Abstract
Many native proteins occasionally form partially unfolded forms (PUFs), which can be detected by hydrogen/deuterium exchange and NMR spectroscopy. Knowledge about these metastable states is required to better understand the onset of folding-related diseases. So far, not much is known about where PUFs reside within the energy landscape for protein folding. Here, four PUFs of the relatively large apoflavodoxin (179 aa) are identified. Remarkably, at least three of them are partially misfolded conformations. The misfolding involves side-chain contacts as well as the protein backbone. The rates at which the PUFs interconvert with native protein have been determined. Comparison of these rates with stopped-flow data positions the PUFs in apoflavodoxin’s complex folding energy landscape. PUF1 and PUF2 are unfolding excursions that start from native apoflavodoxin but do not continue to the unfolded state. PUF3 and PUF4 could be similar excursions, but their rates of formation suggest that they are on a dead-end folding route that starts from unfolded apoflavodoxin and does not continue all of the way to native protein. All PUFs detected thus are off the protein’s productive folding route.
Keywords: hydrogen/deuterium exchange, partially unfolded forms, protein folding
Protein folding can be described by using a free energy landscape model (1, 2). In this model an unfolded protein molecule descends along a funnel describing its free energy until it reaches the state with the lowest free energy, which is the native state. The landscape often contains local minima, which host folding intermediates. Whereas the native state of many proteins is well characterized, little is known about the metastable intermediate states and their positions along the folding funnel. Knowledge about these metastable states is necessary to improve the understanding of protein folding and folding-related diseases, because rarely populated partially folded forms of proteins are known to initiate the formation of amyloids (3).
Detection by nuclear magnetic resonance (NMR) spectroscopy of native state hydrogen/deuterium exchange (H/D exchange), i.e., in the presence of small amounts of a denaturant, allows the characterization of partially unfolded forms (PUFs) of a protein (4). These PUFs are interpreted to reside in high-energy minima in the folding energy landscape (5, 6). PUFs are often undetectable by other techniques, because, for example, they reside behind the highest-energy transition state for folding and because they do not populate significantly at equilibrium. Analysis of H/D exchange data with models that statistically sample the conformational space accessible to a protein can give insight into the high-energy conformations that a protein can adopt (7–9). However, little evidence exists that links PUFs to the folding intermediates that populate during equilibrium unfolding, kinetic unfolding, or kinetic refolding of proteins. This missing link makes it difficult to position PUFs within the energy landscape for protein folding (10).
Recently, both the denaturant-induced equilibrium unfolding and the kinetic folding of Azotobacter vinelandii flavodoxin have been characterized (11–15). Apoflavodoxin kinetic folding is described by (13)
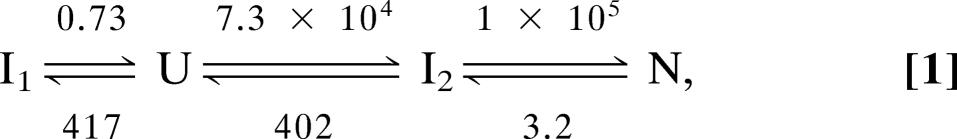 |
where U is unfolded protein, N is native apoflavodoxin, and I1 and I2 are folding intermediates. In the presence of FMN, first apoflavodoxin folds to its native state according to Eq. 1 and then FMN binds to form holoflavodoxin (15). The rate constants shown in Eq. 1 are in s−1 and are obtained by extrapolation to 0 M GuHCl. The rate constants that describe departure from I2, either to N or to U, are not absolute. The kinetic data obtained only allow the determination of their ratio (13). The molten globule-like intermediate I1, which also populates to significant extents at GuHCl concentrations ranging from 1 to 3 M, acts as a trap. It has to unfold before native apoflavodoxin can be formed. Intermediate I2 is highly unstable and thus is not observed during denaturant-induced equilibrium unfolding of apoflavodoxin. The formation of intermediates I1 and I2 during folding is linked to apoflavodoxin’s α-β parallel topology (14).
Here, H/D exchange measurements are used to characterize PUFs of the 179-residue A. vinelandii apoflavodoxin. A detailed picture, which includes the position of apoflavodoxin’s PUFs, is obtained of the folding energy landscape accessible to this relatively large protein. All PUFs turn out to be off the protein’s productive folding route.
Results
Backbone amide proton H/D exchange rates (kex) are determined for apoflavodoxin at deuterated guanidinium chloride (GuDCl) concentrations that range from 0 to 750 mM using NMR spectroscopy and converted into free energy differences for local opening of the protein structure, ΔGop (see Table 2, which is published as supporting information on the PNAS web site). This conversion is valid when exchange takes place according to the EX2 mechanism, which is the case for apoflavodoxin at pD 5.7 to 5.9 (where pD is the pH meter reading of a deuterated solution), the pD range used in this study (see below).
Identification of Cooperatively (Un)folding Clusters Within Apoflavodoxin.
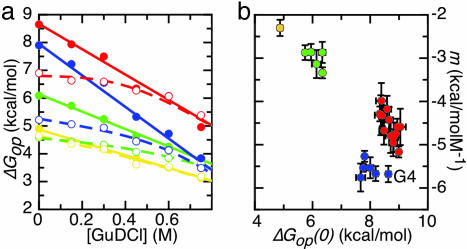
Some apoflavodoxin residues display a linear dependence of ΔGop on GuDCl concentration over the complete concentration range used (Fig. 1a). Their amides exchange either from the globally unfolded state (type 1 residues) or from a PUF of the protein (type 2 residues; see Supporting Experimental Procedures, which is published as supporting information on the PNAS web site). Other residues have ΔGop values that have a curving dependency on GuDCl concentration (Fig. 1a). At low GuDCl concentration, the amides of these residues exchange through local opening of the native conformation (type 3 residues).
Fig. 1.
Native-state H/D exchange data of amino acid residues of apoflavodoxin. (a) Typical examples of ΔGop data that have a linear or curved dependency on GuDCl concentration (filled and open circles, respectively). Cluster 1 within apoflavodoxin (yellow) is represented by V141 (filled circles) and A140 (open circles), Cluster 2 (green) by A150 (filled circles) and K145 (open circles), Cluster 3 (blue) by F6 (filled circles) and V17 (open circles), and Cluster 4 (red) by L93 (filled circles) and F49 (open circles). (b) Clusters of residues identified by similarity of the corresponding ΔGop(0) and m values, respectively (errors are standard fitting errors). Only those residues for which the GuDCl-dependency of their ΔGop data is linear in the GuDCl-range studied are taken into account. Four clusters are identified as follows: Cluster 1 (yellow; V141), Cluster 2 (green; F146, V147, G148, L149, and A150), Cluster 3 (blue; G4, L5, F6, I21, and K22), and Cluster 4 (red; I51, L52, G53, V91, A92, L93, F94, W167, L168, A169, Q170, I171, and A172). The average ΔGop(0) and m values of a particular cluster define the linear GuDCl-dependence of the free energy isotherm of the cluster involved. The isotherms for the four identified clusters are shown in a as solid lines. The curving isotherms in a, which result from a fit of a two-process model (see Eq. S7 in Supporting Experimental Procedures) to the curving data, are dashed and join one of the linear isotherms at GuDCl concentrations >0 M.
Residues for which the ΔGop values depend linearly on GuDCl concentration.
For each residue that displays a linear dependence of ΔGop on GuDCl concentration, the ΔGop value at 0 M GuDCl [i.e., ΔGop(0)] and the corresponding slope (m value) are shown in Fig. 1b. Residues are clustered and colored based on similarities of their ΔGop(0) and m values, respectively. Residues in a particular cluster exchange their amides from the same (partially or globally) unfolded form of the protein (see Supporting Experimental Procedures). Four clusters are thus identified. Each cluster is characterized by a typical average ΔGop(0) and average m value (described below in Features of the four cooperatively unfolding clusters identified). These averaged ΔGop(0) and m values are all smaller than the stability against global unfolding of apoflavodoxin (ΔGN-U) and the corresponding GuHCl-dependence (mN-U), respectively. This finding implies that none of the amides of the identified clusters exchanges through global protein unfolding, the characteristic of type 1 residues. The residues in Fig. 1b are thus all type 2 residues, and each of the four clusters unfolds through a specific subglobal process.
Residues for which the ΔGop values have a curving dependency on GuDCl concentration.
At low GuDCl concentrations, the ΔGop values of the type 3 residues of apoflavodoxin change little with increasing GuDCl concentration, because local opening processes are the dominant mechanism for exchange of type 3 residues. These residues mainly reside in loops and at the edges of elements of secondary structure of apoflavodoxin, where it is reasonable to envisage local unfolding events. In contrast, type 2 residues (Fig. 1b and Table 2) are in the core of secondary-structure elements of apoflavodoxin.
At higher GuDCl concentrations, exchange of type 3 amide protons occurs through subglobal unfolding of the protein. As a consequence, the corresponding ΔGop values curve toward the linear isotherms that characterize the unfolding of the four identified clusters (Fig. 1a). Type 3 residues can be assigned to belong to one of the subglobally unfolding clusters identified in Fig. 1b on the basis of the quality of the fit of a two-process model (see Eq. S7 in Supporting Experimental Procedures) to their curving ΔGop data (assignments are listed in Table 2) (16, 17). In this model, amides can exchange from a (sub)globally unfolded conformation or a locally opened native-like conformation.
Features of the four cooperatively unfolding clusters identified.
Cluster 1 within apoflavodoxin (yellow in Figs. 1 and 2a) is characterized by ΔGop(0) = 4.87 ± 0.03 kcal/mol and m = −2.3 ± 0.1 kcal/mol·M−1 (errors indicated are standard errors). Only one of its residues, V141, has up to 0.75 M a linear dependence of its ΔGop on GuDCl concentration, characteristic for type 2 residues. Neighbor A140 has a somewhat lower ΔGop(0) than V141, and its isotherm curves smoothly toward the one of V141. A140 and also V142 are type 3 residues belonging to Cluster 1. The three residues discussed form one of the two strands of a small β-sheet that is part of a loop typical for long-chain flavodoxins (Fig. 2a). This loop connects β-strands 5A and 5B.
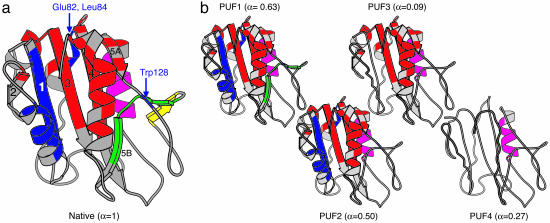
Fig. 2.
Native apoflavodoxin and its partially unfolded forms as detected by native-state H/D exchange and NMR spectroscopy. (a) molscript (18) cartoon drawing of the x-ray structure of A. vinelandii flavodoxin (19), with secondary structure depicted as determined for apoflavodoxin by NMR spectroscopy (20). The clusters of cooperatively unfolding residues are shown in yellow (Cluster 1), green (Cluster 2), blue (Cluster 3), red (Cluster 4), and purple (Cluster 5). The β-sheets are numbered based on the primary sequence of the protein. (b) PUFs of apoflavodoxin presented in increasing order of stability difference with respect to native apoflavodoxin (i.e., PUF1 to PUF4). Elements of secondary structure that are intact in a specific PUF are drawn in cartoon fashion, whereas those that are unfolded and water accessible in a specific PUF are drawn as coils. Secondary-structure elements are assumed to unfold cooperatively unless data opposing this exist. The α values of the individual PUFs are given in brackets.
Cluster 2 consists of the other strand of the small β-sheet discussed and also comprises β-strand 5B. It is colored green in Figs. 1 and 2a and is characterized by ΔGop(0) = 6.14 ± 0.05 kcal/mol and m = −3.1 ± 0.1 kcal/mol·M−1.
Cluster 3 (blue in Figs. 1 and 2a) is characterized by ΔGop(0) = 7.95 ± 0.09 kcal/mol and m = −5.7 ± 0.2 kcal/mol·M−1. It mainly involves a large part of the first 24 N-terminal residues of apoflavodoxin. Of the residues belonging to Cluster 3, the ΔGop(0) of G4 is significantly higher than the ΔGop(0) values of the other residues (Fig. 1b). Because the m value associated with G4 is identical within error to the average m value of the residues belonging to Cluster 3, G4 is considered to be a member of this cluster as well. G4 seems to have residual protection against exchange when Cluster 3 is unfolded; its ΔGop(0) and m value are not used for the determination of the average ΔGop(0) and m value of Cluster 3.
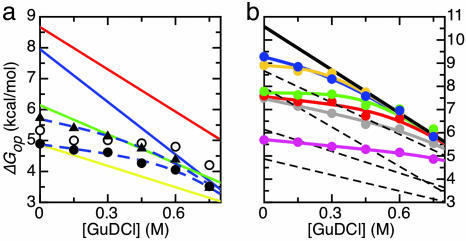
Apparently, the subglobal unfolding of the N-terminal part of apoflavodoxin (i.e., Cluster 3) also affects distant parts within the protein. At least two residues (W128 and E82) that are sequentially and spatially distant from the N terminus (Fig. 2a) have a curvature of their ΔGop isotherm toward the linear one that characterizes the cooperative unfolding of Cluster 3 (Fig. 3a). W128 and E82 thus belong to Cluster 3. The isotherm corresponding to L84 displays a similar curvature. Although at 0.75 M GuDCl the isotherm ends at a somewhat higher ΔGop value than the average ΔGop value for this cluster, L84 most likely belongs to Cluster 3 as well (Fig. 3a).
Fig. 3.
Native-state H/D exchange data of a few specific amino acid residues of apoflavodoxin. (a) Residues E82 (filled circles), L84 (open circles), and W128 (filled triangles) belong to Cluster 3 of apoflavodoxin, as shown by the GuDCl-dependencies of the ΔGop of their backbone amides. The linear isotherms that describe the cooperative unfolding of Clusters 1–4 of apoflavodoxin are drawn in the same colors used in Figs. 1 and 2. The dashed lines are best fits of a two-process model (see Eq. S7 in Supporting Experimental Procedures) to the E82 and the W128 data that join the blue isotherm of Cluster 3. (b) Residues that form Cluster 5 within apoflavodoxin as shown by the GuDCl-dependencies of the ΔGop values of L110, pink; G111, gray; Y114, orange; S115, red; F116, green; and F117, blue. The isotherm that describes the GuDCl-induced global unfolding of apoflavodoxin is shown in black. The best fits of a two-process model (see Eq. S7 in Supporting Experimental Procedures) to the data of the individual residues are shown as colored lines. For comparison, the linear ΔGop isotherms corresponding to the four already-identified clusters within apoflavodoxin are shown by dashed lines.
Cluster 4 (red in Figs. 1 and 2a) is characterized by ΔGop(0) = 8.67 ± 0.05 kcal/mol and m = −4.6 ± 0.1 kcal/mol·M−1. It is the cluster that contains the largest number of residues, and it spans several elements of secondary structure including the core of apoflavodoxin.
Identification of a fifth cluster.
The residues not yet categorized (i.e., L110, G111, Y114, S115, F116, and F117) reside in α-helix 4 of apoflavodoxin (purple in Fig. 2a) and have curved ΔGop isotherms. Interestingly, these curves (except the one for L110) lie at high GuDCl concentrations above the isotherms of the four already-identified clusters (Fig. 3b). These curves bend toward a common linear isotherm at high concentrations of GuDCl (shown in black in Fig. 3b), which represents the GuDCl-dependence of the free energy difference ΔGN-U that is associated with global unfolding of apoflavodoxin (15). In Fig. 3b, ΔGN-U is corrected for prolyl peptide bond isomerizations by adding 0.42 kcal/mol to ΔGN-U (21). These isomerizations do not occur during the short time the unfolded state is visited in the native state H/D exchange experiments but do occur in equilibrium-unfolded apoflavodoxin. Clearly, the results show that the amide protons of the mentioned residues exchange through global unfolding of apoflavodoxin at high GuDCl concentrations. Together these residues form Cluster 5, which is characterized by ΔGop(0) = ΔGN-U = 10.58 ± 0.06 kcal/mol and m = mN-U = −6.22 ± 0.02 kcal/mol·M−1. L110 could be a member of Cluster 4 or 5 (Fig. 3b). It is placed in Cluster 5 because it resides in the same helix as the other residues of Cluster 5, and because the two-process fit (Eq. S7 in Supporting Experimental Procedures) is slightly better when it is assumed to belong to Cluster 5 instead of Cluster 4.
PUFs of Apoflavodoxin.
Native-state H/D exchange data identify five clusters of residues within apoflavodoxin. Four of these clusters unfold subglobally in a cooperative manner. The resulting conformations are thus PUFs of the protein (4). It has been suggested that PUFs are snapshots of productive protein folding, forming sequentially in the order of their stability difference with respect to the native state (6, 22). This order is thought to be caused by the cooperative nature of protein folding (6). Assuming that such sequential unfolding occurs for native apoflavodoxin, conformations of its PUFs are identified (Fig. 2b).
The slope of the GuDCl-dependence of ΔGN-PUF, which is the free energy difference between N and a particular PUF (Figs. 1 and 3), is called mN-PUF. The value of mN-PUF informs about the PUF’s denaturant accessibility relative to the denaturant accessibility of native protein, because m values scale to changes in accessible surface area (23). The denaturant accessibility of a PUF is usually expressed as the α factor, which is the normalized m value of the specific PUF:
where mN-U is the m value associated with global unfolding of apoflavodoxin (13, 15) and mPUF-U is the one associated with unfolding of a specific PUF to unfolded apoflavodoxin. When α = 0, the corresponding PUF is as denaturant-accessible as fully unfolded apoflavodoxin, and when α = 1 it is as denaturant-accessible as native apoflavodoxin. The PUFs α values are listed in Fig. 2b.
The thermodynamic parameters associated with the identified folding states of apoflavodoxin are listed in Table 1.
Table 1.
Thermodynamic parameters of the apoflavodoxin folding states identified
| Species | ΔGN−X(0) | m value | α value | kN−X | kX−N |
|---|---|---|---|---|---|
| N | 0 | 0 | 1 | — | — |
| I1 | 6.69 ± 0.01 | −4.38 ± 0.02 | 0.29 | 7.4 × 10−5 | 0.733 |
| I2 | — | — | 0.8–0.9 | 3.2 | ≈1 × 105 |
| U | 10.58 ± 0.06 | −6.22 ± 0.02 | 0 | 0.013 | 7.3 × 104 |
| PUF1 | 4.87 ± 0.03 | −2.3 ± 0.1 | 0.63 | — | — |
| PUF2 | 6.14 ± 0.05 | −3.1 ± 0.1 | 0.50 | — | — |
| PUF3 | 7.95 ± 0.09 | −5.7 ± 0.2 | 0.09 | (7 ± 4) × 10−5 | 6 ± 4 |
| PUF4 | 8.67 ± 0.05 | −4.6 ± 0.1 | 0.27 | (4 ± 1) × 10−5 | 13 ± 4 |
Listed are the free energy [ΔGN−X(0), kcal/mol] associated with the formation of a particular state starting from native apoflavodoxin in absence of denaturant, the denaturant dependence of ΔGN−X (i.e., the m value, kcal/mol·M−1), the α-value (i.e., the normalized m value), the rate constant kN−X (s−1) at which the species is formed from native apoflavodoxin, and the rate constant kX−N (s−1) at which the species refolds to native apoflavodoxin. Data regarding N, I1, I2, and U are taken from refs. 13 and 15; the relative free energy of the unfolded state is corrected for prolyl peptide bond isomerization, as described in Results.
Folding and Unfolding Rates of the Clusters Identified Within Apoflavodoxin.
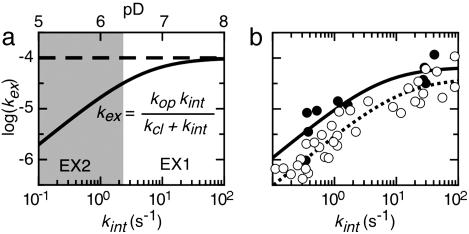
The rate constant for the conversion of native protein to a specific PUF can be determined. Use is made of pD-dependent native-state H/D exchange rates of type 2 residues of the cluster involved (24–27). At elevated pD values, exchange enters the EX1 regime (see Fig. 4a). The fit of the equation shown in Fig. 4a to the curving pD-dependent kex data leads to the determination of kop and kcl of the cluster involved. In the case of type 2 residues, kop equals the rate constant associated with unfolding of native apoflavodoxin to a specific PUF, kN-PUF, and kcl equals the rate constant associated with refolding of the PUF in question (26). Comparison of the latter rate constants with those obtained from stopped-flow folding data of a protein can give clues about the position a PUF has within the energy landscape for folding.
Fig. 4.
Determination of folding and unfolding rates of Clusters 3 and 4 within apoflavodoxin. (a) Simulated pD-dependent exchange curve of a typical residue. The residue has an intrinsic exchange rate constant (kint) of 1 s−1 at pD 6; the rate constant for local opening of the protein (kop) is 1 × 10−4 s−1, and the rate constant for local closing of the protein (kcl) is 5 s−1. An increase of pD by one unit causes kint to increase 10-fold. At low pD values (shaded in gray), the exchange rate constant kex increases accordingly. In this so-called EX2 pD regime, the logarithm of kex depends linearly on pD with a slope of 1. Above a certain pD value, kint is larger than kcl. Exchange then enters the EX1 regime in which kex curves toward a horizontal line defined by kex equaling kop. (b) The exchange rate constants of the backbone amide protons of the type 2 residues L5, F6, I21, and K22 of Cluster 3 (filled circles) and of all type 2 residues of Cluster 4 (open circles) display a pD-dependent curvature from EX2 to EX1 behavior in the pD range studied. Exchange rate constants (kex) are plotted against kint. In this representation, the curves of all residues of a particular cluster coincide. A fit of the equation shown in a to the data of Cluster 3 (solid line) and 4 (dashed line), respectively, allows the extraction of the unfolding and refolding rate constants involved.
H/D exchange rates of backbone amide protons of apoflavodoxin are determined in the absence of GuDCl at pD values of 6.28, 6.80, and 8.23 (a similar change of pH from 6 to 8 marginally destabilizes native apoflavodoxin because the thermal midpoint of its unfolding decreases by only 2.5 K). Unfortunately, the amide protons of the type 2 residues of both Clusters 1 and 2 exchange too rapidly to be tracked at pD 6.80 and 8.23. Their exchange apparently increases significantly with increasing pD because they can be tracked at pD 6.28, and EX2 behavior is likely. Consequently, the closing rates of these residues, which equal the refolding rates of PUF1 and PUF2, respectively, need to be fast compared with their kint values, which are at pD 6.28 between 0.1 and 1 s−1.
In case of Cluster 3, pD-dependent amide proton exchange could be determined for four of its five type 2 residues, which leads to: kop = (7 ± 4) × 10−5 s−1 and kcl = 6 ± 4 s−1 (Fig. 4b). The kop value equals the rate constant at which PUF3 is formed from N, whereas PUF3 refolds at rate constant kcl (26). Analogously, in case of Cluster 4 global fitting of the pD-dependent H/D exchange data of all of its 13 type 2 residues leads to: kop = (4 ± 1) × 10−5 s−1 and kcl = 13 ± 4 s−1 (Fig. 4b).
Cluster 5 only contains type 3 residues. Therefore, the rate constant for global folding and unfolding of apoflavodoxin cannot be determined directly by H/D exchange.
The GuDCl-dependent exchange data, recorded at pD 5.7–5.9, are analyzed assuming that exchange takes place according to the EX2 mechanism. The pD-dependent exchange data validate this assumption. At pD 5.7–5.9, the kint values are all <1.7 s−1. At these kint values the residues shown in Fig. 4b exchange according to the EX2 mechanism. This finding also holds for all other apoflavodoxin residues except for F25 (data not shown).
Discussion
Position of the PUFs Identified Within the Energy Landscape for Apoflavodoxin Folding.
A two-dimensional model of the energy landscape for apoflavodoxin folding has been constructed by using the reported denaturant-induced equilibrium and stopped-flow apoflavodoxin (un)folding results (Fig. 5a) (13). Comparison of the rate constants for interconversion between native apoflavodoxin and its PUFs with the rate constants for apoflavodoxin (un)folding extracted from stopped-flow data allows the positioning of the PUFs within this folding energy landscape.
Fig. 5.
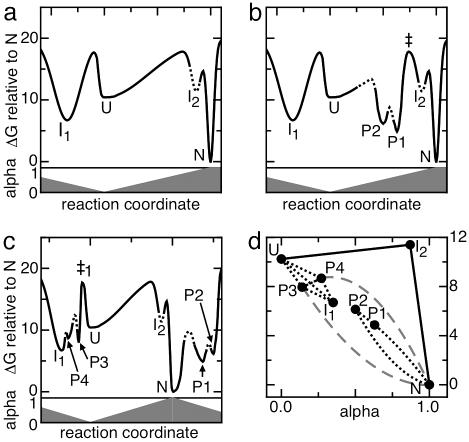
Schematic representation of the energy landscape for folding of apoflavodoxin. (a) Folding-energy landscape derived from denaturant-induced equilibrium and kinetic (un)folding studies (13). The horizontal axis represents the reaction coordinate (i.e., folding state of the protein). The vertical axis in Upper represents the free energy difference between a particular species and native apoflavodoxin. Lower shows the denaturant accessibility of each folding state expressed as the α-value, i.e., the ratio of the m value of a folding species and that of native apoflavodoxin. U (α = 0) and N (α = 1) represent unfolded and native apoflavodoxin, respectively; I1 (α = 0.29) and I2 (α lies between 0.8 and 0.9) are the two folding intermediates presented in Eq. 1. The off-pathway intermediate I1 is represented on the left-hand side of the unfolded state, whereas both the on-pathway intermediate I2 and native apoflavodoxin reside on the right-hand side of the unfolded state. The heights of the barriers ΔGop‡ between the individual species are calculated from the opening (i.e., unfolding) rate constants kop according to ΔGop‡ = −RT ln(kop/k0) using a value of 108 for k0 (28). The depth of the minimum in the free energy landscape in which I2 resides is unknown and therefore is represented by a dashed line. (b) Energy landscape for apoflavodoxin folding in which PUF1 (P1; α = 0.63) and PUF2 (P2; α = 0.50) are assumed to be on the productive folding route that links U and N. The height and position of the transition states that are kinetically important are determined accurately and are shown by continuous lines. The other barriers are created in such a manner that they do not influence the known folding and unfolding kinetics; their position and height are chosen arbitrarily as shown by dashed lines. Note that the energy difference between PUF1 and the transition state marked ‡ is large (12.95 kcal/mol) and causes the rate constant for PUF1 to N conversion to be 0.029 s−1 [calculated using ΔGPUF1–N‡ = −RT ln(kPUF1–N/k0)]. This conversion would be the rate-limiting step for folding. (c) Energy landscape for apoflavodoxin folding in which PUF1 and PUF2 are on a separate unfolding excursion from N. All free energy barriers that are not present in a are chosen such that they do not influence the known (un)folding kinetics. Note that on the right-hand side of N (α = 1) the α-values decrease. PUF3 (P3; α = 0.09) and PUF4 (P4; α = 0.27) are positioned in the region of the landscape where I1 is located, because PUF3 and PUF4 have within error the same barrier ‡1 as I1, according to their opening rate constants as determined from pD-dependent H/D exchange data. The heights of the free energy barriers between these three species are unknown and are represented by dashed lines. (d) Alternative representation of the energy landscape shown in c. Note that the denaturant accessibility axis now starts at α = 0 and ends at α = 1. Productive apoflavodoxin folding follows the solid lines. Dotted lines reflect that no information is available about how the species involved interconvert with one another. Although unlikely, PUF3 and PUF4 could be direct unfolding excursions from N as shown by gray dashes.
Positioning PUF1 and PUF2.
The rate constants at which two apoflavodoxin species interconvert can be calculated from Eq. 1. Unfolded apoflavodoxin (U) forms native apoflavodoxin (N) with a rate constant kU-N of 7.3 × 104 s−1, and N visits U with an unfolding rate constant kN-U of 0.013 s−1, respectively (see Supporting Experimental Procedures). Any folding intermediate that is positioned on the route between U and N must form native apoflavodoxin with a rate constant that is equal to or larger than kU-N. Such a high refolding rate implies that backbone amide protons that exchange from U, I2, or from any other potential apoflavodoxin folding intermediate that lies on the productive folding route between N and U are expected to display EX2 exchange behavior within the pD range studied (see Fig. 4a).
As discussed, EX2 behavior seems likely for the residues of Clusters 1 and 2 of apoflavodoxin. Both PUF1 and PUF2 thus could be on the productive folding route that links U with N, but PUF1 and PUF2 are too accessible for GuDCl to coincide with the kinetic apoflavodoxin folding intermediate I2 (Table 1).
Fig. 5b shows a free energy landscape for apoflavodoxin folding with PUF1 and PUF2 positioned on the productive folding route, taking into account their free energies and denaturant accessibilities. Because both PUFs are significantly more denaturant accessible than the rate-limiting transition state for folding [its α-value is 0.73 (13)], they reside on the unfolded side of this transition state, which is marked ‡. Both PUFs are significantly more stable than U. As a result, the transition from PUF1 via ‡ to N would be the rate-limiting step in folding from U to N. This folding would be much slower (0.029 s−1; see legend of Fig. 5b) than experimentally observed (7.3 × 104 s−1). Such slow folding would cause EX1 exchange behavior for amides that exchange from PUF1 and PUF2 at all pD values used here, which is not observed. Consequently, PUF1 and PUF2 cannot reside on the productive folding route of apoflavodoxin.
PUF1 and PUF2 thus must be excursions from the native state that are separated from the (un)folding route that links native and globally unfolded apoflavodoxin. This result is schematically shown in Fig. 5c.
Positioning PUF3 and PUF4.
None of the backbone amide protons of residues that belong to Clusters 3 and 4 of apoflavodoxin display pure EX2 H/D exchange behavior over the entire pD range studied. PUF3 and PUF4 of apoflavodoxin thus are not on the productive folding route between U and N.
PUF3 and PUF4 could be excursions from the native state separate from those leading to PUF1 and PUF2. Remarkably however, kPUF3-N and kPUF4-N are much smaller than kU-N, whereas kN-PUF3 and kN-PUF4 are within error identical to kN–I1 (Table 1). An explanation for this observation is that PUF3, PUF4, and I1 reside in a free energy region that is separated from unfolded apoflavodoxin by the same high barrier (Fig. 5c). PUF3 and PUF4 do not correspond to I1, because they are too unstable, too accessible for GuDCl, and fold too rapidly (Table 1). Apparently, both PUF3 and PUF4, as well as I1, need to unfold before productive folding occurs.
PUFs Are Not Snapshots of Apoflavodoxin’s Productive Folding.
PUFs were suggested to be snapshots of productive protein folding and to fold one after the other in a strictly hierarchical order determined by their individual free energy values, where the folding of one PUF facilitates the formation of the next one (5, 6, 22). However, apoflavodoxin’s pD-dependent exchange data clearly show that its PUFs do not reside on apoflavodoxin’s productive folding route. Furthermore, note that in Fig. 1a the lines that represent unfolding of native apoflavodoxin to PUF2 (i.e., the green line) and to PUF3 (i.e., the blue line) cross one another at 0.7 M GuDCl. This result implies that above 0.7 M GuDCl PUF3 is visited more often than PUF2, which conflicts with the idea that the stabilities of PUFs represent a sequential (un)folding order. Apoflavodoxin PUFs thus are not snapshots of apoflavodoxin’s productive folding.
Apoflavodoxin PUFs Are Partially Misfolded.
The observation that the PUFs of apoflavodoxin do not develop from one another in a sequential manner means that the representations of the conformations of these PUFs in Fig. 2b are speculative. For example, in the case of PUF2, it is assumed in Fig. 2b that both Cluster 2 and Cluster 1 are unfolded. However, when PUF1 and PUF2 would be separate excursions from the native state, Cluster 1 needs not necessarily be unfolded in PUF2. In addition, the folded parts of a PUF need not necessarily be native-like. In fact, the H/D exchange data presented here show that the PUFs of apoflavodoxin are partially misfolded for the following reasons. (i) In PUF1, one of the two strands of a small β-sheet is unfolded, whereas the second strand remains folded. The residues in the remaining folded strand can only be shielded from water by making nonnative hydrogen bonds with other folded parts of the protein. (ii) In PUF2, both the long loop typical for long-chain flavodoxins and β-strand 5B are unfolded, whereas the backbone amide of a single residue in this part of the protein (i.e., W128) remains protected against exchange. In native apoflavodoxin, the backbone amide of W128 is hydrogen-bonded to the carbonyl group of F146. F146 is water accessible in PUF2. Consequently, in PUF2, W128 must make a nonnative hydrogen bond. (iii) β-Strand 2 in PUF3, a strand that is at the periphery of apoflavodoxin, apparently is natively folded, whereas the neighboring β-strand 1 is unfolded. The backbone amides of β-strand 2 can only be protected against exchange in PUF3 by making nonnative hydrogen bonds.
In conclusion, three PUFs of apoflavodoxin represent partially misfolded conformations. The misfolding does not only involve side-chain contacts, as was recently observed for the PUFs of a four-helix bundle protein (29), but also involves the backbone of secondary structure elements of apoflavodoxin. The apoflavodoxin data thus show that the original idea (5, 6, 22) that PUFs represent a hierarchical folding order in which the parts of the PUFs that are folded are all native-like no longer holds.
It is striking that all four PUFs identified reside on folding and unfolding routes that differ from the one that links native and globally unfolded apoflavodoxin and that at least three of these PUFs are partially misfolded. Both observations probably are related because partially misfolded intermediates need to unfold to continue the route to the native state.
Rugged Folding Energy Landscape of Apoflavodoxin.
The schematic energy landscape presented in Fig. 5c is consistent with the experimental apoflavodoxin equilibrium and kinetic folding data (13) as well as with the H/D exchange data presented here. However, the two-dimensional model is most likely an oversimplification of reality. In Fig. 5d an alternative representation of apoflavodoxin’s folding energy landscape is shown in which the known minima and the likely folding trajectories are presented. Productive apoflavodoxin folding proceeds along the solid lines that link U with N via the high-energy intermediate I2. No direct folding from I1 to N is observed (13). This kinetic mechanism for folding is shared by proteins with the flavodoxin-like topology (14). PUF3 and PUF4 have free energy values and denaturant accessibilities that position these species in a region between U and I1 in Fig. 5d. PUF3 and PUF4 thus could be PUFs of apoflavodoxin’s relatively stable, long-living folding intermediate I1. This hypothesis is supported by kN-PUF3 and kN-PUF4 being identical within error to kN–I1 and by kPUF3-N and kPUF4-N being slightly larger than kI1–N (Table 1). However, although unlikely, PUF3 and PUF4 could be rare unfolding excursions that start directly from native apoflavodoxin (Fig. 5d).
Conclusion
A detailed view has been obtained of the folding energy landscape of A. vinelandii apoflavodoxin. The energy landscape is mapped by denaturant-induced equilibrium unfolding, stopped-flow folding and unfolding, and H/D exchange data under both EX2 and EX1 conditions. The PUFs of apoflavodoxin detected with H/D exchange are partially misfolded and all turn out to be off the protein’s productive folding route. Apoflavodoxin folding involves a search of many local minima in its complex folding-energy landscape. The search includes a population of a stable intermediate. Apparently, this search is characteristic for the folding of the many proteins that share the flavodoxin-like topology.
Methods
Monomeric uniformly 15N-labeled C69A A. vinelandii apoflavodoxin was obtained as described in refs. 11, 13, and 30. Lyophilized apoflavodoxin was dissolved in D2O containing 100 mM potassium pyrophosphate and various GuDCl concentrations ranging from 0 to 750 mM and KCl in various amounts to obtain a final salt concentration (i.e., GuDCl + KCl) of 750 mM. The final concentrations of protein are ±1 mM. Samples were immediately transferred into an AMX500 NMR spectrometer (Bruker, Rheinstetten, Germany) that operates at a proton frequency of 500.13 MHz. Subsequently, a series of gradient-enhanced 1H-15N Heteronuclear single-quantum coherence spectra (31, 32) were recorded to detect H/D exchange. The dead-time was ≈5 min, and the temperature in the probe was 25°C. pD was measured after exchange was complete. The spectra were processed, and a single-exponential function was fitted to the time-dependent peak intensities to obtain kex. Further details are provided in Supporting Experimental Procedures, together with the theory behind H/D exchange and Table 2.
Supplementary Material
Acknowledgments
We thank J. P. Mackay, H. Lill, and I. E. Sánchez for constructive comments.
Abbreviations
- GuDCl
deuterated guanidinium chloride
- pD
pH meter reading of a deuterated solution
- PUF
partially unfolded form
- N
native apoflavodoxin
- U
unfolded protein.
Footnotes
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Bryngelson J. D., Onuchic J. N., Socci N. D., Wolynes P. G. Proteins. 1995;21:167–195. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- 2.Dill K. A., Chan H. S. Nat. Struct. Biol. 1997;4:10–19. doi: 10.1038/nsb0197-10. [DOI] [PubMed] [Google Scholar]
- 3.Dobson C. M. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 4.Bai Y., Sosnick T. R., Mayne L., Englander S. W. Science. 1995;269:192–197. doi: 10.1126/science.7618079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu R., Pei W., Takei J., Bai Y. Biochemistry. 2002;41:7998–8003. doi: 10.1021/bi025872n. [DOI] [PubMed] [Google Scholar]
- 6.Englander S. W., Mayne L., Rumbley J. N. Biophys. Chem. 2002;101–102:57–65. doi: 10.1016/s0301-4622(02)00190-4. [DOI] [PubMed] [Google Scholar]
- 7.Hilser V. J., Dowdy D., Oas T. G., Freire E. Proc. Natl. Acad. Sci. USA. 1998;95:9903–9908. doi: 10.1073/pnas.95.17.9903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sadqi M., Casares S., Abril M. A., Lopez-Mayorga O., Conejero-Lara F., Freire E. Biochemistry. 1999;38:8899–8906. doi: 10.1021/bi990413g. [DOI] [PubMed] [Google Scholar]
- 9.Vendruscolo M., Paci E., Dobson C. M., Karplus M. J. Am. Chem. Soc. 2003;125:15686–15687. doi: 10.1021/ja036523z. [DOI] [PubMed] [Google Scholar]
- 10.Clarke J., Itzhaki L. S., Fersht A. R. Trends Biochem. Sci. 1997;22:284–287. doi: 10.1016/s0968-0004(97)01087-6. [DOI] [PubMed] [Google Scholar]
- 11.van Mierlo C. P. M., van Dongen W. M. A. M., Vergeldt F., van Berkel W. J. H., Steensma E. Protein Sci. 1998;7:2331–2344. doi: 10.1002/pro.5560071110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Mierlo C. P. M., van den Oever J. M. P., Steensma E. Protein Sci. 2000;9:145–157. doi: 10.1110/ps.9.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bollen Y. J. M., Sánchez I. E., Van Mierlo C. P. M. Biochemistry. 2004;43:10475–10489. doi: 10.1021/bi049545m. [DOI] [PubMed] [Google Scholar]
- 14.Bollen Y. J. M., van Mierlo C. P. M. Biophys. Chem. 2005;114:181–189. doi: 10.1016/j.bpc.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 15.Bollen Y. J. M., Nabuurs S. M., van Berkel W. J. H., van Mierlo C. P. M. J. Biol. Chem. 2005;280:7836–7844. doi: 10.1074/jbc.M412871200. [DOI] [PubMed] [Google Scholar]
- 16.Woodward C. K., Hilton B. D. Biophys. J. 1980;32:561–575. doi: 10.1016/S0006-3495(80)84990-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woodward C., Simon I., Tuchsen E. Mol. Cell. Biochem. 1982;48:135–160. doi: 10.1007/BF00421225. [DOI] [PubMed] [Google Scholar]
- 18.Kraulis P. J. J. Appl. Cryst. 1991;24:946–950. [Google Scholar]
- 19.Alagaratnam S., van Pouderoyen G., Pijning T., Dijkstra B. W., Cavazzini D., Rossi G. L., Van Dongen W. M., van Mierlo C. P., van Berkel W. J., Canters G. W. Protein Sci. 2005;14:2284–2295. doi: 10.1110/ps.051582605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steensma E., van Mierlo C. P. M. J. Mol. Biol. 1998;282:653–666. doi: 10.1006/jmbi.1998.2045. [DOI] [PubMed] [Google Scholar]
- 21.Reimer U., Scherer G., Drewello M., Kruber S., Schutkowski M., Fischer G. J. Mol. Biol. 1998;279:449–460. doi: 10.1006/jmbi.1998.1770. [DOI] [PubMed] [Google Scholar]
- 22.Maity H., Maity M., Krishna M. M., Mayne L., Englander S. W. Proc. Natl. Acad. Sci. USA. 2005;102:4741–4746. doi: 10.1073/pnas.0501043102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Myers J. K., Pace C. N., Scholtz J. M. Protein Sci. 1995;4:2138–2148. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arrington C. B., Robertson A. D. J. Mol. Biol. 2000;296:1307–1317. doi: 10.1006/jmbi.2000.3536. [DOI] [PubMed] [Google Scholar]
- 25.Canet D., Last A. M., Tito P., Sunde M., Spencer A., Archer D. B., Redfield C., Robinson C. V., Dobson C. M. Nat. Struct. Biol. 2002;9:308–315. doi: 10.1038/nsb768. [DOI] [PubMed] [Google Scholar]
- 26.Hoang L., Bedard S., Krishna M. M., Lin Y., Englander S. W. Proc. Natl. Acad. Sci. USA. 2002;99:12173–12178. doi: 10.1073/pnas.152439199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yan S., Kennedy S. D., Koide S. J. Mol. Biol. 2002;323:363–375. doi: 10.1016/s0022-2836(02)00882-3. [DOI] [PubMed] [Google Scholar]
- 28.Krieger F., Fierz B., Bieri O., Drewello M., Kiefhaber T. J. Mol. Biol. 2003;332:265–274. doi: 10.1016/s0022-2836(03)00892-1. [DOI] [PubMed] [Google Scholar]
- 29.Feng H., Zhou Z., Bai Y. Proc. Natl. Acad. Sci. USA. 2005;102:5026–5031. doi: 10.1073/pnas.0501372102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steensma E., Nijman M. J. M., Bollen Y. J. M., de Jager P. A., van den Berg W. A. M., van Dongen W. M. A. M., van Mierlo C. P. M. Protein Sci. 1998;7:306–317. doi: 10.1002/pro.5560070210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palmer A. G., III, Cavanagh J., Wright P. E., Rance M. J. Magn. Res. 1991;93:151–170. [Google Scholar]
- 32.Kay L. E., Keifer P., Saarinen T. J. Am. Chem. Soc. 1992;114:10663–10665. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.