays, we have previously demonstrated the ability of HHV-8 full-length gpK8.1A and the recombinant ΔTMgpK8.1A to bind a variety of target cells (HFF, HMVEC-d, BJAB, and CHO-K1) via its interaction with the cell surface HS-like molecules (51). To determine whether gBΔTM binds to the target cells, unlabeled purified gBΔTM and gBΔTM-RGA proteins were allowed to bind the paraformaldehyde-treated suspension of BJAB cells at 4°C. In another method, these proteins were incubated with monolayers of HFF or HMVEC-d cells at 4°C and were then fixed with 4% paraformaldehyde. These cells were washed and tested with rabbit anti-gB or anti-gpK8.1A IgG antibodies in SIFAs. Bright ring-type fluorescence was observed only on cells incubated with gBΔTM, and the results with HFF and BJAB cells are shown in Fig. 2A, panels 1 and 2, respectively. No fluorescence was observed in cells incubated with gBΔTM and anti-gpK8.1A rabbit antibodies (Fig. 2A, panel 3) or in cells incubated only with anti-gB antibodies (Fig. 2A, panel 4). Similar results were also seen with the gBΔTM-RGA protein (data not shown). Binding was not detected when cells were incubated with purified His-tagged HHV-8 latency-associated ORF 73 protein (51; data not shown). These data demonstrated the interaction of gBΔTM with the cell surface and showed that the extracellular domains of gBΔTM mediate this binding.
FIG. 2.
HHV-8-gBΔTM binds to the target cells. (A) Binding of purified gBΔTM protein detected by surface immunofluorescence assay. Panel 1: HFFs in chamber slides were incubated with 16 μg of purified gBΔTM/ml for 90 min at 4°C, washed, fixed with 4% paraformaldehyde for 10 min at room temperature, washed, and incubated with rabbit anti-HHV-8-gB IgG antibodies for 60 min at 4°C. Cells were washed, incubated for 60 min at 4°C with FITC-conjugated goat anti-rabbit IgG antibodies, washed, mounted, and examined under a fluorescence microscope. Panel 2: BJAB cells fixed with 0.1% paraformaldehyde were incubated with 33 μg of purified gBΔTM/ml for 90 min at 4°C, washed, and incubated with rabbit anti-HHV-8-gB IgG antibodies and processed as described for panel 1. Panel 3: BJAB cells incubated with purified gBΔTM, rabbit anti-HHV-8 gpK8.1A IgG antibodies, and FITC-anti-rabbit antibodies. Panel 4: BJAB cells incubated with DMEM, rabbit anti-HHV-8-gB IgG antibodies, and FITC-anti-rabbit antibodies. (B) Morphological changes induced by binding of gBΔTM protein to the target cells. Confluent HFFs in chamber slides were incubated with different concentrations of recombinant gBΔTM, gBΔTM-RGA, or ΔTMgpK8.1A protein (150 μl/well) diluted in serum-free DMEM for 30 min at room temperature, and slides were observed.
Binding of higher concentrations of HHV-8-gBΔTM proteins to the target cells induces morphological changes and detachments.
Since the ability of gBΔTM protein to mediate cell adhesion via its RGD sequence was tested at 37°C, we examined the effect of recombinant gBΔTM protein binding to the target cells. Monolayers of HFF, HMVEC-d, and CV-1 were incubated with different concentrations of recombinant proteins at 37°C or at room temperature for 30 min, washed, and observed for the morphological changes. Subsequently, viability of these cells was tested by trypan blue exclusion test and cytotoxicity assay kit. No significant morphological changes were observed in cells incubated with the ΔTMgpK8.1A and gBΔTM-RGA proteins even at a concentration of 32 μg/ml (Fig. 2B, top and middle panels). In contrast, cells incubated with the gBΔTM protein at a concentration of 8 μg/ml and above exhibited significant morphological changes such as rounding and detachment (Fig. 2B, lower panels). These cells remained viable, and cellular death was not observed. These morphological changes were probably induced by the clumping of integrin molecules and actin cytoskeleton rearrangement and thus by the lifting of cells from the substrate. Nevertheless, these data clearly demonstrated that HHV-8-gBΔTM protein binds cell surface receptors.
Specificity of HHV-8-gBΔTM and gBΔTM-RGA proteins binding to the target cells.
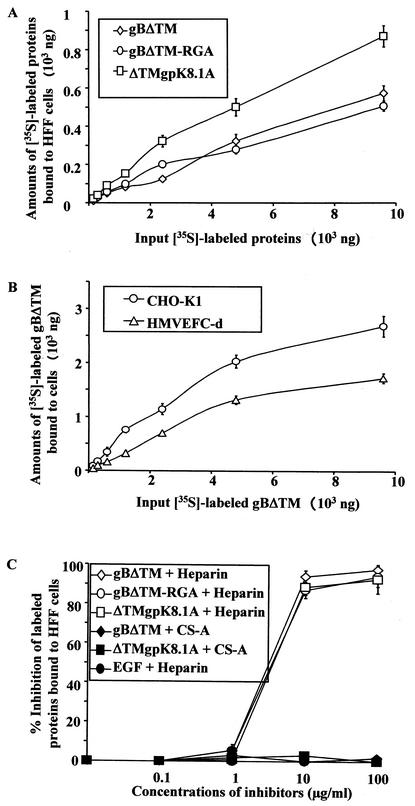
To quantitate the target cell bindings of gBΔTM protein, purified [35S]methionine-labeled His-tagged recombinant gBΔTM, gBΔTM-RGA, and ΔTMgpK8.1A proteins were incubated with HFFs and HMVEC-d, and CHO-K1 cells at 4°C for 90 min. Radiolabeled gBΔTM, gBΔTM-RGA, and ΔTMgpK8.1A proteins bound to HFFs in dose-dependent manners (Fig. 3A). Equal binding was detected with gBΔTM and gBΔTM-RGA proteins. Compared to the gB proteins, more ΔTMgpK8.1A protein was bound to cells. Binding was not detected when cells were incubated with [35S]methionine-labeled purified His-tagged ORF 73 protein (51). We have previously shown that the binding of ΔTMgpK8.1A protein to the target cells occurred in a saturable manner, which was blocked by the preincubation of cells with unlabeled ΔTMgpK8.1A protein (51). Binding of gBΔTM and gBΔTM-RGA protein to HFFs also occurred in a similar saturable manner (data not shown). The gBΔTM protein also bound with equal efficiency to the HMVEC-d and CHO-K1 cells in a dose-dependent and saturable manner (Fig. 3B) and to BJAB cells (data not shown). Binding of gBΔTM-RGA protein to the HFFs (Fig. 3A) and HMVEC-d and CHO-K1 (data not shown) cells occurred with efficiency similar to that of the gBΔTM protein, alleviating the concern that the single amino acid substitution may have altered the conformation of the protein. Binding of gBΔTM to the various human and animal target cells is not surprising, as it probably reflects the broad cellular tropism of HHV-8 (3).
FIG. 3.
Quantitation of HHV-8-gBΔTM binding to the target cells. (A and B) Binding of radiolabeled proteins to target cells. Different concentrations of [35S]methionine-labeled purified gBΔTM, gBΔTM-RGA, or ΔTM gpK8.1A protein were incubated for 90 min at 4°C with HFFs (A) or HMVEC-d (B) or CHO-K1 (B) cells in 24-well plates. After incubation, cells were washed five times and lysed with 1% SDS and 1% Triton X-100, and the cell-bound protein radioactivity was counted. Each reaction was done in triplicate, and each point represents the average plus or minus standard deviation of three experiments. (C) Inhibition of [35S]methionine-labeled purified proteins binding to HFFs by heparin. [125I]-labeled EGF lacking heparin-binding activity was used as the negative control. A predetermined quantity of 125I-labeled EGF (259,272 cpm/ng) or [35S]methionine-labeled purified gBΔTM, gBΔTM-RGA, or ΔTM gpK8.1A protein within the linear range of the dose response-curve (6 μg) (A and B) was mixed with medium alone or with different concentrations of heparin or CS-A and was incubated for 60 min at 4°C. These were then incubated with HFFs for 90 min at 4°C, washed, and lysed with 1% SDS and 1% Triton X-100, and the cell-bound protein radioactivity was counted. The cell-associated protein counts per minute in the presence or absence of heparin and the percentage of inhibition of protein binding were calculated. Each reaction was done in triplicate, and each point represents the average plus or minus standard deviation of three experiments.
We have previously shown the interaction of HHV-8 with host cell surface HS-like molecules via its envelope-associated gB and gpK8.1A (2, 3, 51). HHV-8-gB binding to the target cells probably involves the heparin-binding domain of gB (HBD, aa 108 to 117) and the RGD motif, as well as other domains. Heparin is closely related to HS, and studies showed that HHV-8 interaction with the host cell surface involved HS and that soluble heparin prevented HHV-8 infectivity (3). To determine whether heparin inhibits the binding of gBΔTM, gBΔTM-RGA, or ΔTMgpK8.1A protein to the target cells, a constant quantity of purified radiolabeled protein within the linear range of the dose-response curve (6 μg) (Fig. 3A) was mixed with medium alone or with medium containing different concentrations of heparin or CS-A and was incubated at 4°C for 1 h. These were then added to the HFFs and were incubated at 4°C for 90 min. To further determine the specificity of recombinant proteins binding to the target cells, 125I-labeled EGF lacking heparin-binding activity was used as the negative control. A predetermined quantity of 125I-labeled EGF within the linear range of the dose-response curve was mixed with medium alone or with different concentrations of heparin and incubated for 90 min at 4°C. After incubation, the cells were washed five times and cell-bound radioactivity was counted. Soluble heparin significantly inhibited the binding of labeled gBΔTM, gBΔTM-RGA, and ΔTMgpK8.1A proteins to HFFs in a dose-dependent manner (Fig. 3C). Similar results were also seen with the HMVEC-d cells (data not shown). The specificity of inhibition of recombinant proteins binding to the target cells by heparin was shown by the absence of inhibition by CS-A (Fig. 3C) and by the inability of heparin to block the binding of 125I-labeled EGF to the target cells (Fig. 3C). These results further confirmed the binding of gBΔTM, gBΔTM-RGA, and ΔTMgpK8.1A proteins to the target cells. Inhibition by heparin suggested that these proteins interact with the cell surface HS-like molecules.
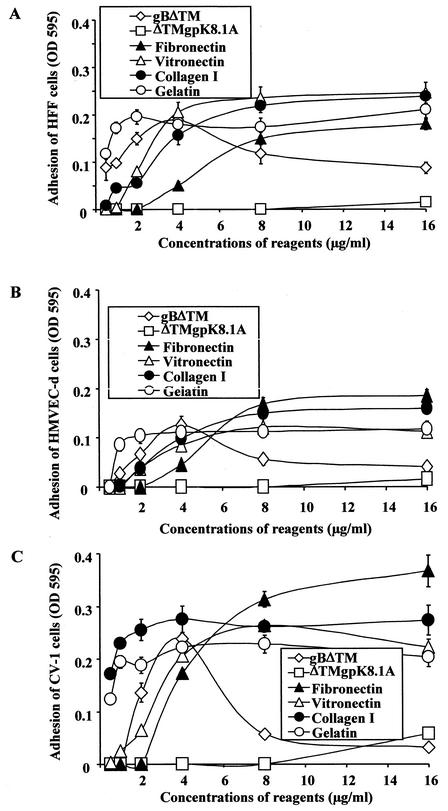
HHV-8-gBΔTM induces the adhesion of cells.
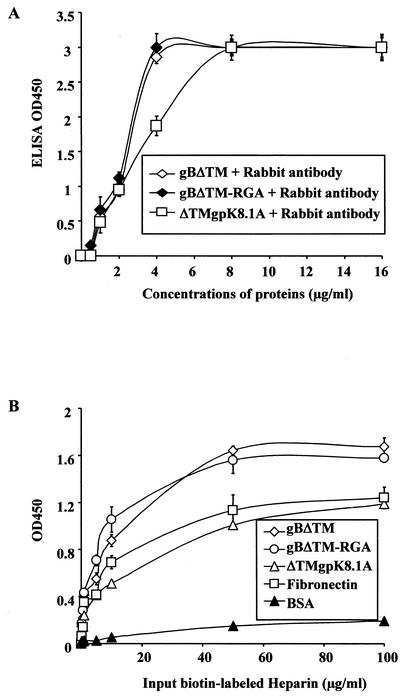
HHV-8-gB possesses the RGD motif, the minimal peptide region of many proteins known to interact with subsets of host cell surface integrins (4, 28). HHV-8 utilizes α3β1 integrin as one of the receptors for its entry into the target cells via its gB interaction and induces the activation of FAK. Since FAK activation is the first step in the outside-in signaling necessary for the integrin-mediated cell adhesion, we next examined whether the recombinant gBΔTM protein could mediate adhesion of human (HFF and HMVEC-d) and monkey (CV-1) cells. ECM proteins such as fibronectin, collagen, vitronectin, and gelatin were used as positive controls, and ΔTMgpK8.1A protein and BSA were used as negative controls. The ability of gBΔTM, gBΔTM-RGA, and ΔTMgpK8.1A to bind the plates was initially examined by coating the plates with different concentrations of these proteins and by testing them with specific antibodies (Fig. 4A). ELISAs demonstrated that the wells were coated with all three proteins with equal efficiency in a dose-dependent manner (Fig. 4A). To determine the ability of Maxisorp plate-bound proteins to bind heparin, plates were coated with a constant quantity of fibronectin, gBΔTM, gBΔTM-RGA, ΔTMgpK8.1A, and BSA (4 μg/ml in PBS, 100 μl/well) overnight at 4°C. Plates were washed, blocked, washed, and incubated with different concentrations of biotin-labeled heparin for 1 h at 4°C and were incubated with streptavidin horseradish peroxidase. Dose-dependent binding of biotin-labeled heparin to plate-bound fibronectin, gBΔTM, gBΔTM-RGA, and ΔTMgpK8.1A proteins but not to BSA was detected (Fig. 4B). The ability of plate-bound recombinant proteins to bind heparin suggested that the conformation of the protein domain(s) involved in the interaction with heparin was probably not altered.
FIG. 4.
HHV-8-gBΔTM bound to the plates retains heparin-binding activity. (A) Binding of purified proteins to the Maxisorp plates. Plates were coated with different concentrations of unlabeled purified gBΔTM, gBΔTM-RGA, and ΔTMgpK8.1A proteins in PBS (4 μg/ml in PBS, 100 μl/well) overnight at 4°C. Plates were washed, blocked with 10% nonfat milk for 1 h at 37°C, washed, and incubated with rabbit anti-gB or anti-gpK8.1A IgG antibodies for 1 h at 37°C, followed by goat anti-rabbit antibodies coupled to horseradish peroxidase. After reacting with substrate, the reaction was stopped by 2 N H2SO4 and the optical density was read at 450 nm (OD450). Each reaction was done in triplicate, and each point represents the average plus or minus standard deviation of three experiments. (B) Heparin binds to the gBΔTM protein-coated plates. Maxisorp plates were coated with BSA, fibronectin, gBΔTM, gBΔTM-RGA, or ΔTMgpK8.1A protein (4 μg/ml, 100 μl/well) overnight at 4°C. Plates were washed, blocked with 3% BSA in PBS for 2 h at 4°C, washed, and incubated with different concentrations of biotin-labeled heparin (100 μl/well) for 1 h at 4°C. Plates were washed and incubated with streptavidin horseradish peroxidase (0.01 μg/well) for 1 h at room temperature. The reaction was stopped by 2 N H2SO4 and read at 450 nm. Each reaction was done in triplicate, and each point represents the average plus or minus the standard deviation of three experiments.
Cell adhesion assays were done by coating Maxisorp ELISA plates with different concentrations of proteins overnight at 4°C in PBS. Plates were washed, blocked with 1% BSA in PBS for 2 h at 4°C, washed, and incubated with fresh suspensions of target cells at a concentration of 2 × 104 cells/well for 45 min to 1 h at 37°C. Cells were washed, adherent cells were fixed with 4% paraformaldehyde, washed, and stained with 0.5% crystal violet-methanol, the dye was extracted with 0.1 M sodium citrate, and the optical density at 595 nm was quantitated. The ECM proteins induced the adhesion of all three cell types examined in a dose-dependent manner (Fig. 5). No cell adhesion was seen with ΔTMgpK8.1A (Fig. 5). In contrast, gBΔTM induced the adhesion of all three cell types in a dose-dependent manner (Fig. 5). Absence of cell adhesion by ΔTMgpK8.1A was not due to its inability to bind the Maxisorp plates, since ELISAs demonstrated that the wells were also coated with ΔTMgpK8.1A (Fig. 4A). When examined under a light microscope, wells coated with 4 μg of gBΔTM/ml showing the highest optical density readings were covered with a monolayer of adherent cells. As the optical density decreased, fewer and fewer cells remained attached to the bottom of the wells. Wells coated with the ΔTMgpK8.1A or BSA were virtually devoid of cells. Thus, the optical density readings correlated with cell adhesion, suggesting that the gBΔTM mediates cell adhesion.
FIG. 5.
HHV-8-gBΔTM induces the adhesion of target cells. Maxisorp plates were coated overnight at 4°C with different concentrations of purified gBΔTM, ΔTMgpK8.1A, fibronectin, vitronectin, collagen I, and gelatin in PBS (100 μl/well). Plates were washed, blocked with 1% BSA in PBS for 2 h at 4°C, washed, and incubated with fresh suspensions of target cells in 0.1% BSA-serum-free DMEM at a concentration of 2 × 104 cells/well for 45 min to 1 h. Cells were washed, and adherent cells were fixed with 4% paraformaldehyde, washed, and stained with 0.5% crystal violet-methanol. The dye was extracted with 0.1 M sodium citrate, and the optical density at 595 nm (OD 595) was quantitated. Each reaction was done in triplicate, and each point represents the average plus or minus standard deviation of three experiments.
Similar to cells adhered to ECM proteins, cells adhering to gBΔTM showed attachment and spread with typical ruffled membranes (lamellipodia) and extended hairlike projections (filopodia) (data not shown). Interestingly, adhesion of cells to higher concentrations of gBΔTM (>4 μg/ml) was remarkably reduced for all three cell types (Fig. 5). Compared to adhesion mediated by 4 μg of gBΔTM/ml, at a concentration of 16 μg of protein/ml, adhesion of only about 10% was observed. In wells coated with gBΔTM at concentrations of 8 μg/ml or higher, most of the cells were rounded and easily washed off even after an extended period of incubation. Adenovirus type 2 penton base protein binds to integrins αVβ3 and αVβ5 via its RGD sequence, and higher concentrations of penton base protein also cause rounding and detachment of cells (8). Detachment of cells by the high concentrations of gBΔTM could be an in vitro phenomenon due to clumping of integrin molecules and thus lifting of the cells from the substrate. Nevertheless, these data clearly demonstrated that HHV-8-gBΔTM protein interactions with cell surface receptors induce cell adhesion.
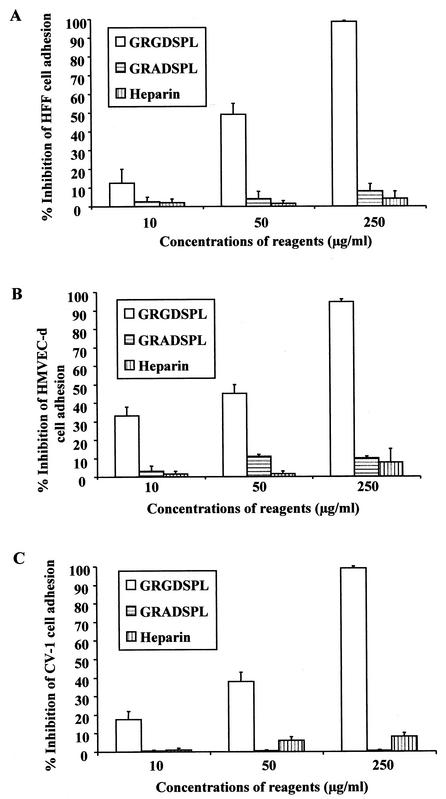
Peptides with RGD amino acids inhibit cell adhesion mediated by gBΔTM.
Since the RGD motif is often essential for the induction of cell adhesion, to determine whether the RGD amino acids in the gBΔTM protein play a role in the observed cell adhesion, target cells were preincubated with different concentrations of synthetic peptides with or without RGD sequences (GRGDSPL and GRADSPL) for 30 min at 4°C. Cells were then added to the plates coated with the gBΔTM protein. To determine whether HS plays a role in cell adhesion, protein-coated plates were also incubated with different concentrations of heparin for 1 h at 4°C before addition of the target cells for adhesion assays. Preincubation of HFF, HMVEC-d, and CV-1 cells with the GRGDSPL peptide significantly inhibited the gBΔTM protein-mediated cell adhesion in a dose-dependent manner and almost completely abolished the cell adhesion at 250-μg/ml concentrations (Fig. 6). In contrast, cell adhesion mediated by gBΔTM protein was unaffected by the GRADSPL peptide even at high concentrations (Fig. 6). These results suggest a role for the RGD amino acids and hence a role for integrins in the cell adhesion induced by the gBΔTM protein.
FIG. 6.
The peptide with RGD amino acids inhibits the cell adhesion induced by the gBΔTM protein. Maxisorp plates were coated with 4 μg of purified gBΔTM protein/ml (100 μl/well) overnight at 4°C, washed, and blocked with 1% BSA-PBS. For testing the effect of RGD amino acids, target cells were preincubated with different concentrations of GRGDSPL or GRADSPL peptides for 30 min at 4°C and were seeded to the protein-coated plates. For testing the effect of heparin, protein-coated plates were incubated with different concentrations of heparin in DMEM for 1 h at 4°C before addition of the target cells. The adhesion assay was carried out as described in the Fig. 3 legend. Data are presented as percentage of inhibition of cell adhesion obtained when cells were preincubated with DMEM only. Each reaction was done in triplicate, and each point represents the average plus or minus standard deviation of three experiments.
Besides the RGD motif at aa 27 to 29, the HHV-8-gB ORF also possesses a putative HBD at aa 108 to 117 (2, 4). It has been previously shown that HHV-8-gB binds to the α3 and β1 integrin subunits to a comparable extent in heparinase I- or III-treated or untreated HFFs (4). This observation suggested that HHV-8-gB interaction with HS via the putative HBD may not be competing with the virus-integrin interaction by the RGD motif. When the gBΔTM protein was preincubated with different concentrations of heparin before adding the target cells, there was no significant change in the adhesion of cells to the plate-bound gBΔTM protein (Fig. 6). Though the preincubation of recombinant proteins with heparin blocked the soluble gBΔTM protein binding to the cell surface HS (Fig. 2C), heparin was not able to block the plate-bound gBΔTM protein RGD motif-dependent mediated cell adhesion. This could be due to two different assay systems being used. However, since heparin binds to the plate-bound gBΔTM protein efficiently (Fig. 4B), it implies that the binding of heparin to gBΔTM protein does not block the binding of the RGD motif to the cellular receptor. These results suggest that adhesion is mediated by the RGD amino acids of gB, which is independent of the HBD domain of gB.
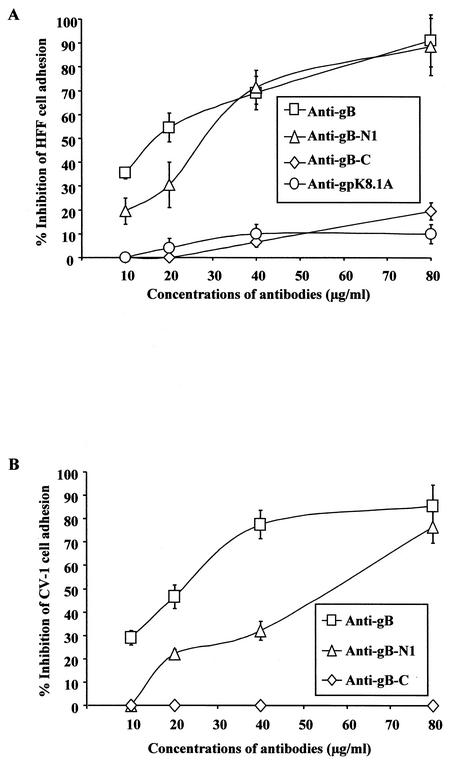
Antibodies to peptides containing RGD amino acids inhibit cell adhesion to gBΔTM.
To ascertain the specificity of cell adhesion to gBΔTM, protein-coated plates were preincubated with different concentrations of IgG antibodies for 1 h at 4°C before seeding of the target cells for adhesion assays. Rabbit IgG antibodies against the full-length gB and the antibodies directed against the RGD-containing gB peptide (gB-N1-RGDTFQTSSSPTPPGSSS) significantly inhibited the gBΔTM-mediated adhesion of HFF and CV-1 cells (Fig. 7). In contrast, rabbit antibodies against the gB peptide lacking the RGD motif but with RGY amino acids (gB-C-RGYKPLTQSLDISPETGE) (Fig. 7) and antibodies against gpK8.1A did not show any significant inhibition of cell adhesion (Fig. 7A). These results demonstrated the specificity of gBΔTM protein-induced cell adhesion and suggested a role for the RGD motif in the cell adhesion induced by HHV-8-gB.
FIG. 7.
Antibodies against gB peptide containing RGD amino acids inhibit the cell adhesion induced by the gBΔTM protein. Maxisorp plates were coated with 4 μg of purified gBΔTM protein/ml (100 μl/well) overnight at 4°C, washed, and incubated for 1 h at 4°C with rabbit IgG antibodies against HHV-8-gB (anti-gB) or against HHV-8-gB peptide with (anti-gB-N1) and without RGD amino acids (anti-gB-C) or against HHV-gpK8.1A (anti-gpK8.1A) in 0.1% BSA-DMEM. HFFs were seeded into the wells, and cell adhesion assays were carried out as described in the Fig. 3 legend. Data are presented as percentage of inhibition of cell adhesion obtained when gBΔTM was preincubated with DMEM only. Each reaction was done in triplicate, and each point represents the average plus or minus standard deviation of three experiments.
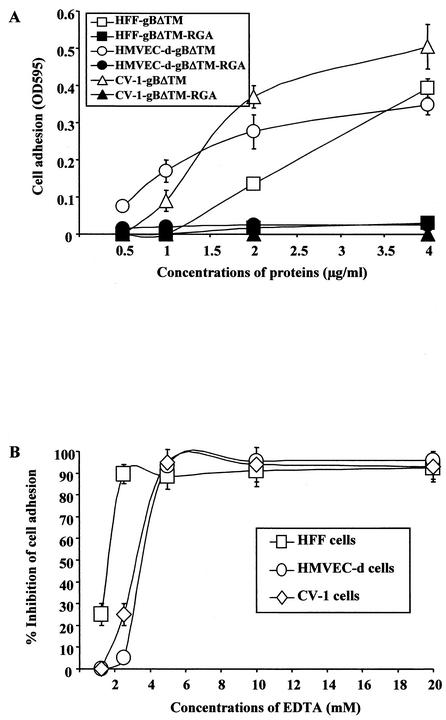
A single amino acid change (RGD to RGA) abolishes the cell adhesion to gBΔTM.
To provide additional proof that the RGD sequence of gBΔTM is responsible for mediating cell adhesion, the aspartic acid at residue 29 was changed to an alanine by site-directed mutagenesis to create the mutant gBΔTM-RGA protein. As seen in our previous experiments, gBΔTM mediated adhesion of HFFs and HMVEC-d and CV-1 cells in a dose-dependent manner (Fig. 8A). In contrast, no adhesion of cells was observed in plates coated with the gBΔTM-RGA mutant protein (Fig. 8A). The similarity of gBΔTM-RGA protein with the gBΔTM protein, in terms of processing and heterodimer formation and the equal binding efficiency to the target cells (Fig. 3A and B) and to the plates (Fig. 4A and B), suggested that the single amino acid substitution may not have altered the conformation of the mutant gBΔTM-RGA protein drastically. These results clearly demonstrated the importance of RGD amino acids in the adhesion mediated by HHV-8-gB.
FIG. 8.
(A) HHV-8-gBΔTM protein-induced cell adhesion was abolished by a single amino acid substitution of the RGD sequence. Maxisorp plates were coated with different concentrations of purified gBΔTM or gBΔTM-RGA mutant proteins overnight at 4°C and washed, and cell adhesion assays were carried out as described in the Fig. 3 legend. Each reaction was done in triplicate, and each point represents the average plus or minus the standard deviation of three experiments. (B) EDTA blocks target cell adhesion induced by HHV-8-gBΔTM. Maxisorp plates were coated with 4 μg of purified HHV-8-gBΔTM/ml (100 μl/well) overnight at 4°C, washed, and blocked with 1% BSA-PBS. Target cells were resuspended in medium with 0.1% BSA and were incubated with different concentrations of EDTA for 15 min on ice before seeding to plates. Adhesion assays were carried out as described in the Fig. 3 legend. Data are presented as percentage of inhibition of cell adhesion obtained when cells were preincubated with DMEM without EDTA. Each reaction was done in triplicate, and each point represents the average plus or minus the standard deviation of three experiments.
Cell adhesion to HHV-8-gBΔTM is dependent on divalent cations.
An important characteristic of integrin-ligand interaction is the dependence on divalent cations, and each integrin heterodimer contains several cation-binding sites. To further verify the specificity of gBΔTM-induced cell adhesion, the requirement for divalent cations for the induced cell adhesion was assessed. Cells were preincubated with different concentrations of EDTA for 15 min on ice before seeding into the protein-coated plates. As shown in Fig. 8B, adhesion of CV-1 cells and HFFs induced by gBΔTM was blocked by EDTA in a dose-dependent manner, and complete inhibition was detected at >4 mM concentrations of EDTA. Lack of cell adhesion in the presence of EDTA clearly demonstrated the divalent cation-dependent gBΔTM-mediated cell adhesion and suggests the role of integrins in the observed adhesions.
HHV-8-gBΔTM activates the FA components.
Based primarily on morphology or method of formation, cell adhesion has been classified into FAs, fibrillar adhesions, focal complexes, and podosomes (41). Over 50 proteins have been found in adhesions, and different classes of adhesions reflect differing molecular compositions (41). The formation and disassembly of adhesions are complex processes and require a coordinated interaction of actin or actin-binding proteins, signaling molecules, and microtubules. FAK is a nonreceptor protein-tyrosine kinase expressed in most tissues. The FAK pathway is activated by most integrin-ligand interactions, and FAK activation is the first step necessary for FA formation and for the outside in signaling by integrins (28). After the interaction with ligands, integrins and numerous signaling molecules, including FAK, assemble into FA aggregates on each side of the membrane and well-developed aggregates of FAs can be detected by immunofluorescence assay (28). FAK is recruited to FAs because it interacts either directly or through the cytoskeletal proteins talin, paxillin, and vinculin with the cytoplasmic tail of integrin subunits (28). FAK also interacts with a number of signaling proteins, including Src, phosphatidylinositol 3-kinase, p130Cas, and Grb2 (28). These interactions link FAK to signaling pathways that modify the cytoskeleton and activate mitogen-activated protein kinase cascades (28). Following integrin-ligand interactions, FAK is autophosphorylated at the Tyr397 residue, which is critical for the subsequent tyrosine phosphorylation of other FA proteins (28). It has been previously shown that HHV-8 induces the FAK phosphorylation as early as 5 min postinfection, which was inhibited by the preincubation of virus with soluble α3β1 integrin and anti-HHV-8-gB antibodies (4).
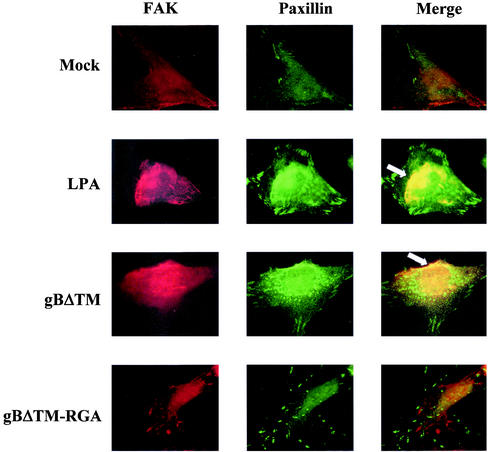
To determine whether the cell adhesion induced by gBΔTM involves FAK activation, paxillin and FAK colocalization was examined in serum-starved HFFs and HMVEC-d cells incubated with various concentrations of recombinant gBΔTM and gBΔTM-RGA proteins and results with HFFs are shown here. In cells mock treated with serum-free DMEM with 50 ng of BSA/ml, the distribution of FAK and paxillin is more diffuse and independent of each other (Fig. 9, top row). In contrast, distinct patchy patterns of FAK and paxillin distribution were observed in cells incubated with 50 ng of gBΔTM/ml for 5 min at 37°C (Fig. 9, third row). Image overlays demonstrated the colocalization of FAK with paxillin, indicating the specificity of altered FAK distribution in gBΔTM-treated cells (Fig. 9, third row). These patterns of FAK colocalization with paxillin were readily observed as early as 5 min after incubation with gBΔTM and were comparable to the patterns observed in 20 ng of LPA-treated cells per ml (Fig. 9, second row). In cells incubated with gBΔTM-RGA protein, similar to the mock-infected cells, the distribution of FAK and paxillin is more diffuse and independent of each other (Fig. 9, fourth row). Similar activation and redistribution of FAK were observed in HMVEC-d cells (data not shown). These results demonstrated that gBΔTM activates FAK presumably by its interaction with integrin molecules and thus indicate that the cell adhesion induced by gBΔTM involves FAK activation.
FIG. 9.
HHV-8-gBΔTM activates the integrin-dependent FAK components. Serum-starved HFFs in chamber slides were incubated with 200 μl of serum-free DMEM (mock) or LPA (20 ng/ml) or gBΔTM protein (50 ng/ml) in serum-free DMEM or gBΔTM-RGA (50 ng/ml) in serum-free DMEM. After incubation for 5 min at 37°C, cells were washed, fixed, permeabilized, and reacted with rabbit anti-p125FAK and mouse MAb to paxillin, followed by anti-rabbit-tetramethyl rhodamine isothiocyanate and anti-mouse-FITC antibodies. Stained cells were examined under a fluorescence microscope with appropriate filters. The arrowheads indicate representative areas of FAK-paxillin colocalization. Magnification, ×1,000.
HHV-8-gBΔTM induces the phosphorylation of FAK in the target cells.
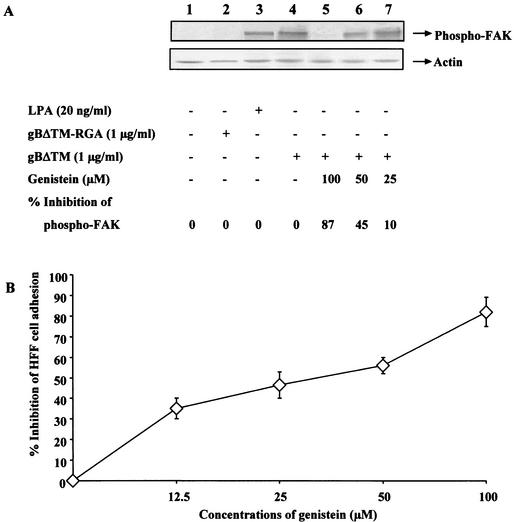
To verify the induction of FAK by gBΔTM, the amount of phosphorylation of FAK at Tyr397 was determined by testing the gBΔTM-incubated cell lysates with anti-Tyr397-FAK antibodies. Induction of cells for 5 min with 20 ng of LPA-treated cells/ml resulted in about a 27-fold increase in FAK phosphorylation over that found in the control cells (Fig. 10A, top panel, lanes 1 and 3). HHV-8-gBΔTM also rapidly induced the phosphorylation of FAK, with about a 23-fold increase at 15 min postincubation (Fig. 10A, top panel, lane 4). Incubation of HFFs with gBΔTM-RGA protein (Fig. 10A, top panel, lane 3) or with ΔTMgpK8.1A protein (data not shown) did not induce any significant levels of FAK phosphorylation. The specificity of anti-phospho-FAK antibodies was demonstrated by the lowering of FAK phosphorylation to undetectable levels by treating infected cell lysates with tyrosine phosphatase 1B (data not shown).
FIG. 10.
FAK activation is critical for the cell adhesion induced by HHV-8-gBΔTM. (A) Induction of FAK phosphorylation by HHV-8-gBΔTM protein and its inhibition by genistein. Top panel: Serum-starved HFFs were uninduced (lane 1) and induced with 1 μg of gBΔTM-RGA protein/ml for 15 min at 37°C (lane 2), with 20 ng of LPA/ml for 5 min at 37°C (lane 3), and with 1 μg of gBΔTM protein/ml for 15 min at 37°C (lane 4) or were pretreated with 100, 50, and 25 μM concentrations of genistein for 1 h at 37°C and were then incubated with 1 μg of gBΔTM protein/ml for 15 min at 37°C in the presence of similar concentrations of genistein (lanes 5 to 7, respectively). Cells were lysed, and equivalent amounts of lysates were subjected to SDS-PAGE, transferred to nitrocellulose membranes, reacted with anti-phospho-p125FAK antibodies, and developed using chemiluminescent reagents. Lower panel: after reactions, membranes were stripped and reprobed with MAb to actin. The bands were scanned, and the band intensities were assessed. Data are presented as percentage of inhibition of FAK phosphorylation obtained when the cells were incubated with the gBΔTM protein only. (B) Kinase inhibitor genistein inhibits HHV-8-gBΔTM mediated cell adhesion. Maxisorp plates were coated with 4 μg of gBΔTM protein (100 μl/well) per ml overnight at 4°C, washed, and blocked with 1% BSA-PBS. Serum-starved HFFs were incubated with different concentrations of genistein in serum-free DMEM for 1 h at 37°C, collected by trypsin, washed, suspended in serum-free DMEM-0.1% BSA with different concentrations of genistein, and added to the protein-coated plates. Adhesion assays were carried out as described in the Fig. 3 legend. Each reaction was done in triplicate, and each point represents the average plus or minus the standard deviation of three experiments. Data are presented as percentage of inhibition of cell adhesion when cells were incubated with DMEM without genistein.
Inhibition of FAK phosphorylation inhibits the cell adhesion induced by gBΔTM.
Genistein (4′,5,7-trihydroxyisoflavone) is a tyrosine kinase-competitive inhibitor of the ATP binding site and inhibits several tyrosine kinases (1, 33). Genistein inhibits serum-induced tyrosine phosphorylation of FAK and the assembly of focal adhesions (18). We used genistein to evaluate the importance of tyrosine phosphorylation of FAK in the process of HHV-8-gBΔTM protein-mediated cell adhesion. Since genistein is cytotoxic at higher concentrations (1, 33), predetermined nontoxic concentrations of genistein were used in these studies. Treatment of cells for 1 h before gBΔTM protein incubation with 25, 50, and 100 μM concentrations of genistein and during the 15-min incubation with the protein lowered the gBΔTM-induced FAK phosphorylation to 10, 45, and 87% of that of control cells, respectively (Fig. 10A, top panel, lanes 4 to 6). The levels of actin did not change in these experiments (Fig. 10A, lower panel), and dimethyl sulfoxide used as a solvent for genistein did not alter the expression of FAK (data not shown). These results further confirmed the activation of FAK by HHV-8-gBΔTM protein in the target cells.
Incubation of HFFs with 140 μM genistein has been shown to inhibit the integrin-induced signaling and cytoskeletal protein complexes without profound cytotoxicity (33). To determine the interlink between FAK activation and cell adhesion induced to gBΔTM, HFFs were incubated with noncytotoxic doses of genistein, harvested by trypsin treatment, suspended in serum-free medium with different concentrations of genistein, and seeded into the plates coated with proteins, and adhesion assays were performed. As shown in Fig. 10B, genistein inhibited HFF adhesion to gBΔTM in a dose-dependent manner, with about 85% inhibition at a 100 μM concentration. These data suggest a direct role for FAK and other tyrosine kinases in the gBΔTM-mediated cell adhesion and the induction of cellular signaling pathways by the gB interactions with integrins.
DISCUSSION
HHV-8 virion envelope-associated glycoprotein gB interacts with the target cell HS and α3β1 integrin molecules, which are critical for virus attachment and entry into the host cells, respectively (2, 3, 4). Several lines of evidence presented in this study demonstrate that HHV-8-gB, like ECM proteins, could also mediate cell adhesion via its RGD motif interactions with integrin molecules. Maintenance of gBΔTM protein in a soluble form throughout the purification procedures and cell adhesion experiments, similarity with the full-length gB in terms of processing, heterodimer formation, binding to the target cells, inhibition of target cell binding by heparin, and the activation of FAK in the target cells suggest that gBΔTM used in this study probably possess a conformation similar to that for the external domain of gB associated with the virion envelope and the infected cell membranes. Moreover, the absence of LPS contamination in our protein preparations, the inhibition of adhesion by preincubating the protein with anti-gB antibodies, and the requirement of gB-RGD motif for the observed adhesion clearly demonstrated the specificity of cell adhesion mediated by the gBΔTM protein.
In results similar to those of our studies, HIV-1 Tat expressed in the prokaryotic system, and baculovirus-expressed adenovirus penton base protein have been successfully used in cell adhesion assays (9, 13, 52). The RGD motif is also present in several viral proteins, such as adenovirus type 2 penton base protein (52), echovirus 9 vp-1 (55), murine and human cytomegalovirus ppM44 (34), foot-and-mouth disease virus vp-1 (25), and HIV-1 Tat protein (13, 27). Some of these viral proteins mediate cell adhesion in an RGD-dependent manner, and HIV-1 Tat protein also mediates cell attachment through its basic domain (9, 10, 13, 34, 50). Our studies show that HHV-8-gB mediates via its RGD sequence but not by its HBD domain. The presence of an RGD motif alone does not guarantee cell adhesive activity, because the flanking sequence often influences its conformation. The availability of appropriate integrin receptor molecules on the cell surface also affects the final outcome. Several studies have shown that for the interactions of proteins with integrins via RGD motif, threonine (T) at the fourth position (RGDT) is also critical (40). It is interesting that HHV-8-gB ORF also possesses a threonine at the fourth position of the RGD motif (43).
Several ECM proteins like laminin can mediate both RGD-dependent and -independent cell adhesion (40). Cell adhesion via binding to integrin molecules can also occur to proteins without the RGD motif. Cyr61, an angiogenic inducer, although devoid of RGD motif, mediates human skin fibroblast cell adhesion via its binding to both HS and integrin α6β1 (16, 17). Cell adhesion mediated by Cyr61 was blocked by heparin as well as by EDTA but not by peptides containing the RGD sequence (16, 17). HIV-1 Tat protein mediates adhesion of several cells through its RGD sequence (13) as well as via its basic domain (9, 10, 50). Cell adhesion to the Tat basic domain is blocked by heparin but resistant to EDTA (50). There is increasing evidence showing that HS proteoglycans can function as coreceptors with integrins in the cell-matrix interactions (53). In addition to the RGD sequence, HHV-8-gB also possesses a heparin-binding basic amino acid domain (HBD aa 108 to 117) (2). However, it is unlikely that the adhesion observed in the present in vitro study is due to the HBD, since adhesion was almost completely blocked by peptides containing RGD and anti-gB-RGD peptide antibodies but not by high concentrations of heparin. Blocking of HHV-8 binding to the target cells by pretreatment of virus with heparin (3), and the inability of soluble integrins, RGD peptides, anti-gB antibodies, and anti-integrin intibodies to block virus binding but ability to neutralize infectivity (4) suggest that the first contact with the target cells by HHV-8 is mediated by its interaction with the cell surface HS-like molecules (3), and that the subsequent entry appears to be mediated by the virus integrin and other molecules (4). Though the preincubation of recombinant proteins with heparin blocked the soluble protein binding to HS, the inability of heparin to block the plate-bound protein-mediated cell adhesion suggests that adhesion mediated by the RGD amino acids is independent of the HBD domain of gB and that cells can still bind to the RGD domain, though the HS-binding site was blocked by heparin. Together with the absence of adhesion by gBΔTM-RGA mutant protein, these results suggest that HHV-8-gB-mediated cell adhesion is through its RGD motif interacting with integrin molecules.
The interaction of HHV-8-gB with α3β1 integrin has been shown previously (4). Ongoing studies show that, in addition to the α3β1 integrin, HHV-8 also utilizes the αvβ3 and αvβ5 integrins for its infectivity (unpublished data). Ongoing studies show that antibodies against αv and β1 block the adhesion mediated by HHV-8-gB. Further studies are in progress to characterize the role of different integrins in the adhesion mediated by HHV-8-gB.
Upon activation, FAK autophosphorylates FAK-Tyr397, creating a binding site for the SH2 domain of Src or Fyn. The Src kinases then phosphorylate a number of FA components. The major targets of Src kinase phosphorylation include the two-cytoskeletal proteins paxillin and tensin, which localize at the FA sites (28, 45). Paxillin is one of the key components of integrin signaling, and tyrosine-phosphorylated paxillin plays a major role in localizing FAK to FAs and FA complexes and is required for integrin-mediated cytoskeletal reorganization (28, 45). Colocalization of FAK and paxillin in HHV-8-gB-incubated cells suggests the activation of paxillin and the onset of integrin-mediated signaling pathway induction in the target cells. Integrins regulate cell adhesion, spreading, and migration through the activation of the Rho family of small guanine nucleotide-binding proteins (28, 29, 35). Among the Rho-related guanosine triphosphatases, Cdc42 induces the hairlike projections (filopodia), Rac induces membrane ruffling (lamellipodia), and Rho induces focal adhesions and the associated stress fibers (28, 29, 35). Each of these Rho-related proteins controls the actin cytoskeleton by interacting with multiple downstream effectors (28, 29, 35). The FAK-Src pathway also plays a role in cell migration, by promoting the disassembly of FAs at the trailing edge of the cell (28, 29, 35). Cells incubated with HHV-8-gBΔTM exhibited filopodia and lamellipodia, suggesting the induction of Rho family kinases. Further work is in progress in defining the signal transduction pathways and cell migration induced by HHV-8-gB.
What role might the adhesion mediated by HHV-8-gB play during virus infection of target cells? HHV-8-gB interaction with integrin and the subsequent activation of FAK and other signaling pathways may promote its entry into the target cells and may generate a cellular state that is more receptive for infection. In a manner similar to that of simian virus 40 (5) and γ1-Epstein-Barr virus (37), the γ2-HHV-8 enters via large endocytic vesicles in the human B-cell line, BJAB (3) and HFFs and 293 and endothelial cells (unpublished data). This is an exciting observation, since HHV-8 utilizes the α3β1 integrin for its infectivity and activates FAK (4), which is intimately associated with the assembly of cytoskeleton and endocytosis (28). Receptor-mediated endocytosis and signaling pathways are interlinked and depend upon one another (35). In contrast to the earlier concept that endocytosis is a way to attenuate ligand-activated responses, recent studies suggest that signaling continues in the endocytic pathway (35). Protein components of signal transduction cascades can assemble at clathrin-coated pits and remain associated with endocytic vesicles following their dynamin-dependent release from the plasma membrane, suggesting that endocytosis plays a role in the activation and propagation of signaling pathways such as phosphatidylinositol 3-kinase and ERK1/2 and vice versa (35). FAK activation and assembly of cytoskeleton have been shown to be critical for the endocytosis of several microbes (21, 48). For example, adenovirus entry into the target cells via endocytosis depends upon the actin cytoskeleton activation by the Rho family of GTPases (32, 52). Further studies are in progress to decipher the interlink between HHV-8-gB interactions with host cells, including adhesion, activation of cellular kinases, and the virus entry and infection of target cells.
What role might the HHV-8-gB activation of FAK and the induction of adhesion play in KS pathogenesis? AIDS KS differs from other tumors in many respects. The disease is characterized by a multifocal, widespread distribution that may involve any location on the skin or mucous membranes, as well as the lymph nodes, gastrointestinal tract, and visceral organs (6, 44). Most human tumors are derived from the clonal outgrowth of a single cell type. In contrast, all KS lesions show a variety of cell types. Advanced lesions include a predominance of spindle-shaped cells, thought to be derived from cells in the endothelial or mesenchymal lineage (6, 44). All KS lesions also display a proliferation of aberrant, slit-like neovascular spaces as well as variable quantities of infiltrating inflammatory cells, including plasma cells, T cells, and monocytes/macrophages (12, 26, 38, 44). In KS tissues, HHV-8 DNA is present in a latent form in the vascular endothelial and spindle cells (6). Only a limited set of HHV-8 genes, LANA1 (ORF 73), cyclin D (vCYC-[ORF 72]), vFLIP (ORF 71), and K12 are expressed in these cells (6, 26, 38, 46). All clinical stages of KS, including patch, plaque, and nodular lesions, were positive for ORF 73 (6, 26, 38, 46). About 1 to 10% of infiltrating monocytic cells in KS lesions express HHV-8 lytic cycle proteins (6, 12, 38, 46).
HHV-8 probably depends upon its proteins expressed during the latent infection to evade the immune system, to overcome host cell restrictions of transcription, and to maintain the latent state. LANA-1 maintains the KSHV episome and tethers the viral genome to chromatin during mitosis (6, 47). In addition, LANA-1 interacts with p53 and represses its transcriptional activity (6, 47). LANA-1 also interacts with pRb. In cooperation with the cellular oncogene Harvey rat sarcoma viral oncogene homologue (Hras), LANA-1 transformed primary rat embryo fibroblasts and rendered them tumorogenic (6, 47). HHV-8-vCYC strongly associates with cdk6, activates cdk6 leading to the phosphorylation of Rb and histone 1 substrates, and evades inhibition by p21 and p16 family members (6, 47). Kaposin is known to interact with cytohesin 1, a guanine nucleotide exchange factor and regulator of integrin-mediated cell adhesion, and induces focus formation, stress fiber dissolution, and activation of the ERK1/2 mitogen-activated protein kinase signal cascade (30).
Despite the expression of these powerful latent viral proteins that can modulate cell growth, cells cultured from KS tumors are not fully transformed, have normal diploid karyotype, grow for a limited number of passages, contain a mixture of cell types (spindle shaped fibroblasts and endothelial cells), depend upon growth factors, fail to grow in soft agar, and do not induce tumors in immunodeficient mice (6, 26, 44, 46). Even though HHV-8 is present in the KS tissues, HHV-8 DNA is lost within a few passages of KS lesion cells. The factors driving the KS tumor spindle cell growth, the inability of KS cells latently infected with HHV-8 to grow in cell culture, and the reason for the loss of HHV-8 DNA within a few passages of KS cells are not known.
Several studies demonstrate an increase in HHV-8 load before KS development and during KS clinical manifestations, suggesting reactivation and lytic HHV-8 replication (6, 24, 26). Together with the detection of the HHV-8 lytic cycle in KS lesion inflammatory cells, reduction of risk of KS by ganciclovir treatment that can inhibit HHV-8 replication, and increased antibodies against lytic cycle proteins in KS patients (6, 24, 26), these data suggest a role for HHV-8 lytic replication in KS. Many of the HHV-8-encoded lytic cycle proteins such as vGPCR, vMIP1, vBcl2, and vIL-6 may contribute to the pathogenesis of KS by their cell growth-altering activities (6, 19, 26, 36, 46). HHV-8-encoded lytic cycle proteins have been shown to interact with several cellular signal transduction proteins. HHV-8 K1 protein has been shown to interact with Vav, p85, and Syk kinase (31). The cytoplasmic domain of K15 is tyrosine phosphorylated at the putative SH2 binding motif by cellular tyrosine kinases (19). Constitutive expression of ORF 74 has been shown to induce activation of several cellular signal transduction pathways, leading to activation of cellular immediate-early response genes such as NF-κB and activator protein 1 via mitogenic cascades (39).
In addition to the HHV-8 lytic cycle proteins mentioned above, HHV-8-gB's interaction with integrin, activation of FAK and the induction of cell adhesion may also have important implications in the unique biology of KS lesions and in HHV-8's role in KS pathogenesis. Productive infection of monocytic cells resulting in cell lysis could release virion particles, gB, and other regulatory proteins that could have a profound growth-altering effect on the latently infected endothelial cells and neighboring cells. For example, activation of FAK by HHV-8-gB may induce cytokines and growth factors controlling cell migration and proliferation. HHV-8 interactions with β1 integrin, a molecule known to induce inflammatory cytokines and vascular endothelial growth factor, may also stimulate the production of cytokines and growth factors in latently infected endothelial and adjacent cells, which in turn may stimulate the endothelial growth. Adhesions, migration, invasion, and proliferation are interlinked dynamic processes of tumor cell invasion, involve homotypic or heterotypic cellular interactions, cell interactions with ECM proteins and local stroma, and dissolution of these contacts (41). Adhesion can function in a positive way to stabilize tissue structure and organization under normal conditions or negatively to facilitate dissemination (41). In the context of invasion, adhesion can be broken into a continuous circle of attachment, detachment, and spreading. Cells also require anchorage to ECM to proliferate, and integrins activate growth-promoting signaling pathways such as FAK/Src and Fyn/Shc pathways that are responsible for the anchorage requirement (28, 41). The interaction between HHV-8-gB RGD sequence and integrins activating FAK may contribute in the homing and adhesion of HHV-8-infected cells to endothelial cells and their proliferation. Interactions with latently infected endothelial cells may contribute to the detachment and spread of infected cells and thus contribute to the multifocal, widespread distribution of KS lesions. Further studies are needed to examine the consequences of HHV-8-gB interactions with cells that are already programmed by HHV-8 latency-associated proteins and to show whether such interactions play a role in the establishment and/or maintenance of latent infection and/or cellular proliferation of latently infected endothelial cells.
Acknowledgments
This study was supported in part by Public Health Service grants CA 75911 and CA 82056 to B.C. and P01 RR16443 to S.M.A. and by a University of Kansas Medical Center Biomedical Research Training program postdoctoral fellowship to S.M.A.
We thank Clark Bloomer and Bo Wisdom, Biotechnology Center, University of Kansas Medical Center, Kansas City, for DNA sequencing and for the synthetic peptides. We thank Marilyn Smith for critically reading the manuscript.
REFERENCES
- 1.Akiyama, T., J. Ishida, S. Nakagawa, H. Ogawara, S. Watanabe, N. Itoh, M. Shibuya, and Y. Fukami. 1987. Genistein, a specific inhibitor of tyrosine-specific protein kinases. J. Biol. Chem. 262:5592-5595. [PubMed] [Google Scholar]
- 2.Akula, S. M., N. P. Pramod, F. Z. Wang, and B. Chandran. 2001. Human herpesvirus 8 envelope-associated glycoprotein B interacts with heparan sulfate-like moieties. Virology 284:235-249. [DOI] [PubMed] [Google Scholar]
- 3.Akula, S. M., F. Z. Wang, J. Vieira, and B. Chandran. 2001. Human herpesvirus 8 interaction with target cells involves heparan sulfate. Virology 282:245-255. [DOI] [PubMed] [Google Scholar]
- 4.Akula, S. M., N. P. Pramod, F. Z. Wang, and B. Chandran. 2002. Integrin alpha3beta1 (CD 49c/29) is a cellular receptor for Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) entry into the target cells. Cell 108:407-419. [DOI] [PubMed] [Google Scholar]
- 5.Anderson, H. A., Y. Chen, and L. C. Norkin. 1996. Bound simian virus 40 translocates to caveolin-enriched membrane domains, and its entry is inhibited by drugs that selectively disrupt caveolae. Mol. Biol. Cell 7:1825-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Antman, K., and Y. Chang. 2000. Kaposi's sarcoma. N. Engl. J. Med. 342:1027-1038. [DOI] [PubMed] [Google Scholar]
- 7.Baghian, A., M. Luftig, J. B. Black, Y. X. Meng, C. P. Pau, T. Voss, P. E. Pellett, and K. G. Kousoulas. 2000. Glycoprotein B of human herpesvirus 8 is a component of the virion in a cleaved form composed of amino- and carboxyl-terminal fragments. Virology 269:18-25. [DOI] [PubMed] [Google Scholar]
- 8.Bai, M., B. Harfe, and P. Freimuth. 1993. Mutations that alter an Arg-Gly-Asp (RGD) sequence in the adenovirus type 2 penton base protein abolish its cell-rounding activity and delay virus reproduction in flat cells. J. Virol. 67:5198-5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barillari, G., R. Gendelman, R. C. Gallo, and B. Ensoli. 1993. The Tat protein of human immunodeficiency virus type 1, a growth factor for AIDS Kaposi sarcoma and cytokine-activated vascular cells, induces adhesion of the same cell types by using integrin receptors recognizing the RGD amino acid sequence. Proc. Natl. Acad. Sci. USA 90:7941-7945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barillari, G., C. Sgadari, V. Fiorelli, F. Samaniego, S. Colombini, V. Manzari, A. Modesti, B. C. Nair, A. Cafaro, M. Sturzl, and B. Ensoli. 1999. The Tat protein of human immunodeficiency virus type-1 promotes vascular cell growth and locomotion by engaging the α5β1 and αVβ3 integrins and by mobilizing sequestered basic fibroblast growth factor. Blood 94:663-672. [PubMed] [Google Scholar]
- 11.Birkmann, A., A. Mahr, A. Ensser, S. Yaguboglu, F. Titgemeyer, B. Fleckenstein, and F. Neipel. 2001. Cell surface heparan sulfate is a receptor for human herpesvirus 8 and interacts with envelope glycoprotein K8.1. J. Virol. 75:11583-11593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blasig, C., C. Zietz, B. Haar, F. Neipel, S. Esser, N. H. Brockmeyer, E. Tschachler, S. Colombini, B. Ensoli, and M. Sturzl. 1997. Monocytes in Kaposi's sarcoma lesions are productively infected by human herpesvirus 8. J. Virol. 71:7963-7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brake, D. A., C. Debouck, and G. Biesecker. 1990. Identification of an Arg-Gly-Asp (RGD) cell adhesion site in human immunodeficiency virus type 1 transactivation protein, tat. J. Cell Biol. 111:1275-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cerimele, F., F. Curreli, S. Ely, A. E. Friedman-Kien, E. Cesarman, and O. Flore. 2001. Kaposi's sarcoma-associated herpesvirus can productively infect primary human keratinocytes and alter their growth properties. J. Virol. 75:2435-2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 16.Chen, N., C. C. Chen, and L. F. Lau. 2000. Adhesion of human skin fibroblasts to Cyr61 is mediated through integrin α6β1 and cell surface heparan sulfate proteoglycans. J. Biol. Chem. 275:24953-24961. [DOI] [PubMed] [Google Scholar]
- 17.Chen, C. C., N. Chen, and L. F. Lau. 2001. The angiogenic factors Cyr61 and connective tissue growth factor induce adhesive signaling in primary human skin fibroblasts. J. Biol. Chem. 276:10443-10452. [DOI] [PubMed] [Google Scholar]
- 18.Chrzanowska-Wodnicka, M., and K. Burridge. 1994. Tyrosine phosphorylation is involved in reorganization of the actin cytoskeleton in response to serum or LPA stimulation. J. Cell Sci. 107:3643-3654. [DOI] [PubMed] [Google Scholar]
- 19.Damania, B., J.-K. Choi, and J. U. Jung. 2000. Signaling activities of gammaherpesvirus membrane proteins. J. Virol. 74:1593-1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dezube, B. J. 2000. The role of human immunodeficiency virus-I in the pathogenesis of acquired immunodeficiency syndrome-related Kaposi's sarcoma: the importance of an inflammatory and angiogenic milieu. Semin. Oncol. 27:420-423. [PubMed] [Google Scholar]
- 21.Dramsi, S., and P. Cossart. 1998. Intracellular pathogens and the actin cytoskeleton. Annu. Rev. Cell Dev. Biol. 14:137-166. [DOI] [PubMed] [Google Scholar]
- 22.Ensoli, B., R. Gendelman, P. Markham, V. Fiorelli, S. Colombini, M. Raffeld, A. Cafaro, H. K. Chang, J. N. Brady, and R. C. Gallo. 1994. Synergy between basic fibroblast growth factor and HIV-1 Tat protein in induction of Kaposi's sarcoma. Nature 371:674-680. [DOI] [PubMed] [Google Scholar]
- 23.Ensoli, B., and M. Sturzl. 1998. Kaposi's sarcoma: a result of the interplay among inflammatory cytokines, angiogenic factors and viral agents. Cytokine Growth Factor Rev. 9:63-83. [DOI] [PubMed] [Google Scholar]
- 24.Ensoli, B., M. Sturzl, and P. Monini. 2001. Reactivation and role of HHV-8 in Kaposi's sarcoma initiation. Adv. Cancer Res. 81:161-200. [DOI] [PubMed] [Google Scholar]
- 25.Fox, G., N. R. Parry, P. V. Barnett, B. McGinn, D. J. Rowlands, and F. Brown. 1989. The cell attachment site on foot-and-mouth disease virus includes the amino acid sequence RGD (arginine-glycine-aspartic acid). J. Gen. Virol. 70:625-637. [DOI] [PubMed] [Google Scholar]
- 26.Ganem, D. 1998. Human herpesvirus 8 and its role in the genesis of Kaposi's sarcoma. Curr. Clin. Top. Infect. Dis. 18:237-251. [PubMed] [Google Scholar]
- 27.Ganju, R. K., N. Munshi, B. C. Nair, Z.-Y. Liu, P. Gill, and J. E. Groopman. 1998. Human immunodeficiency virus Tat modulates the Flk-1/KDR receptor, mitogen-activated protein kinases, and components of focal adhesion in Kaposi's sarcoma cells. J. Virol. 72:6131-6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giancotti, F. G., and E. Ruoslahti. 1999. Integrin signaling. Science 285:1028-1032. [DOI] [PubMed] [Google Scholar]
- 29.Hall, A. 1998. Rho GTPases and the actin cytoskeleton. Science 279:509-514. [DOI] [PubMed] [Google Scholar]
- 30.Kliche, S., W. Nagel, E. Kremmer, C. Atzler, A. Ege, T. Knorr, U. Koszinowski, W. Kolanus, and J. Haas. 2001. Signaling by human herpesvirus 8 kaposin A through direct membrane recruitment of cytohesin-1. Mol. Cell 7:833-843. [DOI] [PubMed] [Google Scholar]
- 31.Lagunoff, M., R. Majeti, A. Weiss, and D. Ganem. 1999. Deregulated signal transduction by the K1 gene product of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 96:5704-5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, E., D. Stupack, G. M. Bokoch, and G. R. Nemerow. 1998. Adenovirus endocytosis requires actin cytoskeleton reorganization mediated by Rho family GTPases. J. Virol. 72:8806-8812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Linassier, C., M. Pierre, J. B. Le Pecq, and J. Pierre. 1990. Mechanisms of action in NIH-3T3 cells of genistein, an inhibitor of EGF receptor tyrosine kinase activity. Biochem. Pharmacol. 39:187-193. [DOI] [PubMed] [Google Scholar]
- 34.Loh, L. C., D. Locke, R. Melnychuk, and S. Laferte. 2000. The RGD sequence in the cytomegalovirus DNA polymerase accessory protein can mediate cell adhesion. Virology 272:302-314. [DOI] [PubMed] [Google Scholar]
- 35.McPherson, P. S., B. K. Kay, and N. K. Hussain. 2001. Signaling on the endocytic pathway. Traffic 2:375-384. [DOI] [PubMed] [Google Scholar]
- 36.Neipel, F., J. C. Albrecht, and B. Fleckenstein. 1997. Cell-homologous genes in the Kaposi's sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J. Virol. 71:4187-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nemerow, G. R., and N. R. Cooper. 1984. Early events in the infection of human B lymphocytes by Epstein-Barr virus: the internalization process. Virology 132:186-198. [DOI] [PubMed] [Google Scholar]
- 38.Parravicini, C., B. Chandran, M. Corbellino, E. Berti, M. Paulli, P. S. Moore, and Y. Chang. 2000. Differential viral protein expression in Kaposi's sarcoma-associated herpesvirus-infected diseases: Kaposi's sarcoma, primary effusion lymphoma, and multicentric Castleman's disease. Am. J. Pathol. 156:743-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pati, S., M. Cavrois, H.-G. Guo, J. S. Foulke, Jr., J. Kim, R. A. Feldman, and M. Reitz. 2001. Activation of NF-κB by the human herpesvirus 8 chemokine receptor ORF74: evidence for a paracrine model of Kaposi's sarcoma pathogenesis. J. Virol. 75:8660-8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Plow, E. F., T. A. Haas, L. Zhang, J. Loftus, and J. W. Smith. 2000. Ligand binding to integrins. J. Biol. Chem. 275:21785-21788. [DOI] [PubMed] [Google Scholar]
- 41.Price, J. T., M. T. Bonovich, and E. C. Kohn. 1997. The biochemistry of cancer dissemination. Crit. Rev. Biochem. Mol. Biol. 32:175-253. [DOI] [PubMed] [Google Scholar]
- 42.Renne, R., D. Blackbourn, D. Whitby, J. Levy, and D. Ganem. 1998. Limited transmission of Kaposi's sarcoma-associated herpesvirus in cultured cells. J. Virol. 72:5182-5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Safai, B., and J. J. Schwartz. 1992. Kaposi's sarcoma and the acquired immunodeficiency syndrome, p. 209-223. In V. T. DeVitta, S. Helllman, and S. A. Rosenberg (ed.), AIDS: etiology, diagnosis, treatment and prevention. Lippincott Co., Philadelphia, Pa.
- 45.Schaller, M. D. 2001. Paxillin: a focal adhesion-associated adaptor protein. Oncogene 20:6459-6472. [DOI] [PubMed] [Google Scholar]
- 46.Schulz, T. F., Y. Chang, and P. S. Moore. 1998. Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8), p. 87-134. In D. J. McCance (ed.), Human tumor viruses. American Society for Microbiology, Washington, D.C.
- 47.Swanton, C., and N. Jones. 2001. Strategies in subversion: de-regulation of the mammalian cell cycle by viral gene products. Int. J. Exp. Pathol. 82:3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Triantafilou, K., Y. Takada, and M. Triantafilou. 2001. Mechanisms of integrin-mediated virus attachment and internalization process. Crit. Rev. Immunol. 21:311-322. [PubMed] [Google Scholar]
- 49.Vieira, J., P. O'Hearn, L. Kimball, B. Chandran, and L. Corey. 2001. Activation of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) lytic replication by human cytomegalovirus. J. Virol. 75:1378-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vogel, B. E., S. J. Lee, A. Hildebrand, W. Craig, M. D. Pierschbacher, F. Wong-Staal, and E. Ruoslahti. 1993. A novel integrin specificity exemplified by binding of the αVβ5 integrin to the basic domain of the HIV Tat protein and vitronectin. J. Cell Biol. 121:461-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, F.-Z., S. M. Akula, N. P. Pramod, L. Zeng, and B. Chandran. 2001. Human herpesvirus 8 envelope glycoprotein K8.1A interaction with the target cells involves heparan sulfate. J. Virol. 75:7517-7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wickham, T. J., P. Mathias, D. A. Cheresh, and G. R. Nemerow. 1993. Integrins αVβ3 and αVβ5 promote adenovirus internalization but not virus attachment. Cell 73:309-319. [DOI] [PubMed] [Google Scholar]
- 53.Woods, A., E. S. Oh, and J. R. Couchman. 1998. Syndecan proteoglycans and cell adhesion. Matrix Biol. 17:477-483. [DOI] [PubMed] [Google Scholar]
- 54.Zhu, L., V. Puri, and B. Chandran. 1999. Characterization of human herpesvirus-8 K8.1A/B glycoproteins by monoclonal antibodies. Virology 262:237-249. [DOI] [PubMed] [Google Scholar]
- 55.Zimmermann, H., H. J. Eggers, and B. Nelsen-Salz. 1997. Cell attachment and mouse virulence of echovirus 9 correlate with an RGD motif in the capsid protein VP1. Virology 233:149-156. [DOI] [PubMed] [Google Scholar]