Abstract
The oncoproteins of the DNA tumor viruses, adenovirus E1A, simian virus 40 T antigen, and papillomavirus E7, each interact with the retinoblastoma family of tumor suppressors, leading to cell cycle stimulation, apoptosis induction, and cellular transformation. These proteins utilize a conserved LXCXE motif, which is also found in cellular proteins, to target the retinoblastoma family. Here, we describe a herpesvirus protein that shares a subset of the properties of the DNA tumor virus oncoproteins but maintains important differences as well. The human cytomegalovirus pp71 protein employs an LXCXD motif to attack the retinoblastoma family members and induce DNA synthesis in quiescent cells. pp71 binds to and induces the degradation of the hypophosphorylated forms of the retinoblastoma protein and its family members p107 and p130 in a proteasome-dependent manner. However, pp71 does not induce apoptosis and fails to transform cells. Thus, the similarities and differences in comparison to E1A, T antigen, and E7 make pp71 an interesting new tool with which to further dissect the role of the retinoblastoma/E2F pathway in cellular growth control and carcinogenesis.
Viruses are reliant upon their host cells for their replication, and thus, they have evolved methods to efficiently alter the cellular processes that are essential to their duplication and dissemination. Since the repertoire of proteins expressed by viruses is constrained by their relatively small genomes, they generally target important cellular processes, and the means by which they attack them must be effective. Thus, the analysis of how individual viral proteins commandeer and disrupt cellular proteins and networks has been a powerful approach for investigating fundamental mechanisms in cell biology. For example, adenovirus E1A, simian virus 40 (SV40) T antigen, and papillomavirus E7 continue to be invaluable tools in the molecular dissection of the processes governed by the retinoblastoma (Rb) family of tumor suppressors. Here, we demonstrate that a herpesvirus protein, the human cytomegalovirus (HCMV) phosphoprotein 71 (pp71), shares some, but not all, of the properties of these three oncoproteins from the DNA tumor viruses and thus represents a new means by which a viral protein alters cell biology.
HCMV is a ubiquitous human pathogen, and a new infection or the reactivation of a latent infection in immunocompromised individuals can result in life-threatening disease. HCMV infection may also play a role in proliferative diseases such as atherosclerosis (27) and restenosis (38). Thus, we and others have been exploring how HCMV alters cell cycle progression (reviewed in reference 23). Infection of cells brought to quiescence either by contact inhibition or serum starvation results in an abortive mitogenic response with an increase in the level of cyclin E protein and kinase activity (4, 18). However, cyclin A is not induced, and cells do not synthesize host genomic DNA (4, 24). Thus, infection of quiescent cells stimulates their reentry into the cell cycle and progression through the G1 phase, with an eventual cell cycle arrest at the G1/S border. This cell cycle compartment is presumably favorable for viral replication, since the precursors for DNA replication are available but are not being consumed in the synthesis of the host cell's genome.
Here, we show that the HCMV UL82 gene product pp71 stimulates quiescent cells to enter the cell cycle. pp71 is a component of HCMV virions and is delivered to the cell at the very start of infection, a time when altering the host's cell cycle may be important to establishing a successful infection. pp71 targets the hypophosphorylated forms of the Rb family for proteasome-dependent degradation. Like the DNA tumor virus proteins E1A, T antigen, and E7, which contain LXCXE motifs (reviewed in reference 30), pp71 employs an LXCXD motif to attack the Rb pathway and induce cell cycle progression. However, unlike the DNA tumor virus oncoproteins, pp71 does not transform cells and does not induce apoptosis. pp71 sends a proliferative signal to cells that is strong enough to induce entry into the cell cycle and progression into S phase in the absence of other viral proteins but mild enough not to override the eventual cell cycle arrest imposed by the virus at the G1/S border during an infection. Thus, pp71 provides a new means with which to study the complexities of progression through the G1 phase of the cell cycle and the Rb/E2F pathway.
MATERIALS AND METHODS
Cell culture and analysis of infectivity, apoptosis, and cell cycle progression.
Primary human foreskin fibroblasts, primary rat embryo fibroblasts, U-2 OS, 293, Rat-1, and baby rat kidney cells were maintained in Dulbecco's modification of Eagle's medium supplemented with 10% fetal bovine serum (FBS; HyClone) in an atmosphere of 5% CO2 at 37°C. For serum starvation, Rat-1 cells were washed twice with and then cultured in Dulbecco's modification of Eagle's medium plus 0.1% FBS for 72 h. The ability of pp71 to increase the infectivity of transfected viral genomic DNA was assayed as described previously (1). For cell cycle and apoptosis analysis by flow cytometry, floating and attached cells were harvested, fixed in ethanol, stained with propidium iodide and analyzed as described previously (22). For determination of bromodeoxyuridine (BrDU) incorporation, cells grown on glass coverslips were pulse-labeled for 2 h with 25 μM BrDU beginning at 22 h after infection, immediately fixed with paraformaldehyde, treated with 2 N HCl, and stained with biotinylated anti-BrDU (Zymed) and streptavidin-Alexa 546 (Molecular Probes). Nuclear DNA was counterstained with yoyo-1 (Molecular Probes) and RNase A. At least 500 nuclei were scored by fluorescence microscopy.
Plasmids.
pCGN71 (C. J. Baldick and T. Shenk, unpublished data) encodes an XbaI-BamHI PCR fragment corresponding to the HCMV strain AD169 UL82 gene cloned in frame with the hemagglutinin (HA) epitope tag of the vector pCGN (41). This N-terminal HA-tagged pp71 allele was employed as the template to make all of the additional clones by PCR amplification. BamHI PCR fragments encoding N-terminal HA- and His-tagged pp71 alleles were cloned into pSG5 (Stratagene) to generate pSG5-pp71 and pSG5-His-pp71, respectively. The HA-pp71 point mutants were created by PCR amplification with mutagenic primers. pGSTpp71 was constructed by inserting a PCR fragment containing amino acids 1 to 407 of pp71 into pGTK (H. Zhu and T. Shenk, unpublished data). All clones generated by PCR amplification were confirmed by complete sequencing. pSG5L-HApRb (provided by J. DeCaprio) encodes HA-tagged Rb. pCMV-Rb, pCMV-p107, pHAp107, pCMV-p130, pHAp130, pCMVE2F-1, pCGNpp65, pSG5-T, pGSTpRb, pGSTp107, pGSTp130, pCMVE1A, pCMV19K, pCGNIE1, pCGNIE2, pCBSFlag-myc, and pH-rasEJ have been described previously (1, 8, 10, 19, 20, 25, 26, 34, 40, 43, 45, 46, 47).
Recombinant adenoviruses.
Recombinant adenoviruses were generated by using the AdEasy system (16) with pADEasy-1 and either pADtrack-CMV (16) or pACIC. ADTrack viruses express the indicated protein and enhanced green fluorescent protein (GFP), each from its own CMV promoter. To make pADIC, a PstI-BamHI internal ribosome entry site (IRES)-containing fragment from pIRES-EYFP (Clontech) was inserted into pECFP-N1 (Clontech) to make pIR-CFP. The IRES-enhanced cyan fluorescent protein (CFP) cassette was removed with XhoI and XbaI and cloned into the corresponding sites of pADtrack-CMV (the polylinker XhoI and a methylation-sensitive XbaI site after the enhanced GFP gene) to create pADIC. Adenovirus-IRES-CFP (ADIC) viruses express the gene of interest from the CMV promoter and the enhanced CFP gene from an IRES element. BamHI fragments encoding the desired gene were cloned into the BglII site of pADtrack-CMV or pADIC, and viruses were created as described previously (16). pADtrack-CMV and pADIC without inserts were used to make control viruses that express only the fluorescent protein. The ADtrack recombinant adenovirus expressing E2F-1 has been previously described (42). Purified adenovirus stocks were prepared as described previously (28), diluted with an equal volume of 2× storage buffer (50% glycerol, 4 mM MgCl2, 20 mM Tris [pH 8.0]), and stored at −20°C. Virus titers were determined by optical absorbance (28). Infections were at ∼10,000 particles/cell.
Degradation analysis.
For cotransfection assays, U-2 OS cells were transfected with the indicated plasmids by the calcium phosphate method. The DNA precipitate was removed 12 h later, and cells were rinsed and cultured in either 10% FBS-containing medium for 48 h or 0.1% FBS-containing medium for 24 h before protein lysates were harvested. Protein levels were quantitated by the Bradford assay, lysates separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and Western blots were probed with the indicated antibodies. For the pulse-chase analysis, at 24 h after transfection U-2 OS cells cultured in 10% FBS-containing medium were incubated in cysteine- and methionine-free medium containing 10% dialyzed FBS for 1 h prior to a 30 min pulse-label period with 4 ml of medium containing 0.2 mCi of 35S-express (NEN)/ml. Cultures were chased in medium containing 10% FBS, and lysates were immunoprecipitated with the HA-specific antibody. Blots were quantitated with a phosphorimager. For the plots shown in Fig. 4A, arbitrary units for each time point were divided by the arbitrary units for the vector time-zero sample and the ratio is presented.
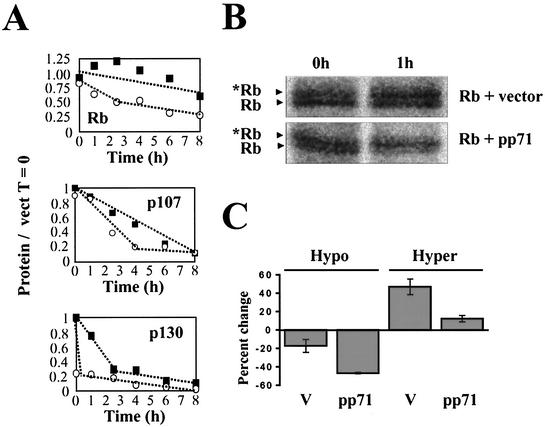
FIG. 4.
pp71 degrades the Rb family. (A) U-2 OS cells cotransfected with pSG5L-HApRb, pHAp107, or pHAp130 and either pSG5 (filled squares) or pSG5-pp71 (open circles) were subjected to a pulse-chase analysis as described in Materials and Methods. The relative amounts of total protein (hypo- and hyperphosphorylated forms) are shown. (B) Phosphorimages of the 0- and 1-h time points of a representative Rb pulse-chase experiment are shown. The asterisk indicates the phosphorylated Rb species. (C) The relative amount of each individual form of Rb (lower, hypophosphorylated band; upper, hyperphosphorylated band) in the presence and absence of pp71 was determined after chase times of 0 and 1 h for two independent experiments, and the percent change during that time period is shown for each species.
Analysis of protein interactions in vitro and in vivo.
Glutathione S-transferase (GST) and GST fusion proteins of Rb, p107, and p130 were prepared in Escherichiacoli as described previously (12) and captured on glutathione-Sepharose 4B. This and all subsequent manipulations were performed at 4°C. The loaded beads were washed twice with PBS and then blocked with 0.5% bovine serum albumin and 5% goat serum for 30 min prior to incubation for 2 h with lysates of U-2 OS cells transfected with pCGN71. The beads were washed 4 times with lysis buffer (50 mM HEPES [pH 7.9], 75 mM KCl, 75 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, 5 mM dithiothreitol, 5 μg of aprotinin/ml, 25 μg of leupeptin/ml, 25 μg of trypsin inhibitor/ml) and once with PBS, and bound proteins were separated by SDS-PAGE. pp71 was visualized on Western blots with the HA-specific antibody. GST-pp71 was prepared as described above and incubated with lysates from U-2 OS cells transfected with pSG5L-HApRb, and bound proteins were analyzed as described above. For immunoprecipitations, cells were lysed in radioimmunoprecipitation assay buffer (50 mM Tris [pH 8.0], 5 mM EDTA, 150 mM NaCl, 10% glycerol, 0.1% SDS, 1% NP-40, 1% sodium deoxycholate, 1% Triton X-100, 2 mM dithiothreitol, 1 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride, 25 mM NaF, 10 μg of pepstatin A/ml, 10 μg of aprotinin/ml, 25 μg of leupeptin/ml, 25 μg of trypsin inhibitor/ml) with mild sonication, and all subsequent incubations were at 4°C. Lysates were precleared by the addition of goat serum to a final concentration of 5% for 30 min prior to incubation with a preparation of heat-killed protein A-positive Staphylococcus aureus (Roche) for 30 min. The cleared lysate was incubated with the appropriate antibody for 2 h. Immune complexes were captured on protein A-agarose beads (Pharmacia) and washed six times with lysis buffer and once with PBS. Bound proteins were separated by SDS-PAGE and visualized by either autoradiography or Western blotting.
Transformation assay.
Primary rat embryo fibroblasts were a gift from M. Cole (Princeton University). Baby rat kidney cells were prepared essentially as described previously (37). Cells cultured on 10-cm-diameter plates were transfected with 3 μg of each of the indicated plasmids by the calcium phosphate method. Cultures were fed fresh media every 4 to 5 days, and transformed foci were counted after 18 to 21 days.
Antibodies and inhibitors.
The following antibodies were from commercial sources: tubulin (for Western blots, DM 1A [Sigma]; for immunoprecipitations, H-300 [Santa Cruz]); HA (16B12; Babco); pp65 (1025; Rumbaugh-Goodwin Institute for Cancer Research, Inc., Plantation, Fla.); and E2F-1 (KH95), RB (C-20), p107(C-18), and p130 (C-20) (all from Santa Cruz). The E1A antibody was M73. The pp71 antibodies 10G11, 1E-233, 2H10-9, and 4F7 were identified in a screen of antibodies raised against HCMV virion proteins by P. Robinson. Mice were immunized with purified HCMV virions, and individual hybridomas that reacted in Western blots to virion proteins of approximately 70 kDa were screened for pp71 reactivity by Western blotting and immunoprecipitation of lysates from cells transfected with pCGN71. The pp71 antibody CMV355 has been described previously (31). For immunoprecipitations, a cocktail containing equal volumes of each of the five pp71 antibody tissue culture supernatants was employed. The following compounds were purchased from Calbiochem, dissolved in dimethyl sulfoxide (DMSO), and employed at the concentrations indicated in parentheses: lactacystin (20 μM), E64 (50 μM), proteasome inhibitor no. 1 (1 μM), ALLN (50 μM), epoxomicin (10 μM), and clasto-lactacystin β-lactone (20 μM).
RESULTS
HCMV pp71 induces DNA synthesis in quiescent cells and binds to the Rb family of tumor suppressors.
We employed a transient transfection assay (22) to identify pp71 as a cell cycle regulator (23a). To determine if pp71 could drive quiescent cells into the cell cycle, we infected them with recombinant adenoviruses that express either pp71 or another HCMV virion protein, pp65. These two proteins are members of a viral gene family (36). pp71 was able to induce DNA synthesis in quiescent cells, but pp65 was not (Fig. 1). The increased DNA content induced by pp71 appears to result from true DNA replication since it was inhibited by mimosine (data not shown), an inhibitor of DNA replication (21). We were also able to demonstrate with BrDU uptake that pp71, but not pp65 or the GFP, was able to induce DNA synthesis in quiescent cells (data not shown) (see Fig. 5C).
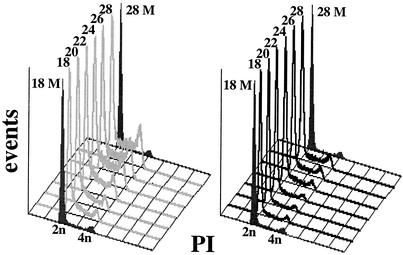
FIG. 1.
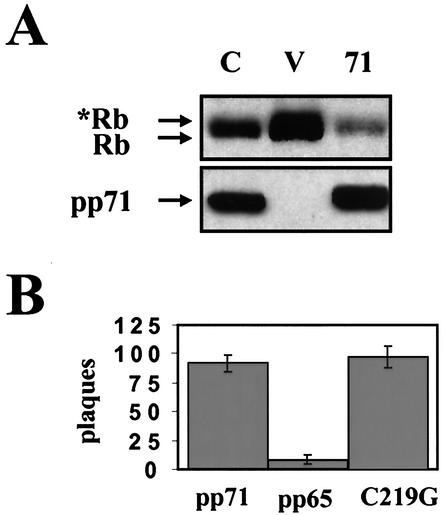
HCMV pp71 induces DNA synthesis in quiescent cells. Quiescent Rat-1 cells were mock infected (filled histograms) or infected with ADtrack-based recombinant adenoviruses expressing either pp71 (gray) or pp65 (black), harvested at the indicated time after infection (in hours) and analyzed for DNA content by flow cytometry. Similar expression levels of pp71 and pp65 were confirmed by Western blotting (data not shown). M, mock; PI, propidium iodide.
FIG. 5.
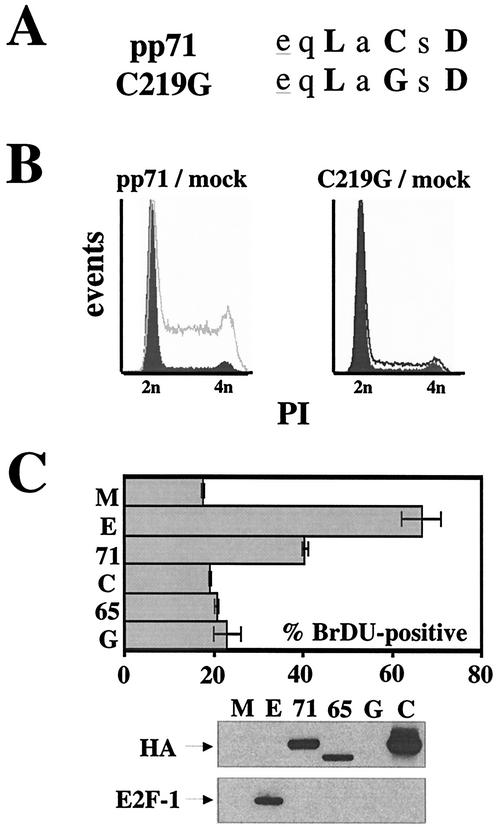
The LXCXD motif is required for the induction of S phase by pp71. (A) Sequences of wild-type pp71 and the C219G mutant in which a single cysteine residue was changed to a glycine. Conserved residues are in boldface, and acidic residues preceding the motif are underlined. (B) Quiescent Rat-1 cells were mock infected (filled histograms) or infected with ADtrack-based recombinant adenoviruses expressing either pp71 (gray) or the C219G mutant (black) and harvested at 24 h postinfection, and their DNA contents were analyzed by flow cytometry. Similar expression levels were confirmed by Western blotting (data not shown). (C) Quiescent Rat-1 cells grown on coverslips were infected with ADtrack-based recombinant adenoviruses expressing the indicated protein as described above and pulse-labeled with BrDU prior to harvesting. The coverslips were stained as described in Materials and Methods, and the number of BrDU-positive nuclei was counted by fluorescence microscopy and is presented as a percentage of the total number of nuclei counted (top). The cells remaining attached to the plates were processed for Western blot analysis of equal amounts of protein lysate with the indicated antibody (bottom). M, mock; E, E2F-1; 71, pp71; 65, pp65; G, GFP; C, C219G; HA, HA-specific antibody; E2F-1, E2F-1-specific antibody.
We identified an LXCXD sequence in pp71 that is similar to the LXCXE sequence (5) in adenovirus E1A, SV40 T antigen, and papillomavirus E7 (Fig. 2A). This motif mediates interactions with the Rb family of tumor suppressors, Rb, p107, and p130 (reviewed in reference 6). The hypophosphorylated forms of these proteins bind to the E2F family of transcription factors. These complexes repress transcription from E2F-responsive promoters and, since E2F controls the transcription of many genes required for entry into the S phase, arrest the cell cycle in G1 (reviewed in reference 11). Normal cell cycle progression is achieved through cyclin-dependent kinase phosphorylation of the Rb family members, which disrupts their interaction with E2F. This association is also disrupted upon the binding of E1A, T antigen, or E7 to the Rb protein, resulting in cell cycle stimulation (reviewed in reference 30).
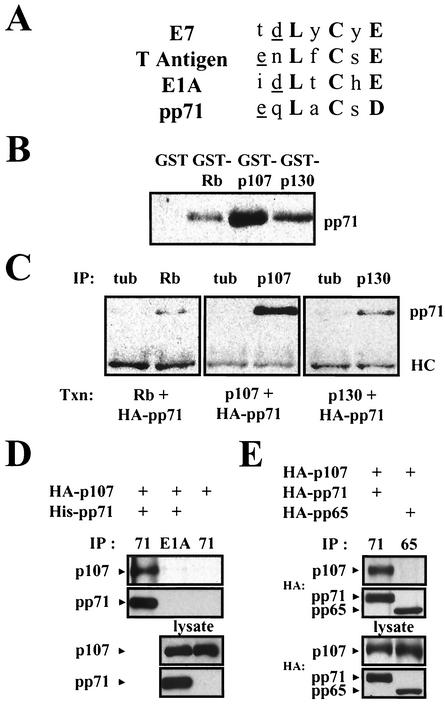
FIG. 2.
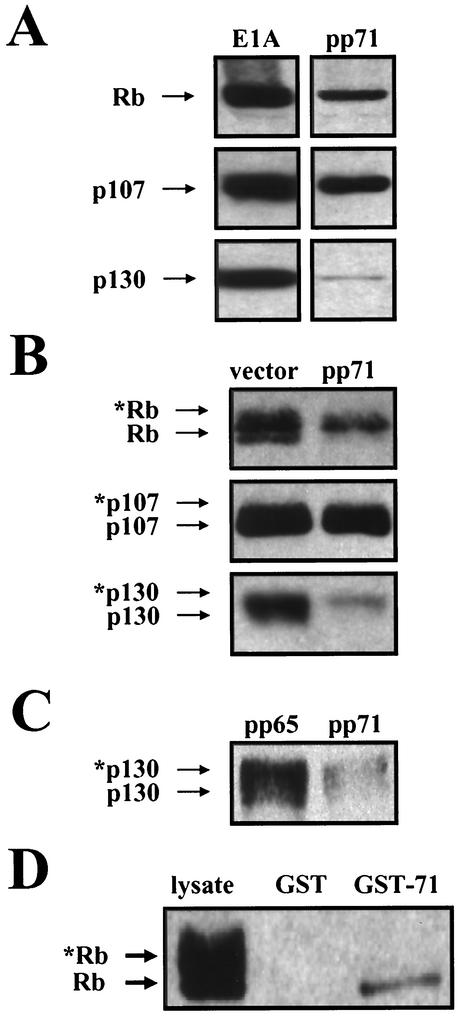
pp71 binds to the Rb family in vitro and in vivo. (A) Comparison of viral LXCXE sequences. Conserved residues are in bold. This motif is generally preceded by an acidic residue (underlined). (B) Glutathione-Sepharose beads preloaded with GST or GST fusions to the indicated Rb family members were incubated with lysates of U-2 OS cells transfected with pCGN71 and then processed for immunoblotting with an HA-specific antibody. (C) U-2 OS cells were transfected (Txn) with expression plasmids (pSG5-pp71, pCMV-Rb, pCMV-p107, and pCMV-p130) for the indicated proteins. After 24 h in 0.1% serum-containing medium, lysates were prepared and subjected to immunoprecipitation (IP) with the indicated rabbit polyclonal antisera. Bound pp71 was detected with the HA antibody. HC, antibody heavy chain; tub, tubulin. (D) U-2 OS cells were transfected (+) with expression plasmids (pSG5-His-pp71 and pHAp107) for the indicated proteins. After 24 h in 0.1% serum-containing medium, lysates were prepared and subjected to immunoprecipitation (IP) with either a cocktail of the five pp71 mouse monoclonal antibodies (71) or the M73 E1A antibody (E1A). Proteins on Western blots were detected with either the HA antibody (p107) or the pp71 antibody 4F7. (E) Transfection-immunoprecipitation experiments were performed as described above with pHAp107, pCGN71, and pCGNpp65. The pp65 antibody for immunoprecipitation was 1025, and proteins on Western blots were detected with the HA antibody.
When the LXCXE motif in E1A was converted to LXCXD, the mutant protein still bound to Rb, activated E2F-mediated transcription, and transformed cells, although at reduced levels compared to the wild-type protein (7). Consequently, we tested for an interaction between pp71 and the Rb family members in vitro and observed an interaction between pp71 from transfected cell lysates and GST fusions of each of the Rb family members but not with GST alone (Fig. 2B). We also observed an in vivo interaction of pp71 with all of the Rb family members (Fig. 2C). pp71 was found in immunoprecipitates from transfected cells with antibodies specific for each Rb family member but not with equal amounts of a control antibody to tubulin. In both circumstances, p107 appeared to interact most strongly with pp71, p130 displayed moderate binding, and Rb displayed a weak association.
We were also able to demonstrate a pp71-p107 in vivo interaction by immunoprecipitating pp71 and probing Western blots for bound p107 (Fig. 2D). p107 was captured by a pp71 antibody cocktail only when pp71 was coexpressed in cells, and it was not captured in pp71-coexpressing cells by an antibody to the adenovirus E1A protein. We were unable to detect an in vivo interaction between pp71 and either Rb or p130 by immunoprecipitation with the pp71 antibodies (data not shown). Although we could detect this complex by immunoprecipitation with antibodies to the Rb family member (Fig. 2C), the half-life of these complexes (see below) as well as the higher steady-state levels of pp71 in comparison to those of the Rb family members in these experiments (data not shown) probably prevent us from detecting all but the most-abundant pp71-p107 complexes with antibodies to pp71.
We also employed pp65 as a control in these binding studies since it is similar in sequence to pp71 but does not induce cell cycle progression in quiescent cells (Fig. 1). pp65 and pp71 are 26% identical and 44% homologous over the central 57% of the protein (data not shown) that contains the LXCXD motif of pp71. However, this sequence is altered to LXCXM in pp65, and despite their overall similarity, pp65 is unable to bind p107 in vivo (Fig. 2E). This finding agrees well with the inability of pp65 to stimulate the cell cycle and the importance of a proper LXCXE/D motif for interaction with the Rb family.
pp71 degrades the hypophosphorylated Rb family members.
When we performed Western blots to analyze the lysates that were employed in the in vivo association experiments described above, we observed a decrease in the level of the Rb family proteins in cells expressing pp71 compared to E1A (Fig. 3A). We reexamined the effect of pp71 expression on the steady-state levels of each member of the Rb family in cotransfection assays by performing Western blot analysis with equal amounts of protein lysates that were separated on low-percentage polyacrylamide gels to distinguish between the differentially phosphorylated forms of the Rb family members. Under these conditions, hypophosphorylated forms migrate farther in the gels than hyperphosphorylated forms. Although only a small change in the levels of p107 was detected, we once again observed a dramatic decrease in the levels of Rb and p130 in the presence of pp71 (Fig. 3B). Interestingly, the growth-suppressive hypophosphorylated forms of the two proteins were preferentially missing. We also compared the effect of pp71 and pp65 expression on the levels of p130 (Fig. 3C) and found that, compared to pp65, which does not bind the Rb family (Fig. 2E) and does not stimulate the cell cycle (Fig. 1), pp71 expression caused a drop in the level of the p130 protein, with the hypophosphorylated forms preferentially missing. Thus, we speculated that pp71 might specifically bind to and subsequently degrade the hypophosphorylated forms of the Rb family members. Another possibility was that pp71 could phosphorylate the Rb family, but since it contains no consensus kinase or ATP binding motifs, we considered this unlikely.
FIG. 3.
pp71 targets the hypophosphorylated forms of the Rb family. (A) U-2 OS cells were transfected with pCMV-Rb, pCMV-p107, or pCMV-p130 and either pCMVE1A or pCGN71. Lysates were harvested 48 h later, separated on 10% gels, and probed with antibodies to the indicated Rb family members. (B) U-2 OS cells were transfected with pSG5L-HApRB, pHAp107, or pHAp130 and either the empty pSG5 vector or pSG5-pp71. Lysates were harvested 48 h later, separated on 6% gels, and probed with the HA antibody. Asterisks represent phosphorylated protein species. (C) U-2 OS cells were transfected with pHAp130 and either pCGNpp65 or pCGN71. Lysates were harvested 48 h later, separated on 6% gels, and probed with the HA antibody. Asterisks represent phosphorylated protein species. (D) Glutathione-Sepharose beads preloaded with GST or a GST-pp71 fusion protein were incubated with lysates of U-2 OS cells transfected with pSG5L-HApRb and then processed for immunoblotting with an HA-specific antibody. Asterisks indicate phosphorylated Rb proteins.
To test for a specific interaction with the hypophosphorylated form of Rb, we employed an in vitro interaction assay and found that only the hypophosphorylated form of Rb was captured by GST-pp71 (Fig. 3D). This is not a result of dephosphorylation during the binding reaction since the spectrum of phosphorylated forms of Rb present in the unbound fraction was identical to that in the lysate (data not shown). No binding was observed to GST alone.
To test for degradation, we performed a pulse-chase analysis (Fig. 4), which demonstrated that the half-lives of Rb, p107, and p130 are all decreased by pp71. The half-life of Rb drops from >8 h to 3.9 ± 1.2 h, the half-life of p107 drops from 4.3 ± 0.3 h to 2.4 ± 0.4 h, and the half-life of p130 drops from 1.8 ± 0.1 h to <30 min. For p130, a significant amount of the protein was degraded during the 30-min pulse period before the chase was initiated. The proteins in labeled immunoprecipitates from the time course exhibited biphasic degradation curves in the presence of pp71 (Fig. 4A). The Rb family members are extremely susceptible to degradation by pp71 during the initial chase period, but at later time points they are more stable. This is the expected result if pp71 is degrading the hypophosphorylated forms of the proteins shortly after they are made, and once phosphorylated, they become more resistant to degradation. An autoradiograph of the first two time points of the Rb pulse-chase analysis supports this hypothesis (Fig. 4B). A qualitative examination indicates that, without pp71, there is more hypophosphorylated Rb at time zero than of the slower-migrating, hyperphosphorylated band. After 1 h of the chase period, a significant conversion of hypophosphorylated to hyperphosphorylated form is observed. In contrast, in the presence of pp71, there are similar amounts of hypo- and hyperphosphorylated Rb at time zero, and by 1 h, the hypophosphorylated form is substantially reduced with very little increase in the hyperphosphorylated form. This argues for a mechanism in which the hypophosphorylated form is lost by degradation not by conversion to the phosphorylated species. A quantitative examination of two independent experiments (Fig. 4C) confirms this interpretation. During the first hour of chase, a higher percentage of hypophosphorylated Rb is lost in the presence of pp71 than in its absence. However, only a small minority is converted to the hyperphosphorylated form in the presence of pp71 (much less than in its absence), and therefore, it must be degraded.
The pp71 LXCXD motif is required for the induction of DNA synthesis in quiescent cells and for degradation of the Rb family.
We generated a mutation in the pp71 LXCXD motif (Fig. 5A) modeled after one that disrupted the ability of E1A to bind Rb and transform cells (7). This mutant protein (C219G), which contains a single cysteine-to-glycine change in the LXCXD motif, accumulates to levels similar to those of wild-type pp71 (Fig. 5C, bottom panel) and correctly localizes to the nucleus (data not shown). We made a recombinant adenovirus that expresses the C219G mutant and demonstrated by flow cytometry (Fig. 5B) and BrDU uptake (Fig. 5C, top panel) that it fails to induce DNA synthesis in quiescent cells.
Since the C219G mutant failed to induce DNA synthesis in quiescent cells, we reasoned that it would also be incapable of degrading the Rb family members. Western blotting of lysates from cotransfection assays demonstrated that the C219G mutant was indeed unable to degrade Rb (Fig. 6A) and p130 (see below) to the same extent as the wild-type protein. However, the C219G protein retained an unrelated function of pp71. When transfected into permissive cells, viral DNA produces live virus. This happens at a low efficiency that can be dramatically increased by cotransfection with a pp71 expression plasmid but not with plasmids expressing other tegument or immediate-early proteins (1). The C219G protein was able to enhance the infectivity of viral DNA as efficiently as wild-type pp71 (Fig. 6B). Consequently, we can conclude that the C219G mutation does not lead to a global inactivation of pp71. Rather, it specifically abolishes its cell cycle regulatory function.
FIG. 6.
The pp71-mediated degradation of Rb requires a wild type LXCXD motif. (A) Cotransfection assays were performed as described in the legend to Fig. 3B with pSG5L-HApRb. C, pSG5-pp71-C219G; V, vector (pSG5); 71, pSG5-pp71. Asterisks indicate phosphorylated forms of the Rb protein. (B) Primary human foreskin fibroblasts were transfected with infectious HCMV DNA isolated from virions and either pCGN71, pCGNpp65, or pCGN71-C219G. The cells were overlaid with agarose, and plaques were stained and counted 18 days after transfection.
pp71 directs the proteasome-dependent degradation of the Rb family.
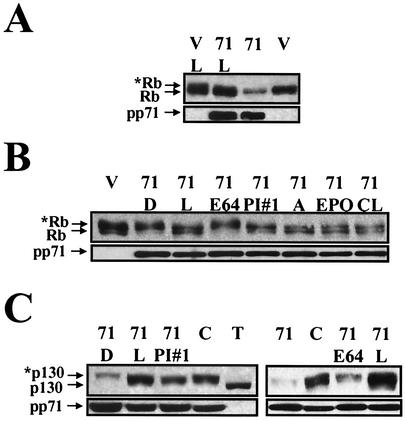
The proteasome, a large, multisubunit protease, is responsible for degrading the majority of cellular proteins that are specifically targeted for degradation (reviewed in reference 44). To determine if the pp71-mediated degradation of Rb required proteasome function, we added different proteasome inhibitors to cells cotransfected with Rb and pp71 prior to harvesting protein lysates. We initially tested lactacystin and found that treatment of cells with this drug prevented the pp71-mediated degradation of Rb (Fig. 7A). We repeated this experiment with additional proteasome inhibitors and negative controls. All of the five proteasome inhibitors tested were able to stabilize the hypophosphorylated form of Rb in the presence of pp71 (Fig. 7B). This degradation was not affected by the addition of an equal volume of DMSO or by E64, a cell-permeable inhibitor of nonproteasomal cysteine proteases such as calpain. Since pp71 is present for the duration of the experiment (48 h), but the proteasome inhibitors are only added for the final 12 h, the amount of the Rb protein detected in the presence of pp71 and the proteasome inhibitors is not expected to reach the level achieved in the absence of pp71. However, because the hypophosphorylated form is stabilized in the presence of different proteasome inhibitors, we conclude that proteasome function is required for the pp71-mediated degradation of Rb.
FIG. 7.
The pp71-mediated degradation of Rb and p130 requires proteasome function. (A) Cotransfection assays with pSG5L-HApRb were performed as described in the legend to Fig. 3B. Where indicated, lactacystin (L) was added 14 h before harvesting lysates. V, vector; 71, pp71. (B) Cotransfection assays were preformed as described above. Where indicated, the proteasome inhibitors lactacystin (L), proteasome inhibitor no. 1 (PI#1), ALLN (A), epoxomicin (EPO), and clasto-lactacystin β-lactone (CL), the cysteine protease inhibitor E64, or, as a control, the solvent DMSO (D) were added as described above. (C) Cotransfection assays were performed with pHAp130 as described above. The indicated additions were as described above. Also analyzed in these experiments were the pp71 C219G mutant (C) and SV40 T antigen (T).
We obtained similar results when we examined the pp71-mediated degradation of p130. Proteasome inhibitors, but not DMSO or E64, were able to prevent the degradation of hypophosphorylated p130 by pp71 (Fig. 7C). In these experiments, we also showed that the C219G mutant, which does not induce quiescent cells to enter the cell cycle and does not degrade Rb, also fails to degrade p130 (Fig. 7C). Furthermore, in one of the experiments (Fig. 7C, lane T), we included a cotransfection with T antigen, which prevents phosphorylation of p130 (39). By comparing the T antigen and pp71 lanes, it is clear that pp71 is degrading the hypophosphorylated form of p130. Since it is difficult to observe the pp71-mediated degradation of p107 by Western blotting, even though pp71 appears to bind most strongly to p107 (Fig. 2B and C) and the pp71-mediated degradation of p107 was detected by the pulse-chase analysis (Fig. 4A), we have not examined p107 degradation in these experiments. Since pp71 only targets the hypophosphorylated forms of the Rb family members, the difficulty in separating the differentially phosphorylated forms of p107 probably prevents us from detecting its degradation by Western blotting.
Unlike the DNA tumor virus oncoproteins, pp71 does not induce apoptosis and does not cooperate to transform cells.
In many respects, pp71 appears to function in a fashion essentially identical to E1A, T antigen, and E7. These DNA tumor virus oncoproteins also share the ability to induce apoptosis (13, 32, 35) and to transform cells (reviewed in reference 30). We reasoned that if pp71 were to function exactly like these proteins, it would also retain these activities. Thus, we asked whether pp71 could induce apoptosis and transform cells.
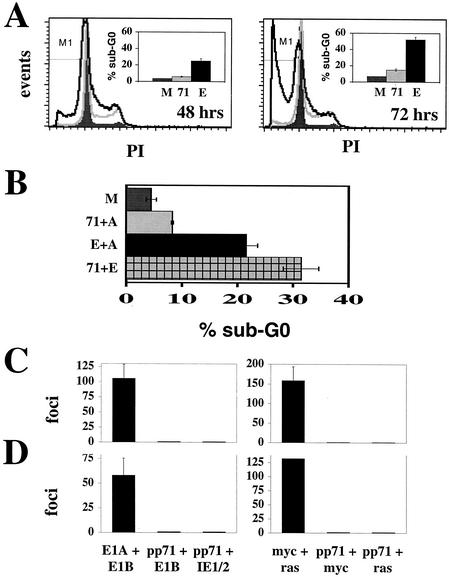
We infected quiescent cells with recombinant adenoviruses expressing either pp71 or E2F-1 and assayed for apoptosis by determining the percentage of cells with a sub-G0 DNA content. Overexpression of E2F-1 has been shown to induce apoptotic cell death (9). Infection of quiescent cells with the recombinant adenovirus expressing E2F-1 led to apoptosis, as expected (Fig. 8A). However, pp71 induced little apoptosis at either time point examined, even though, in these same experiments, we observed significant cell cycle stimulation by pp71 (Fig. 8A). Although it is possible that pp71 induces and subsequently blocks apoptosis much like T antigen, we find this unlikely since pp71 was unable to prevent E2F-1-induced apoptosis (Fig. 8B). In this respect, the interaction of pp71 with the cellular pathways leading to apoptosis, and thus presumably the mechanism through which it induces cell cycle progression, is different from that of E1A, T antigen, and E7.
FIG. 8.
pp71 neither induces apoptosis nor transforms cells. (A) Quiescent Rat-1 cells were infected with the indicated ADIC-based recombinant adenovirus. Floating and attached cells were collected at 48 and 72 h postinfection and analyzed by flow cytometry. Quantitation of sub-G0 cells indicated by the M1 gate in the histograms was determined with CELLQuest software (Becton Dickinson). M, mock, dark gray filled histograms; 71, pp71, light gray; E, E2F-1, black. (B) Quiescent Rat-1 cells were infected with the indicated viruses and harvested at 72 h postinfection as described above. Because cells were coinfected with two viruses, the amount of each individual virus employed was half that used in the experiments for which results are shown in panel A so as to keep the total amount of input virus the same as in all other experiments. Where pp71 and E2F-1 were expressed individually, a control virus without an insert (ADIC [A]) was coinfected to maintain the proper virus level. (C) Primary rat embryo fibroblasts were transfected with the indicated plasmids and cultured for 18 to 21 days, at which time transformed colonies were counted. Error bars represent standard deviations. (D) Transformation assays were performed with baby rat kidney cells as described above.
Unlike the DNA tumor virus oncoproteins, pp71 does not induce apoptosis. Likewise, we found that pp71 does not transform cells. When either primary rat embryo fibroblasts (Fig. 8C) or baby rat kidney cells (Fig. 8D) were employed in standard transformation assays, we were able to count numerous transformed foci when either adenovirus E1A and E1B or myc and activated ras were transfected, but no pp71-transformed foci were observed. pp71 did not transform cells alone or in combination with adenovirus E1B, HCMV IE1 and IE2 (which cooperate with E1A to transform cells) (37), myc, or activated ras (Fig. 8C and D). In addition, pp71 does not cooperate with cowpox virus CrmA, Bcl2, or two different dominant-negative p53 alleles to transform cells (data not shown). The inability of pp71 to induce cellular transformation is another indication that this protein, while maintaining a similar approach to stimulate cell cycle progression as the DNA tumor virus proteins, must have some important mechanistic differences that may be exploited to learn more about the cell cycle and cancer.
DISCUSSION
We have shown that pp71 interacts with the hypophosphorylated forms of Rb, p107, and p130 (Fig. 2B to E and 3D), inducing their degradation (Fig. 3A to C, 4, 6A, and 7) and thus stimulating cell cycle progression (Fig. 1, 5B and C, and 8A). Since pp71 stimulates cells to enter the cell cycle, where hypophosphorylated forms of the Rb proteins would be less populous than in quiescent cells, it was important to differentiate between degradation of these proteins by pp71 and their phosphorylation either directly by pp71 or as an indirect result of the induced cell cycle progression. The pulse-chase analysis demonstrated that, in the presence of pp71, the hypophosphorylated form of Rb is dramatically reduced after 1 h of chase while at the same time the hyperphosphorylated form of the protein is not substantially increased (Fig. 4B and C). This argues against the loss of the hypophosphorylated form of Rb by phosphorylation and strongly supports the model in which pp71 induces the degradation of hypophosphorylated Rb.
Furthermore, proteasome inhibitors arrest cells in the G2 phase (reviewed in reference 29), where kinases that phosphorylate the Rb family are active and hyperphosphorylated forms of these proteins accumulate. If pp71 were to induce the phosphorylation of the Rb family, one would predict that proteasome inhibitors would either have no effect or perhaps enhance the ability of pp71 to direct the loss of hypophosphorylated Rb. However, these inhibitors stabilize the hypophosphorylated forms of the Rb family in the presence of pp71 (Fig. 7), opposing its function. Once again, this supports a model where pp71 works to degrade the Rb family members and not to directly or indirectly induce their phosphorylation. Furthermore, the E1A protein, which stimulates the cell cycle, does not affect the level of the Rb proteins in experiments in transiently transfected, asynchronous cells (Fig. 3A), where we have shown pp71 to have a dramatic effect (Fig. 3A to C, 4, 6A, and 7). Thus, the instability of the hypophosphorylated forms of the Rb proteins in these experiments is unique to pp71 and thus cannot be the result of cell cycle stimulation-induced phosphorylation.
pp71 and the DNA tumor virus oncoproteins E1A, T antigen, and E7 all contain an LXCXE/D motif that mediates binding in the pocket domains of the Rb family members. However, beyond this conserved motif, there are significant differences in the mechanisms with which these viral proteins attack the Rb pathway, in the eventual fate of the targeted Rb family member, and in the consequences of the cell cycle stimulation induced by these proteins.
E1A requires a CR1 domain that helps disrupt Rb-E2F complexes by competing for the E2F binding site on the Rb proteins (17). E1A does not degrade the Rb family members (2), but it does prevent the phosphorylation of p107 and p130, but not Rb, independently of its ability to bind them (33). T antigen prevents the phosphorylation of p107 and p130 through an unknown mechanism that requires binding (39). T antigen also degrades p130, but not Rb or p107 (40). The degradation is mediated by the proteasome and requires the DNA J domain of T antigen (40). E7 degrades all of the differentially phosphorylated forms of Rb in a reaction that requires proteasome function (3). Although this degradation was originally thought to be specific for Rb, recent evidence suggests that E7 also degrades p107 and p130 (2, 3, 14). An amino-terminal domain of unknown function is required for the E7-mediated degradation of Rb. We have no evidence that pp71 effects the phosphorylation status of the Rb family. It degrades Rb, p107, and p130 and has a strict preference for their hypophosphorylated forms (Fig. 3B and C, 4, and 7). As in each of the examples above, it is likely that a domain in addition to the LXCXD motif of pp71 is required for the degradation of the Rb family, and we are actively searching for such a sequence.
In addition to the subtle differences between pp71, E1A, T antigen, and E7 with regards to the effects on the phosphorylation and stability of the targeted Rb family member, as well as on the protein domains other than the LXCXD/E motif employed by these proteins, a more-profound difference separates pp71 from the others. Whereas E1A, T antigen, and E7 each induce apoptosis and transform cells, pp71 can perform neither of these functions (Fig. 8). Thus, the mechanism and consequences of the pp71-mediated degradation of the Rb family differs in important aspects from the way in which the DNA tumor virus proteins function.
There are several possible models for how pp71 attacks the Rb family and stimulates cell cycle progression without inducing apoptosis or transforming cells. For example, while conversion of the LXCXE in E1A to LXCXD did not abolish that protein's ability to bind Rb and transform cells (7), it did attenuate it. Perhaps because of the altered consensus sequence of the Rb-binding motif, pp71 is not as efficient as the DNA tumor virus oncoproteins in targeting the Rb family. If this were true, pp71 may deliver a signal that, while sufficient to induce cell cycle progression, may not be strong enough to trigger apoptosis or result in transformation. Alternatively, in addition to degrading the Rb family member, pp71 may degrade the E2F component of the Rb-E2F complex as well. This may serve to derepress the E2F-responsive genes but not activate them to a high degree, thus promoting an attenuated proliferative response.
A speculative yet provocative model is that pp71 may interact with Rb-E2F complexes in a manner that is fundamentally different from the way in which the DNA tumor virus proteins are thought to function. Although pp71 induces cell cycle progression, it does so less efficiently than E2F-1 (Fig. 5C and 8A and data not shown). Another member of the E2F family, E2F-4, also induces cell cycle progression, but it does so to a lesser extent than E2F-1 (9). Furthermore, E2F-1 is the only member of that family of transcription factors that induces apoptosis upon overexpression (9). Since the efficiency of cell cycle and apoptosis induction by pp71 more-closely mimics overexpression of E2F-4 than E2F-1, a mechanism in which pp71 were to target E2F-4-bound Rb family members more efficiently than those bound to other E2F family members is consistent with our cell cycle and apoptosis data. If this model is correct, then perhaps more E2F-4 is released than other family members, such as E2F-1. This in turn sends a moderate proliferative signal to the cell, one that HCMV can efficiently block before the infected cells enter the S phase (4, 24) and one that might not lead to apoptosis or transformation. This model is consistent with our data, since E2F-4 can bind to each of the Rb family members but is most often found in complexes with p130 (which we find to be degraded by pp71 with very rapid kinetics) and p107 (which we find binds most strongly to pp71). A comprehensive series of experiments will be required to test each of these possible models and to determine the molecular details of the mechanism employed by pp71 to disrupt the Rb pathway.
The inability of pp71 to induce apoptosis partially explains why inhibitors of apoptosis do not cooperate with pp71 to transform cells (Fig. 8C and D). However, unlike E1A, pp71 was also unable to cooperate with an activated ras allele in transformation assays. If pp71 does not interact as strongly with the Rb family members or in a way fundamentally different from that described above, this may explain why pp71 fails to transform cells. Alternatively pp71 may target Rb efficiently but lack an additional function required for transformation. For example, in addition to targeting the Rb family, E1A and T antigen also disrupt the function of the p300 tumor suppressor (reviewed in reference 15), and this activity is required for cellular transformation. Perhaps the ability to target p300 has not been conserved in pp71, and thus, it cannot transform cells. Further experiments are required to determine if pp71 fails to transform cells because of the way it targets Rb-E2F complexes or because it lacks an additional required activity, such as antagonizing p300 function.
Finally, pp71 represents the first example of a herpesvirus protein to utilize an LXCXD/E motif to target the Rb family. Although it shares many properties with the oncoproteins from the DNA tumor viruses, we believe the unique features of this interesting protein will provide an opportunity to further investigate the roles of the Rb proteins not only in HCMV infection but also in cellular growth control and carcinogenesis.
Acknowledgments
We thank A. Beavis for flow cytometry assistance; C. Baldick, M. Cole, D. Cress, J. DeCaprio, E. Harlow, W. Krek, M. Murphy, B. Vogelstein, and H. Zhu for reagents; and P. Robinson for generating and identifying candidate pp71 antibodies.
This work was supported by a grant from the NIH (CA82396).
REFERENCES
- 1.Baldick, C. J., A. Marchini, C. E. Patterson, and T. Shenk. 1997. Human cytomegalovirus protein pp71 (ppUL82) enhances the infectivity of viral DNA and accelerates the infectious cycle. J. Virol. 71:4400-4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berezutskaya, E., B. Yu, A. Morozov, P. Raychaudhuri, and S. Bagchi. 1997. Differential regulation of the pocket domains of the retinoblastoma family proteins by the HPV16 E7 oncoprotein. Cell Growth Differ. 8:1277-1286. [PubMed] [Google Scholar]
- 3.Boyer, S. N., D. E. Wazer, and V. Band. 1996. E7 protein of human papillomavirus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 56:4620-4624. [PubMed] [Google Scholar]
- 4.Bresnahan, W. A., I. Boldogh, E. A. Thompson, and T. Albrecht. 1996. Human cytomegalovirus inhibits cellular DNA synthesis and arrests productively infected cells in G1. Virology 224:150-160. [DOI] [PubMed] [Google Scholar]
- 5.Chellappan, S., V. B. Kraus, B. Kroger, K. Munger, P. M. Howley, W. C. Phelps, and J. R. Nevins. 1992. Adenovirus E1A, simian virus 40 tumor antigen, and human papillomavirus E7 protein share the capacity to disrupt the interaction between transcription factor E2F and the retinoblastoma gene product. Proc. Natl. Acad. Sci. USA 89:4549-4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Classon, M., and N. Dyson. 2001. p107 and p130: versatile proteins with interesting pockets. Exp. Cell Res. 264:135-147. [DOI] [PubMed] [Google Scholar]
- 7.Corbeil, H. B., and P. E. Branton. 1994. Functional importance of complex formation between the retinoblastoma tumor suppressor family and adenovirus E1A proteins as determined by mutational analysis of E1A conserved region 2. J. Virol. 68:6697-6709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cress, W. D., D. G. Johnson, and J. R. Nevins. 1993. A genetic analysis of the E2F1 gene distinguishes regulation by Rb, p107, and adenovirus E4. Mol. Cell. Biol. 13:6314-6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeGregori, J., G. Leone, A. Miron, L. Jakoi, and J. R. Nevins. 1997. Distinct roles for E2F proteins in cell growth control and apoptosis. Proc. Natl. Acad. Sci. USA 94:7245-7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong, F., W. D. Cress, D. Agrawal, and W. J. Pledger. 1998. The role of cyclin D3-dependent kinase in the phosphorylation of p130 in mouse BALB/c 3T3 fibroblasts. J. Biol. Chem. 273:6190-6195. [DOI] [PubMed] [Google Scholar]
- 11.Dyson, N. 1998. The regulation of E2F proteins. Genes Dev. 12:2245-2262. [DOI] [PubMed] [Google Scholar]
- 12.Frangioni, J. V., and B. G. Neel. 1993. Solubilization and purification of enzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal. Biochem. 210:179-187. [DOI] [PubMed] [Google Scholar]
- 13.Fromm, L., W. Shawlot, K. Gunning, J. S. Butel, and P. A. Overbeek. 1994. The retinoblastoma protein-binding region of simian virus 40 large T antigen alters cell cycle regulation in lenses of transgenic mice. Mol. Cell. Biol. 14:6743-6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzalez, S. L., M. Stremlau, X. He, J. R. Basile, and K. Munger. 2001. Degradation of the retinoblastoma tumor suppressor by the human papillomavirus type 16 E7 oncoprotein is important for functional inactivation and is separable from proteasomal degradation of E7. J. Virol. 75:7583-7591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodman, R. H., and S. Smolik. 2000. CBP/p300 in cell growth, transformation and development. Genes Dev. 14:1553-1577. [PubMed] [Google Scholar]
- 16.He, T.-C., S. Zhou, L. T. DaCosta, J. Yu, K. W. Kinzler, and B. Vogelstein. 1998. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA 95:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ikeda, M. A., and J. R. Nevins. 1993. Identification of distinct roles for separate E1A domains in disruption of E2F complexes. Mol. Cell. Biol. 13:7029-7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jualt, F. M., J. M. Jualt, F. Ruchti, E. A. Fortunato, C. Clark, J. Corbeil, D. D. Richman, and D. H. Spector. 1995. Cytomegalovirus infection induces high levels of cyclins, phosphorylated Rb, and p53, leading to cell cycle arrest. J. Virol. 69:6697-6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaelin, W. G., D. C. Pallas, J. A. DeCaprio, F. J. Kaye, and D. M. Livingston. 1991. Identification of cellular proteins that can interact specifically with the T/E1A-binding region of the retinoblastoma gene product. Cell 64:521-532. [DOI] [PubMed] [Google Scholar]
- 20.Kaelin, W. G., W. Krek, W. R. Sellers, J. A. DeCaprio, F. Ajchenbaum, C. S. Fuchs, T. Chittenden, Y. Li, P. J. Farnham, and M. A. Blanar. 1992. Expression cloning of a cDNA encoding a retinoblastoma-binding protein with E2F-like properties. Cell 70:351-364. [DOI] [PubMed] [Google Scholar]
- 21.Kalejta, R. F., and J. L. Hamlin. 1997. The dual effect of mimosine on DNA replication. Exp. Cell Res. 231:173-183. [DOI] [PubMed] [Google Scholar]
- 22.Kalejta, R. F., A. D. Brideau, B. W. Banfield, and A. J. Beavis. 1999. An integral membrane green fluorescent protein marker, Us9-GFP, is quantitatively retained in cells during propidium iodide-based cell cycle analysis by flow cytometry. Exp. Cell Res. 248:322-328. [DOI] [PubMed] [Google Scholar]
- 23a.Kalejta, R. F., and T. Shenk. 2003. The human cytomegalovirus UL82 gene product (pp71) accelerates progression through the G1 phase of the cell cycle. J. Virol. 77:3451-3459. [DOI] [PMC free article] [PubMed]
- 23.Kalejta, R. F., and T. Shenk. 2002. Manipulation of the cell cycle by human cytomegalovirus. Front. Biosci. 7:d295-d306. [DOI] [PubMed] [Google Scholar]
- 24.Lu, M., and T. Shenk. 1996. Human cytomegalovirus infection inhibits cell cycle progression at multiple points, including the transition from G1 to S. J. Virol. 70:8850-8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magae, J., C. L. Wu, S. Illenye, E. Harlow, and N. H. Heintz. 1996. Nuclear localization of DP and E2F transcription factors by heterodimeric partners and retinoblastoma protein family members. J. Cell Sci. 109:1717-1726. [DOI] [PubMed] [Google Scholar]
- 26.McMahon, S. B., H. A. Van Buskirk, K. A. Dugan, T. D. Copeland, and M. D. Cole. 1998. The novel ATM-related protein TRAP is an essential cofactor for the c-MYC and E2F oncoproteins. Cell 94:363-374. [DOI] [PubMed] [Google Scholar]
- 27.Melnick, J. L., E. Adam, and M. E. DeBakey. 1995. Cytomegalovirus and atherosclerosis. Bioessays 17:899-903. [DOI] [PubMed] [Google Scholar]
- 28.Mittereder, N., K. L. March, and B. C. Trapnell. 1996. Evaluation of the concentration and bioactivity of adenovirus vectors for gene therapy. J. Virol. 70:7498-7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Myung, J., K. B. Kim, and C. M. Crews. 2001. The ubiquitin-proteasome pathway and proteasome inhibitors. Med. Res. Rev. 21:245-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nevins, J. R. 2002. Cell transformation by viruses, p. 245-283. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 31.Nowak, B., C. Sullivan, P. Sarnow, R. Thomas, F. Bricout, J. C. Nicolas, B. Fleckenstein, and A. J. Levine. 1984. Characterization of monoclonal antibodies and polyclonal immune sera directed against human cytomegalovirus virion proteins. Virology 132:325-338. [DOI] [PubMed] [Google Scholar]
- 32.Pan, H., and A. E. Griep. 1994. Altered cell cycle regulation in the lens of HPV-16 E6 or E7 transgenic mice: implications for tumor suppressor gene function in development. Genes Dev. 8:1285-1299. [DOI] [PubMed] [Google Scholar]
- 33.Parreno, M., J. Garriga, A. Limon, X. Mayol, G. R. Beck, E. Moran, and X. Grana. 2000. E1A blocks hyperphosphorylation of p130 and p107 without affecting the phosphorylation status of the retinoblastoma protein. J. Virol. 74:3166-3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qin, X.-Q., T. Chittenden, D. M. Livingston, and W. G. Kaelin. 1992. Identification of a growth suppression domain within the retinoblastoma gene product. Genes Dev. 6:953-964. [DOI] [PubMed] [Google Scholar]
- 35.Rao, L., M. Debbas, P. Sabbatini, D. Hockenbery, S. Korsmeyer, and E. White. 1992. The adenovirus E1A proteins induce apoptosis, which is inhibited by the E1B 19-kDa and Bcl-2 proteins. Proc. Natl. Acad. Sci. USA 89:7742-7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruger, B., S. Klages, B. Walla, J. Albrecht, B. Fleckenstein, P. Tomlinson, and B. Barrell. 1987. Primary structure and transcription of the genes coding for the two virion phosphoproteins pp65 and pp71 of human cytomegalovirus. J. Virol. 61:446-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shen, Y., H. Zhu, and T. Shenk. 1997. Human cytomegalovirus IE1 and IE2 proteins are mutagenic and mediate “hit and run” oncogenic transformation in cooperation with the adenovirus E1A proteins. Proc. Natl. Acad. Sci. USA 94:3341-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Speir, E., R. Modali, E. S. Huang, M. B. Leon, F. Shawl, T. Finkel, and S. E. Epstein. 1994. Potential role of human cytomegalovirus and p53 interaction in coronary restenosis. Science 265:391-394. [DOI] [PubMed] [Google Scholar]
- 39.Stubdal, H., J. Zalvide, and J. A. DeCaprio. 1996. Simian virus 40 large T antigen alters the phosphorylation state of the Rb-related proteins p130 and p107. J. Virol. 70:2781-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stubdal, H., J. Zalvide, K. S. Campbell, C. Schweitzer, T. M. Roberts, and J. A. DeCaprio. 1997. Inactivation of pRB-related proteins p130 and p107 mediated by the J domain of simian virus 40 large T antigen. Mol. Cell. Biol. 17:4979-4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka, M., and W. Herr. 1990. Differential transcriptional activation by Oct-1 and Oct-2: interdependent activation domains induce Oct-2 phosphorylation. Cell 60:375-386. [DOI] [PubMed] [Google Scholar]
- 42.Tao, W., D. Pennica, L. Xu, R. F. Kalejta, and A. J. Levine. 2001. Wrch-1, a novel member of the Rho gene family that is regulated by Wnt-1. Genes Dev. 15:1796-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vairo, G., D. M. Livingston, and D. Ginsberg. 1995. Functional interaction between E2F-4 and p130: evidence for distinct mechanisms underlying growth suppression by different retinoblastoma protein family members. Genes Dev. 9:869-881. [DOI] [PubMed] [Google Scholar]
- 44.Voges, D., P. Zwickl, and W. Baumeister. 1999. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 68:1015-1068. [DOI] [PubMed] [Google Scholar]
- 45.White, E., R. Cipriani, P. Sabbatini, and A. Denton. 1991. Adenovirus E1B 19-kilodalton protein overcomes the cytotoxicity of E1a proteins. J. Virol. 65:2968-2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu, H., Y. Shen, and T. Shenk. 1995. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J. Virol. 69:7960-7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu, L., S. van den Heuvel, K. Helin, A. Fattaey, M. Ewen, D. Livingston, N. Dyson, and E. Harlow. 1993. Inhibition of cell proliferation by p107, a relative of the retinoblastoma protein. Genes Dev. 7:1111-1125. [DOI] [PubMed] [Google Scholar]