Abstract
Transfected siRNAs regulate numerous transcripts sharing limited complementarity to the RNA duplex. This unintended (“off-target”) silencing can hinder the use of RNAi to define gene function. Here we describe position-specific, sequence-independent chemical modifications that reduced silencing of partially complementary transcripts by all siRNAs tested. Silencing of perfectly matched targets was unaffected by these modifications. The chemical modification also reduced off-target phenotypes in growth inhibition studies. Key to the modification was 2′-O-methyl ribosyl substitution at position 2 in the guide strand, which reduced silencing of most off-target transcripts with complementarity to the seed region of the siRNA guide strand. The sharp position dependence of 2′-O-methyl ribosyl modification contrasts with the broader position dependence of base-pair substitutions within the seed region, suggesting a role for position 2 of the guide strand distinct from its effects on pairing to target transcripts.
Keywords: siRNA, 2′-O-methyl, off-target, RNAi
INTRODUCTION
Off-target transcript silencing (Jackson et al. 2003) limits the specificity of siRNAs for functional genomic and therapeutic applications. Off-target transcript silencing is widespread and mediated largely by limited target sequence complementarity to the seed region of the siRNA guide strand (Jackson et al. 2006). Our studies show that off-target silencing cannot be easily eliminated by siRNA sequence selection. Furthermore, they cannot be distinguished from on-target silencing by reducing the siRNA concentration. Taken together, these findings suggest that off-target transcript silencing is a fundamental feature of RNAi-mediated transcript silencing. In agreement with these findings, several false-positive hits resulting from an siRNA screen showed sequence complementarity to the siRNA guide strand (Lin et al. 2005). The high frequency of such off-target or false-positive phenotypes demonstrates their impact on functional genetic studies.
We hypothesized that modifications to siRNAs that weaken or disrupt RISC–mRNA interaction in the seed region could reduce off-target silencing by further disrupting interaction with transcripts containing only partial complementarity. Chemical modification of RNA can alter RNA–RNA and RNA–protein interactions (Miller et al. 1982; Blake et al. 1985). O-methyl groups added to the 2′ position of the ribosyl ring are commonly used for RNA modification to alter key thermodynamic and binding properties of modified duplexes (Monia et al. 1993; Lubini et al. 1994; Cummins et al. 1995; Nishizaki et al. 1997; Adamiak et al. 2001). Here, we show that 2′-O-methyl modifications to specific positions within the siRNA seed region reduce both the number of off-target transcripts and the magnitude of their regulation, without significantly affecting silencing of the intended targets. Furthermore, these modifications reduce off-target phenotypes in functional studies.
RESULTS
In a separate study, we showed that siRNAs can silence unintended transcripts with sequence complementarity to seed regions of siRNAs (Jackson et al. 2006). Furthermore, we showed that base substitutions in the siRNA seed region disrupt regulation of unintended transcripts, just as mismatches disrupt miRNA target regulation (Doench and Sharp 2004; Lim et al. 2005). However, while base substitutions in siRNA seed regions reduce silencing of off-target transcripts complementary to the wild-type siRNA sequence, they trigger regulation of new off-target transcripts complementary to the mismatch sequence (Jackson et al. 2006).
To determine whether backbone modifications to the siRNA similarly impact transcript regulation, we analyzed the effect of 2′-O-methyl seed region modifications on off-target silencing by an siRNA targeting MAPK14. For these studies, the 5′-end of the antisense strand was phosphorylated, as previous studies suggested that phosphorylation is required for association of siRNAs with RISC (Nykanen et al. 2001; Schwarz et al. 2003). Furthermore, the bases in positions 1 and 2 of the sense strand were modified with 2′-O-methyl groups to reduce the contribution of this strand to transcript silencing (data not shown). This allowed us to concentrate on the impact of 2′-O-methyl modifications within the seed region of the guide strand. We transfected HeLa cells with duplexes having single or paired 2′-O-methyl modifications of the first 12 nucleotides of the guide strand and analyzed gene expression profiles. No single modified position negatively affected target transcript silencing (Fig. 1A; data not shown), confirming previous observations that no single 2′-hydroxyl group in the siRNA duplex is indispensable in RNAi (Amarzguioui et al. 2003). Likewise, paired modifications did not reduce target silencing. However, paired 2′-O-methyl modifications of positions 1–5 reduced the number of transcripts regulated, with the strongest effect seen with paired modification of positions 1 and 2 (Fig. 1A). This modification reduced both the number of off-target transcripts regulated and the magnitude of the effects, while regulation of the intended target was unaffected (Fig. 1A). siRNAs with all other 2′-O-methyl modification pairs produced expression signatures similar to the unmodified siRNA.
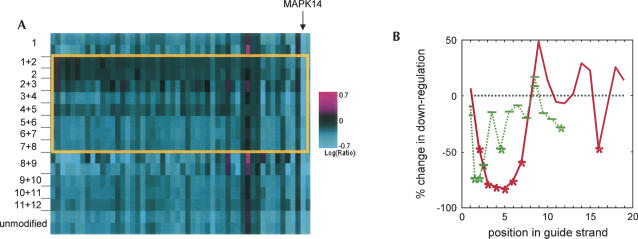
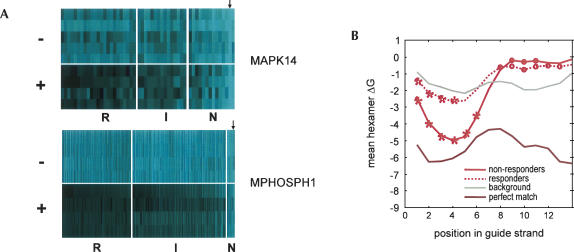
FIGURE 1.
Position-specific impact of chemical modification on siRNA specificity. (A) Position-specific effect of 2′-O-methyl modifications on silencing of off-target transcripts. siRNAs to MAPK14-193 were synthesized to contain either a single 2′-O-methyl modification or paired 2′-O-methyl modifications at overlapping consecutive pairs of nucleotides on the antisense (guide) strand. Chemically modified duplexes were phosphorylated on the antisense strand. siRNAs were transfected into HeLa cells, and changes in transcript regulation were analyzed by microarray profiling (Jackson et al. 2003). Shown is a heat map representing the entire signature of transcripts down-regulated by the wild-type MAPK14 siRNA (52 genes, X-axis) in 29 experiments (Y-axis). The transcripts shown were regulated with p ≤ 0.01, with no cuts placed on fold regulation. siRNA transcript regulations were analyzed using a consensus signature list for the unmodified MAPK14-193 duplex. Transcripts down-regulated in siRNA-transfected cells are shown in light blue, and transcripts up-regulated in siRNA-transfected cells are shown in magenta. Black indicates no change in regulation. Two or three independent experiments are shown for each modified or unmodified siRNA. The gold box indicates the location of the siRNA seed region (positions 2–8 of the guide strand). Transcripts are ordered by percent change in down-regulation (normalized mlratio change) across the wild-type signature. The arrow indicates the location of the target transcript MAPK14. (B) Comparison of MAPK14-193 modification walks. The average percent change in down-regulation (normalized mean log ratio change) for off-target transcripts relative to the wild-type siRNA sequence was calculated from the microarray for both base mismatches (Jackson et al. 2006) and chemical modification, analyzed as in A, and plotted as a function of position in the siRNA guide strand, 5′–3′ orientation. Green dashes indicate chemical modification; longer dashes represent paired modifications, and shorter dashes represent single residue modifications. The red line indicates base substitutions. Positions are marked with asterisks where modification reduced down-regulation significantly more than the random reductions seen between repeats of unaltered transfections (p < 1e − 4 with Bonferroni correction, Kolmogorov-Smirnov goodness-of-fit test, negative tail).
We further explored the position dependence of 2′-O-methyl modifications on off-target silencing by comparing the expression signatures of duplexes with 2′-O-methyl modifications of position 1, position 2, or positions 1 + 2 of the seed region. None of these modifications affected silencing of the intended target. However, modification of position 2 alone of a MAPK14 siRNA reduced off-target transcript down-regulation to a similar extent as positions 1 + 2 (Fig. 1A). In contrast, modification of position 1 alone did not decrease off-target transcript silencing. Similar results were observed for siRNAs to two additional targets (data not shown.) The 2′-O-methyl modification reduced silencing of the original off-target transcripts without inducing a new off-target signature, a consequence of base mismatches (Jackson et al. 2006). The effects of 2′-O-methyl modification on off-target transcript silencing showed sharp position dependence, peaking at position 2. In contrast, the effects of seed region mismatches showed a broader position dependence among positions 2–7 (Fig. 1B). We conclude that 2′-O-methyl modifications are therefore superior to mismatched siRNAs for improving siRNA specificity. We performed subsequent studies with siRNAs (“modified” duplexes) containing 2′-O-methyl modifications of positions 1 + 2 of the sense strand and position 2 of the guide strand, with 5′ phosphorylation of the guide strand.
We next examined the effect of modification on seven additional siRNAs (Fig. 2). In total, 10 of 10 modified siRNAs tested showed full silencing of the intended targets but reduced off-target transcript silencing (Fig. 2; data not shown). Overall, the modification reduced silencing of ∼80% of off-target transcripts. The magnitude of reduction of off-target transcript regulation by different siRNAs tested at a single concentration was relatively uniform, averaging ∼66% (Fig. 2B).
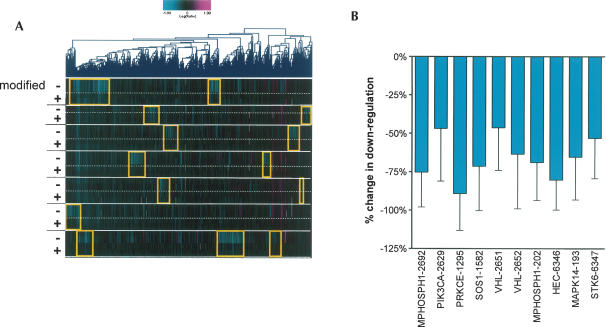
FIGURE 2.
Chemical modification reduces off-target silencing for all siRNAs tested. (A) Seven siRNAs were synthesized with (+) and without (−) the 2′-O-methyl modification as described in the text. Transcripts regulated by siRNAs with p ≤ 0.01 in at least one experiment are shown. Replicate experiments are shown for transfections when available. siRNA transcript regulations were clustered using a combined consensus signature list for repeats of each unmodified duplex. A common signature unrelated to duplex sequence was observed and removed from further analysis. Gold boxes indicate the unique transcript signature for each wild-type siRNA duplex. siRNAs from top to bottom: MPHOSPH1-2692, PIK3CA-2629, PRKCE-1295, SOS1-1582, VHL-2651, VHL-2652, MPHOSPH1-202. (B) Quantitation of the effects of chemical modification on off-target transcript regulation. Show is the mean decrease (±SD) of regulation of consensus off-target transcripts by chemical modification for the siRNAs shown in A. Also included are data for three additional siRNAs not shown in A: MAPK14-193, HEC-6346, and STK6–6347.
We next tested whether the preferential reduction in off-target silencing was maintained over a range of siRNA concentrations. The effects of chemical modification for on- and off-target gene silencing were determined by microarray for siRNAs targeting two different transcripts. The extent of MAPK14 or MPHOSPH1 target gene silencing was minimally affected by modification over a wide dose range (Fig. 3A), but off-target silencing was significantly reduced at all doses (p = 0.004, Wilcoxon rank-sum).
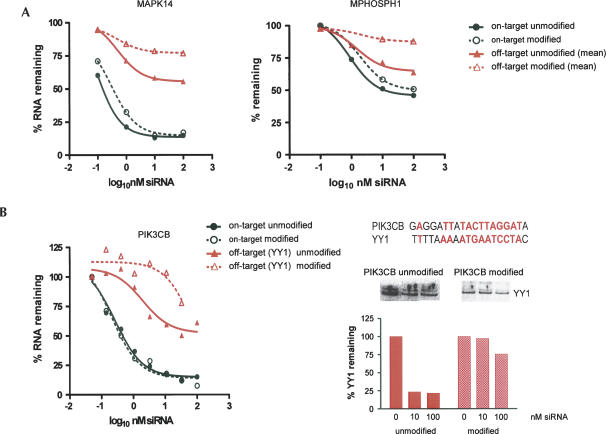
FIGURE 3.
Chemical modification preferentially reduces silencing of off-target transcripts and proteins. (A) siRNA dose titration analysis of on-target and off-target transcript silencing. Target transcript silencing was quantitated from the microarray. Off-target silencing was quantitated as the mean silencing of all off-target events, defined as those transcripts down-regulated with p < 0.01. No cuts were placed on fold down-regulation. In all cases, there was a greater reduction in off-target gene silencing relative to on-target silencing with the modified siRNA (p = 0.004, Wilcoxon rank-sum). (B) siRNA dose titration analysis of silencing of off-target RNA and protein levels. Transcript silencing was quantitated by TaqMan analysis 24 h following transfection of the PIK3CB siRNA at the indicated concentrations. YY1 protein levels were quantitated by Western analysis 48 h following transfection of the PIK3CB siRNA at the indicated concentrations. The extent of sequence complementarity between the PIK3CB siRNA and the YY1 off-target transcript are indicated. Red text indicates complementary nucleotides.
Unintended transcript silencing leads to corresponding reductions in the encoded proteins (Jackson et al. 2006). It was important to determine whether off-target silencing of proteins was also reduced by modification. We selected an siRNA for which the on-target protein and an off-target protein could be measured using commercial antibodies. Expression profiling of an siRNA targeting PIK3CB revealed many off-target transcript regulations. From these, we selected for off-target protein analysis YY1, which shares sequence complementarity to the seed region of the PIK3CB siRNA (Fig. 3A). We performed a dose titration of modified and unmodified PIK3CB siRNAs, and measured transcript and protein levels for both PIK3CB and YY1 at each siRNA concentration. Modification had no impact on the potency or maximal extent of silencing of the intended target transcript (PIK3CB, Fig. 3B, p-value = 0.8 for potency comparison). In contrast, modification reduced both the potency and maximal extent of silencing of the off-target transcript YY1 (Fig. 3B, p-value = 0.006 for potency comparison). Modification did not affect regulation of the target protein PIK3CB (data not shown), but reduced regulation of the off-target protein YY1 (Fig. 3B). Thus, modification preferentially reduced silencing of both the off-target transcript and its corresponding protein.
It was also important to determine whether modification could reduce false-positive phenotypes triggered by siRNA silencing. We had observed that some siRNAs caused growth inhibition in short-term assays without significantly silencing their intended target transcripts (data not shown). Thus, these growth-inhibition phenotypes must be false positives. To assess the impact of the chemical modification on these phenotypes, we measured target silencing and growth inhibition by these siRNAs with and without modification. In all cases, growth inhibition was reduced by modification, while silencing of the intended target was either improved or unaffected (Fig. 4). The extent to which modification reduced growth inhibition correlated with the extent to which it reduced average off-target transcript down-regulation. This experiment demonstrates that off-target transcript silencing can have significant phenotypic impact, and off-target phenotypes can be reduced by the 2′-O-methyl modification.
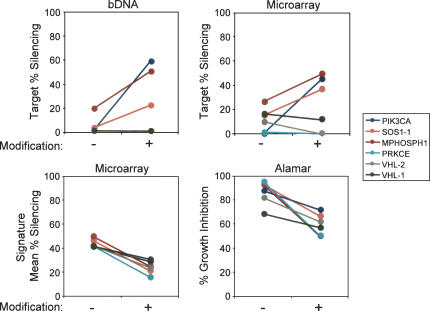
FIGURE 4.
2′-O-Methyl modification reduces off-target phenotypes. The impact of the modification on target transcript silencing was measured by bDNA for the six siRNAs indicated (upper left panel). The effect of the modification on target transcript silencing was also measured by microarray analysis (upper right panel). The effect of the modification on off-target transcript silencing was measured by microarray, quantitated as the mean percent silencing for the entire off-target signature for each siRNA (lower left panel). The effect of the modification on growth inhibition phenotype was measured for the same six siRNAs by Alamar Blue analysis 72 h post-transfection (lower right panel). The extent of on-target silencing showed no correlation with growth inhibition (R 2 = 0.05), while the extent of off-target silencing was highly correlated with growth inhibition (R 2 = 0.73).
While modification greatly reduced off-target transcript silencing by siRNAs, not all transcripts were affected equally. To understand better the mechanisms by which chemical modification reduced off-target silencing, we compared properties of the most-responsive and least-responsive transcripts. The most-responsive transcripts (“responders”) were defined as transcripts significantly down-regulated (p < 0.01) by an unmodified siRNA, and either not regulated (p > 0.1) or up-regulated by a modified siRNA. The least-responsive transcripts (“nonresponders”) were defined as transcripts showing significant down-regulation (p < 0.01) with both unmodified and modified siRNAs. Transcripts whose responses were between these extremes were labeled “intermediate.” The distributions of down-regulation of responder, intermediate, and nonresponder transcripts for two siRNA duplexes are shown in Figure 5A. Although intermediate transcripts were clearly affected by modification, these were excluded from further comparisons so that factors distinguishing the extremes of the response spectrum could be more easily discerned.
FIGURE 5.
Chemical modification preferentially reduces silencing of off-target transcripts with weaker free energy in the seed region. (A) Responder and nonresponder transcripts were identified for siRNAs MAPK14-193 and MPHOSPH1-202. Responders (R) and nonresponders (N) were defined as described in the text. Transcripts regulated with p-values between these extremes are indicated as intermediate (I). (−) Indicates transcript regulation in the absence of modification, (+) indicates transcripts regulated in the presence of modification. Within each class (R, I, N), transcripts are sorted by percent change in down-regulation (normalized mlratio change) between modified and unmodified duplex, high (left) to low (right). Five (MAPK14) or four (MPHOSPH1) independent experiments are shown for unmodified and modified duplexes. The arrows indicate the presence of the target transcripts. (B) Free energy analysis of responder and nonresponder transcripts for four siRNAs: MAPK14-193, MPHOSPH1-202, KNTC2, STK6. Shown is the average ΔG for the four siRNAs. Asterisks mark positions where the signature alignment hexamer ΔG is more negative than the background alignment hexamer ΔG with p ≤ 0.01 by Kolmogorov-Smirnov goodness-of-fit test with Bonferroni correction for the number of positions examined.
To explore how transcript response to modification is related to transcript binding to siRNAs, we took several approaches based on sequence alignment. We used FASTA alignment to predict binding sites for four siRNAs to their responder and nonresponder off-target transcripts. Nonresponder transcript alignments showed significantly more complementarity to the seed regions of targeting siRNAs than responder transcript alignments did (p = 0.008 by Wilcoxon sign-rank; data not shown). In addition, more nonresponder than responder transcripts contained hexamers perfectly complementary to the siRNA seed regions (72% vs. 32%, respectively; data not shown). We predicted binding properties of siRNA/transcript pairs by calculating RNA:RNA duplex free energies of the aligned sequences, over 6-base windows. Both responder and nonresponder transcript alignments showed stronger average binding energy to the siRNA seed regions than to the 3′-end of the siRNA guide strands (Fig. 5B). However, nonresponder transcripts showed significantly stronger binding energy than responder transcripts to siRNA seed regions. Seed region binding energy for the nonresponders approximated the free energies of perfectly matched duplexes. Taken together, these findings show that weakly hybridizing off-target transcripts are more responsive to modification than more strongly hybridizing ones.
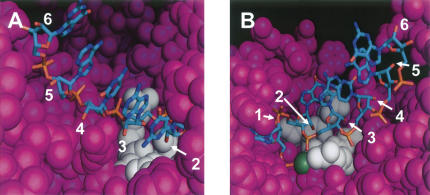
To gain insight into how 2′-O-methyl modification at position 2 affects off-target silencing, we examined published structures of siRNA guide strands bound to components of RISC (Song et al. 2003; Ma et al. 2005; Parker et al. 2005). These structures indicate that position 2 has limited space in which to accommodate a methyl group (Fig. 6). Position 2 is unique in this regard, in that it is the only position in the seed region for which the 2′ oxygen is positioned within 4 Å of any Piwi residue (Ma et al. 2005). This suggests that conformational adjustment of RISC and/or the guide strand is required to accommodate a methyl moiety at position 2.
FIGURE 6.
Position of the 5′ nucleotides of the siRNA guide strand in the complex with Piwi protein from Archaeoglobus fulgidus. (A) Position of the 5′ nucleotides of the siRNA guide (antisense) strand in the complex with Piwi protein from A. fulgidus. The crystal structure coordinates are from Ma et al. (2005). Piwi residues within 4 Å of the 2′-OH of base G2 are shown in white (Asn 155, Leu 156, Gln 159); all other Piwi residues are in purple. The sense RNA strand is not shown. White numbers and arrows show the 2′-OH positions of nucleotides 2–6 of the guide strand where methyl moieties were added in this study. Images were created using PyMol (DeLano Scientific). (B) View of the binding pocket rotated roughly 180° from the view in A. Nucleotides 1–6 are indicated. The divalent cation is shown as a green sphere.
DISCUSSION
The position-specific chemical modification of siRNA duplexes described here reduced silencing of off-target transcripts by all siRNAs tested (N = 10), without compromising silencing of the intended targets. The preferential reduction of off-target silencing was maintained over a dose titration of the siRNA and was observed for both transcript and protein. The modification also reduced false-positive phenotypes in growth-inhibition studies. The modified siRNAs described here reduced silencing of off-target transcripts by 66% on the average, and reduced false-positive phenotypes proportionally. The use of modified siRNAs will aid functional analyses and/or microarray profiling experiments.
Previous studies of chemical modification of siRNAs demonstrated that siRNAs containing full 2′-O-methyl substitutions on the sense strand, antisense strand, or both strands were inactive in transcript silencing (Elbashir et al. 2001; Amarzguioui et al. 2003; Braasch et al. 2003; Chiu and Rana 2003; Czauderna et al. 2003). A more recent study demonstrated that siRNAs with fully 2′-O-methyl-substituted sense strands were functional in transcript silencing for duplexes with 20-bp blunt construction but not for canonical duplexes with 19-bp constructs with 3′-overhangs (Kraynack and Baker 2006). In contrast, siRNAs with alternating 2′-O-methyl and unmodified nucleotides (Czauderna et al. 2003), or alternating 2′-O-methyl and 2′-O-fluoro nucleotides (Allerson et al. 2005) had activity equivalent to unmodified duplexes, suggesting that minimal chemical modifications are compatible with siRNA function. In the present study, siRNA duplexes contained 2′-O-methyl substitutions on only three nucleotides. This minimal modification was compatible with full silencing of on-target transcripts, but preferentially reduced silencing of seed sequence-complementary off-target transcripts. While modification improves siRNA specificity, it does not eliminate all sequence-dependent off-target silencing. Therefore, inclusion of multiple independent siRNAs to the target gene of interest is still helpful for distinguishing true positives from false positives in functional genomic studies.
In another study (Jackson et al. 2006), we demonstrated that much of the off-target transcript silencing of siRNAs is mediated through miRNA-like mechanism(s). Here we show that 2′-O-methyl modification distinguishes off-target silencing from silencing of perfectly matched targets. Our findings therefore suggest that miRNA-like transcript silencing is differentially sensitive to 2′-O-methyl modification. This may reflect mechanistic differences between miRNA-like target degradation and silencing of perfectly matched targets. Consistent with this possibility, Bagga et al. (2005) showed that degradation of miRNA target transcripts does not involve slicer activity.
Alternatively, the differential effects of 2′-O-methyl modification may reflect step(s) in which the silencing of partially complementary and perfectly matched targets differ only quantitatively. Supporting this possibility is our observation that the free energy of target complementarity to the siRNA seed determines sensitivity to chemical modification. More strongly hybridizing targets may be resistant to chemical modification because their binding is strong enough to overcome disruptive effects of modification. However, other considerations suggest that chemical modification likely does not exert its effects by directly affecting the strength of siRNA seed region:target interactions. Current evidence suggests that 2′-O-methyl modifications decrease the free energy of hybridization, which would tend to compensate for, rather than impair, weaker base-pairing (Inoue et al. 1987; Lesnik et al. 1993). Additionally, the effects on off-target silencing of the 2′-O-methyl modification show a sharp position dependence, in contrast to the broader position dependence of base substitutions. Thus, the strength of seed region binding modulates target sensitivity to position 2 modification, but the modification most likely affects a step(s) distinct from RNA:RNA binding in target recognition.
One possibility is that 2′-O-methyl modification may render the RISC complex incapable of cleaving targets with suboptimal siRNA binding. Brown et al. (2005) suggested a model for recognition of perfectly matched targets by an siRNA in association with RISC (RISC*). They argue for a diffusion-controlled mechanism in which many target sites are sampled nonspecifically until the correct one is found. Once the correct target RNA is found, conformational changes occur that enhance RISC* catalytic efficiency, leading to slicer activity. While degradation of imperfectly matched targets may not involve slicer activity (Bagga et al. 2005), our findings suggest a model for their recognition similar to that of Brown et al. 2′-O-Methyl modification of siRNAs does not detectably affect silencing of perfectly matched targets, arguing that modification does not prevent siRNA incorporation into RISC, target scanning by RISC*, or slicer activity. However, degradation of imperfectly matched targets with weaker seed region binding is sensitive to modification, suggesting that the modification affects another step(s). This second step may become rate limiting for formation of a stable and catalytically active RISC* with modified siRNAs. Structural analysis of an RNA guide strand associated with Piwi (Fig. 6) suggests that conformational alterations of RISC and/or the guide strand are required to accommodate a methyl moiety at position 2. These conformational changes may reduce the rate of RISC* formation so that weaker binding imperfectly matched targets dissociate from guide strands before they can be cleaved.
A separate study by Federov et al. (2006) reports effects of 2′-O-methyl modification on non-seed-region-mediated off-target siRNA activity. Together with our studies, these findings suggest that multiple distinct steps in the silencing process are affected by 2′-O-methyl modification of position 2. Further structural studies will clarify the key role of position 2 in regulating the specificity of siRNA-mediated silencing.
MATERIALS AND METHODS
siRNA synthesis
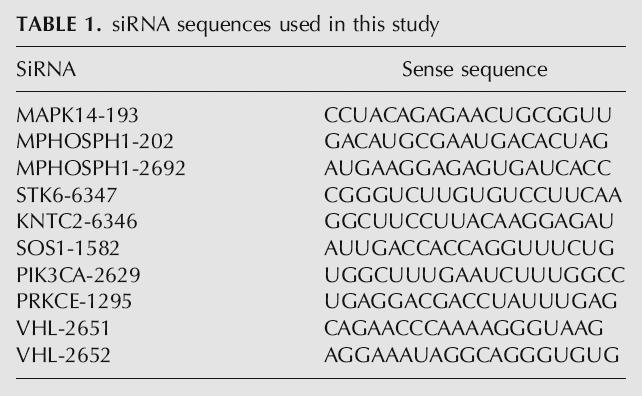
All siRNAs were synthesized and chemically modified by Dharmacon Inc. All nucleotide modifications on the guide and passenger strands of siRNAs are proprietary chemical modification patterns (ON-TARGET, ON-TARGET PLUS, patent pending). See Table 1 for siRNA sequences.
TABLE 1.
siRNA sequences used in this study
Microarray analysis
HeLa cells were transfected in six-well plates using Oligofectamine (Invitrogen) and the indicated doses of siRNA duplex. Where not specified, the concentration of siRNA was 100 nM. RNA was isolated 24 h following transfection. RNA from siRNA-transfected cells was hybridized against RNA from mock-transfected cells (treated with transfection reagent in the absence of RNA duplex). Total RNA was purified by a QIAGEN RNeasy kit, and processed as described previously (Hughes et al. 2001) for hybridization to microarrays containing oligonucleotides corresponding to ∼21,000 human genes. Ratio hybridizations were performed with fluorescent label reversal to eliminate dye bias. The data shown are signature genes that display a difference in expression level (p<0.01) relative to mock-transfected cells. No cuts were placed on fold change in expression. Blue indicates decreased expression; magenta indicates increased expression. The data were analyzed using Rosetta Resolver software. Differences in transcript regulation between unmodified and modified duplexes were calculated individually for each transcript. Transcript regulation was calculated as the error-weighted mean log10 ratio for each transcript across the fluor-reversed pair (ratio hybridizations were performed with fluorescent label reversal to eliminate dye bias). Differences in regulation between unmodified and modified duplex were then divided by the log10 ratio for the unmodified duplex for that transcript to result in the normalized mlratio change.
RNA and protein quantitation
RNA was harvested 24 h following siRNA transfection. RNA was quantitated using TaqMan qPCR or bDNA (branched DNA; Genospectra) and was normalized to hGUS RNA levels. Protein lysates were harvested 48 h following transfection of the siRNAs into HeLa cells. Antibodies were from Upstate Biotechnology (PIK3CB), Santa Cruz (YY1), or Abcam (Actin). Protein levels were normalized to actin levels at each siRNA concentration.
Free energy calculation
Off-target transcripts (p < 0.01) with mapped CDS were selected as having sufficiently complete sequence available for alignment. Each strand of the siRNA or miRNA duplex was aligned with each off-target transcript using FASTA 3.4 (Pearson and Lipman 1988) with parameters:
ktup: 2
display up to 50,000 alignments and alignment scores
gap opening penalty: −10
gap extension penalty: −10
match reward/mismatch penalty: +5/−5′
Hexamer duplexes connected by an “LLL” loop were constructed in rolling-window fashion from the aligned sequences, and RNA:RNA duplex binding energies were calculated using Mfold (Zuker 2003). Mfold treats sequences linked by “LLL” as two strands of a bimolecular RNA:RNA duplex.
ACKNOWLEDGMENTS
We thank members of the Rosetta biology team for advice and critical reading of the manuscript, Rosetta Gene Expression Lab for microarray hybridizations, and the Dharmacon production team for siRNA synthesis.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.30706.
REFERENCES
- Adamiak D.A., Rypniewski W.R., Milecki J., Adamiak R.W. The 1.19 Å X-ray structure of 2′-O-Me(CGCGCG)(2) duplex shows dehydrated RNA with 2-methyl-2,4-pentanediol in the minor groove. Nucleic Acids Res. 2001;29:4144–4153. doi: 10.1093/nar/29.20.4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allerson C.R., Sioufi N., Jarres R., Prakash T.P., Naik N., Berdeja A., Wanders L., Griffey R.H., Swayze E.E., Bhat B. Fully 2′-modified oligonucleotide duplexes with improved in vitro potency and stability compared to unmodified small interfering RNA. J. Med. Chem. 2005;48:901–904. doi: 10.1021/jm049167j. [DOI] [PubMed] [Google Scholar]
- Amarzguioui M., Holen T., Babaie E., Prydz H. Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res. 2003;31:589–595. doi: 10.1093/nar/gkg147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagga S., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A.E. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Blake K., Murakami A., Miller P. Hybridization arrest of globin synthesis in rabbit reticulocyte lysates and cells by oligodeoxyribonucleoside methylphosphonates. Biochemistry. 1985;24:6139–6145. doi: 10.1021/bi00343a016. [DOI] [PubMed] [Google Scholar]
- Braasch D.A., Jensen S., Liu Y., Kaur K., Arar K., White M.A., Corey D.R. RNA interference in mammalian cells by chemically-modified RNA. Biochemistry. 2003;42:7967–7975. doi: 10.1021/bi0343774. [DOI] [PubMed] [Google Scholar]
- Brown K.M., Chu C.Y., Rana T.M. Target accessibility dictates the potency of human RISC. Nat. Struct. Mol. Biol. 2005;12:469–470. doi: 10.1038/nsmb931. [DOI] [PubMed] [Google Scholar]
- Chiu Y.L., Rana T.M. siRNA function in RNAiA chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins L.L., Owens S.R., Risen L.M., Lesnik E.A., Freier S., McGee D., Guinosso C., Cook P.D. Characterization of fully 2′-modified oliogoribonucleotide hetero- and homoduplex hybridization and nuclease sensitivity. Nucleic Acids Res. 1995;23:2019–2024. doi: 10.1093/nar/23.11.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czauderna F., Fechtner M., Dames S., Aygun H., Klippel A., Pronk G.J., Giese K., Kaufmann J. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 2003;31:2705–2716. doi: 10.1093/nar/gkg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J.G., Sharp P.A. Specificity of microRNA target selection in translational repression. Genes & Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir S.M., Martinez J., Patkaniowska A., Lendeckel W., Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov Y., Anderson E.M., Birmingham A., Reynolds A., Karpilow J., Robinson K., Leake D., Marshall W.S., Khvorova N. Off-target effects by siRNA can induce toxic phenotype. RNA. 2006 doi: 10.1261/rna.28106. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes T.R., Mao M., Jones A.R., Burchard J., Marton M.J., Shannon K.W., Lefkowitz S.M., Ziman M., Schelter J.M., Meyer M.R., et al. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat. Biotechnol. 2001;19:342–347. doi: 10.1038/86730. [DOI] [PubMed] [Google Scholar]
- Inoue H., Hayase Y., Imura A., Iwai S., Miura K., Ohtsuka E. Synthesis and hybridization studies on two complementary nona(2′-O-methyl)ribonucleotides. Nucleic Acids Res. 1987;15:6131–6148. doi: 10.1093/nar/15.15.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A.L., Bartz S.R., Schelter J., Kobayashi S.V., Burchard J., Mao M., Li B., Cavet G., Linsley P.S. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- Jackson A.L., Burchard L., Schelter J., Chau B.N., Cleary M., Lim L., Linsley P. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA. 2006 doi: 10.1261/rna.25706. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraynack B.A., Baker B.F. Small interfering RNAs containing full 2′-O-methylribonucleotide-modified sense strands display Argonaute2/eIF2C2-dependent activity. RNA. 2006;12:163–176. doi: 10.1261/rna.2150806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesnik E.A., Guinosso C.J., Kawasaki A.M., Sasmor H., Zounes M., Cummins L.L., Ecker D.J., Cook P.D., Freier S.M. Oligodeoxynucleotides containing 2′-O-modified adenosine: Synthesis and effects on stability of DNA:RNA duplexes. Biochemistry. 1993;32:7832–7838. doi: 10.1021/bi00081a031. [DOI] [PubMed] [Google Scholar]
- Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lin S., Ruan S., Anderson M.G., McDowell J.A., Kroeger P.E., Fesik S.W., Shen Y. siRNA-mediated off-target gene silencing triggered by a 7nt complementation. Nucleic Acids Res. 2005;33:4527–4535. doi: 10.1093/nar/gki762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubini P., Zurcher W., Egli M. Stabilizing effects of the RNA 2′-substituent: crystal structure of an oligodeoxynucleotide duplex containing 2′-O-methylated adenosines. Chem. Biol. 1994;1:39–45. doi: 10.1016/1074-5521(94)90039-6. [DOI] [PubMed] [Google Scholar]
- Ma J.-B., Yuan Y.-R., Meister G., Pei Y., Tuschl T., Patel D.J. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller P., Annan N., McParland K., Pulford S. Oligothymidylate analogues having stereoregular, alternating methylphosphonate/phosphodiester backbones as primers for DNA polymerase. Biochemistry. 1982;21:2507–2512. doi: 10.1021/bi00539a033. [DOI] [PubMed] [Google Scholar]
- Monia B., Lesnick E., Gonzalez C., Lima W., McGee D., Guinosso C., Kawasaki A., Cook P., Freier S. Evaluation of 2′-modified oligonucleotides containing 2′-deoxy gaps as antisense inhibitors of gene expression. J. Biol. Chem. 1993;268:14514–14522. [PubMed] [Google Scholar]
- Nishizaki T., Iwai S., Ohtsuka E., Nakamura H. Solution structure of an RNA·2′-O-methylated RNA hybrid duplex containing an RNA·DNA hybrid segment at the center. Biochemistry. 1997;36:2577–2585. doi: 10.1021/bi962297c. [DOI] [PubMed] [Google Scholar]
- Nykanen A., Haley B., Zamore P.D. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- Parker J.S., Roe M., Barford D. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature. 2005;434:663–666. doi: 10.1038/nature03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W.R., Lipman D.J. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz D.S., Hutvagner G., Du T., Xu Z., Aronin N., Zamore P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Song J.J., Liu J., Tolia N.H., Schneiderman J., Smith S.K., Martienssen R.A., Hannon G.J., Joshua-Tor L. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat. Struct. Biol. 2003;10:1026–1032. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]