Abstract
To determine the physiological functions of the mammalian double-stranded RNA-binding protein PACT, the single-copy mouse Pact gene was disrupted and expression of the protein was completely ablated. The most notable phenotypes of the Pact−/− mouse were reduced size and severe microtia. As a result of the congenital abnormality of both outer and middle ears, these mice were hearing impaired. In situ hybridization revealed that PACT mRNA was expressed in specific regions of all three parts of the ear in adult and embryonic wild-type mice. Our study demonstrated an essential role of PACT in mammalian ear development and produced the first animal model for studying human microtia.
Keywords: gene disruption, microtia, antiviral, innate immunity, PKR
The human protein PACT and its murine counterpart RAX are almost identical in sequence. These proteins were discovered by virtue of their ability to activate the latent protein kinase PKR (1, 2). They and PKR share another property, namely, the ability to bind double-stranded RNA (dsRNA). Many, but not all, dsRNA-binding proteins contain one or more dsRNA-binding motif (dsRBM), which assumes an α-β-β-β-α structure and binds to the A-form of dsRNA in a sequence-independent fashion (reviewed in ref. 3). The same motif also mediates direct protein–protein interaction, thus providing opportunities for homomeric and heteromeric interactions between proteins of this family. PACT contains two dsRBMs, each of which can bind dsRNA. The same domains of PACT, domains 1 and 2, mediate strong interaction with the two dsRBMs of PKR, but that interaction does not cause PKR activation. Another domain of PACT, domain 3 consisting of 66 residues, is responsible for activating PKR by binding to its kinase domain (4).
Although different dsRNA-binding proteins share the ability to bind dsRNA, they have distinct biochemical, cellular, and physiological properties that are mediated by the specific “executor” parts of these proteins, such as the kinase domain of PKR (reviewed in ref. 5). These proteins participate in a quite diverse array of cellular processes. For example, PKR, PACT, and TRBP can regulate translation (1, 6, 7), RHA can regulate transcription (8), Staufen and NF90 can regulate mRNA localization (9, 10), and RNaseIII, ADAR, Dicer, and Drosha can regulate RNA processing (reviewed in ref. 5). Functions of these proteins, in the context of a whole organism, have been explored in only a few cases. In Drosophila, disruption of the Staufen gene causes lethal developmental defects (11). In mice, ablation of PKR causes defects in innate immune response to virus infections and cytokine signaling (12), and ablation of ADARI is embryonic lethal (13). The current study was designed to investigate the role of PACT in mouse physiology by disrupting the Pact gene. Because this protein is expressed in many tissues, although in varying amounts, we anticipated that disruption of the Pact gene would cause noticeable phenotypes in the knockout mice. Indeed, we observed that the Pact−/− mice were smaller in size and exhibited severe microtia.
Results
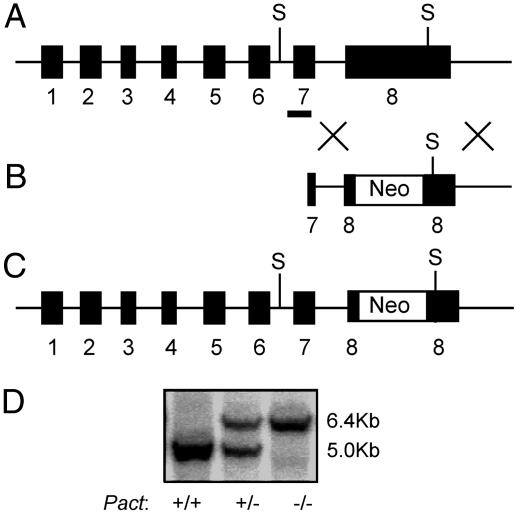
For probing functions of a mouse protein, disruption of the corresponding gene is often used as a tool. This approach is quite feasible for single-copy genes, as is the case for the Pact gene. However, it is often difficult to decide which exon of the gene to disrupt. Disruption of the exon containing the translation start site is the popular choice, but in many such cases, truncated proteins are expressed from the disrupted allele through the use of cryptic internal translation initiation sites. To avoid this problem for ablating Pact expression in mice, we targeted exon 8, which encodes its domain 3, because this domain is necessary and sufficient for activating PKR (4). Thus, even if the disrupted PACT allele were expressed in the knockout mice, the truncated protein containing only domains 1 and 2 would not be able to activate PKR. To the contrary, experiments in cultured cells suggested that such a protein should act as an inhibitor of PKR activation (14). With the above reasons in mind, we constructed a targeting vector containing a Neo cassette flanked by a part of exon 7, intron 7, and a part of exon 8 on the 5′ side. Part of exon 8 and the downstream genomic sequence were at the 3′ side, followed by a TK cassette. Moreover, we introduced a translation stop codon and a polyadenylylation signal in exon 8 just 5′ to the Neo cassette (Fig. 1B). Homologous recombination of this targeting vector with the resident gene should delete ≈500 bp from exon 8 and introduce the Neo gene in its place (Fig. 1C). Digestion of the disrupted Pact gene with SacI should produce a 6.4-kb DNA fragment, instead of a 5-kb fragment, that can hybridize with an exon 7-specific probe. An ES cell clone with a disrupted allele was isolated, and chimeric mice were generated from this clone. Successful propagation of the disrupted allele produced the founder Pact+/− mice. After several generations, these mice of mixed genetic background were bred with pure C57BL/6 mice, and the Pact+/− progenies were successively back-crossed with C57BL/6 partners for 10–14 successive generations. This crossing led to the establishment of a Pact+/− line in the C57BL/6 genetic background. The genotyping of wild-type, Pact+/−, and Pact−/− mice is shown in Fig. 1D.
Fig. 1.
Generation of the Pact−/− mouse. (A) Genomic structure of the Pact gene. The black boxes represent the eight exons present in the gene. S indicates the location of the SacI restriction sites used for genotype determination by Southern blot analysis. The black bar covering portions of intron 6 and exon 7 denotes the location of the probe used for Southern hybridization. (B) Structure of the targeting vector. Neo denotes the PGK-neomycin resistance cassette. A stop codon and a poly(A) signal were engineered into the targeting vector in exon 8 just 5′ to the PGK-Neo resistance gene as described in materials and methods. (C) Targeted Pact allele. Through homologous recombination, the neomycin resistance gene was inserted into exon 8, removing ≈500 bp of exon 8 sequence encoding the PKR activation domain. (D) Southern blot analysis of the various Pact genotypes in mice. The upper band (6.4 kb) is the disrupted allele. The lower band (5.0 kb) is the endogenous wild-type allele.
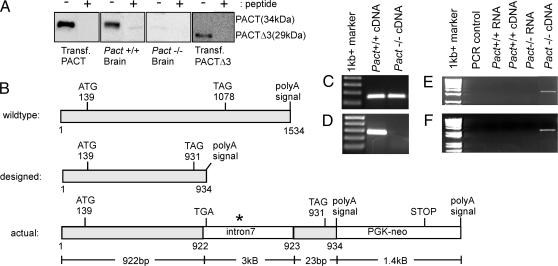
Unexpectedly, no truncated protein was expressed from the disrupted Pact allele (Fig. 2A). For detecting Pact expression, Western blot analysis was performed with extracts of several tissues in which PACT is expressed at a high level; the antibody used in these experiments recognizes specifically domain 2 of PACT. As positive controls, we used extracts of tissue culture cells expressing transfected wild-type PACT or Pact Δ3, which was the expected product of the targeted allele in mice. As shown in Fig. 2A, the expected full-length PACT was expressed in the brain of wild-type mice, but not in Pact−/− mice. The authenticity of the protein band was established by competing it out with the peptide against which the antibody was raised. No PACTΔ3 protein was detectable in the brain extracts of either mouse, although it was readily detected in the transfected cells. The observed absence of PACT-related protein expression in Pact−/− mice prompted us to investigate whether the corresponding mRNA was expressed. For mRNA analyses, we used a combination of RT-PCR, 3′ RACE, and cDNA sequencing. When the structures of the designed truncated mRNA and the actual mRNA from the disrupted allele were compared (Fig. 2B), it was apparent that the additional polyadenylylation signal of the targeting vector (at base 934) was not used for generating the mRNA; instead, the polyadenylylation signal of the Neo cassette was used. Moreover, intron 7 was retained in this aberrant PACT mRNA that was ≈5.5 kb long, instead of the expected 1-kb length. Thus, this PACT/neo hybrid mRNA contained 1–922 bases of authentic PACT mRNA, intron 7, 923–934 of the disrupted PACT exon 8, and all of the Neo cassette. This mRNA contained multiple translation termination signals in all three reading frames in the 5′ end of intron 7. The PACT ORF is terminated at a TGA codon right in the beginning of intron 7, and hence the encoded protein, if expressed, would have been almost identical to the one designed. Although we did not investigate the reason for the lack of translation of the aberrant mRNA, enhanced degradation, inhibition of cytoplasmic export, or inhibition of translation initiation due to the presence of the unnatural 3′ sequences could all have contributed to this effect. Regardless of the mechanism, because no PACT-related protein was expressed in the PACT−/− mouse, a true null strain of mouse line had been established.
Fig. 2.
PACT protein and mRNA expression in the Pact−/− mice. (A) Western blot for detecting PACT-related proteins in brains of wild-type and Pact−/− mice. The peptide from PACT domain 2 that was used as the antigen for raising the antibody was used as the competitor where indicated (+). (B) mRNA structure from both the wild-type and disrupted Pact alleles. The wild-type mRNA is ≈1.5 kb. The disrupted mRNA was designed to be ≈0.93 kb. The actual mRNA size from the disrupted allele is ≈5.4 kb. The asterisk denotes that there are 61 stop codons within intron 7. (C–F) RT-PCR analysis on Pact+/+ and Pact−/− RNA to determine message length. Primer sets used were: 599 sense and 743 antisense (144 bp) (C), 836 sense and 1096 antisense (260 bp) (D), 329 sense and intron 7 antisense (3.5 kb) (E), and intron 7 sense and 1415 neo antisense (2 kb) (F). All RNA was DNase treated and tested to ensure that all genomic DNA contamination had been removed before RT-PCR.
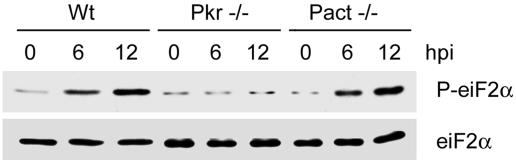
Pact−/− MEF lines were generated and used for examining PACT's role in PKR activation in cells infected with vesicular stomatitis virus (Fig. 3). In wild-type MEF, virus infection caused increasing eIF-2α phosphorylation. Because this is mediated by activated PKR, it did not occur in Pkr−/− cells; however, Pact−/− cells behaved like wild-type cells. These results suggest that dsRNA, not PACT, is the activator of PKR in vesicular stomatitis virus-infected MEF.
Fig. 3.
Lack of a need of PACT in PKR-mediated eIF-2a phosphorylation. MEF lines of the noted genotypes were infected with vesicular stomatitis virus at an MOI of 10. Cell extracts were made after the indicated hours of infection and Western blotted for phospho-eIF-2α (Upper) and total eIF-2α (Lower).
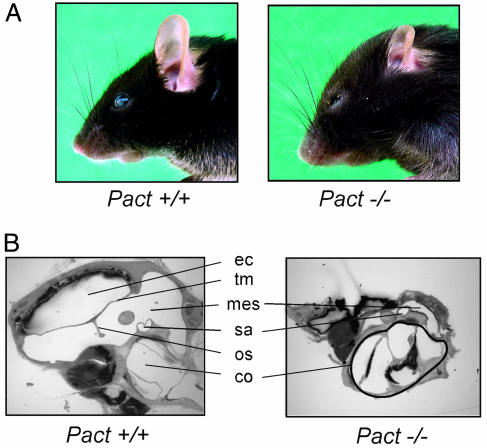
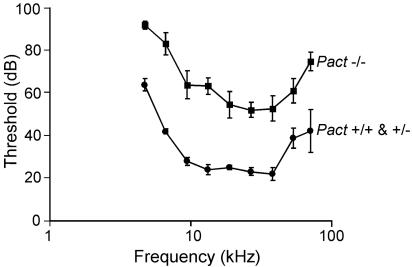
When Pact+/− male and female mice were mated, Pact−/− mice were born at the expected frequency, indicating normal Mendelian inheritance of the disrupted allele. The Pact−/− mice, both of the original mixed genetic background and pure C57BL/6 background, were smaller in size than wild-type mice. The difference in size was less noticeable in the first two weeks after birth, but after 4–6 weeks of age, the Pact−/− mice weighed ≈40% less than the wild-type mice, whereas Pact+/− mice weighed the same as wild-type mice. This difference in weight did not change over the whole life span of the Pact−/− mice. Despite the smaller size, anatomy of the Pact−/− mice is relatively normal except for the craniofacial areas: the rostrum is rounded, the nose is shortened, the turbinates are hypoplastic, and most remarkably, there is severe reduction of the size of the outer ear (Fig. 4A). Microscopic examination of the ear tissues revealed smaller pinna and external auditory canals and malformed ossicles, as well as a very small middle ear space and bulla (Fig. 4B). In contrast to the defects of the outer and middle ear, the cochlea of the Pact−/− mice is normal. The noted microtia of the Pact−/− mice was accompanied by hearing defects. When auditory brainstem responses were performed, hearing thresholds were elevated by 30–45 dB across all frequencies (4–60 kHz) in experimental mice as compared to the Pact+/+ and Pact+/− littermates (Fig. 5). This threshold shift did not change with age up to mice of 6 months. Pact−/− mice are clearly hearing impaired as a result of the congenital anomaly of the ear.
Fig. 4.
Differences in both external and internal ear structures of the Pact−/− mouse. (A) External ear and head morphology of a Pact+/+ and Pact−/− mouse. Note the extremely reduced size of the pinna as well as the shortened nose and prominent bossing of the forehead in the Pact−/− mouse as compared to the Pact+/+ mouse. (B) Internal ear structure of a Pact+/+ and Pact−/− mouse. The most abnormal ear structures in the Pact−/− mouse as seen above are the ear canal (ec), tympanic membrane (tm), ossicles (os), and middle ear space (mes). There is no impairment in the size of the cochlea (co) in the Pact−/− mouse. Also shown is the stapedial artery (sa).
Fig. 5.
Auditory brainstem response (ABR) of Pact+/+, Pact+/−, and Pact−/− mice. The above graph denotes data from mice tested at 8 weeks of age. n = 6 for Pact+/+ and Pact+/−, n = 7 for Pact−/−. For the study, mice were tested at 4, 8, and 12 weeks of age and demonstrated no sign of changes in hearing sensitivity during the first 12 weeks of life.
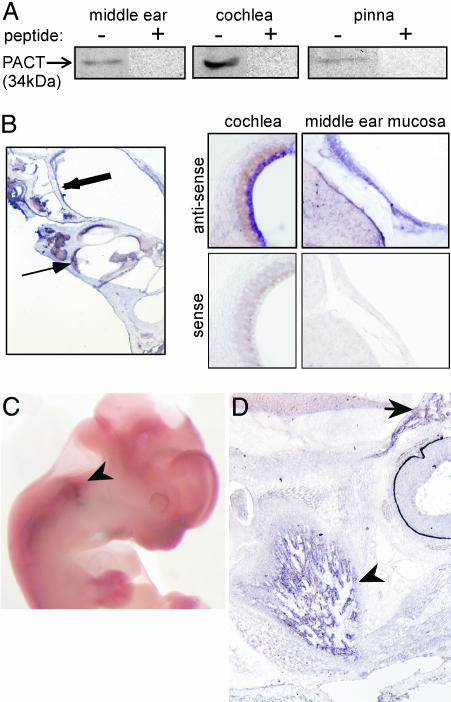
The above observations suggested that PACT might be expressed in the ear tissues of wild-type mice and required for their normal development. To examine PACT expression in different regions of the ears of 3-month-old adult C57BL/6 mice, the middle ear, the cochlea, and the pinna were dissected, and their extracts were analyzed for the presence of PACT protein by Western blotting. PACT was expressed in all three parts of the ear (Fig. 6A). To further localize the sites of PACT expression in the ear, we used in situ hybridization for detecting PACT mRNA expression in middle and inner ear sections. PACT mRNA was clearly detectable in the middle ear and the cochlea. There were strong signals with the antisense probe and no signals with the sense probe (Fig. 6B). In the middle ear, PACT mRNA was detected in the middle ear mucosa throughout the bulla in the respiratory epithelium. In the cochlea, PACT mRNA was observed in the marginal cell layer of the stria vascularis (Fig. 6B) and in the spiral prominence (data not shown). PACT mRNA was also expressed in the supporting cell layer of the sensory epithelium (Claudius and Hensen cells), but it was not expressed in the spiral limbus or the spiral ganglion (not shown).
Fig. 6.
Pact expression in the ear structures of wild-type adult mouse and embryos. (A) Western blot analysis of protein isolated from the pinna, middle ear and cochlea of a 3-month-old mouse. (B) Pact mRNA expression as revealed by in situ hybridization in the middle ear mucosa (thick arrow) and the lateral cochlear wall (thin arrow). Higher magnification (×20) reveals Pact mRNA in the marginal cell layer of the stria vascularis and in the respiratory epithelial cells of the middle ear mucosa. Hybridization using a sense probe in these regions served as a negative control. (C) Whole mount in situ hybridization of a wild-type E12 embryo. PACT mRNA expression is seen in the developing ear (arrowhead). (D) Parasagittal section of wild-type E16 embryo head. PACT mRNA is highly expressed in Meckel's cartilage (arrowhead), which is undergoing condensation to form the mandible. Additional signal is seen in the region of the developing anterior skull base (arrow).
In situ hybridization for PACT mRNA was performed in mouse embryos to determine the timing and location of PACT expression during development. PACT mRNA was present in the developing ear at embryonic day 12 (E12) as seen in the whole-mount embryo (Fig. 6C). Although the signal was weak, it was specific; no signal was obtained with the sense probe (data not shown). Strong signals were obtained in the mandible and anterior skull base at E16 in parasagittal sections (Fig. 6D). There were no signals when the sense probe was used (data not shown). Both the mandible and external ear are derived from the first branchial arch and PACT mRNA was expressed in both. These results demonstrated that PACT is expressed early in mouse development in structures derived from the first branchial arch (external ear and mandible) and the anterior skull base, and it is clearly important for normal development of the ear, mandible, and craniofacial skeleton. Additionally, PACT expression in the anterior skull base is likely responsible for normal development of the nose, as the PACT knockout mouse demonstrates a foreshortened nose and hypoplastic nasal turbinates.
Discussion
Genes encoding several dsRNA-binding proteins have been ablated in mice, and every ablation has caused a major change in their phenotypes. Mice lacking spermatid perinuclear RNA-binding protein show a high rate of mortality, reduced weight, and defects in spermatogenesis (15). Those lacking Prbp, encoded by Tarbp2, are sterile because of a failure to synthesize protamine (16). Disruption of the gene encoding RNA helicase A led to early embryonic lethality because of apoptotic death of embryonic ectodemal cells (17). The RNA-editing enzyme ADAR1 was shown to be required for embryonic erythropoiesis, and disruption of the corresponding gene caused embryonic lethality (13). On the other hand, the ADAR2 mice were viable, but prone to seizure and early death (18). These reports clearly indicated the critical importance of members of this family of proteins in mouse development.
PACT was not required for PKR activation and the resultant eIF-2α phosphorylation caused by vesicular stomatitis virus infection of MEF, indicating that, in the context of the specific cell type and the virus, dsRNA, not PACT, is the activator of PKR. It remains to be seen whether the same is true for other viruses and cells of other lineages. The observations presented here on Pact−/− mice demonstrate a critical function of PACT in ear development. Because Pkr−/− mice (12) do not share any of the defects of Pact−/− mice reported here, it is fair to assume that the corresponding functions of PACT are not mediated by PKR activation. Because PACT is a strong RNA-binding protein as well, we postulate that the missing functions are mediated by that property of PACT. This idea is strongly supported by what is known about several other dsRNA-binding proteins, most notably Staufen. In Drosophila, Staufen functions as an mRNA chaperon and transports and regulates the translation respectively of bicoid mRNA and Oskar mRNA at the anterior and the posterior poles of oocytes (9, 19, 20). Consequently, disruption of the Staufen gene is lethal to Drosophila development (11). In mammals, the protein is ubiquitously expressed and has been implicated in mRNA transport and translational regulation in hippocampal neurons (21, 22). It also interacts with telomerase RNA affecting telomerase functions (23, 24). Staufen promotes decay of specific mRNAs as well (25), thus down-regulating their cellular expression. It remains to be seen whether PACT functions in the above fashion, but the observed expression of PACT in different parts of the ear, which are developmentally affected in Pact−/− mouse, is consistent with this notion. This effect is obviously tissue specific and developmental lineage specific, because, although PACT was expressed in all three parts of the ear, the structure of the cochlea was not affected by the absence of PACT, whereas those of the pinna and the middle ear were profoundly affected. It should be noted that the outer and middle ears have a different embryologic origin than the inner ear (26). The Pact−/− mice must have additional developmental anomalies that cause the observed dwarfism. However, despite both of these defects, the mice have normal life span.
The Pact−/− mice may represent the first animal model for human microtia, a congenital otologic malformation of unknown etiology. People with microtia have small malformed pinnae, narrow or atretic ear canals, malformed ossicles, and associated conductive hearing loss (27), all of which are present in the Pact−/− mouse. Because ear development is intimately connected to craniofacial development, microtia is often accompanied by craniofacial abnormalities, such as in Treacher Collins syndrome or Goldenhar's syndrome (28). In the same vein, Pact−/− mice displays flattening of the midface, shortening of the nose, and hypoplastic nasal turbinates. In humans, microtia is relatively common (1 in 6,000 births). Although many cases are sporadic, others are clearly inherited, but the pattern of inheritance remains unclear (29). Our study suggests that some patients with microtia may have defects in PACT expression, a hypothesis that can be tested experimentally in the future.
Materials and Methods
Generation of the Pact Knockout Mouse.
The targeting vector (mPACT KO Domain 3) was constructed in two steps. Initially, a 3-kb genomic fragment of Pact containing the last five amino acids of exon 7, intron 7, the first 3 amino acids of exon 8 followed by a stop codon and poly(A) signal was PCR amplified from mouse BAC clone 68D6 (30) using the following primer set: 5′-CCGCTCGAGACGTATTTGGATATAGGTATGC-3′ and 5′-CCCAAGCTTTTTATTCTACAGCTCCTCTGTAATGACG-3′. This fragment was subcloned into the XhoI and HindIII sites of a pBSII KS plasmid already containing both a PGK-neo and a PGK-tk cassette. The second fragment was inserted into the previously mentioned vector by blunt-end ligation into a BamHI site located between the PGK-neo and PGK-tk cassette. This fragment consists of genomic DNA beginning from ≈150 bp 3′ to the natural poly(A) signal of Pact and continuing ≈3 kb into the genome. This fragment of DNA was removed from a plasmid containing a 7-kb EcoRI genomic fragment of Pact (from BAC clone 68D6) by BamHI and HindIII digestion. The construct was designed to remove 486 bp of exon 8 in the knockout mouse, which comprises domain 3, the PKR-activating region. The targeting vector was used for generation of the knockout mouse by standard protocol. The Pact−/− mice were backcrossed into the C57BL/6 background by mating with wild-type C57BL/6 mice (Charles River Breeding Laboratories) for a minimum of 10 generations. These backcrossed mice then became the experimental model.
DNA Extraction and Southern Blot Hybridization.
DNA extraction and Southern blot hybridization were performed as described (31). Genotyping of the mice was determined by using an [α-32P]dCTP-radiolabeled pact probe containing ≈750 bp of Pact intron 7 sequence located 5′ to the site of homologous recombination.
RNA Isolation and RT-PCR.
RNA was isolated from tissues by using RNA-Bee according to manufacturer's instructions (Tel-Test). Reverse transcription was performed with random hexamers on DNase treated RNA by using the Superscript III Random Prime Synthesis kit for RT-PCR (Invitrogen/Life Science) according to manufacturer's instructions. PCR was performed by using various primer sets located within the coding region of Pact and the region of the targeting vector that had undergone homologous recombination.
Protein Isolation and Western Blot Analysis.
Protein was isolated in lysis buffer containing 20 mM Tris·HCl (pH 7.5), 50 mM KCl, 100 mM NaCl, 1% Triton X-100, 1 mM EDTA, 20% glycerol, 0.2 mM PMSF, 1 mM DTT, and complete EDTA-free protease inhibitor tablet (Roche). The three areas of the ear were harvested separately from freshly killed mice. The pinna was sharply excised from the skull, followed by degloving of the soft tissues of the scalp. The middle ear was then fragmented and harvested under binocular microscopy. The cochlea was then dislodged and removed. Tissue was dounce-homogenized multiple times, allowed to undergo a freeze/thaw cycle, and spun down to remove debris. A total of 100 μg of protein lysate was run on a SDS/12% PAGE gel and Western blotted with a PACT domain 2-specific polyclonal antibody 1414 (1:1,000) that was custom made by Biosynthesis (Lewisville, TX). For the peptide competition assays, 20 μl of PACT antibody was mixed with 100 μg of peptide in PBS, incubated at 37°C for 2 h, and then incubated at 4°C for 16 h. The mixture was then spun down to remove any immune complexes that had formed and used for Western blotting.
Generation of MEF and Assay for eIF-2α Phosphorylation After Vesicular Stomatitis Virus Infection.
Primary MEF cultures were established from E14 embryos as described (12). Cells were genotyped as above. Cells were grown to 90–100% confluence and infected with vesicular stomatitis virus in serum-free Dulbecco's modified Eagle's medium at a multiplicity of 10 plaque-forming units per cell in a small volume (500 μl per well for a six-well dish). At 1 h after infection, virus was removed and cells were washed with PBS. Complete medium containing 10% FBS was added to the wells, and the cells were incubated for an additional 5 or 11 h. Cells were lysed in modified RIPA buffer (10 mM Tris·HCl, pH 7.5/100 mM NaCl/1 mM EDTA/1 mM EGTA/1 mM NaF/20 mM Na4P2O7/2 mM Na3VO4/0.1% SDS/1% Triton X-100/10% glycerol/1 mM PMSF/1× protease inhibitor mixture, Sigma). The extracts were electrophoresed and Western blotted with antibodies for phopho-eIF-2α (BioSource) and eIF-2α (Cell Signaling Technology).
Auditory Brainstem Response (ABR).
ABRs were elicted through tone pips and recorded via s.c. electrodes placed in the ipsilateral pinna, vertex, and ground electrode placed in the back by the tail. The stimulus was 5-ms tone pips (delivered at 40 per s). The response was amplified (10,000×), filtered (100–3,000 Hz), and averaged by using computer software (Tucker-Davis Technologies, Gainesville, FL). The sound level was raised in 5-dB steps from 5 dB SPL up to 99 dB SPL. At each sound level, 1,024 responses were averaged with stimulus polarity alternated. Response waveforms were discarded as artifact if the peak-to-peak voltage exceeded 15 μV. Stimulus frequencies ranged from 4 to 60 kHz for threshold testing. Threshold was determined by a single observer who noted the lowest sound level at which a recognizable waveform was seen on a screen of tracings stacked from lowest to highest sound level.
Histologic Preparation.
Anesthetized mice were perfused via intracardiac route with 4% paraformaldehyde for 1 min. Temporal bones were dissected, and the round and oval windows opened to allow for intralabyrinthine perfusion of fixative. After 1-h fixation, cochleas were decalcified in 1 M EDTA for 3 days at 4°C.
For plastic embedding, after decalcification, cochleas were osmicated (1% OsO4 in dH2O) for 1 h and dehydrated in serial ethanols and propylene oxide, and then embedded in Araldite resin and baked for 3 days at 60°C. The cochleas were serial sectioned (40-μm sections) with a carbide steel knife.
In Situ Hybridization.
All aqueous, non-Tris-containing solutions were treated with diethylpyrocarbonate (DEPC) to a concentration of 0.1% and RNase free conditions were strictly maintained. Antisense and sense riboprobes were generated by transcription of a vector containing bases 795-1100 of Pact cDNA using digoxigenin-labeled UTP. Fragments obtained were stored at −80°C in 50% deionized formamide, 5× SSC, 50 μg/ml yeast tRNA, 1% SDS, and 50 μg/ml heparin.
Adult C57BL/6 mice were anesthetized via i.p. infusion of ketamine (50 mg/kg) and xylazine (25 mg/kg) and perfused by intracardiac infusion of 4% paraformaldehyde/PBS. Mice were decapitated, and temporal bones were rapidly dissected, keeping the bulla intact. The specimens were fixed overnight in 4% paraformaldehyde at 4°C, washed with PBS, and decalcified in 0.5 M EDTA/PBS at 4°C for 3 days. Tissues were placed in 30% sucrose/PBS for 24 h and embedded in 30% sucrose on dry ice. Sections (30 μm) were placed onto Superfrost Plus slides (Fisher Scientific) and stored at −80°C.
For in situ hybridization, slides were fixed in 4% paraformaldehyde/PBS, bleached in 6% H2O2 and permeabilized in 2 μg/ml proteinase K/PBT for 20 min at 37°C. Slides were immersed in 2 mg/ml glycine/PBT and fixed in 4% PFA/0.2% glutaraldehyde/PBT. Prehybridization was carried out in 50% deionized formamide, 5× SSC, 50 μg/ml yeast tRNA, 1% SDS, and 50 μg/ml heparin in DEPC-treated H2O at 55°C for 1 h. Slides were hybridized with 250 ng of riboprobe in prehybridization solution at 55°C overnight, washed in Solution 1 (50% formamide/5× SSC/1% SDS) at 62°C, in solution 2 (50% formamide/5× SSC) at 58°C, and then blocked in 5% sheep serum. Sheep anti-digoxigenin Fab fragments conjugated to alkaline phosphatase (Roche Diagnostics) were preadsorbed in 1% sheep serum/TBST at a concentration of 1:1,000 and exposed to the tissue sections overnight at 4°C. Slides were washed and placed in NBT/BCIP solution until a blue color was noted. Postfixation was carried out in 4% paraformaldehyde/0.1% glutaraldehyde.
Embryos were prepared by harvesting the uteri of timed pregnant CBA/CaJ mice. After fixing embryos in 4% paraformaldehyde and dehydrating in methanol, some embryos were prepared as whole-mount preparations, whereas the larger embryos were sectioned on a cryostat. In situ hybridization was then performed on the specimens as described above.
Acknowledgments
We thank Sean Kessler for helpful discussion and Christina Gaughan for technical assistance. This work was partially supported by National Institutes of Health Grants CA62220 and CA68782.
Abbreviation
- En
embryonic day n.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Patel R. C., Sen G. C. EMBO J. 1998;17:4379–4390. doi: 10.1093/emboj/17.15.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ito T., Yang M., May W. S. J. Biol. Chem. 1999;274:15427–15432. doi: 10.1074/jbc.274.22.15427. [DOI] [PubMed] [Google Scholar]
- 3.Tian B., Bevilacqua P. C., Diegelman-Parente A., Mathews M. B. Nat. Rev. 2004;5:1013–1023. doi: 10.1038/nrm1528. [DOI] [PubMed] [Google Scholar]
- 4.Peters G. A., Hartmann R., Qin J., Sen G. C. Mol. Cell. Biol. 2001;21:1908–1920. doi: 10.1128/MCB.21.6.1908-1920.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saunders L. R., Barber G. N. FASEB J. 2003;17:961–983. doi: 10.1096/fj.02-0958rev. [DOI] [PubMed] [Google Scholar]
- 6.Meurs E., Chong K., Galabru J., Thomas N. S., Kerr I. M., Williams B. R., Hovanessian A. G. Cell. 1990;62:379–390. doi: 10.1016/0092-8674(90)90374-n. [DOI] [PubMed] [Google Scholar]
- 7.Gatignol A., Buckler-White A., Berkhout B., Jeang K. T. Science. 1991;251:1597–1600. doi: 10.1126/science.2011739. [DOI] [PubMed] [Google Scholar]
- 8.Nakajima T., Uchida C., Anderson S. F., Lee C. G., Hurwitz J., Parvin J. D., Montiminy M. Cell. 1997;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 9.St. Johnston D., Beuchle D., Nusslein-Volhard C. Cell. 1991;66:51–63. doi: 10.1016/0092-8674(91)90138-o. [DOI] [PubMed] [Google Scholar]
- 10.Shim J., Lim H., Yates R., Jr., Karin M. Mol. Cell. 2002;10:1331–1344. doi: 10.1016/s1097-2765(02)00730-x. [DOI] [PubMed] [Google Scholar]
- 11.Winslow G. M., Carroll S. B., Scott M. P. Dev. Biol. 1988;129:72–83. doi: 10.1016/0012-1606(88)90162-5. [DOI] [PubMed] [Google Scholar]
- 12.Yang Y. L., Reis L. F., Pavlovic J., Aguzzi A., Schafer R., Kumar A., Williams B. R., Aguet M., Weissmann C. EMBO J. 1995;14:6095–6106. doi: 10.1002/j.1460-2075.1995.tb00300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Q., Khillan J., Gadue P., Nishikura K. Science. 2000;290:1765–1768. doi: 10.1126/science.290.5497.1765. [DOI] [PubMed] [Google Scholar]
- 14.Li S., Sen G. C. J. Interferon Cytokine Res. 2003;23:689–697. doi: 10.1089/107999003772084806. [DOI] [PubMed] [Google Scholar]
- 15.Pires-daSilva A., Nayernia K., Engel W., Torres M., Stoykova A., Chowdhury K., Gruss P. Dev. Biol. 2001;233:319–328. doi: 10.1006/dbio.2001.0169. [DOI] [PubMed] [Google Scholar]
- 16.Zhong J., Peters A. H. F. M., Lee K., Braun R. E. Nat. Genet. 1999;22:171–174. doi: 10.1038/9684. [DOI] [PubMed] [Google Scholar]
- 17.Lee C.-G., Da Costa Soares V., Newberger C., Manova K., Lacy E., Hurwitz J. Proc. Natl. Acad. Sci. USA. 1998;95:13709–13713. doi: 10.1073/pnas.95.23.13709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higuchi M., Maas S., Single F. N., Hartner J., Rozov A., Burnashev N., Feldmeyer D., Sprengel R., Seeburg P. H. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 19.St. Johnston D., Driever W., Berleth T., Richstein S., Nusslein-Volhard C. Development (Cambridge, U.K.) 1989;107:13–19. doi: 10.1242/dev.107.Supplement.13. [DOI] [PubMed] [Google Scholar]
- 20.Micklem D. R., Adams J., Grunert S., St. Johnston D. EMBO J. 2000;19:1366–1377. doi: 10.1093/emboj/19.6.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiebler M. A., Hemraj I., Verkade P., Kohrmann M., Fortes P., Marion R. M., Ortin J., Dotti C. G. J. Neurosci. 1999;19:288–297. doi: 10.1523/JNEUROSCI.19-01-00288.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kohrmann M., Luo M., Kaether C., DesGroseillers L., Dotti C. G., Kiebler M. A. Mol. Biol. Cell. 1999;10:2945–2953. doi: 10.1091/mbc.10.9.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bachand F., Triki I., Autexier C. Nucleic Acids Res. 2001;29:3385–3393. doi: 10.1093/nar/29.16.3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le S., Sternglanz R., Greider C. W. Mol. Biol. Cell. 2000;11:999–1010. doi: 10.1091/mbc.11.3.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim Y. K., Furic L., DesGroseillers L., Maquat L. E. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 26.Fekete D. Trends Neurosci. 1999;22:263–269. doi: 10.1016/s0166-2236(98)01366-6. [DOI] [PubMed] [Google Scholar]
- 27.Tanzer R. C. Clin. Plast. Surg. 1978;5:317–336. [PubMed] [Google Scholar]
- 28.Marszalek B., Wojcicki P., Kobus K., Trzeciak W. H. J. Appl. Genet. 2002;43:223–233. [PubMed] [Google Scholar]
- 29.Gupta A., Patton M. A. Am. J. Med. Genet. 1995;59:238–241. doi: 10.1002/ajmg.1320590223. [DOI] [PubMed] [Google Scholar]
- 30.Rowe T. M., Sen G. C. Gene. 2001;273:215–225. doi: 10.1016/s0378-1119(01)00588-1. [DOI] [PubMed] [Google Scholar]
- 31.Kessler S. P., Rowe T. M., Gomos J. B., Kessler P. M., Sen G. C. J. Biol. Chem. 2000;275:26259–26264. doi: 10.1074/jbc.M004006200. [DOI] [PubMed] [Google Scholar]