Abstract
The versatility of RNA-dependent RNA polymerases (RDRPs) in eukaryotic gene silencing is perhaps best illustrated in the kingdom Fungi. Biochemical and genetic studies of Schizosaccharomyces pombe and Neurospora crassa show that these types of enzymes are involved in a number of fundamental gene-silencing processes, including heterochromatin regulation and RNA silencing in S. pombe and meiotic silencing and RNA silencing in N. crassa. Here we show that Aspergillus nidulans, another model fungus, does not require an RDRP for inverted repeat transgene (IRT)-induced RNA silencing. However, RDRP requirements may vary within the Aspergillus genus as genomic analysis indicates that A. nidulans, but not A. fumigatus or A. oryzae, has lost a QDE-1 ortholog, an RDRP associated with RNA silencing in N. crassa. We also provide evidence suggesting that 5′ → 3′ transitive RNA silencing is not a significant aspect of A. nidulans IRT-RNA silencing. These results indicate a lack of conserved kingdom-wide requirements for RDRPs in fungal RNA silencing.
RNA silencing refers to a group of very similar post-transcriptional gene-silencing mechanisms that have been discovered in a diverse range of eukaryotes (Pickford et al. 2002; Denli and Hannon 2003; Tang et al. 2003). The core processes of RNA silencing are highly conserved and involve double-stranded RNA (dsRNA) processing by an RNAse III domain-containing enzyme (Dicer) into 21- to 26-nt small interfering RNAs (siRNAs; Bernstein et al. 2001), which are then incorporated into a ribonucleoprotein complex (RNA-induced silencing complex, RISC). RISC recognizes and degrades target mRNAs by complementary base pairing to the incorporated siRNA (Hammond et al. 2000; Elbashir et al. 2001). An essential protein member of RISC is an argonaute family protein with a PAZ and PIWI domain (PPD; Carmell et al. 2002). Examples include Rde-1 in Caenorhabditis elegans (Tabara et al. 1999), dAgo2 in Drosophila melanogaster (Hammond et al. 2001), Ago1 in Arabidopsis thaliana (Fagard et al. 2000), Ago1 in Schizosaccharomyces pombe (Volpe et al. 2002), and QDE-2 in Neurospora crassa (Catalanotto et al. 2002). Recent evidence suggests that the PAZ domain of argonaute proteins facilitates transfer of siRNAs to the RISC complex (Lingel et al. 2003; Yan et al. 2003) and that the PIWI domain contains the nuclease activity responsible for siRNA-guided mRNA cleavage (Song et al. 2004).
In some organisms, RNA-dependent RNA polymerases (RDRPs) are essential components of RNA silencing (e.g., protists, nematodes; Smardon et al. 2000; Sijen et al. 2001; Martens et al. 2002; Simmer et al. 2002), while in others RDRPs appear to be dispensable for this process (e.g., flies, mammals; Schwarz et al. 2002; Stein et al. 2003). In plants and fungi, the roles of RDRPs in RNA silencing are not as well defined. For example, the model plant A. thaliana encodes six putative RDRPs and thus far only two have been partially investigated. Of these two RDRPs, SGS2/SDE1 is required for RNA silencing activated by sense transgenes (Beclin et al. 2002), but not for RNA silencing activated by inverted repeat transgenes (IRTs) or RNA viruses (Dalmay et al. 2000; Beclin et al. 2002; Muangsan et al. 2004), and AtRdRP1 is involved in viral defense (Yu et al. 2003; Yang et al. 2004).
Studies of fungal RDRPs suggest that these enzymes are involved in RNA silencing and a number of other gene-silencing-related processes in fungi. For example, the S. pombe RDRP, Rdp1, is required for RNA silencing induced by IRTs (IRT-RNA silencing) and for RNAi-dependent heterochromatin formation at centromeric regions, mating-type loci, and euchromatic regions (Volpe et al. 2002, 2003; Schramke and Allshire 2003; Jia et al. 2004; Verdel et al. 2004). While it is currently unknown why the process of IRT-RNA silencing requires an RDRP in S. pombe, current models suggest that RNAi-dependent heterochromatin formation requires Rdp1 to create, directly or indirectly, small RNAs used to direct a complex of proteins, referred to as RNA-induced initiation of transcriptional gene silencing (RITS) proteins, to specific chromatin regions (Volpe et al. 2002, 2003; Schramke and Allshire 2003; Verdel et al. 2004).
In the filamentous fungus N. crassa, there are two gene silencing processes that require two of three N. crassa RDRPs (Galagan et al. 2003). The first is N. crassa quelling, a type of RNA silencing that is thought to be related to high transgene number (Pickford et al. 2002; Forrest et al. 2004). This process requires the RDRP QDE-1 (Cogoni and Macino 1999a). In vitro studies of QDE-1 activity indicate that it produces both full-length complementary RNA (cRNA) and 9- to 21-nt cRNAs along the length of single-stranded RNA templates (Makeyev and Bamford 2002), suggesting the possibility that QDE-1 creates dsRNA for processing by Dicer or directly forms siRNAs for incorporation into RISC during quelling (Makeyev and Bamford 2002). Such activities may be unnecessary when RNA silencing is activated by IRTs, which may explain the recent finding that QDE-1 is dispensable for IRT-RNA silencing (Catalanotto et al. 2004). The second N. crassa gene-silencing process requiring an RDRP is meiotic silencing by unpaired DNA (MSUD; Shiu et al. 2001; Shiu and Metzenberg 2002). This process requires the RDRP SAD-1 (Shiu et al. 2001; Lee et al. 2003). A third N. crassa RDRP, RRP-3, has not yet been attributed with a function. Phylogenetic analysis suggests RRP-3 is not part of the quelling or MSUD pathways (Galagan et al. 2003; Borkovich et al. 2004) and biochemical studies suggests that it is not involved in DNA methylation or heterochromatin formation (Freitag et al. 2004b).
The zygomycete Mucor circinelloides may encode an RDRP with an important role in transitive RNA silencing (Nicolas et al. 2003). This process, more thoroughly investigated in plants (Vaistij et al. 2002; Van Houdt et al. 2003) and nematodes (Sijen et al. 2001), forms dsRNA/siRNAs from sequences upstream (3′ → 5′) and/or downstream (5′ → 3′) of primary target sequences on targeted mRNA, leading to the creation of secondary siRNAs and the spreading of RNA silencing (Denli and Hannon 2003). In M. circinelloides these secondary siRNAs have been detected, but a specific RDRP has yet to be identified (Nicolas et al. 2003).
Recently, a clear dissimilarity in fungal RDRP function became apparent when examination of a N. crassa strain devoid of all its RDRPs showed that, unlike S. pombe Rdp1 mutants, it was not affected in DNA methylation or heterochromatin silencing (Freitag et al. 2004b). Here, in addition to reporting that IRTs efficiently silence homologous mRNAs in the model filamentous fungus Aspergillus nidulans, we report that dissimilarity in fungal RDRP function is also observed in the process of IRT-RNA silencing. Comparative analysis of all predicted RDRPs in the three sequenced Aspergilli revealed that A. nidulans encodes two RDRPs and, in contrast to the related species A. fumigatus and A. oryzae, has lost an ortholog of N. crassa QDE-1. Deletion of the remaining two A. nidulans RDRPs had no detectable effect upon IRT-RNA silencing while deletion of a putative PPD protein, named RsdA, disrupted this process. Possible reasons to account for the apparent difference in a RDRP requirement for IRT-RNA silencing in S. pombe and A. nidulans are discussed.
MATERIALS AND METHODS
Strains, growth conditions, and transformation conditions:
All strains used in this study are listed in Table 1. A. nidulans RJH0128 was transformed with aflR-specific IRTs and control transgenes using the method described by Yu and Adams (1999). Standard crossing techniques (Pontecorvo 1953) were then used to introduce different auxotrophic markers from A. nidulans RDIT1.1 into the aflR(IRT) and aflR single-sense transgene (SST) transformants. Gene replacements were confirmed by Southern blotting with probes specific for the internal deleted region and at least one flanking region of the deleted gene. A. nidulans RTMH13.C5 (for rsdA) and A. nidulans RTMH13.F5 (for rrpB and rrpC) were used as the hosts for gene replacements. A. nidulans cultures were grown in 25 ml of appropriate supplemented liquid or solid minimal media or oatmeal agar as previously described (Butchko et al. 1999). All strains were cultured under dark, stationary conditions at 37° in standard Petri dishes. Liquid cultures were inoculated with ∼1 × 106 spores/ml and solid cultures were point inoculated with freshly harvested conidia.
TABLE 1.
A. nidulans strains used in this study
| Fungal straina | Genotype | Source |
|---|---|---|
| RJH0128 | biA1; trpC801; ΔstcE::argB; wA2 | J. K. Hicks and N. P. Keller (unpublished data) |
| TTMH13.1 | biA1; aflR(IRT1300)::trpC; ΔstcE::argB; wA2 | This study |
| TTMH13.2 | biA1; ΔstcE::argB; wA2 | This study |
| TTMH20.8 | biA1; aflR(IRT900)::trpC; ΔstcE::argB; wA2 | This study |
| TTMH20.9 | biA1; ΔstcE::argB; wA2 | This study |
| TTMH16.9 | biA1; aflR(SST1300)::trpC; ΔstcE::argB; wA2 | This study |
| RDIT1.1 | pyrG89; argB2; metG1 | Tsitsigiannis et al. (2004) |
| RTMH13.B1 | aflR(IRT1300)::trpC; ΔstcE::argB | This study |
| RTMH13.B3 | ΔstcE::argB | This study |
| RTMH13.C5 | aflR(IRT1300)::trpC; ΔstcE::argB; pyrG89; wA | This study |
| RTMH13.F5 | aflR(IRT1300)::trpC; ΔstcE::argB; pyrG89; metG1 | This study |
| TTMH65.1 | aflR(IRT1300)::trpC; ΔstcE::argB; ΔrsdA::pyrG; pyrG89; wA | This study |
| TTMH74.12 | aflR(IRT1300)::trpC; ΔstcE::argB; ΔrrpB::pyrG; pyrG89; metG1 | This study |
| TTMH75.17 | aflR(IRT1300)::trpC; ΔstcE::argB; pyrG89; ΔrrpC::metG; metG1 | This study |
| RTMH7475.1A | aflR(IRT1300)::trpC; ΔstcE::argB; ΔrrpB::pyrG; ΔrrpC::metG | This study |
| RTMH7475.3A | ΔstcE::argB; ΔrrpB::pyrG; ΔrrpC::metG | This study |
| AAH16 | ΔmusN::pyrG; pabaA1; yA2 | Hofmann and Harris (2001) |
| RTMH13.D9 | aflR(IRT1300)::trpC; ΔstcE::argB; ΔmusN::pyrG | This study |
| RTMH13.D16 | ΔstcE::argB; ΔmusN::pyrG | This study |
Strains starting with T are original transformants. Strains starting with R are recombinants. All strains contain the veA1 allele (Mooney et al. 1990).
Vector construction:
Oligonucleotides used in vector construction are listed in Table 2.
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence | Purpose |
|---|---|---|
| aflh5-Mod | 5′-CATATGCAAGCTTCATGGAGC-3′ | aflR(IRT) |
| aflr3-BamHI | 5′-AAGGATCCGAGGCGTGGCGG-3′ | aflR(IRT/SST) |
| afln5-NcoI | 5′-TTTCCATGGAGCCCCAGCGA-3′ | aflR(IRT/SST) |
| 5gfp | 5′-ATGACCTAGGGCTATGTGCAGGAG-3′ | aflR(IRT) |
| 3gfp-BamHI | 5′-GCACGGATCCATCCTCAATGTTG-3′ | aflR(IRT) |
| 5gfp-SacII | 5′-CATGACCGCGGACTATGTGCAG-3′ | aflR(IRT) |
| 3gfp | 5′-GCACGGCGCCATCCTCAATG-3′ | aflR(IRT) |
| AnQDE2-5fl5 | 5′-TTGTGGTACCGAGTCTGCCGACCGTGAAATC-3′ | rsdA replacement vector |
| AnQDE2-5fl3 | 5′-CTGATCCAAGCTTCCACCACCACCG-3′ | rsdA replacement vector |
| AnQDE2-3fl5 | 5′-CTGAGCGAATTCACTCATTTCATTAGCTC-3′ | rsdA replacement vector |
| AnQDE2-3fl3 | 5′-CCTTTGGATCCCCTAACTCGGCTTCCCTGG-3′ | rsdA replacement vector |
| AnRDRP1-5f5 | 5′-CGTCAGGTACCTCCAAGTGTTTATACTGCG-3′ | rrpB replacement vector |
| AnRDRP1-5f3 | 5′-TCAAAAAGCTTCCTTGCCTGCACCTC-3′ | rrpB replacement vector |
| AnRDRP1-3f5 | 5′-AAACGAATTCCGGCACAGCACGAGATC-3′ | rrpB replacement vector |
| AnRDRP1-3f3 | 5′-TTTGGATCCAGTTCCTTGCGAAAACCCATATCCC-3′ | rrpB replacement vector |
| AnRDRP2-5f5 | 5′-CAAAGCAACTCGAGCCACCGCAAACC-3′ | rrpC replacement vector |
| AnRDRP2-5f3 | 5′-TAAGTAAAGCTTCTCCCCGTTCTCATCGCAG-3′ | rrpC replacement vector |
| AnRDRP2-3f5 | 5′-CCCCGGGATCCCTAGCCAAGATC-3′ | rrpC replacement vector |
| AnRDRP2-3f3 | 5′-GTCTTCTAGATACACCCACCGCCTAGTCTG-3′ | rrpC replacement vector |
| aflR3END5 | 5′-GTCGCCATGGCTCGGGATAGGTC-3′ | Sense (S) oligo (Figure 3A) |
| aflR3END3B | 5′-GGCGACGGCTTACCTGAGTCACCAG-3′ | Antisense (AS) oligo (Figure 3B) |
pTMH13.7 [also referred to as aflR(IRT1300)]:
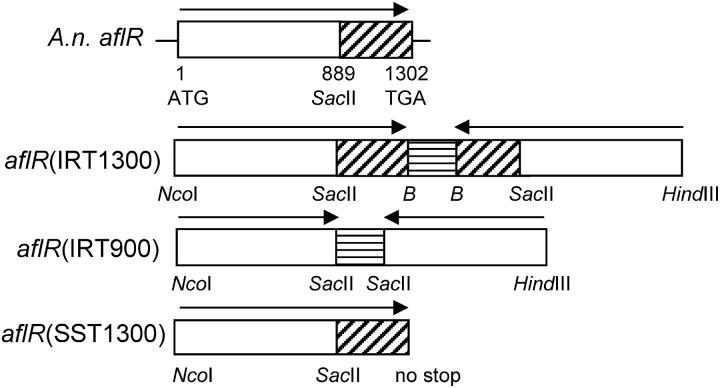
This transformation vector consists of an inverted repeat of two ∼1300-bp aflR fragments (Figure 1) separated by an ∼280-bp spacer, driven by the A. nidulans gpdA promoter and terminated by the A. nidulans trpC terminator (Punt et al. 1991). It also contains a truncated A. nidulans trpC selectable marker for targeted integration of the IRT next to the A. nidulans trpC locus (Mullaney et al. 1985). Oligonucleotides aflh5-mod and aflr3-BamHI were used to amplify an ∼1300-bp aflR fragment (5′ HindIII-aflR), containing the full-length A. nidulans aflR coding sequence. This PCR product was cloned into the EcoRV site of pBluescript II SK− (pBS, Stratagene, La Jolla, CA) to create pTMH2.3. Oligonucleotides afln5-NcoI and aflr3-BamHI were then used to amplify a similar aflR fragment with a different restriction site in the 5′ primer (5′ NcoI-aflR). This PCR product was cloned into the SmaI site of pBS to create pTMH3.3. Using multiple cloning steps, these two ∼1300-bp aflR fragments were then placed in an inverted orientation on opposite sides of a ∼280-bp spacer (gf1), creating plasmid pTMH8.7. The gf1 spacer was amplified from pPRgf-T4 (Zolotukhin et al. 1996) with oligonucleotides 5gfp and 3gfp-BamHI. The aflR IRT was then released from pTMH8.7 and cloned between the NcoI and HindIII sites of the high-expression vector pAN52-3 (gi:474929), creating pTMH10.1. Finally, a ∼2400-bp 5′ EcoRI fragment of A. nidulans trpC was cloned into the EcoRI site of pTMH10.1 to give the aflR(IRT1300) transgene (Figure 1).
Figure 1.—
IRT and SST constructs of A. nidulans aflR. (Top) A diagram of the 1302-bp A. nidulans aflR coding sequence from the ATG start site to the TGA stop site is shown. A SacII site at position 889 is indicated. The region 3′ to the SacII site was used to make riboprobes for aflR mRNA and siRNA analysis (diagonal hatching). The aflR(IRT1300) transgene consists of two complete aflR coding regions in an inverted orientation, separated by an ∼280-bp spacer fragment (horizontal hatching). B, BamHI site. The aflR(IRT900) transgene consists of two identical truncated fragments of aflR coding region in an inverted orientation, separated by an ∼280-bp spacer fragment. This is essentially the same IRT as aflR(IRT1300), except that aflR sequences between the SacII sites have been removed. The aflR(SST1300) transgene contains the complete open reading frame of aflR with a mutated stop codon. All three aflR(IRT) and aflR(SST) transgenes are flanked by the A. nidulans gpdA promoter and trpC terminator (not shown).
pTMH20.7 [also referred to as aflR(IRT900)]:
This transformation vector differs from pTMH13.7 only in that it consists of two ∼900-bp aflR fragments instead of two ∼1300-bp aflR fragments. Oligonucleotides 5gfp-SacII and 3gfp were used to amplify an ∼280-bp fragment of GFP from pPRgf-T4. This fragment (gf2) was then cloned into the SmaI site of pBS, giving a plasmid containing a SacII-gf2-SacII fragment (pTMH14.8). Plasmid pTMH10.1 was then digested with SacII, thereby releasing ∼1100 bp from the center of the aflR IRT, including ∼400 bp from the 3′ end of each aflR fragment. This fragment was replaced with the SacII-gf2-SacII fragment from pTMH14.8 to give pTMH17.2. As described above, a trpC selectable marker was cloned into the EcoRI site of pTMH17.2 to give the aflR(IRT900) transgene (Figure 1).
pTMH16.3 [also referred to as aflR(SST1300)]:
This transformation vector contains a single full-length aflR ORF, in a sense orientation between the gpdA promoter and trpC terminator of pAN52-3; thus it is referred to as a SST to distinguish it from the inverted repeat nature of the IRTs. Plasmid pTMH10.1 was digested with BamHI and HindIII to release an ∼1600-bp BamHI-gf1-Rfla-HindIII fragment. The BamHI and HindIII ends of the digested plasmid were filled in with Klenow DNA polymerase (New England Biolabs, Beverly, MA) and ligated together to give pTMH12.1. As described above, a trpC selectable marker was cloned into the EcoRI site of pTMH12.1 to complete the aflR(SST1300) transgene (Figure 1).
Gene replacement vectors:
DNA flanking regions were cloned from A. nidulans genomic DNA for creation of rsdA, rrpB, and rrpC gene replacement vectors, using the oligonucleotides listed in Table 2. Oligonucleotide restriction sites were used to place the flanking regions into the matching sites in pBS. A. parasiticus pyrG (Skory et al. 1990; for rsdA and rrpB) or A. nidulans metG (Sienko and Paszewski 1999; for rrpC) selectable markers were placed between the flanking DNA and the resulting plasmids were used in A. nidulans transformations.
Northern hybridizations:
Total RNA analysis:
Trizol reagent (Invitrogen, Carlsbad, CA) was used to isolate total RNA from lyophilized A. nidulans cultures per manufacturer's instructions. RNA was then blotted to Hybond-XL (Amersham, San Francisco). Ambion's (Austin, TX) Maxiscript kit was used to make a 3′ aflR sense-specific α-32P-labeled riboprobe (Figure 1, diagonal hatching) for hybridization to total RNA.
Low-molecular-weight (MW) RNA analysis:
Low-MW RNA was isolated as described (Catalanotto et al. 2002). This was separated in a denaturing gel, blotted to a Hybond-XL nylon membrane, and hybridized to a riboprobe as described (Nicolas et al. 2003), except that Denhardt's reagent was excluded from the hybridization buffer. The 3′ sense and antisense aflR-specific riboprobes (Figure 1) were prepared as described above, except that before hybridization the probe was hydrolyzed into ∼50-nt fragments as described (Hamilton and Baulcombe 1999).
Norsolorinic acid analysis:
From liquid cultures:
Twenty-five-milliliter A. nidulans cultures were mixed with an equal volume of acetone. The mixture was slightly agitated for 1 hr and then 7.5 ml was transferred to a new container, which was then shaken vigorously with an equal volume of CHCl3. The mixture was allowed to separate and a 5-ml aliquot of the CHCl3 layer was transferred to a new tube, evaporated, and redissolved in 200 μl of CHCl3. Five-microliter aliquots from each sample were then loaded on a TLC plate (no. 4410221; Whatman, Brentford, UK). Compounds were separated using a toluene:ethyl-acetate:acetic acid (80:10:10) solvent system.
From solid media cultures:
A single 1.4-cm-diameter core was removed from the center of a 6-day-old colony, ground in 3 ml of 50% acetone, and then mixed with 1.5 ml of CHCl3. A 1-ml aliquot of the CHCl3 layer was then transferred to a new tube and evaporated. The residual compounds were redissolved in 20 ul of CHCl3 and analyzed by TLC as described above.
Computer-based analyses and database searches:
Table 4 lists accession numbers for N. crassa, S. pombe, Magnaporthe grisea, and Aspergillus genes used in this study. Putative Aspergillus RDRPs were identified by a search (blastp and tblastn) of the A. nidulans, A. fumigatus, and A. oryzae genome databases with RDRP sequences from N. crassa and the conserved domain for eukaryotic RDRPs (pfam no. 05183.5). Putative A. nidulans PPD proteins and RecQ DNA helicases were identified by searching the same databases (blastp) with N. crassa QDE-2 and QDE-3 sequences. Specific database websites are listed in the acknowledgments section. Similarity comparisons (Clustal W), phylogenetic trees (DrawTree), and ORF analysis (SixFrame) were performed on the SDSC Biology Workbench (http://workbench.sdsc.edu). A search for a conserved RDRP motif (DbDGD) in the putative degenerated A. nidulans rrpA locus was performed by pasting amino acid translations of this locus in all six reading frames into Microsoft Word and visually scanning the document with the aid of the program for the conserved motif.
TABLE 4.
Identification numbers for protein and nucleotide sequences used in this study
| Branch no. | Gene | Identification no.a |
|---|---|---|
| 1 | M.g. rrp1 | MG07682.4 |
| 2 | N.c. qde-1 | 28920432,b 4803727c |
| 3 | A.n. degen. RrpA | See belowd |
| 4 | A.o. rrpA | 20113.m00115 |
| 5 | A.f. rrpA | 71.m15624 |
| 6 | A.n. actin | 168004 |
| 7 | A.o. rrpB | 20166.m00248 |
| 8 | A.n. rrpB | An4790.2 |
| 9 | A.f. rrpB | 59.m09193 |
| 10 | M.g. rrp2 | MG02748.4 |
| 11 | N.c. sad-1 | 28926016,b 13699900c |
| 12 | A.o. rrpC | 20139.m00205 |
| 13 | A.n. rrpC | An2717.2 |
| 14 | N.c. rrp-3 | 32423631,b 32418006c |
| 15 | M.g. rrp3 | MG06205.4 |
| 16 | A.n. dewA | 533424 |
| 17 | A.n. stcA | 1235618 |
| 18 | A.n. pkaR | 3170247 |
| 19 | A.n. cyaA | 21326181 |
| 20 | A.n. nimP | 4102989 |
| 21 | A.n. ppoA | 40715887 |
| 22 | A.n. yA | 2425 |
| 23 | A.n. aflR | 1235618 |
| 24 | A.n. actin | 168004 |
| 25 | N.c. qde-2 | 7248732 |
| 26 | N.c. qde-3 | 6934277 |
| 27 | A.n. rsdA | 40744619 |
| 28 | S.p. rdp1 | 2330856 |
| 29 | A.n. musN | 14039839 |
| 30 | N.c. sms-2 | 23452214 |
| 31 | A.n. smsA | 40741781 |
| 32 | N.c. recQ-2 | 32403964 |
Aspergillus and Magnaporthe RDRP nucleotide and amino acid data were obtained from the Broad Institute, TIGR, and the National Institute of Advanced Industrial Science and Technology (see acknowledgements for web site information) and the sequences are accessible using the given identification numbers. All other genes are listed with their GenBank accession numbers.
Nucleotide sequence.
Amino acid sequence.
The ∼4.0-kb A. nidulans degenerate rrpA sequence can be obtained from the A. nidulans genome database (www.broad.mit.edu) contig 1.161 (position 256,699–260,649).
RESULTS
IRTs correlate with inhibition of gene expression in A. nidulans:
To test if RNA silencing exists in A. nidulans, two different IRTs were designed with A. nidulans aflR sequences (Figure 1, top). AflR is a transcription factor required for the biosynthesis of a bright orange compound, norsolorinic acid (NOR), in A. nidulans ΔstcE strains (Butchko et al. 1999). Thus loss of aflR expression leads to loss of NOR production. Transformation of A. nidulans RJH0128 with aflR(IRT1300) or aflR(IRT900) (Figure 1) resulted in a few transformants that did not produce NOR (Figure 2A). Southern analysis indicated that the NOR− phenotype correlated 100% with the successful integration of an aflR(IRT) into the genome and that in each case the endogenous aflR locus was not affected (Figure 2, A and B). The NOR− phenotype was stable in time course experiments (Figure 2C) and through sexual crosses (data not shown). In contrast, all 23 transformants resulting from transformation of A. nidulans RJH0128 with aflR(SST1300) (Figure 1) were NOR+ (data not shown).
Figure 2.—
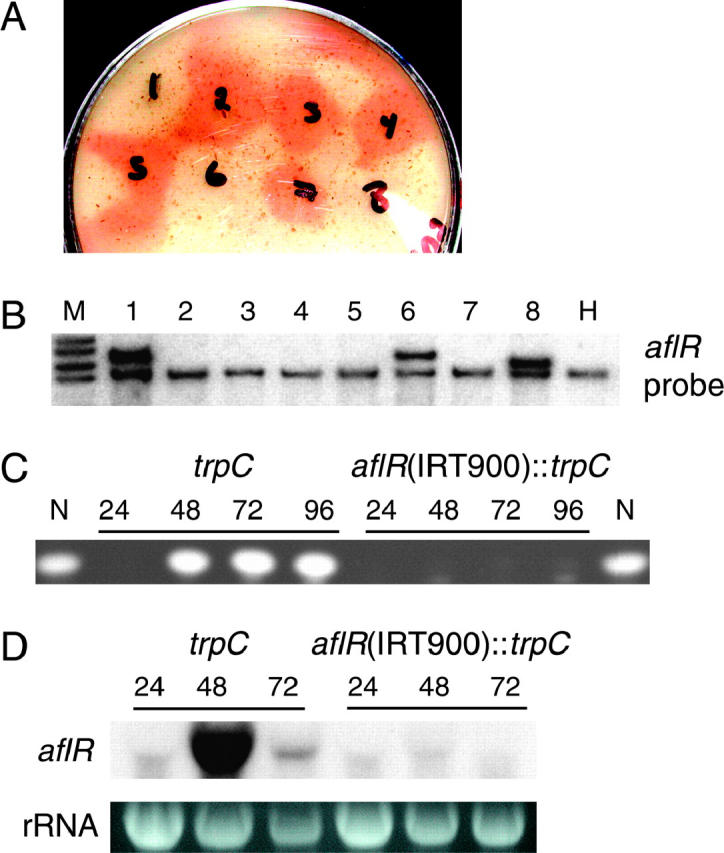
A. nidulans aflR IRTs suppress NOR production and aflR expression. (A) A random selection of NOR+ and NOR− transformants were point inoculated onto oatmeal medium and cultured for ∼5 days to assay NOR production; aflR(IRT1300) transformants are numbered 1–6 and aflR(IRT900) transformants are numbered 7 and 8. (B) Southern analysis of HindIII-digested genomic DNA from the eight transformants depicted in A and the transformation host strain are shown (lanes 1–H). Lane M shows nonspecific hybridization of the probe to the DNA ladder (top band, 10 kb; bottom band, 6 kb). (C and D) A trpC control transformant (TTMH20.9) and an aflR(IRT900) transformant (TTMH20.8) were cultured in liquid minimal media and analyzed for (C) NOR production and (D) aflR expression over 96 and 72 hr, respectively. The riboprobe was specific for aflR sequences not present in the aflR(IRT900) transgene to detect aflR transcripts derived from the endogenous aflR locus only. N, NOR standard.
To determine if the NOR− phenotype was a result of decreased aflR transcript levels, total RNA extracts were analyzed from two strains resulting from the transformation of RJH0128 with the aflR(IRT900) transgene. A. nidulans TTMH20.9 is a NOR+ transformant that integrated the trpC-selectable marker without the aflR(IRT900) inverted repeat, while A. nidulans TTMH20.8 is a NOR− transformant that integrated both the trpC-selectable marker and the aflR(IRT900) inverted repeat. Analysis of aflR mRNA levels with a riboprobe specific for the 3′ ∼400 bp of aflR (Figure 1, diagonal hatching), a segment of aflR not contained in the aflR(IRT900) transgene, indicated that aflR transcript levels were significantly decreased in TTMH20.8 relative to TTMH20.9 (Figure 2D).
RNA silencing in A. nidulans is characterized by a single class of 25 nt siRNAs:
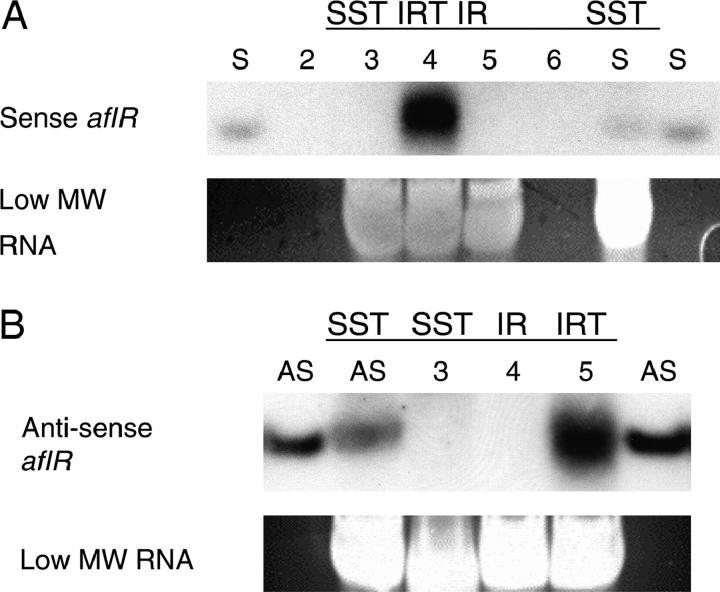
To confirm that RNA silencing is the mechanism responsible for the IRT-induced decrease in aflR mRNA, low-molecular-weight RNAs were analyzed for the presence of aflR-specific siRNAs. Using sense- and anti-sense-specific riboprobes for the 3′ end of aflR (Figure 1), siRNAs of 25 nt in length were detected in a NOR− strain containing the aflR(IRT1300) transgene (TTMH13.1) but not in a NOR+ strain containing the aflR(SST1300) transgene (TTMH16.9), or in a NOR− strain containing the aflR(IRT900) transgene (TTMH20.8; Figure 3). The absence of siRNAs in TTMH20.8 indicates that A. nidulans RDRPs do not form significant levels of dsRNA or secondary siRNAs from the 3′ region of targeted aflR mRNA during IRT-RNA silencing.
Figure 3.—
A. nidulans IRT-RNA silencing is characterized by a single class of ∼25-nt siRNAs. The following strains were cultured for 72 hr in liquid minimal medium: TTMH16.9, aflR(SST1300); TTMH13.1, aflR(IRT1300); and TTMH20.8, aflR(IRT900); designated SST, IRT, and IR, respectively. A riboprobe specific for (A) aflR sense sequences or (B) antisense aflR sequences (see Figure 1 for aflR region specificity) was hybridized to ∼30 μg of low-molecular-weight (MW) RNAs. Approximately 20 pmol of sense (S) and antisense (AS) aflR oligonucleotides, 23 and 25 nt, respectively (Table 2), was used as a control for the probe. It was also mixed with ∼30 μg of low-MW RNAs from TTMH16.9 as a migration control to more accurately determine the size of A. nidulans siRNAs. Lanes are labeled with the letter of the oligonucleotide used in the particular lane or with the appropriate lane number if an oligonucleotide was not added to the lane. Ethidium bromide staining of the highest-concentration RNA species is shown to demonstrate relative amounts of RNA between lanes.
RNA silencing in A. nidulans requires RsdA, a putative argonaute family protein:
Two putative PPD proteins (e−139 and e−31) were identified in the A. nidulans genome database by searching (blastp) with the predicted N. crassa QDE-2 amino acid sequence. Deleting the highest-matching gene (e−139) resulted in a loss of aflR silencing (Figure 4 and supplementary Figure S1 at http://www.genetics.org/supplemental/) in the aflR(IRT1300) genetic background (Figure 5). This gene was therefore named rsdA, for RNA-silencing-deficient A, and it is likely a RISC complex protein required for RNA silencing. The second match (e−31) may be an ortholog of N. crassa SMS-2, a QDE-2 paralog required for meiotic silencing (Lee et al. 2003). This putative protein (referred to as smsA in Table 4) was also identified during a search of the A. nidulans genome with the predicted N. crassa SMS-2 sequence (e−15). SmsA was not investigated further during this study.
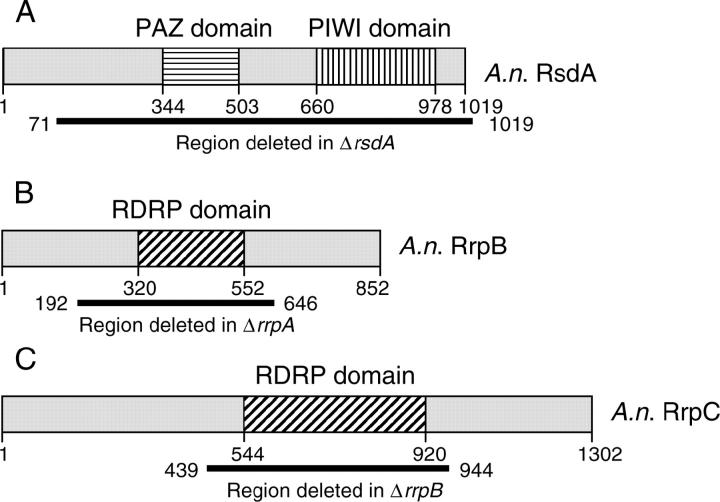
Figure 4.—
A. nidulans rsdA, rrpB, and rrpC. The predicted amino acid sequences of RsdA, RrpB, and RrpC were used to search NCBI's conserved domain database. (A–C) The identified conserved domains are indicated along with the predicted starting and stopping points for each domain, the predicted length of the protein, and the codons deleted in these studies. (A) Predicted PAZ and PIWI domains of A. nidulans RsdA. (B and C) Predicted RDRP domains of RrpB and RrpC, respectively.
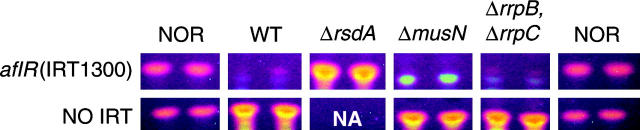
Figure 5.—
Genetic analysis of IRT-RNA silencing in A. nidulans. A. nidulans mutants with (top) or without (bottom) the aflR(IRT1300) transgene were point inoculated onto 25 ml of solid minimal medium and incubated for 6 days. NOR production was analyzed by TLC. Bright orange spots are NOR. NA, not analyzed. Top row, strains are: WT, RTMH13.B1; ΔrsdA, RTMH65.1; ΔmusN, RTMH13.D9; and ΔrrpB ΔrrpC, RTMH7475.1A. Bottom row, strains are: WT, RTMH13.B3; ΔmusN, RTMH13.D16; and ΔrrpB ΔrrpC, RTMH7475.3A.
RNA silencing in A. nidulans does not require A. nidulans MusN:
N. crassa quelling requires QDE-3, a RecQ DNA helicase (Cogoni and Macino 1999b). Therefore we searched for an A. nidulans QDE-3 ortholog in the A. nidulans genome database using the predicted QDE-3 sequence. The most likely ortholog identified during this search is a previously characterized A. nidulans RecQ DNA helicase involved in the DNA damage response, MusN (e−174; Hofmann and Harris 2001). The second-best match (An5092.2) had a relatively low expect value (e−23) and was not investigated further during this study. However, the role of MusN in IRT-RNA silencing was investigated. A significant portion of musN was deleted from the A. nidulans genome (Hofmann and Harris 2001) and the resulting ΔmusN strain (AAH16) was crossed to an aflR(IRT1300) strain (TTMH13.1) to give progeny with ΔmusN in the aflR(IRT1300) genetic background. NOR analysis indicated that loss of musN had no effect upon IRT-RNA silencing (Figure 5). This finding is in agreement with that of Catalanotto et al. (2004), who recently reported that QDE-3 is not required for N. crassa IRT-RNA silencing.
Loss of a QDE-1 ortholog in A. nidulans:
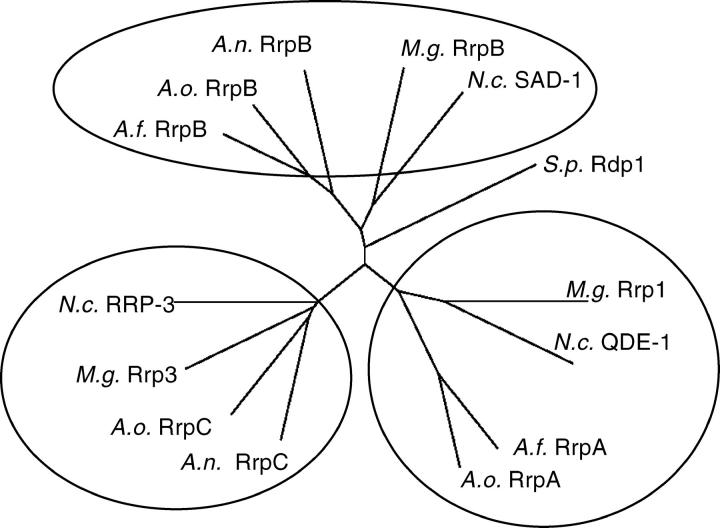
Comparative analysis of all putative RDRPs identified through literature searches (N. crassa and S. pombe) and database searches (A. nidulans, A. fumigatus, A. oryzae, and M. grisea) suggests that these enzymes fall into three distinct classes in filamentous fungi represented by N. crassa QDE-1, SAD-1, and RRP-3 (Figure 6). This is in contrast to the single RDRP in S. pombe, which is most similar to the SAD-1 RDRP class (Galagan et al. 2003 and Figure 6). Surprisingly, while A. oryzae has conserved members of all three classes, there are no orthologs of QDE-1 in the A. nidulans genome database or orthologs of RRP-3 in the A. fumigatus genome database. Analysis of the Aspergilli genomes suggests that the A. nidulans QDE-1 ortholog was lost during evolution (see below).
Figure 6.—
Three classes of RDRPs exist in N. crassa, M. grisea, and Aspergillus species. The neighbor-joining method was used with the predicted amino acid sequences of all known RDRPs and RDRPs identified in this study from N. crassa, S. pombe, M. grisea, and the three sequenced Aspergilli (Table 4) to create this noniterated, unrooted tree. A. nidulans is missing a QDE-1-like RDRP and A. fumigatus is missing a RRP-3-like RDRP. N.c., N. crassa; A.n., A. nidulans; A.o., A. oryzae; A.f., A. fumigatus; M.g., M. grisea; S.p., S. pombe.
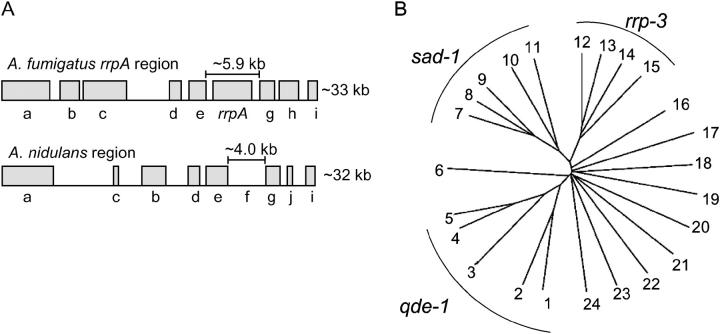
Comparison of the chromosomal region adjacent to the putative A. fumigatus QDE-1 ortholog (RrpA; Galagan et al. 2003) indicates that a high level of synteny exists between this region and a specific chromosomal region in A. nidulans. A section of this synteny is depicted in Figure 7. These two chromosomal fragments are nearly identical except for the presence of a unique ORF in each, a rearrangement of two ORFs, and the absence of a predicted rrpA ortholog in A. nidulans (Figure 7A and Table 3).
Figure 7.—
Genome analysis indicates that A. nidulans rrpA has been lost during evolution. (A) Synteny exists between the A. fumigatus rrpA region and an analogous region in A. nidulans. Generic gene-type predictions were obtained from the draft annotation of the A. nidulans and A. fumigatus genomes and are listed in Table 3. Expect values were obtained by searching (blastp) the A. fumigatus genome database with the predicted amino acid sequences of the corresponding A. nidulans ORF depicted in this diagram. The nucleotide length designations above the A. fumigatus rrpA locus and the analogous region in A. nidulans indicate the approximate numbers of bases between the stop and start sites of the flanking genes. (B) The ∼4.0-kb region of A. nidulans genomic DNA depicted in A was compared to genomic sequences of all known and predicted fungal RDRPs from N. crassa, A. fumigatus, A. oryzae, A. nidulans, and M. grisea and 10 random genes from A. nidulans. Genomic sequences (including introns) between the known or predicted start and stop sites of the genes listed in Table 4 were compared using the neighbor-joining method and are depicted in a noniterated, unrooted tree. Numbers on the branches correspond to the genes listed in Table 4.
TABLE 3.
Gene types near theA. fumigatus rrpA locus and the corresponding locus inA. nidulans
| Letter code | Gene type | Expect valuea |
|---|---|---|
| a | Transporter | e0 |
| c | Transporter | e−57 |
| b | Permease | e−216 |
| d | Reductase | e−187 |
| e | Permease | e−64 |
| rrpA | QDE-1 like RDRP | NA |
| f | No predicted gene | NA |
| g | Clock controlled | e−142 |
| i | Sugar utilization | e−117 |
| h and j | (unique ORFs) | NA |
Expect values were determined by searching (blastp) the A. fumigatus peptide database with the predicted A. nidulans ORFs depicted in Figure 7. NA, not applicable.
Guessing that an A. nidulans QDE-1 ortholog might be undetected in the ∼4.0-kb genomic region corresponding to A. fumigatus rrpA (Figure 7A, f), a search (blastx) of NCBI's protein database was performed with this sequence. No significant matches were identified during this process. Contrastingly, a similar search with the corresponding ∼5.9-kb region spanning the A. fumigatus rrpA locus (Figure 7A, rrpA) identified N. crassa QDE-1 as the most significant match (e−96). This suggests that a RDRP is not present in the ∼4.0-kb region of A. nidulans genomic DNA. Next, the ∼4.0-kb A. nidulans sequence was aligned (Clustal W) with the genomic sequences of 23 predicted or known genes from A. nidulans, A. fumigatus, A. oryzae, and N. crassa and M. grisea. The results indicated that this region is more similar to the sequences of the QDE-1-like RDRPs of all included fungi than to any other sequence included in the analysis, including the genomic DNA of the two predicted A. nidulans RDRPs (Figure 7B and Table 4). This finding is supportive of the hypothesis that this ∼4.0-kb region of DNA constitutes the remnants of an Aspergillus QDE-1 ortholog that has degenerated during evolution. Further support for the hypothesis that this region actually contains a relic RDRP and not a functional RDRP comes from a detailed six-frame analysis of the ∼4.0-kb region. This analysis indicated that a conserved RDRP motif (DbDGD, b is a bulky residue) found in all known RDRPs (Iyer et al. 2003), including the two putative A. nidulans RDRPs, is not present in any of the six frames (data not shown).
IRT-RNA silencing in A. nidulans is independent of RDRPs:
We next tested whether or not the two A. nidulans RDRPs, RrpB and RrpC, are essential for IRT-RNA silencing in A. nidulans. Replacement vectors were designed to eliminate the conserved RDRP domains in both genes (Figure 4 and Figure S1). Transformation of A. nidulans RTMH13.F5 with the RDRP gene replacement vectors resulted in the RDRP single knockouts, TTMH74.12 and TTMH75.17 (Figure S1). Double-RDRP knock-out strains were then obtained by crossing TTMH74.12 and TTMH75.17 (data not shown). Our results indicated that neither the single nor double mutants were compromised in their ability to silence aflR (Figure 5). This finding indicates that A. nidulans RrpB and RrpC are not required for IRT-RNA silencing in A. nidulans.
DISCUSSION
Here we demonstrate that IRTs efficiently induce RNA silencing in A. nidulans in a process that is independent of the two A. nidulans RDRPs, RrpB and RrpC. A third RDRP of the QDE-1 class of fungal RDRPs has apparently degenerated over time through changes in the DNA code. In addition, we have shown that RNA silencing is stable in A. nidulans, is characterized by a significant decrease in mRNA expression, results in the accumulation of a single class of 25-nt siRNA molecules, and requires a PPD protein, RsdA.
Before this report, RDRPs had not been investigated in the Aspergilli or in any other fungi with the exception of S. pombe and N. crassa. Recent studies indicate that RDRPs are involved in various gene-silencing processes in these two fungi (Cogoni and Macino 1999a; Shiu et al. 2001; Volpe et al. 2002; Schramke and Allshire 2003; Verdel et al. 2004), protists (Martens et al. 2002), nematodes (Smardon et al. 2000; Sijen et al. 2001; Simmer et al. 2002), and plants (Dalmay et al. 2000; Beclin et al. 2002; Vaistij et al. 2002; Van Houdt et al. 2003; Muangsan et al. 2004). On the other hand, studies with Drosophila and mammals (Schwarz et al. 2002; Roignant et al. 2003; Stein et al. 2003) suggest RDRPs have no role in gene-silencing processes and, furthermore, are not evolutionary conserved in these species (Stein et al. 2003). Our current findings do not support a required role for an A. nidulans RDRP in IRT-RNA silencing but do not negate a role for these enzymes in other gene-silencing processes. For example, N. crassa SAD-1 is required for MSUD and successful progression through the meiotic cycle (Shiu et al. 2001) and our preliminary results suggest that A. nidulans RrpB may be required for some aspect of outcrossing (T. M. Hammond and N. P. Keller, unpublished results). Certainly, the surprising finding that the three currently sequenced Aspergilli contain three different combinations of RDRP classes has identified a unique opportunity to investigate how these different RDRPs might affect fundamental gene-silencing processes.
The biological significance of possessing a QDE-1 type RDRP is currently unknown. It is possible that the loss of a QDE-1 ortholog in A. nidulans may account for the lack of reported quelling phenotypes in the A. nidulans literature. Furthermore, our use of an aflR(SST) in this and previous studies (Shimizu and Keller 2001; Shimizu et al. 2003) suggests this form of RNA silencing does not exist in A. nidulans. However, given the small number of aflR(SST) transformants examined in this study, the tendency of the transformants to acquire only a single copy of the transgene (data not shown), and the small percentage of transformants that stably display cosuppression phenotypes in N. crassa (Cogoni et al. 1996; Cogoni and Macino 1997), our analysis of “quelling” like RNA silencing in A. nidulans was not thorough enough to firmly conclude that this process does not occur in this fungus.
In vitro analysis of N. crassa QDE-1 indicates that this enzyme can make full-length dsRNA from a single-stranded template but that it preferentially makes small 9- to 21-nt single-stranded RNAs from RNA templates (Makeyev and Bamford 2002). Such a finding suggests that QDE-1 may amplify siRNA levels during RNA silencing. Such amplification could lead to transitive RNA silencing, which seems to occur in the fungus M. circinelloides (Nicolas et al. 2003) but has not been reported in N. crassa. The fact that the low-molecular-weight RNA fraction in an A. nidulans aflR(IRT900)-carrying strain did not contain aflR siRNAs from the 3′-untargeted region of aflR suggests that 5′ → 3′ transitive RNA silencing does not occur in this organism. Given the in vitro studies of QDE-1, it is possible that the absence of these siRNAs in A. nidulans could be due to the loss of a QDE-1 ortholog. Alternatively, the lack of secondary siRNAs could be the result of an inefficient amplification of dsRNA by RrpB or RrpC from endogenous aflR transcripts. Future studies with A. fumigatus or A. oryzae QDE-1 orthologs and more sensitive secondary siRNA detection methods should help test these hypotheses.
The ability of A. nidulans RDRP double mutants and the inability of S. pombe Rdp1 mutants to perform IRT-RNA silencing highlights a fundamental difference in how RDRP mutations affect the RNA-silencing pathway in these model fungi. Theoretically IRTs should bypass RDRPs in RNA silencing because siRNAs should be derived from hpRNA in an RDRP-independent process. Examples of RNA silencing working independently of RDRPs when dsRNA is introduced into the cell in an RDRP-independent manner are present in the literature (Dalmay et al. 2000; Beclin et al. 2002; Muangsan et al. 2004) and the direct formation of hpRNA by an IRT is a logical reason to explain why N. crassa QDE-1 is not required for IRT-RNA silencing even though it is required for quelling. Therefore, why S. pombe, but not A. nidulans, requires an RDRP for IRT-RNA silencing is a perplexing question.
In S. pombe, the RNA-silencing machinery is responsible for RNAi-dependent heterochromatin regulation (Volpe et al. 2002; Schramke and Allshire 2003; Verdel et al. 2004). Deleting any of the core RNA-silencing proteins or a histone methyltransferase (Clr4) disrupts this process (Volpe et al. 2002; Schramke and Allshire 2003). Surprisingly, IRT-RNA silencing is also eliminated by a Clr4 mutation (Schramke and Allshire 2003). These results suggest that mutations in the RNAi-dependent heterochromatin regulation pathway disrupt IRT-RNA silencing and vice versa. Therefore it is possible that Rdp1 involvement in RNAi-dependent heterochromatin regulation contributes to the collapse of the IRT-RNA-silencing pathway. Such a collapse may not be encountered in A. nidulans because this fungus may lack RNAi-dependent heterochromatin regulation.
Although evidence for RNAi-dependent heterochromatin regulation has been reported in eukaryotes other than S. pombe, such as Drosophila (Pal-Bhadra et al. 2004) and Arabidopsis (Aufsatz et al. 2002), recent results have shown that N. crassa does not use either its RDRPs or its RNA-silencing machinery for transcriptional silencing by heterochromatin formation (Chicas et al. 2004; Freitag et al. 2004b). It is unknown if heterochromatin regulation requires RDRPs or any component of the RNA-silencing machinery in A. nidulans. Currently, the Aspergilli appear to be “hybrid” with regard to gene-silencing equipment found in other fungi. For example, in contrast to the filamentous fungi studied so far, e.g., N. crassa (Kouzminova and Selker 2001), Ascobolus immerses (Goyon and Faugeron 1989), and Coprinus cinereus (Freedman and Pukkila 1993), but like Saccharomyces cerevisiae and S. pombe, Aspergillus species lack significant DNA methylation and concomitant methylation-dependent gene inactivation (Gowher et al. 2001). Also, analysis of their genomes shows that the Aspergilli lack a homolog of N. crassa DNA methylase DIM-2. Yet the Aspergilli contain heterochromatin-maintenance homologs (e.g., HP1/Swi6) similar to those found in N. crassa (Freitag et al. 2004a). Given that removal of functional RDRPs does not inhibit IRT-RNA silencing in A. nidulans as it does in S. pombe, we propose that A. nidulans does not commingle its RNA-silencing machinery with heterochromatin and post-transcriptional silencing components. Therefore, we further speculate that RDRPs will be required for IRT-RNA silencing only in those fungi that use the RNA-silencing machinery for both processes.
Acknowledgments
We thank Steve Harris for the A. nidulans ΔmusN strain and Mark Caddick for preliminary analysis of the degenerate rrpA (qde-1) locus in A. nidulans. Genomic data for A. fumigatus was provided by the Institute for Genomic Research (www.tigr.org/tdb/e2k1/afu1) and the Wellcome Trust Sanger Institute (www.sanger.ac.uk/Projects/A_fumigatus); genomic data for A. nidulans was provided by the Broad Institute (www.broad.mit.edu/annotation/fungi/ aspergillus/); and genomic data for A. oryzae was provided by the National Institute of Advanced Industrial Science and Technology (http://oryzae.cbrc.jp/ and www.bio.nite.go.jp/dogan/Top). Coordination of analyses of these data was enabled by an international collaboration involving more than 50 institutions from 10 countries and coordinated from Manchester, United Kingdom (www.cadre.man.ac.uk and www.aspergillus.man.ac.uk). This work was supported by Hatch funds and the graduate school of the University of Wisconsin-Madison.
References
- Aufsatz, W., M. F. Mette, J. Van Der Winden, A. J. Matzke and M. Matzke, 2002. RNA-directed DNA methylation in Arabidopsis. Proc. Natl. Acad. Sci. USA 99(Suppl 4): 16499–16506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beclin, C., S. Boutet, P. Waterhouse and H. Vaucheret, 2002. A branched pathway for transgene-induced RNA silencing in plants. Curr. Biol. 12: 684–688. [DOI] [PubMed] [Google Scholar]
- Bernstein, E., A. A. Caudy, S. M. Hammond and G. J. Hannon, 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366. [DOI] [PubMed] [Google Scholar]
- Borkovich, K. A., L. A. Alex, O. Yarden, M. Freitag, G. E. Turner et al., 2004. Lessons from the genome sequence of Neurospora crassa: tracing the path from genomic blueprint to multicellular organism. Microbiol. Mol. Biol. Rev. 68: 1–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butchko, R. A., T. H. Adams and N. P. Keller, 1999. Aspergillus nidulans mutants defective in stc gene cluster regulation. Genetics 153: 715–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell, M. A., Z. Xuan, M. Q. Zhang and G. J. Hannon, 2002. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 16: 2733–2742. [DOI] [PubMed] [Google Scholar]
- Catalanotto, C., G. Azzalin, G. Macino and C. Cogoni, 2002. Involvement of small RNAs and role of the qde genes in the gene silencing pathway in Neurospora. Genes Dev. 16: 790–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalanotto, C., M. Pallotta, P. Refalo, M. S. Sachs, L. Vayssie et al., 2004. Redundancy of the two dicer genes in transgene-induced posttranscriptional gene silencing in Neurospora crassa. Mol. Cell. Biol. 24: 2536–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chicas, A., C. Cogoni and G. Macino, 2004. RNAi-dependent and RNAi-independent mechanisms contribute to the silencing of RIPed sequences in Neurospora crassa. Nucleic Acids Res. 32: 4237–4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni, C., and G. Macino, 1997. Isolation of quelling-defective (qde) mutants impaired in posttranscriptional transgene-induced gene silencing in Neurospora crassa. Proc. Natl. Acad. Sci. USA 94: 10233–10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni, C., and G. Macino, 1999. a Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature 399: 166–169. [DOI] [PubMed] [Google Scholar]
- Cogoni, C., and G. Macino, 1999. b Posttranscriptional gene silencing in Neurospora by a RecQ DNA helicase. Science 286: 2342–2344. [DOI] [PubMed] [Google Scholar]
- Cogoni, C., J. T. Irelan, M. Schumacher, T. J. Schmidhauser, E. U. Selker et al., 1996. Transgene silencing of the al-1 gene in vegetative cells of Neurospora is mediated by a cytoplasmic effector and does not depend on DNA-DNA interactions or DNA methylation. EMBO J. 15: 3153–3163. [PMC free article] [PubMed] [Google Scholar]
- Dalmay, T., A. Hamilton, S. Rudd, S. Angell and D. C. Baulcombe, 2000. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101: 543–553. [DOI] [PubMed] [Google Scholar]
- Denli, A. M., and G. J. Hannon, 2003. RNAi: an ever-growing puzzle. Trends Biochem. Sci. 28: 196–201. [DOI] [PubMed] [Google Scholar]
- Elbashir, S. M., W. Lendeckel and T. Tuschl, 2001. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15: 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard, M., S. Boutet, J. B. Morel, C. Bellini and H. Vaucheret, 2000. AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc. Natl. Acad. Sci. USA 97: 11650–11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest, E. C., C. Cogoni and G. Macino, 2004. The RNA-dependent RNA polymerase, QDE-1, is a rate-limiting factor in post-transcriptional gene silencing in Neurospora crassa. Nucleic Acids Res. 32: 2123–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman, T., and P. J. Pukkila, 1993. De novo methylation of repeated sequences in Coprinus cinereus. Genetics 135: 357–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag, M., P. C. Hickey, T. K. Khlafallah, N. D. Read and E. U. Selker, 2004. a HP1 is essential for DNA methylation in Neurospora. Mol. Cell 13: 427–434. [DOI] [PubMed] [Google Scholar]
- Freitag, M., D. W. Lee, G. O. Kothe, R. J. Pratt, R. Aramayo et al., 2004b DNA methylation is independent of RNA interference in Neurospora. Science 304: 1939. [DOI] [PubMed]
- Galagan, J. E., S. E. Calvo, K. A. Borkovich, E. U. Selker, N. D. Read et al., 2003. The genome sequence of the filamentous fungus Neurospora crassa. Nature 422: 859–868. [DOI] [PubMed] [Google Scholar]
- Gowher, H., K. C. Ehrlich and A. Jeltsch, 2001. DNA from Aspergillus flavus contains 5-methylcytosine. FEMS Microbiol. Lett. 205: 151–155. [DOI] [PubMed] [Google Scholar]
- Goyon, C., and G. Faugeron, 1989. Targeted transformation of Ascobolus immersus and de novo methylation of the resulting duplicated DNA sequences. Mol. Cell. Biol. 9: 2818–2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, A. J., and D. C. Baulcombe, 1999. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286: 950–952. [DOI] [PubMed] [Google Scholar]
- Hammond, S. M., E. Bernstein, D. Beach and G. J. Hannon, 2000. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404: 293–296. [DOI] [PubMed] [Google Scholar]
- Hammond, S. M., S. Boettcher, A. A. Caudy, R. Kobayashi and G. J. Hannon, 2001. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293: 1146–1150. [DOI] [PubMed] [Google Scholar]
- Hofmann, A. F., and S. D. Harris, 2001. The Aspergillus nidulans musN gene encodes a RecQ helicase that interacts with the PI-3K-related kinase UVSB. Genetics 159: 1595–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer, L. M., E. V. Koonin and L. Aravind, 2003. Evolutionary connection between the catalytic subunits of DNA-dependent RNA polymerases and eukaryotic RNA-dependent RNA polymerases and the origin of RNA polymerases. BMC Struct. Biol. 3: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, S., K. Noma and S. I. Grewal, 2004. RNAi-independent heterochromatin nucleation by the stress-activated ATF/CREB family proteins. Science 304: 1971–1976. [DOI] [PubMed] [Google Scholar]
- Kouzminova, E., and E. U. Selker, 2001. dim-2 encodes a DNA methyltransferase responsible for all known cytosine methylation in Neurospora. EMBO J. 20: 4309–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, D. W., R. J. Pratt, M. Mclaughlin and R. Aramayo, 2003. An argonaute-like protein is required for meiotic silencing. Genetics 164: 821–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingel, A., B. Simon, E. Izaurralde and M. Sattler, 2003. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature 426: 465–469. [DOI] [PubMed] [Google Scholar]
- Makeyev, E. V., and D. H. Bamford, 2002. Cellular RNA-dependent RNA polymerase involved in posttranscriptional gene silencing has two distinct activity modes. Mol. Cell 10: 1417–1427. [DOI] [PubMed] [Google Scholar]
- Martens, H., J. Novotny, J. Oberstrass, T. L. Steck, P. Postlethwait et al., 2002. RNAi in Dictyostelium: the role of RNA-directed RNA polymerases and double-stranded RNase. Mol. Biol. Cell 13: 445–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooney, J. L., D. E. Hassett and L. N. Yager, 1990. Genetic analysis of suppressors of the veA1 mutation in Aspergillus nidulans. Genetics 126: 869–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muangsan, N., C. Beclin, H. Vaucheret and D. Robertson, 2004. Geminivirus VIGS of endogenous genes requires SGS2/SDE1 and SGS3 and defines a new branch in the genetic pathway for silencing in plants. Plant J. 38: 1004–1014. [DOI] [PubMed] [Google Scholar]
- Mullaney, E. J., J. E. Hamer, K. A. Roberti, M. M. Yelton and W. E. Timberlake, 1985. Primary structure of the trpC gene from Aspergillus nidulans. Mol. Gen. Genet. 199: 37–45. [DOI] [PubMed] [Google Scholar]
- Nicolas, F. E., S. Torres-Martinez and R. M. Ruiz-Vazquez, 2003. Two classes of small antisense RNAs in fungal RNA silencing triggered by nonintegrative transgenes. EMBO J. 22: 3983–3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal-Bhadra, M., B. A. Leibovitch, S. G. Gandhi, M. Rao, U. Bhadra et al., 2004. Heterochromatic silencing and HP1 localization in Drosophila are dependent on the RNAi machinery. Science 303: 669–672. [DOI] [PubMed] [Google Scholar]
- Pickford, A. S., C. Catalanotto, C. Cogoni and G. Macino, 2002. Quelling in Neurospora crassa. Adv. Genet. 46: 277–303. [DOI] [PubMed] [Google Scholar]
- Pontecorvo, G., 1953 The genetics of Aspergillus nidulans, pp. 142–235 in Advances in Genetics, edited by M. Demerec. Academic Press, New York.
- Punt, P. J., N. D. Zegers, M. Busscher, P. H. Pouwels and C. A. Van Den Hondel, 1991. Intracellular and extracellular production of proteins in Aspergillus under the control of expression signals of the highly expressed Aspergillus nidulans gpdA gene. J. Biotechnol. 17: 19–33. [DOI] [PubMed] [Google Scholar]
- Roignant, J. Y., C. Carre, B. Mugat, D. Szymczak, J. A. Lepesant et al., 2003. Absence of transitive and systemic pathways allows cell-specific and isoform-specific RNAi in Drosophila. RNA 9: 299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramke, V., and R. Allshire, 2003. Hairpin RNAs and retrotransposon LTRs effect RNAi and chromatin-based gene silencing. Science 301: 1069–1074. [DOI] [PubMed] [Google Scholar]
- Schwarz, D. S., G. Hutvagner, B. Haley and P. D. Zamore, 2002. Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Mol. Cell 10: 537–548. [DOI] [PubMed] [Google Scholar]
- Shimizu, K., and N. P. Keller, 2001. Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics 157: 591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu, K., J. K. Hicks, T. P. Huang and N. P. Keller, 2003. Pka, Ras and RGS protein interactions regulate activity of AflR, a Zn(II)2Cys6 transcription factor in Aspergillus nidulans. Genetics 165: 1095–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu, P. K., and R. L. Metzenberg, 2002. Meiotic silencing by unpaired DNA: properties, regulation and suppression. Genetics 161: 1483–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu, P. K., N. B. Raju, D. Zickler and R. L. Metzenberg, 2001. Meiotic silencing by unpaired DNA. Cell 107: 905–916. [DOI] [PubMed] [Google Scholar]
- Sienko, M., and A. Paszewski, 1999. The metG gene of Aspergillus nidulans encoding cystathionine beta-lyase: cloning and analysis. Curr. Genet. 35: 638–646. [DOI] [PubMed] [Google Scholar]
- Sijen, T., J. Fleenor, F. Simmer, K. L. Thijssen, S. Parrish et al., 2001. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107: 465–476. [DOI] [PubMed] [Google Scholar]
- Simmer, F., M. Tijsterman, S. Parrish, S. P. Koushika, M. L. Nonet et al., 2002. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr. Biol. 12: 1317–1319. [DOI] [PubMed] [Google Scholar]
- Skory, C. D., J. S. Horng, J. J. Pestka and J. E. Linz, 1990. Transformation of Aspergillus parasiticus with a homologous gene (pyrG) involved in pyrimidine biosynthesis. Appl. Environ. Microbiol. 56: 3315–3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smardon, A., J. M. Spoerke, S. C. Stacey, M. E. Klein, N. Mackin et al., 2000. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr. Biol. 10: 169–178. [DOI] [PubMed] [Google Scholar]
- Stein, P., P. Svoboda, M. Anger and R. M. Schultz, 2003. RNAi: mammalian oocytes do it without RNA-dependent RNA polymerase. RNA 9: 187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, J. J., S. K. Smith, G. J. Hannon and L. Joshua-Tor, 2004. Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305: 1434–1437. [DOI] [PubMed] [Google Scholar]
- Tabara, H., M. Sarkissian, W. G. Kelly, J. Fleenor, A. Grishok et al., 1999. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99: 123–132. [DOI] [PubMed] [Google Scholar]
- Tang, G., B. J. Reinhart, D. P. Bartel and P. D. Zamore, 2003. A biochemical framework for RNA silencing in plants. Genes Dev. 17: 49–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsitsigiannis, D. I., R. Zarnowski and N. P. Keller, 2004. The lipid body protein, PpoA, coordinates sexual and asexual sporulation in Aspergillus nidulans. J. Biol. Chem. 279: 11344–11353. [DOI] [PubMed] [Google Scholar]
- Vaistij, F. E., L. Jones and D. C. Baulcombe, 2002. Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell 14: 857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houdt, H., A. Bleys and A. Depicker, 2003. RNA target sequences promote spreading of RNA silencing. Plant Physiol. 131: 245–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdel, A., S. Jia, S. Gerber, T. Sugiyama, S. Gygi et al., 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303: 672–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe, T., V. Schramke, G. L. Hamilton, S. A. White, G. Teng et al., 2003. RNA interference is required for normal centromere function in fission yeast. Chromosome Res. 11: 137–146. [DOI] [PubMed] [Google Scholar]
- Volpe, T. A., C. Kidner, I. M. Hall, G. Teng, S. I. Grewal et al., 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837. [DOI] [PubMed] [Google Scholar]
- Yan, K. S., S. Yan, A. Farooq, A. Han, L. Zeng et al., 2003. Structure and conserved RNA binding of the PAZ domain. Nature 426: 468–474. [DOI] [PubMed] [Google Scholar]
- Yang, S. J., S. A. Carter, A. B. Cole, N. H. Cheng and R. S. Nelson, 2004. A natural variant of a host RNA-dependent RNA polymerase is associated with increased susceptibility to viruses by Nicotiana benthamiana. Proc. Natl. Acad. Sci. USA 101: 6297–6302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, D., B. Fan, S. A. Macfarlane and Z. Chen, 2003. Analysis of the involvement of an inducible Arabidopsis RNA-dependent RNA polymerase in antiviral defense. Mol. Plant-Microbe Interact. 16: 206–216. [DOI] [PubMed] [Google Scholar]
- Yu, J. H., and T. H. Adams, 1999 Filamentous fungi, pp. 417–434 in Manual of Industrial Microbiology and Biotechnology, edited by A. L. Demain and J. E. Davies. American Society for Microbiology Press, Washington, DC.
- Zolotukhin, S., M. Potter, W. W. Hauswirth, J. Guy and N. Muzyczka, 1996. A “humanized” green fluorescent protein cDNA adapted for high-level expression in mammalian cells. J. Virol. 70: 4646–4654. [DOI] [PMC free article] [PubMed] [Google Scholar]