Abstract
To understand the roles of heptad repeat 1(HR1) and HR2 of the spike (S) protein of the severe acute respiratory syndrome coronavirus (SARS-CoV) in virus-cell interactions, the conserved Leu or Ile residues located at positions 913, 927, 941, and 955 in HR1 and 1151, 1165, and 1179 in HR2 were individually replaced with an α-helix-breaker Pro residue. The 913P mutant was expressed mainly as a faster-migrating, lower-molecular-weight SL form, while the wild type and all other mutants produced similar levels of both the SL form and the slower-migrating, higher-molecular-weight SH form. The wild-type SL form was processed to the SH form, whereas the SL form of the 913P mutant was inefficiently converted to the SH form after biosynthesis. None of these mutations affected cell surface expression or binding to its cognate ACE2 receptor. In a human immunodeficiency virus type 1/SARS S coexpression study, all mutants except the 913P mutant incorporated the SH form into the virions as effectively as did the wild-type SH form. The mutation at Ile-1151 did not affect membrane fusion or viral entry. The impaired viral entry of the 927P, 941P, 955P, and 1165P mutants was due to their inability to mediate membrane fusion, whereas the defect in viral entry of the 1179P mutant occurred not at the stage of membrane fusion but rather at a postfusion stage. Our study demonstrates the functional importance of HR1 and HR2 of the SARS-CoV spike protein in membrane fusion and viral entry.
The severe acute respiratory syndrome coronavirus (SARS-CoV), the etiologic agent of the outbreak of atypical pneumonia known as SARS, is a membrane-enveloped virus which encodes a 29,727-nucleotide, polyadenylated, positive-strand RNA. The genome of this virus contains five major open reading frames that encode the replicase polyprotein: the spike (S), envelope (E), and membrane (M) glycoproteins and the nucleocapsid (N) protein in the same order and of approximately the same sizes as those of other CoVs (34, 39). In CoVs, the structural proteins, S, E, M, and N, play crucial roles during host cell entry and virion morphogenesis, assembly, and budding. The S protein is incorporated into the viral envelope by interaction with the M protein and forms large spikes on virion surfaces, and mature virions are released from smooth vesicles (20).
The S protein of CoVs, which is oligomerized in the endoplasmic reticulum, is N glycosylated and has an apparent molecular mass of 150 to 180 kDa (reviewed in reference 28). The S1 domain is required for binding to specific cellular receptors, thus defining the host range of the virus. Angiotensin-converting enzyme 2 (ACE2) has been shown to be a physiologically relevant, functional receptor for the SARS-CoV (31). SARS-CoV S protein-mediated viral infection can be effectively blocked by either a soluble form of ACE2, anti-ACE2 antibodies, or an antibody recognizing the ACE2-binding domain of the S protein (31, 44). The S2 domain forms the membrane-anchored stalk region and mediates the fusion between the viral and cellular membranes (39, 55). The S protein-mediated entry of the SARS-CoV into host cells occurs via a pH-dependent endocytic pathway (24, 41, 59), although S-mediated cell-to-cell fusion can occur at a neutral pH (31, 41, 55).
Membrane-enveloped viruses have evolved a similar mode of entry to ensure that their viral genomes are properly translocated into host cells. Class I fusion proteins, including hemagglutinin 2 (HA2) in influenza viruses, gp41 in the human immunodeficiency virus (HIV)/simian immunodeficiency virus (SIV), GP2 in the Ebola virus, and F2 in paramyxoviruses (17, 23), are characterized by an N-terminal heptad repeat 1 (HR1) adjacent to the fusion peptide, which is just C-terminal to the cleavage site or is near the N terminus of the fusion protein, and a C-terminal HR2, which is close to another short spacer and the transmembrane domain. These HR sequences play important roles in membrane fusion and viral entry (reviewed in references 17, 23, and 42). For viruses such as the influenza virus (6), virions are internalized after HA1-induced receptor binding and receptor-mediated endocytosis; the low pH within the endosomes induces a major structural rearrangement of HA, and the transition from the metastable (nonfusogenic) to the active (fusogenic), prehairpin intermediate of HA triggers membrane fusion. Such structural switching also includes the conversion of a part of the HR region of HA2, which forms a nonhelical loop in its native state (53), into a pH-induced fusogenic coiled-coil structure (6). This conformational change is also the basis of the “spring-loaded mechanism” for activation of viral fusion (8). Formation of the six-helix bundle positions both the N-terminal fusion peptide and C-terminal transmembrane domain on the same site of a stable protein rod and exposes the fusion peptide to target cell membranes, placing the cellular endosomal and viral membranes in close proximity to promote formation of a fusion pore. In both the HIV and SIV, binding to CD4 and its coreceptor triggers conformational changes in the noncovalent gp120-gp41 oligomeric complex present on the viral envelope, which then activate the pH-independent viral fusion process (3, 7, 9, 33, 46, 50-52).
Despite the fact that the SARS-CoV S protein shares little amino acid similarity with other CoVs, the putative S2 domain of the SARS-CoV also contains two predicted amphipathic α-helical, 4-3 heptad repeat sequences characteristic of coiled coils. The sequences most homologous to the SARS-CoV are the mouse hepatitis virus and bovine CoV. Because it shares several features with other class I fusion protein-mediated membrane fusion elements, the six-stranded coiled-coil structure of the mouse hepatitis virus (5, 56) can be used as a model to study the SARS-CoV S protein. Recent studies based on peptide modeling have shown that the HR1 and HR2 fragments of the SARS-CoV S protein form a stable six-helix bundle structure (4, 26, 32, 47, 58, 60, 61). The three-dimensional structure revealed by X-ray crystallography confirms that the fusion core of the SARS-CoV S protein is a six-helix bundle with three HR2 helices packed against the hydrophobic grooves in an oblique antiparallel manner on the surface of a central coiled coil formed by three parallel HR1 helices (45, 57). The hairpin trimer structure of the SARS-CoV S protein suggests that these regions are involved in the membrane fusion process and that this structure may represent a postfusion conformation of the core. The observations that synthetic peptides corresponding to the HR motifs possess inhibitory activity against viral fusion (4, 32, 57, 60, 61) support this hypothesis. Follis et al. recently reported that Ser substitutions for the hydrophobic residues at the a and d positions within the short α-helical segment of HR2, but not in the outlying extended chain in the core structure, affect the cell-to-cell fusion ability of the S protein (18). Nevertheless, the precise roles of HR1 and HR2 motifs in the context of virus infection remain to be determined.
To elucidate the functional basis of the HR1 and HR2 motifs in S protein-mediated virus-cell interactions, in the present study, we conducted mutagenesis analyses by substituting a Pro residue, which disrupts the α-helical structure more severely than any other amino acids, for the HR-conserved Leu or Ile residues located at positions 913, 927, 941, and 955 in HR1 and 1151, 1165, and 1179 in HR2, based on a codon-optimized S gene. We developed sensitive membrane fusion assays based on the loxP-Cre recombination system of bacteriophage P1 and the HIV-1 Tat long-terminal-repeat (LTR) transactivation system to monitor membrane fusion of these mutants. We also employed an HIV/SARS S phenotypic infectivity assay to determine mutant S-mediated viral entry. The results revealed that the defect in viral entry of the 927P, 941P, 955P, and 1165P mutants is due to their impaired membrane fusion ability and that the impaired viral entry of the 1179P mutant is not due to a blockage in membrane fusion but rather to a defect at a postfusion stage. Our study demonstrates for the first time the direct role of α-helices of HR1 and HR2 in membrane fusion and viral entry.
MATERIALS AND METHODS
Cells and antibodies.
Human embryonic kidney 293T and Vero E6 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% heat-inactivated fetal bovine serum. Hybridoma 183 (clone H12-5C) was described previously (29). Rabbit anti-SP4 was generated by immunizing rabbits with residues 435 to 454 of the S protein (12). Rabbit anti-S protein antiserum was prepared by immunizing rabbits with a bacterially expressed recombinant soluble S protein (residues 511 to 1255) (25). A monoclonal antibody (MAb) directed against ACE2 was purchased from R&D (Minneapolis, MN).
Construction of plasmids.
To construct S plasmids encoding mutations in the HR1 and HR2 sequences, site-directed mutagenesis based on a codon-optimized S gene, pCDNA3.1(−)S, was performed using a QuikChange site-directed mutagenesis kit (Stratagene). Since the S gene was cloned at the XbaI and BamHI sites in pCDNA3.1(−) to facilitate the subcloning steps, the KpnI site in multiple cloning sites of the vector and 3′ to the S gene was first deleted by replacing a 0.15-kb PCR fragment. This DNA fragment, which was flanked by the HindIII and BamHI sites and contained the sequence for transmembrane and cytoplasmic domains of the vesicular stomatitis virus (VSV) glycoprotein G protein, was substituted for the corresponding sequence of pCDNA3.1(−)S. Introduction of this DNA fragment did not affect the coding sequence of the S gene. In order to generate HR2 mutants, the 0.85-kb EcoRI-BamHI fragment of the S gene was subcloned in a pBluescript SKII(+) vector at the corresponding sites, and the plasmid obtained was used as a template to generate HR2 mutations. The mutated fragments were then subcloned back into pCDNA3.1(−)S to generate HR2 mutants. To construct the HR1 mutants, the 3-kb XbaI-EcoRV fragment from pCDNA3.1(−)S was subcloned in the corresponding sites of the pBluescript SKII(+) vector, and the resultant plasmid was used as the template to generate HR1 mutations. The 2-kb KpnI-EcoRI fragments containing HR1 mutations isolated from these mutated plasmids were then subcloned back into the corresponding sites in pCDNA3.1(−)S to yield various HR1 mutants. The following oligonucleotide pairs were used to generate the HR1 and HR2 mutants: 913P, 5′-TTCAACAAGGCCCCTAGCCAGATCCAGGAGAGCCTGACC-3′ (sense) and 5′-CTGGATCTGGCTAGGGGCCTTGTTGAACTGGTTGGC-3′ (antisense); 927P, 5′-ACCAGCACGGCCCCAGGCAAGCTGCAGGACGTGG-3′ (sense) and 5′-CTGCAGCTTGCCTGGGGCCGTGCTGGTCGTGG-3′ (antisense); 941P, 5′-AACGCCCAGGCCCCTAACACCCTGGTGAAGCAGC-3′ (sense) and 5′-CACCAGGGTGTTAGGGGCCTGGGCGTTCTGGTTG-3′ (antisense); 955P, 5′-AACTTCGGCGCCCCAAGCTCCGTGCTGAACGATATC-3 (sense) and 5′-CAGCACGGAGCTTGGGGCGCCGAAGTTGCTGGAC-3′ (antisense); 1151P, 5′-GACCTGGGCGACCCTAGCGGCATCAACGCCAGCG-3′ (sense) and 5′-GTTGATGCCGCTAGGGTCGCCCAGGTCCACGTCC-3′ (antisense); 1165P, 5′-ATCCAGAAGGAGCCAGACCGCCTGAACGAGGTGG-3′ (sense) and 5′-GTTCAGGCGGTCTGGCTCCTTCTGGATGTTCACG-3′ (antisense); and 1179P, 5′-CTGAACGAGAGCCCTATCGACCTCCAGGAGCTGGG-3′ (sense) and 5′-CTGGAGGTCGATAGGGCTCTCGTTCAGGTTCTTGG-3′ (antisense). The underlined nucleotides mark the respective sense and antisense primer Pro substitutions. Primers OptiS2743 (5′-CAACAACGTGTTCCAGACCC-3′) and OptiS3210 (5′-CTTCAACTTCAGCCAGATCC-3′) were used to sequence the DNA fragments containing HR1 mutations, and primer OptiS4154 (5′-TCATCACCACGGACAACACC-3′) was used to sequence the DNA fragments containing HR2 mutations.
pCAGGS-MCS was constructed by replacing the 1-kb PstI-to-EcoRV coding region of pCAGGS-NLS-Cre with a synthetic oligonucleotide linker, 5′-CTGCAGGAATTCTCGAGCTCACGCGTGCATGCGATATC-3′, which contained the PstI/EcoRI/XhoI/SacI/MluI/Sph/EcoRV recognition sequence. The SalI site at the 5′ boundary of the cytomegalovirus immediate-early 1 enhancer was then replaced by an oligonucleotide containing the SalI/NotI/XbaI sites, and the HindIII site at the 3′ boundary of the poly(A) signal was replaced by the NotI site. Therefore, the entire expression cassette could be easily isolated using NotI digestion. ploxP1, derived from pBluescript SKII, contained two tandem repeats of the 34-bp loxP sequence in multiple cloning sites. To generate ploxP-EGFP-loxP, ploxP1 was codigested with SalI and NheI, followed by Klenow fill-in. The 1.15-kb KpnI-to-SspI fragment encoding the enhanced fluorescence green protein (EGFP) and poly(A) sequences of pIRES-EGFP (Clontech) was blunt ended by T4 DNA polymerase and then inserted into the treated ploxP1 vector by blunt-end ligation. The XhoI-EcoRV fragment containing the EGFP gene flanked at both ends by loxP sequences was then isolated and inserted into the same restriction enzyme sites in pCAGGS-MCS to create pCAGGS-loxP-EGFP-loxP. The 0.7-kb BamHI-to-NotI fragment of pDsRed2 N1 (Clontech) was blunt ended by fill-in with the Klenow fragment and was then inserted into the EcoRV site of pCAGGS-loxP-EGFP-loxP to obtain pCAGGS-loxP-EGFP-loxP-DsRed2.
pCDNA3.1(−)ACE2-Ig was generated by replacing the EcoRI-to-KpnI fragment in pCDNA3.1(−)ACE2 with a PCR product encoding the Fc domain of a human immunoglobulin γ1 (Igγ1) heavy chain. The insert was generated using 5′-TGAATTCACCAGCACCTGAACTCCTGG-3′ and 5′-TGGTACCTCATTTACCCGGGGACAGGGAGAGG-3′ as the primers and p2C11-γ1, which encodes a human IgG1 Fc domain, as the template in PCR. The resultant plasmid encoded a fusion protein containing the CH2 and CH3 sequences of the Fc domain, beginning with Pro-Ala-Pro-Glu and ending with Gly-Lys-Val-Asp, attached at the C terminus of Ser-602 within the ectodomain of ACE2. To construct pCAGGS-ACE2-Ig, pCDNA3.1(−)ACE2-Ig was digested with XbaI, blunt ended with the Klenow fragment, and cut by KpnI to obtain the ACE2-CH2-CH3 fusion gene. The pCAGGS-MCS vector was digested by EcoRI and treated with the Klenow enzyme to fill in the staggered end before KpnI digestion. Then, the ACE2-CH2-CH3 DNA fragment was inserted into the treated pCAGGS-MCS vector to obtain pCAGGS-ACE2-Ig.
Transfection of DNA plasmids.
For expression of the SARS S protein, 293T cells grown on 6- or 10-cm petri dishes were transfected with 3 or 10 μg of the codon-optimized wild-type (wt) and mutant pCDNA3.1(−)S constructs by a standard calcium phosphate coprecipitation method (14). For the one-cycle viral replication assay, 293T cells grown in 10-cm petri dishes were cotransfected with 10 μg of pNL4-3R−E−Luc and 12 μg of either the wt or mutant pCDNA3.1(−)S plasmids. Alternatively, cells were cotransfected with 10 μg of pHXBΔBglCAT and 7.5 μg of either the wt or mutant S plasmids. pCDNA3, which was added to the transfection mixture that did not contain S expression plasmids, was used as a negative control.
Virus preparation and Western blot analysis.
Two days after transfection, cell-free virions were isolated through a 20% sucrose cushion as described previously (13), and equal volumes of cell and viral lysates were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting. The membrane blots were incubated with the appropriate primary and secondary antibodies, followed by enhanced chemiluminescence detection.
Assessment of the ACE2-binding ability of the S proteins.
293T cells grown on 6-cm dishes were cotransfected with 2 μg of either the wt or mutant pCDNA3.1(−)S plasmids together with 2 μg of pCAGGS-ACE2-Ig using the Lipofectamine transfection reagent (Invitrogen, Carlsbad, CA) according to the procedures provided by the manufacturer. Two days after transfection, cells were washed with phosphate-buffered saline (PBS) and then lysed in 200 μl of PBS containing 1% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS) and protease inhibitor cocktail (Roche) on ice for 30 min, followed by centrifugation to remove cell debris. Equal volumes of cell lysates were incubated with protein A-Sepharose 4B beads. After extensive washes with PBS containing 0.5% CHAPS, the bound proteins were analyzed by Western blotting using an ACE2 MAb and rabbit anti-S antiserum to detect ACE2 and the S protein, respectively. Alternatively, culture supernatants collected from 293T cells transfected with pCDNA3.1(−)ACE2-ecto were concentrated by Amicon Centriprep YM-10 membranes (Millipore, Bedford, MA). 293T cells transfected with each of the wt and mutant S plasmids were lysed with PBS containing 1% 3-[(3-cholamidopropyl)-dimethylammonio]-2-hydroxy-1-propanesulfonate (CHAPSO) and protease inhibitor cocktail. The cell lysates were then incubated with concentrated soluble ACE2 at 4°C for 2 h, followed by incubation with rabbit anti-S-coated protein A-Sepharose 4B beads at 4°C overnight. After being washed extensively with PBS containing 0.5% CHAPSO, the immune complexes were analyzed by SDS-PAGE, followed by Western blotting using rabbit anti-S and anti-ACE2 MAb to detect precipitated S and ACE2 proteins.
SARS S protein-mediated membrane fusion.
To prepare effector cells for qualitative analysis of S-mediated membrane fusion, 293T cells grown on 6-cm petri dishes were cotransfected by the Lipofectamine transfection method with 3 μg of either the wt or mutant pCDNA3.1(−)S plasmids and 1 μg of pCAGGS-NLS-Cre. For preparation of target cells, 293T cells grown on two 10-cm dishes were cotransfected with 6 μg of pCDNA3.1(−)ACE2 and 4 μg of pCAGGS-loxP-EGFP-loxP-DsRed2. One day after transfection, PBS-washed transfected cells were trypsinized, and 6 × 105 (each) of the effector and target cells were seeded onto 6-cm dishes and then cocultured for 2 days. Cocultured cells were examined under a Zeiss model 135 inverted fluorescence microscope (Carl Zeiss AG, Germany), and green and red fluorescent images were overlaid. For quantitative membrane fusion analysis, effector cells were cotransfected with 4 μg of the wt or mutant S expression plasmids and 2.5 μg of a cytomegalovirus promoter-driven HIV-1 Tat expression plasmid, pCEP4-tat, and target cell were cotransfected with 10 μg of pCDNA3.1(−)ACE2 and 5 μg pIIIexcat, a chloramphenicol acetyltransferase (cat) gene linked to the HIV-1 LTR, by the calcium phosphate coprecipitation method. On the following day, effector and target cells were trypsinized, and 6 × 105 of each kind of cell were cocultured. Two days after coculturing, cells were harvested for CAT activity measurement.
HIV/SARS S pseudotype infection assay.
One day before pseudotype infection, 3 ×104 or 1.5 × 105 Vero E6 cells were seeded onto 24-well plates (for the luciferase assay) or 6-well dishes (for the CAT assay), respectively. Cell-free NL4-3R−E−Luc or HXBΔBglCAT pseudotypes containing 6 × 104 or 105 cpm of reverse transcriptase (RT) activity in 0.6 or 2 ml, respectively, of Dulbecco's modified Eagle's medium containing 2.5% fetal bovine serum in the presence of 8 μg/ml of Polybrene were added to the wells. After centrifugation at 2,200 rpm for 2 h at room temperature, the cultures were incubated at 37°C. Two days after transfection, cells were harvested and prepared for the luciferase or CAT assay.
RT, luciferase, and CAT assays.
Cell-free, SARS S protein-pseudotyped viruses were assayed for virion-associated RT activity as described previously (13, 14). Cell lysates (25 μl) prepared from NL4-3R−E−Luc pseudotype infection were assayed for firefly luciferase activity using 50 μl of luciferase assay substrate reagent (Promega). Cell extracts prepared from an HXBΔBglCAT pseudotype infection were assayed for CAT activity using a Packard Instant Imager (model A202401; Packard Instrument Company, Meriden, CT) as described previously (14).
Binding of HIV/SARS S pseudotypes to ACE2-expressing cells.
The wt and mutant pseudotypes produced as described above were concentrated by an Amicon Ultra 15 filter with a nominal molecular mass limit of 100 kDa (Millipore), and comparable amounts of wt and mutant pseudotypes (containing 106 cpm of RT activity) were incubated with 293T cells transfected with pCDNA3.1(−)ACE2 at 4°C for 1 h. After being washed three times with PBS containing 0.5% bovine serum albumin (BSA), cells were blocked with PBS containing 2% BSA on ice for 30 min. The cells were successively incubated with rabbit anti-S and fluorescein isothiocyanate-conjugated anti-rabbit IgG on ice. After each antibody incubation, the cells were washed three times with ice-cold PBS containing 0.5% BSA. After fixation with 4% paraformaldehyde for 30 min, the immunostained cells were quantitated by fluorescence-activated cell sorter analysis according to the procedure described previously (10).
Pulse-chase and immunoprecipitation.
Two days after transfection, 293T cells expressing the wt or 913P mutant S proteins were metabolically labeled with [35S]methionine for 30 min and chased with excess cold methionine as described previously (10, 29) for different time periods. Equal amounts of cell lysates were incubated with rabbit anti-S preadsorbed onto protein A-Sepharose 4B beads at 4°C overnight, extensively washed, and separated by SDS-PAGE followed by fluorography.
Cell surface biotinylation of SARS spike proteins.
Transfected 293T cells grown in 6-cm petri dishes were metabolically labeled with [35S]methionine overnight, and labeled cells were surface biotinylated with 0.25 mg/ml of membrane-impermeable sulfo-N-hydroxysuccinimide-biotin on ice for 30 min as described previously (10, 11, 29). Cell lysates were immunoprecipitated with rabbit anti-S preadsorbed onto protein A-Sepharose 4B beads. The precipitated S proteins were released from the beads by being boiled in sample loading buffer without bromophenol blue. A fraction of samples was directly resolved by SDS-PAGE, and another fraction of samples was precipitated with neutravidin-agarose beads (Pierce) prior to SDS-PAGE.
RESULTS
Construction and expression of the HR1 and HR2 mutants.
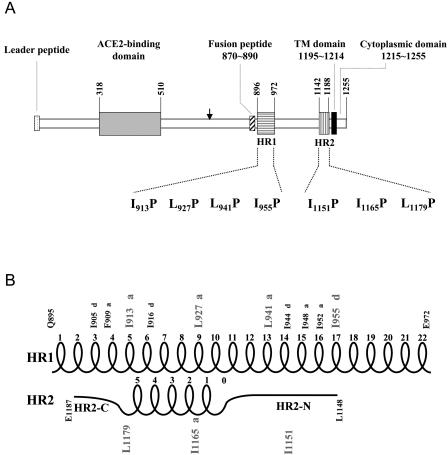
To understand the functional involvement of the two HR1 and HR2 motifs in the SARS-CoV life cycle, the HR residues Ile-913, Leu-927, Leu-941, and Ile-955 within HR1 and Ile-1151, Ile-1165, and Leu-1179 within HR2 (Fig. 1A) were individually replaced by a Pro residue via site-directed mutagenesis based on pCDNA3.1(−)S, a codon-optimized S expression plasmid (31, 36). The relative positions of these substituted residues in the α-helical, hairpin structure are indicated in Fig. 1B. Pro is believed to drastically disrupt the α-helical structures of HR motifs in envelope glycoproteins. pCDNA3.1(−)S encodes the signal sequence of CD5, the entire TOR2 strain S protein but without its signal sequence (residues 12 to 1255), and a nine-residue C-terminal tag, TETSQVAPA, derived from the C terminus of bovine rhodopsin.
FIG. 1.
(A) Construction of the HR1 and HR2 motif mutants. A schematic diagram of the spike protein encoded by the SARS-CoV is shown. The vertical arrow represents the putative cleavage site which separates the S protein into two putative S1 and S2 domains. The HR1 and HR2 motifs are marked by horizontally and vertically hatched bars, respectively. Site-directed mutagenesis was performed to replace the HR-conserved Leu or Ile located at residues 913, 927, 941, and 955 within HR1 and residues 1151, 1165, and 1179 within HR2 with Pro residues. The leader peptide, the ACE2-binding domain, a putative fusion peptide domain, the transmembrane region, and a short cytoplasmic domain are also indicated. (B) The hairpin structure formed by HR1 and HR2. The schematic diagram is based on the ribbon structures presented by Supekar et al. (45) and Duquerroy et al. (16). The numbering of mutated HR1 residues is the same as that indicated in the model of Supekar et al. (45). However, Ile-1151, Ile-1165, and Leu-1179 are numbered 1150, 1164, and 1178, respectively, in the model of Duquerroy et al. (16). HR1 is a canonical, heptad-repeated amphipathic α-helical structure with Leu or Ile residues periodically occurring in a 3-4-3-4 (or a-d-a-d) manner. Two 3-4-4-3 (or d-a-a-d) shifts were noted around the Phe-909 and Ile-948 residues. HR2 comprises extended N- and C-terminal domains, marked as HR2-N and HR2-C, respectively, as well as five and a half turns in the α-helix core. Ile-1151 and Ile-1165 are located in the HR2-N and α-helix, respectively, while Leu-1179 is located at the junction of the helix core and the extended HR2-C.
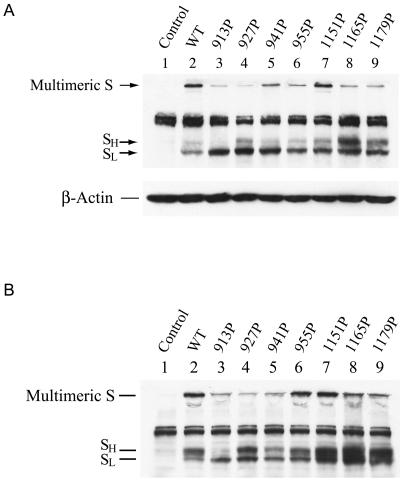
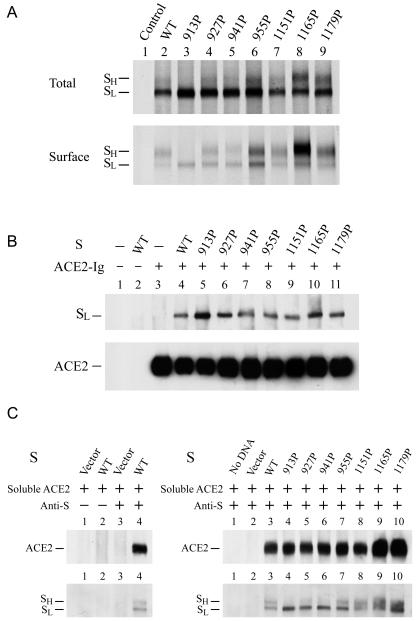
To assess the expression of S proteins, 293T cells were transfected with each of the wt and mutant constructs, and cell lysates prepared at 24 h after transfection were analyzed by Western blotting using a rabbit monospecific anti-SP4 polyclonal antibody, which is directed against residues 435 to 454 of the S protein. The expressed wt S protein migrated as a tightly spaced doublet, marked as SL and SH, around 180 to 200 kDa in SDS-PAGE, with the faster-migrating, lower-molecular-mass SL form being predominant over the slower-migrating, higher-molecular-mass SH form (Fig. 2A, lane 2). Similar results were obtained when a rabbit polyclonal antibody directed against a recombinant soluble S protein was used in Western blotting (data not shown). Consistent with other results (55), no significant cleavage products of the S protein were observed in addition to these two forms. Furthermore, a species migrating between the separating and stacking gels was noted (Fig. 2A, lane 2). This high-molecular-weight species represented aggregates or high-ordered multimeric structures of the S protein that were not dissociated by treatment with SDS and the reducing agent during the boiling of samples (2, 43). When cell lysates prepared at 48 h posttransfection were analyzed, the SH form of the wt S protein was more predominant than the SL form (Fig. 2B, lane 2). These results together suggest that SL is converted to SH during biosynthesis and maturation of the S protein. The nature of this doublet may represent differential glycosylation of the wt S protein (2, 43). An endoglycosidase H digestion study also showed that the faster-migrating, lower-molecular-weight species contains high-mannose N-glycans while the slower-migrating, higher-molecular-weight species contains the complex-type N-glycans (41).
FIG. 2.
Expression of HR1 and HR2 mutant spike proteins. 293T cells grown on 6-cm dishes were transfected with 3 μg each of the wt and mutant pCDNA3.1(−)S plasmids. Cell lysates were prepared 24 (A) or 48 (B) h after transfection, and equal volumes of cell lysates were analyzed by Western blotting using the rabbit anti-SP4 antibody.
When the expressions of the mutant proteins were analyzed, with the exception of the 913P mutant, all mutants produced SL and SH levels comparable to those produced by the wt S construct (Fig. 2A and B). Nevertheless, the 913P mutant produced a smaller amount of the SH form than did the wt and other mutants regardless of whether lysates prepared at 24 or 48 h were analyzed (Fig. 2A and B, lane 3). Moreover, high-ordered multimeric structures of all mutants were also observed (Fig. 2A and B).
Altered processing of the 913P mutant S protein.
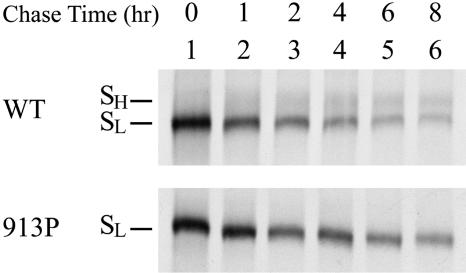
To understand the relationship between the SL and SH forms of the wt and 913P mutant proteins, a pulse-chase study was performed. Cell lysates were precipitated with rabbit anti-S antiserum, and the precipitated proteins were resolved by SDS-PAGE. The intensity of the wt SL form decreased while that of the SH form increased as the chase proceeded (Fig. 3, top panel), indicating that wt SL is converted to SH during protein maturation. Although the intensity of the 913P mutant SL gradually decreased at a rate slower than that of wt SL as the chase proceeded, the SH form of this mutant could not be efficiently detected even after 8 h of chasing (Fig. 3, bottom panel). These results indicate that a mutation at Ile-913 may affect the glycosylation process of the S protein.
FIG. 3.
Pulse-chase of the wt and 913P mutant proteins. 293T cells expressing the wt or 913P mutant proteins were metabolically labeled with [35S]methionine and chased with excess cold methionine. Cell lysates harvested at different times after the chase were immunoprecipitated with rabbit anti-S antiserum preadsorbed onto protein A-Sepharose 4B beads. The precipitated proteins were subjected to SDS-PAGE followed by fluorography.
Cell surface expression of the HR1 and HR2 mutants.
To understand whether the SL and SH forms of the wt and mutants are expressed on the cell surface, 293T cells transfected with the wt or each of the mutant S expression plasmids were metabolically labeled with [35S]methionine, and cell surface biotinylation using membrane-impermeable sulfo-N-hydroxysuccinimide-biotin was performed. Cell lysates were first immunoprecipitated with rabbit anti-S, and the immune complexes were directly separated by SDS-PAGE or precipitated with neutravidin-agarose beads prior to SDS-PAGE. As shown by the Western blot analyses (Fig. 2), with the exception of the 913P mutant in which only the SL form was observed, all mutants produced comparable or even slightly greater amounts of the SL and SH forms than did the wt construct when the total protein expression was assessed (Fig. 4A, top panel). The 913P mutant showed only the SL form on the cell surface, whereas all of the other mutants produced similar or even slightly greater amounts of the SL and SH forms on the cell surface than the wt protein (Fig. 4A, bottom panel). This result indicated that mutations in the HR1 and HR2 sequences do not affect cell surface expression of the S protein.
FIG. 4.
Characterization of the HR1 and HR2 mutant S proteins. (A) Cell surface expression of mutant proteins. 293T cells were transfected with each of the wt and mutant S plasmids as indicated. Cells were metabolically labeled with [35S]methionine and surface biotinylated with sulfo-N-hydroxysuccinimide-biotin. Cell lysates were analyzed as described in Materials and Methods. (B and C) ACE2-binding abilities of HR1 and HR2 mutants. In panel B, 293T cells were transfected with or without pCAGGS-ACE2-Ig in the presence or absence of wt or mutant S plasmids. Equal volumes of cell lysates were then precipitated with protein A-Sepharose 4B beads, and the bound proteins were analyzed by Western blotting using an ACE2 MAb and rabbit anti-S antiserum. On the left side of panel C, lysates from mock-transfected or wt S plasmid-transfected 293T cells were incubated with concentrated soluble ACE2. After incubation with rabbit control serum or anti-S-coated protein A-Sepharose beads, the precipitated proteins were separated by SDS-PAGE, followed by Western blotting using rabbit anti-S and ACE2 MAb. On the right side of panel C, soluble ACE2 was added to lysates containing wt or mutant S proteins. After incubation with rabbit anti-S-coated protein A-Sepharose 4B beads, the immune complexes were analyzed by Western blotting using rabbit anti-S and ACE2 MAb.
ACE2-binding ability of the HR1 and HR2 mutants.
The ACE2-binding site in the S protein has been mapped to residues 270 to 510 (1) or 318 to 510 (54), as shown by the binding of S fragments to Vero E6 cells, followed by flow cytometry or by immunoprecipitation of 35S-labeled ACE2 with S fragments fused to the Fc domain of human IgG1, respectively. It was expected that mutations in the HR1 or HR2 motif would not affect the receptor-binding ability of the S protein. To confirm this notion, 293T cells were cotransfected with or without the wt or mutant S expression plasmids in the presence or absence of a plasmid encoding soluble ACE2 fused to the Fc domain of human IgG1. Cell lysates were precipitated with protein A-Sepharose beads, and the bound proteins were analyzed by Western blotting using rabbit anti-S and MAb anti-ACE2. In the absence of ACE2-Ig coexpression, the wt S protein did not bind to the protein A-Sepharose 4B beads (Fig. 4B, lane 2). Comparison of the mobility of the precipitated S protein to that of the SL and SH forms shown in Fig. 2, 3, and 4A revealed that SL but not SH was bound to ACE2-Ig (Fig. 4B, lane 4). The wt S and ACE2-Ig may rapidly interact with each other in the endoplasmic reticulum after biosynthesis, resulting in coprecipitation of the wt SL form with ACE2-Ig-coated protein A-Sepharose beads. However, the wt S molecules which do not bind to ACE2 are transported to the Golgi for further processing into SH. A similar or even slightly greater amount of mutant SL proteins than that of wt SL was bound to ACE2-Ig (Fig. 4B, lanes 4 to 11).
To further determine the ACE2-binding ability of these S mutants, concentrated soluble ACE2 collected from culture supernatants of 293T cells transfected with pCDNA3.1(−)ACE2-ecto (31) was incubated with lysates of cells transfected with each of the wt and mutant S plasmids. After incubation with rabbit anti-S-coated protein A-Sepharose 4B beads, the precipitated complexes were resolved by SDS-PAGE, followed by Western blotting using rabbit anti-S and ACE2 MAb. This assay determined the specific binding of wt S with ACE2 since incubation with rabbit anti-S, but not with a rabbit control serum, coprecipitated ACE2 (Fig. 4C, left panel). Levels of soluble ACE2 coprecipitated with mutant proteins were comparable to or even greater than that of ACE2 coprecipitated with the wt protein (Fig. 4C, right panel). Also, the levels of soluble ACE2 coprecipitated paralleled the levels of mutant proteins precipitated (Fig. 4C, right panel). These results taken together indicated that mutations in the HR1 or HR2 motif do not affect the receptor-binding ability of the S protein.
Assessment of the membrane fusion abilities of the HR1 and HR2 mutants.
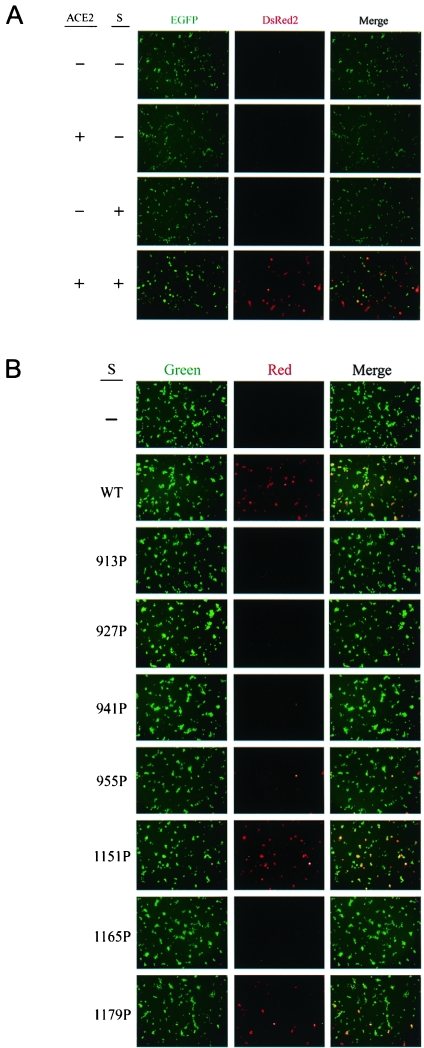
To examine whether these S mutants expressed on the cell surface are able to mediate membrane fusion, we first developed a sensitive cell-to-cell fusion assay based on the loxP-Cre recombination system of bacteriophage P1 (27, 48). Effector cells cotransfected with the S- and Cre-expressing plasmids were cocultured with target cells cotransfected with the loxP-EGFP-loxP-DsRed2 gene and an ACE2 expression plasmid. If membrane fusion between the effector and target cells occurred, Cre recombinase would catalyze the reciprocal site-specific recombination between the two specific 34-bp sites of loxP. The intramolecular recombination of the two loxP sites positioned head to tail resulted in deletion of the intervening EGFP sequences, and an intermolecular recombination caused integration at the loxP site, converting the target gene from EGFP expression to DsRed2 expression. The wt S-mediated membrane fusion was strictly dependent on the S protein expressed on effector cells and ACE2 on target cells (Fig. 5A). The 913P, 927P, 941P, 955P, and 1165P mutants displayed significantly impaired membrane fusion phenotypes compared to the wt S protein (Fig. 5B). However, the 1151P and 1179P mutants still exhibited effective membrane fusion (Fig. 5B).
FIG.5.
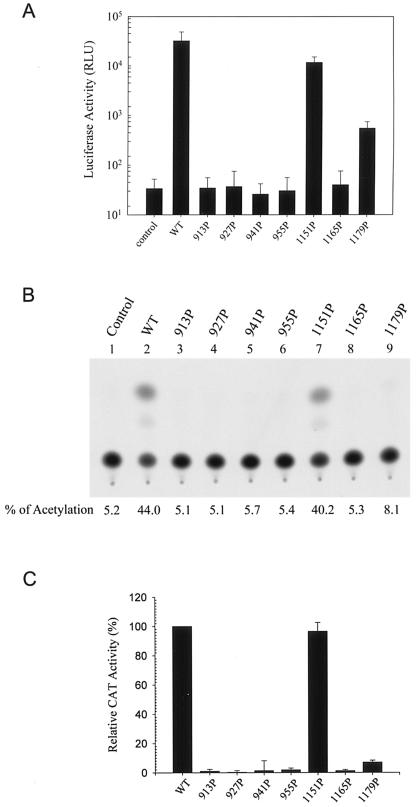
Assessment of the membrane fusion abilities of the HR1 and HR2 mutants. (A) SARS S- and ACE2-dependent membrane fusion based on the loxP-Cre recombination system. 293T effector cells were cotransfected with pCDNA3 or the wt pCDNA3.1(−)S plasmid together with pCAGGS-NLS-Cre. 293T target cells were cotransfected with pCDNA3 or pCDNA3.1(−)ACE2 in the presence of pCAGGS-loxP-EGFP-loxP-DsRed2. One day after transfection, effector and target cells were trypsinized, mixed, and cocultured for 2 days, and cells were examined under a fluorescence microscope for expression of fluorescent proteins. (B) Membrane fusion mediated by mutant S proteins. 293T effector cells cotransfected with pCAGGS-NLS-Cre along with either the wt or mutant S plasmids were cocultured with 293T target cells cotransfected with pCDNA3.1(−)ACE2 and pCAGGS-loxP-EGFP-loxP-DsRed2 and then examined for expression of fluorescent proteins. (C and D) Quantification of S protein-mediated membrane fusion based on the HIV-1 Tat LTR system. 293T cells expressing HIV-1 Tat along with the wt or mutant S proteins were cocultured with 293T cells cotransfected with ACE2 and pIIIexcat plasmids, and the CAT activity of the cell lysates was determined. A representative result is shown in panel C. The background CAT level detected in the absence of the S protein was subtracted from the CAT activity detected in the presence of wt or mutant proteins. The relative membrane fusion activities of the mutant S proteins are expressed as percentages of that of the wt S protein. Panel D represents the results from three independent experiments (means ± standard deviations).
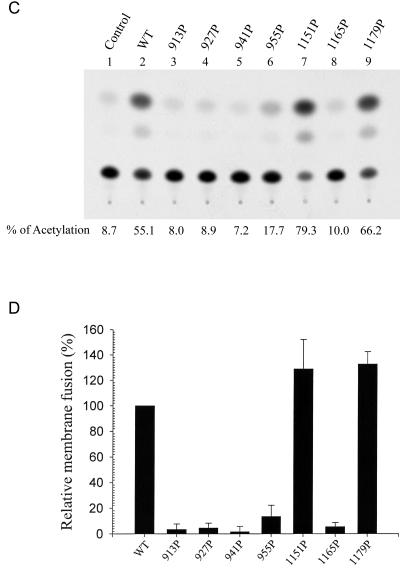
To quantify membrane fusion mediated by the wt and mutant S proteins, 293T cells cotransfected with wt or mutant S and HIV-1 Tat expression plasmids were cocultured with 293T cells cotransfected with the ACE2 expression plasmid along with pIIIexcat, in which a cat gene is driven by the HIV-1 LTR. This assay is based on the ability of Tat expressed in S-synthesizing effector cells to transactivate HIV-1 LTR-linked cat gene expression in ACE2-expressing target cells upon membrane fusion. Consistent with the membrane fusion determined by the Cre-loxP system (Fig. 5B), the 1151P and 1179P mutant proteins effectively mediated membrane fusion as did the wt S protein, whereas the membrane fusion abilities of the other five mutants were significantly reduced (Fig. 5C). When the results from three individual experiments were averaged, the 1151P and 1179P mutants were found to slightly enhance membrane fusion whereas the 913P, 927P, 941P, 955P, and 1165P mutants strikingly decreased membrane fusion, compared to the wt S protein (Fig. 5D).
Incorporation of the HR1 and HR2 mutant proteins into the HIV-1 pseudotypes.
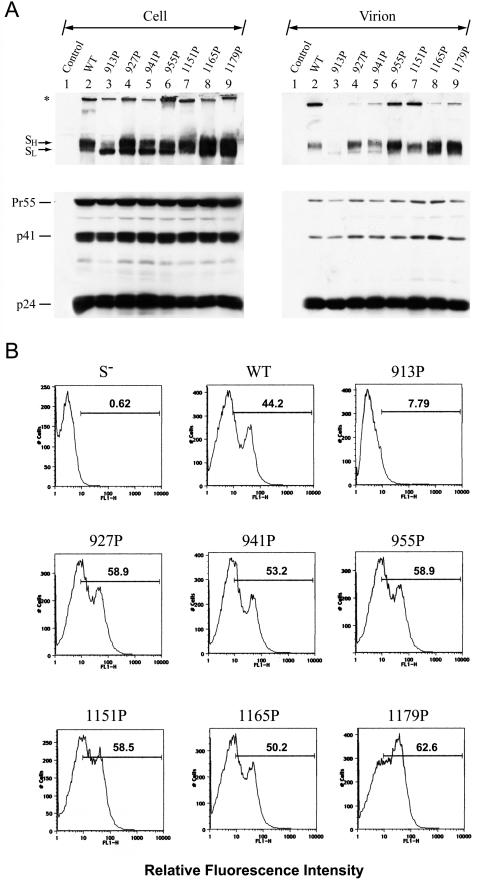
The SARS-CoV S pseudotypes based on the vector particles of VSV, murine leukemia virus, HIV, and SIV (19, 21, 24, 30, 35, 36, 41, 59) have been demonstrated, and infection with these pseudotypes is restricted to target cells expressing ACE2 (30, 31, 36). These retroviral and VSV pseudotyping systems therefore provide a quantitative assay for monitoring SARS-CoV S protein-mediated viral infection, while concomitantly avoiding the need for specialized biosafety containment for handling the live SARS-CoV. Since all of the mutant S proteins are expressed on the cell surface, we then determined whether these mutant S proteins could be assembled and incorporated into HIV-1-like particles using an env-defective, luciferase gene-containing HIV-1 reporter viral expression system. 293T cells were cotransfected with pNL4-3R−E−Luc (15) and each of the wt and mutant S plasmids. Forty-eight hours after transfection, lysates of cells and virions were subjected to SDS-PAGE, followed by Western blotting using MAb 183, which specifically recognizes HIV-1 capsid protein p24, and rabbit anti-S. Similar amounts of the Gag precursor, Pr55, and its cleaved products, p41 and p24, were detected in cells as well as virions obtained from each transfection (Fig. 6A). Consistent with the Western blotting results for cell lysates obtained 48 h after transfection (Fig. 2B), the slower-migrating SH form of the wt S protein was present at a level higher than that of the faster-migrating SL form (Fig. 6A, left panel, lane 2). With the exception of the 913P mutant, all mutant S constructs produced comparable or even slightly greater amounts of the cell-associated SH form than the wt construct (Fig. 6A, left panel). The S protein of the 913P mutant was present mainly in SL, with SH expressed at a level lower than those in the wt and other mutants (Fig. 6A, left panel), confirming that the processing of SL to SH in the 913P mutant is impaired. When virions were analyzed for S protein incorporation, the SH forms of all mutants except for the 913P mutant were sufficiently incorporated into the virions compared to wt SH (Fig. 6A, right panel). Moreover, the SL forms of the wt and all mutants were less efficiently incorporated into the virions compared to their SH counterparts (Fig. 6A, right panel).
FIG. 6.
Analyses of wt and mutant HIV/SARS S pseudotypes. (A) Incorporation of wt and mutant S proteins into the HIV-1 pseudotypes. 293T cells were cotransfected with pNL4-3R−E−Luc and either the wt or mutant S plasmids as indicated. Equal volumes of cell and virion lysates were analyzed by Western blotting using rabbit anti-S (top panels) and MAb 183 (bottom panels), respectively. The asterisk marks the high-ordered multimeric structures of S proteins. (B) ACE2-binding ability of S-pseudotyped viruses. ACE2-expressing 293T cells were incubated with RT-normalized wt or mutant pseudotypes, and the cell surface-bound S proteins were quantified by flow cytometry using rabbit anti-S. HIV-1 particles produced from pNL4-3R−E−Luc and pCDNA3 cotransfection, marked as S−, were used as a negative control.
Binding of pseudotypes to ACE2-expressing cells.
To further determine the specific interactions of mutant proteins with ACE2, binding of wt and mutant pseudotypes to ACE2-expressing 293T cells was determined by fluorescence-activated cell sorter analysis using rabbit anti-S. With the exception of the 913P mutant, which did not show effective ACE2 binding, mutant protein levels similar to or even greater than that of the wt S protein bound to the surface of ACE2-expressing target cells (Fig. 6B), indicating that mutations in HR1 and HR2 motifs do not affect the binding of the S protein present on the viral envelope to ACE2.
Effects of mutations in the HR1 and HR2 motifs on viral entry.
Since all of the mutant SH proteins, except the 913P mutant, were effectively incorporated into the HIV-1-like viral particles, we then determined whether these mutant S proteins were functional for viral entry by an env trans-complementation assay based on pNL4-3R−E−Luc, which measures viral replication in a single-cycle mode. Pseudotyped viruses produced from each of the four HR1 mutants expressed significantly reduced luciferase activity upon challenge to target Vero E6 cells compared to the wt S pseudotype (Fig. 7A). Among the HR2 mutants, the 1151P mutant mediated one-cycle viral transmission as effectively as did the wt S protein, whereas the 1179P mutant still mediated a low degree of viral entry (Fig. 7A). In contrast, viral entry mediated by the 1165P mutant was significantly impaired (Fig. 7A).
FIG. 7.
Examination of viral entry mediated by HR1 and HR2 mutant S proteins. (A) The wt and mutant pseudotypes were prepared by cotransfecting 293T cells with either the wt or mutant S plasmids along with pNL4-3R−E−Luc. Pseudotyped viruses normalized by RT activity were used to challenge Vero E6 cells, and cell lysates were assayed for luciferase activity. Results from two separate experiments, each with triplicate infection assays, were averaged, and the standard deviation was calculated. (B and C) The wt and mutant S pseudotypes based on the HIV-1 vector, pHXBΔBglCAT, were used to challenge Vero E6 cells, and cell lysates were then assayed for CAT activity. Results from a representative analysis are shown in panel B. The background CAT level detected for the defective virus produced in the absence of the S protein was subtracted from the CAT activity of the wt and mutant S pseudotypes. The relative infectivities of the mutant S pseudotypes are expressed as percentages of that of the wt S pseudotype. The results from three separate studies are shown with the means ± standard deviations indicated (C).
To confirm that the 1179P mutant still mediates a low level of viral entry activity, the pseudotyping system based on another env-deficient, cat gene-containing pHXBΔBglCAT reporter virus (22) was employed. As shown in a representative analysis, the 1179P mutant still mediated one-cycle viral infectivity compared to the other HR1 and HR2 mutants; nevertheless, viral infectivity mediated by this mutant was strikingly reduced compared to that mediated by the wt S protein (Fig. 7B). Quantitation from three individual studies clearly demonstrated that virus-to-cell transmission mediated by the 1179P mutant S protein was severely impaired but not completely blocked (Fig. 7C).
DISCUSSION
Although the trimeric hairpin structure formed by the HR1 and HR2 sequences in the SARS-CoV spike protein is believed to play a crucial role in membrane fusion, the precise roles of the HR1 and HR2 motifs in the context of virus infection have not been documented. In the present study, we provide genetic evidence to show that the α-helical structures of HR1 and HR2 motifs play a crucial role in S-mediated membrane fusion and viral entry.
Many studies have shown that Env glycoproteins often degrade intracellularly if these proteins are improperly assembled or misfolded. All mutant S proteins except the 913P mutant protein were effectively processed to yield the SH forms (Fig. 2), which presumably contain complex-type N-glycans, indicating that all of these mutant S proteins except the 913P mutant are transported to the trans-Golgi network, where complex N-glycans are added to the backbone of the S protein. Furthermore, except the 913P mutant, all mutants produced slower-migrating SH and faster-migrating SL levels similar to those of the wt S protein on the cell surface (Fig. 4A). The SL form of the 913P mutant was also effectively expressed on the cell surface compared to that of the wt and other mutants (Fig. 4A). The cell surface biotinylation method we employed to evaluate the cell surface expression of S proteins has been widely used to examine cell surface expression of the HIV envelope glycoprotein (10, 11, 37, 40). The fidelity of this method for detecting cell surface glycoproteins was provided in our previous documentation (10, 11). These results indicate that mutations in the HR1 and HR2 motifs do not significantly affect the intracellular transport of the S protein to the cell surface. The retention of a normal ACE2-binding ability by all mutants (Fig. 4B, 4C, and 6B) suggests that the S1 domain of these mutant proteins may still fold into an ACE2-binding domain even in the absence of a correctly folded S2 domain.
The kinetics of the conversion of wt SL into SH showed that a significant fraction of the SL form can be processed into the SH form within 8 h (Fig. 3). Only a small fraction of the wt S protein was present as SH when cells were analyzed 24 h after transfection (Fig. 2A), but the SH form became more dominant when cells were analyzed 48 h posttransfection (Fig. 2B). It should be noted that the S bands shown in Fig. 3 were labeled for 30 min and then chased, which merely measures the rate of processing of SL into SH. However, the Western blotting analysis reflects both the rates of de novo synthesis and processing of the S protein. It is likely that in the first 24 h after transfection, the rate of de novo synthesis of SL may far exceed the rate of its processing into SH, resulting in the SL form being dominant. However, the rate of protein synthesis may decrease due to confluence of cell cultures at later times of transfection, resulting in the accumulation of SH rather than SL.
It was noted that some of the mutant proteins, particularly the 1165P mutant, are expressed at higher levels than the wt protein (Fig. 2, Fig. 4A, top panel, and Fig. 6A, left panel), which may result in greater cell surface expression and incorporation into the HIV-1 particles than those of the wt S protein (Fig. 4A, bottom panel, and Fig. 6A, right panel). The higher expression levels of these mutants may also result in seemingly more ACE2 being coprecipitated with these mutant proteins than with the wt protein in the soluble S and ACE2-binding study (Fig. 4C). The greater incorporation of these mutant proteins than that of the wt S protein into the virus may also in turn result in apparently more binding of these mutant pseudotypes to ACE2-expressing cells (Fig. 6B). It is likely that these particular mutations may somehow stabilize the S protein, resulting in expression levels higher than that of the wt protein. Alternatively, the wt and mutant proteins may differentially assemble into high-ordered multimeric forms; the lower degree of assembly into high-ordered structures of these mutants may somehow account for seemingly greater accumulation of their SL and SH forms than those of the wt protein in cells.
It should be pointed out that Pro substitution for a conserved Leu or Ile located in the HR1 or HR2 motif does not necessarily result in impaired membrane fusion and/or viral entry. A mutation at Ile-1151 did not affect membrane fusion or viral entry, and a mutation at Leu-1179 affected only viral entry but not membrane fusion (Fig. 5 and 7). The S protein incorporation study revealed that except for the 913P mutant, which did not show incorporation of the SH form into the virus, similar levels of SH of the wt and all mutants were associated with the virions (Fig. 6A). Although the SL forms of the wt and all mutants were expressed on the cell surface as SH (Fig. 4A, bottom panel), the SL forms of the wt and all mutants were insufficiently incorporated into the HIV-1 particles (Fig. 6A, right panel). These results suggest a mechanism that selects SH but not SL for efficient incorporation into the virus. Therefore, the inability of the 913P mutant to mediate membrane fusion and viral entry (Fig. 5 and 7) is due to the low efficiency of its SH form being expressed on the cell surface and thus incorporation into the virus. Moreover, all mutant S-pseudotyped viruses except the 913P mutant pseudotype bound to ACE2-expressing cells as effectively as the wt pseudotype (Fig. 6B). Thus, the strikingly reduced viral entry of the 927P, 941P, 955P, and 1165P mutants (Fig. 7) is due to their inability to mediate membrane fusion (Fig. 5). Since the 1179P mutant was still able to mediate membrane fusion (Fig. 5), the inhibited ability of this mutant to mediate one-cycle viral entry (Fig. 7) suggests that this mutant has a defect after membrane fusion.
The envelope formation of CoVs is believed to be dominated by the laterally interacting M molecules, which form a two-dimensional lattice in intracellular membranes (38). Interaction of the E protein with the M lattice was shown to induce curving and budding of the membrane (49). These results indicate that the M and E proteins play a crucial role in CoV budding. Although the S protein is not necessary for CoV particle formation, the S protein can be incorporated into the particles through its specific interactions with the M protein (for a review, see reference 20). It is unknown whether these S mutants can still interact with the M protein and whether these mutants can be assembled into the viruses in natural SARS-CoV infection. However, the present study showed that with the exception of the 913P mutant, all mutant SH forms can be effectively transported to the cell surface and incorporated into the HIV-1 pseudotypes (Fig. 4A and 6A). Thus, our study provides a system to dissect the bona fide role of the α-helical structures of the HR1 and HR2 motifs in membrane fusion, which may not be unambiguously addressed if a real SARS-CoV infection system is employed.
Ingallinella et al. previously showed that the specific boundary of the HR1 and HR2 interaction sites can be mapped to residues 914 to 949 in HR1 and 1148 to 1185 in HR2 (26). Tripet et al. also mapped the site of interaction to residues 916 to 950 in HR1 and 1151 to 1185 in HR2 (47). Examining the fusion core structure formed by the shorter HR1 peptide and the longer HR2 one which extends up to the transmembrane region, Supekar et al. reported that the majority of the HR1 amino acid side chains in the hydrophobic a and d positions of the HRs assume a classical “knobs-into-holes” close packing typical of coiled coils despite some noted irregularities (45). In their model, the N-terminal Thr-1142 to Asn-1160 of HR2 has an extended conformation, which matches HR1 residues 962 to 929 (Fig. 1B). Ile-1161 to Ser-1178 of HR2 forms an amphipathic α-helix, which packs against the HR1 trimer grooves of residues 928 to 909 (Fig. 1B). The HR2 again assumes an extended conformation after residue 1178 for another two residues (Fig. 1B). Xu et al. also showed that residues 1161 to 1177 form a typical α-helical conformation, which packs against a deep groove consisting of residues 909 to 927 in the HR1 region, and that the other regions in HR2 form extended structures packing against the relatively shallow grooves on the surface of the central HR1 coiled coil (58).
Our mutagenesis studies showed that all HR1 mutants except for the 913P mutant, which has a defect in SH maturation (Fig. 3), exhibited an impaired membrane fusion phenotype and that a mutation at Ile-1165, which is situated in the amphipathic α-helix (Fig. 1B), also impaired membrane fusion. However, mutations at residues 1151 and 1179, which are located in the extended structure of HR2 (Fig. 1B), do not affect S-mediated membrane fusion despite the 1179P mutant possibly having a defect at a postfusion stage. These results suggest that the α-helical structure of the HR1 and HR2 motifs is critical for S-mediated membrane fusion. Replacement in these α-helical sequences with a Pro residue may severely alter the conformation of these regions, thus interfering with the formation of the six-helix bundle during membrane fusion.
Our study helps elucidate the molecular basis underlying the membrane fusion process mediated by the S protein of the SARS-CoV. Identification and characterization of the functional domains in the S protein will certainly reveal potential viral fusion targets that may have antiviral therapeutic implications.
Acknowledgments
We are indebted to the National Taiwan University SARS Research Team for providing Vero E6 cells. pNL4-3R−E−Luc was obtained from Nathaniel Landau through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. We are also grateful to Michael Farzan (Harvard Medical School, Boston, MA) for providing pCDNA3.1(−)S, pCDNA3.1(−)ACE2, and pCDNA3.1(−)ACE2-ecto, Lung-Ji Chang (University of Florida, Gainesville, FL) for providing pCEP4-tat, Andras Nagy (University of Toronto, Toronto, Canada) for providing pCAGGS-NLS-Cre, and Shie-Liang Hsieh (National Yang-Ming University, Taipei, Taiwan, Republic of China) for providing a plasmid encoding the Fc domain of human IgG1.
This work was supported by grants (NSC92-2751-B-001-022-Y and NSC94-2320-B-001-005) from the National Science Council and Academia Sinica, Taipei, Taiwan, Republic of China.
REFERENCES
- 1.Babcock, G. J., D. J. Esshaki, W. D. Thomas, Jr., and D. M. Ambrosino. 2004. Amino acids 270 to 510 of the severe acute respiratory syndrome coronavirus spike protein are required for interaction with receptor. J. Virol. 78:4552-4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bisht, H., A. Roberts, L. Vogel, A. Bukreyev, P. L. Collins, B. R. Murphy, K. Subbarao, and B. Moss. 2004. Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc. Natl. Acad. Sci. USA 101:6641-6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blacklow, S. C., M. Lu, and P. S. Kim. 1995. A trimeric subdomain of the simian immunodeficiency virus envelope glycoprotein. Biochemistry 34:14955-14962. [DOI] [PubMed] [Google Scholar]
- 4.Bosch, B. J., B. E. Martina, R. Van Der Zee, J. Lepault, B. J. Haijema, C. Versluis, A. J. Heck, R. De Groot, A. D. Osterhaus, and P. J. Rottier. 2004. Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proc. Natl. Acad. Sci. USA 101:8455-8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosch, B. J., R. van der Zee, C. A. M. de Haan, and P. J. M. Rottier. 2003. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 77:8801-8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bullough, P. A., F. M. Hughson, J. J. Skehel, and D. C. Wiley. 1994. Structure of influenza hemagglutinin at the pH of membrane fusion. Nature (London) 371:37-43. [DOI] [PubMed] [Google Scholar]
- 7.Caffrey, M., M. Cai, J. Kaufman, S. J. Stahl, P. T. Wingfield, D. G. Covell, A. M. Gronenborn, and G. M. Clore. 1998. Three-dimensional solution structure of the 44 kDa ectodomain of SIV gp41. EMBO J. 17:4572-4584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carr, C. M., and P. S. Kim. 1993. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell 73:823-832. [DOI] [PubMed] [Google Scholar]
- 9.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 10.Chan, W.-E., Y.-L. Wang, H.-H. Lin, and S. S.-L. Chen. 2004. Effect of extension of the cytoplasmic domain of human immunodeficiency type 1 virus transmembrane protein gp41 on virus replication. J. Virol. 78:5157-5169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan, W.-E., H.-H. Lin, and S. S.-L. Chen. 2005. Wild-type-like viral replication potential of human immunodeficiency virus type 1 envelope mutants lacking palmitoylation signals. J. Virol. 79:8374-8387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang, M. S., Y. T. Lu, S. T. Ho, C. C. Wu, T. Y. Wei, C. J. Chen, Y. T. Hsu, P. C. Chu, C. H. Chen, J. M. Chu, Y. L. Jan, C. C. Hung, C. C. Fan, and Y. C. Yang. 2004. Antibody detection of SARS-CoV spike and nucleocapsid protein. Biochem. Biophys. Res. Commun. 314:931-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, S. S.-L., A. A. Ferrante, and E. F. Terwilliger. 1996. Characterization of an envelope mutant of HIV-1 that interferes with viral infectivity. Virology 226:260-268. [DOI] [PubMed] [Google Scholar]
- 14.Chen, S. S.-L., S.-F. Lee, H.-J. Hao, and C.-K. Chuang. 1998. Mutations in the leucine zipper-like heptad repeat sequence of human immunodeficiency virus type 1 gp41 dominantly interfere with wild-type virus infectivity. J. Virol. 72:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conor, R. L., B. K. Chen, and N. R. Landau. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935-944. [DOI] [PubMed] [Google Scholar]
- 16.Duquerroy, S., A. Vigouroux, P. J. Rottier, F. A. Rey, and B. J. Bosch. 2005. Central ions and lateral asparagine/glutamine zippers stabilize the post-fusion hairpin conformation of the SARS coronavirus spike glycoprotein. Virology 335:276-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 18.Follis, K. E., J. York, and J. H. Nunberg. 2005. Serine-scanning mutagenesis studies of the C-terminal heptad repeats in the SARS coronavirus S glycoprotein highlight the important role of the short helical region. Virology 341:122-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukushi, S., T. Mizutani, M. Saijo, S. Matsuyama, N. Miyajima, F. Taguchi, S. Itamura, I. Kurane, and S. Morikawa. 2005. Vesicular stomatitis virus pseudotyped with severe acute respiratory syndrome coronavirus spike protein. J. Gen. Virol. 86:2269-2274. [DOI] [PubMed] [Google Scholar]
- 20.Garoff, H., R. Hewson, and D. J. Opstelten. 1998. Virus maturation by budding. Microbiol. Mol. Biol. Rev. 62:1171-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giroglou, T., J. Cinatl, Jr., H. Rabenau, C. Drosten, H. Schwalbe, H. W. Doerr, and D. von Laer. 2004. Retroviral vectors pseudotyped with severe acute respiratory syndrome coronavirus S protein. J. Virol. 78:9007-9015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Helseth, E., M. Kowalski, D. Gabuzda, U. Olshevsky, W. Haseltine, and J. Sodroski. 1990. Rapid complementation assays measuring replicative potential of human immunodeficiency virus type 1 envelope glycoprotein mutants. J. Virol. 64:2416-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hernandez, L. D., L. R. Hoffman, T. G. Wolfsberg, and J. M. White. 1996. Virus-cell and cell-cell fusion. Annu. Rev. Cell Dev. Biol. 12:627-661. [DOI] [PubMed] [Google Scholar]
- 24.Hofmann, H., K. Hattermann, A. Marzi, T. Gramberg, M. Geier, M. Krumbiegel, S. Kuate, K. Uberla, M. Niedrig, and S. Pohlmann. 2004. S protein of severe acute respiratory syndrome-associated coronavirus mediates entry into hepatoma cell lines and is targeted by neutralizing antibodies in infected patients. J. Virol. 78:6134-6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang, L. R., C. M. Chiu, S. H. Yeh, W. H. Huang, P. R. Hsueh, W. Z. Yang, J. Y. Yang, I. J. Su, S. C. Chang, and P. J. Chen. 2004. Evaluation of antibody responses against SARS coronaviral nucleocapsid or spike proteins by immunoblotting or ELISA. J. Med. Virol. 73:338-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ingallinella, P., E. Bianchi, M. Finotto, G. Cantoni, D. M. Eckert, V. M. Supekar, C. Bruckmann, A. Carfi, and A. Pessi. 2004. Structural characterization of the fusion-active complex of severe acute respiratory syndrome (SARS) coronavirus. Proc. Natl. Acad. Sci. USA 101:8709-8714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kos, C. H. 2004. Cre/loxP system for generating tissue-specific knockout mouse models. Nutr. Rev. 62:243-246. [DOI] [PubMed] [Google Scholar]
- 28.Lai, M. M. C., and K. V. Holmes. 2001. Coronaviridae: the viruses and their replication, p. 1163-1185. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 29.Lee, S.-F., C.-Y. Ko, C.-T. Wang, and S. S.-L. Chen. 2002. Effect of point mutations in the N terminus of the lentivirus lytic peptide-1 sequence of human immunodeficiency virus type 1 transmembrane glycoprotein gp41 on Env stability. J. Biol. Chem. 277:15363-15375. [DOI] [PubMed] [Google Scholar]
- 30.Li, W., T. C. Greenough, M. J. Moore, N. Vasilieva, M. Somasundaran, J. L. Sullivan, M. Farzan, and H. Choe. 2004. Efficient replication of severe acute respiratory syndrome coronavirus in mouse cells is limited by murine angiotensin-converting enzyme 2. J. Virol. 78:11429-11433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, W., M. J. Moore, N. Vasilieva, J. Sui, S. K. Wong, M. A. Berne, M. Somasundaran, J. L. Sullivan, K. Luzuriaga, T. C. Greenough, H. Choe, and M. Farzan. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature (London) 426:450-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, S., G. Xiao, Y. Chen, Y. He, J. Niu, C. R. Escalante, H. Xiong, J. Farmar, A. K. Debnath, P. Tien, and S. Jiang. 2004. Interaction between heptad repeat 1 and 2 regions in spike protein of SARS-associated coronavirus: implications for virus fusogenic mechanism and identification of fusion inhibitors. Lancet 363:938-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu, M., S. C. Blacklow, and P. S. Kim. 1995. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat. Struct. Biol. 2:1075-1082. [DOI] [PubMed] [Google Scholar]
- 34.Marra, M. A., S. J. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 35.Marzi, A., T. Gramberg, G. Simmons, P. Moller, A. J. Rennekamp, M. Krumbiegel, M. Geier, J. Eisemann, N. Turza, B. Saunier, A. Steinkasserer, S. Becker, P. Bates, H. Hofmann, and S. Pohlmann. 2004. DC-SIGN and DC-SIGNR interact with the glycoprotein of Marburg virus and the S protein of severe acute respiratory syndrome coronavirus. J. Virol. 78:12090-12095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moore, M. J., T. Dorfman, W. Li, S. K. Wong, Y. Li, J. H. Kuhn, J. Coderre, N. Vasilieva, Z. Han, T. C. Greenough, M. Farzan, and H. Choe. 2004. Retroviruses pseudotyped with the severe acute respiratory syndrome coronavirus spike protein efficiently infect cells expressing angiotensin-converting enzyme 2. J. Virol. 78:10628-10635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murakami, T., and E. O. Freed. 2000. Genetic evidence for an interaction between human immunodeficiency virus type 1 matrix and α-helix 2 of the gp41 cytoplasmic tail. J. Virol. 74:3548-3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Opstelten, D. J., M. J. Raamsman, K. Wolfs, M. C. Horzinek, and P. J. Rottier. 1995. Envelope glycoprotein interactions in coronavirus assembly. J. Cell Biol. 131:339-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 40.Salzwedel, K., J. T. West, and E. Hunter. 1999. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J. Virol. 73:2469-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simmons, G., J. D. Reeves, A. J. Rennekamp, S. M. Amberg, A. J. Piefer, and P. Bates. 2004. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl. Acad. Sci. USA 101:4240-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skehel, J. J., and D. C. Wiley. 2000. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu. Rev. Biochem. 69:531-569. [DOI] [PubMed] [Google Scholar]
- 43.Song, H. C., M. Y. Seo, K. Stadler, B. J. Yoo, Q. L. Choo, S. R. Coates, Y. Uematsu, T. Harada, C. E. Greer, J. M. Polo, P. Pileri, M. Eickmann, R. Rappuoli, S. Abrignani, M. Houghton, and J. H. Han. 2004. Synthesis and characterization of a native, oligomeric form of recombinant severe acute respiratory syndrome coronavirus spike glycoprotein. J. Virol. 78:10328-10335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sui, J., W. Li, A. Murakami, A. Tamin, L. J. Matthews, S. K. Wong, M. J. Moore, A. S. Tallarico, M. Olurinde, H. Choe, L. J. Anderson, W. J. Bellini, M. Farzan, and W. A. Marasco. 2004. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc. Natl. Acad. Sci. USA 101:2536-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Supekar, V. M., C. Bruckmann, P. Ingallinella, E. Bianchi, A. Pessi, and A. Carfi. 2004. Structure of a proteolytically resistant core from the severe acute respiratory syndrome coronavirus S2 fusion protein. Proc. Natl. Acad. Sci. USA 101:17958-17963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tan, K., J.-H. Liu, J.-H. Wang, S. Shen, and M. Lu. 1997. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc. Natl. Acad. Sci. USA 94:12303-12308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tripet, B., M. W. Howard, M. Jobling, R. K. Holmes, K. V. Holmes, and R. S. Hodges. 2004. Structural characterization of the SARS-coronavirus spike S fusion protein core. J. Biol. Chem. 279:20836-20849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Duyne, G. D. 2001. A structural view of Cre-loxp site-specific recombination. Annu. Rev. Biophys. Biomol. Struct. 30:87-104. [DOI] [PubMed] [Google Scholar]
- 49.Vennema, H., G. J. Godeke, J. W. Rossen, W. F. Voorhout, M. C. Horzinek, D. J. Opstelten, and P. J. Rottier. 1996. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 15:2020-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weissenhorn, W., L. Calder, A. Dessen, T. Laue, J. J. Skehel, and D. C. Wiley. 1997. Assembly of a rod-shaped chimera of a trimeric GCN4 zipper and the HIV-1 gp41 ectodomain expressed in Escherichia coli. Proc. Natl. Acad. Sci. USA 94:6065-6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Willey. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature (London) 387:426-430. [DOI] [PubMed] [Google Scholar]
- 52.Weissenhorn, W., S. A. Wharton, L. J. Calder, P. L. Earl, B. Moss, E. Aliprandis, J. J. Skehel, and D. C. Wiley. 1996. The ectodomain of HIV-1 env subunit gp41 forms a soluble, alpha-helical, rod-like oligomer in the absence of gp120 and the N-terminal fusion peptide. EMBO J. 15:1507-1514. [PMC free article] [PubMed] [Google Scholar]
- 53.Wilson, I. A., J. J. Skehel, and D. C. Wiley. 1981. Structure of the hemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature (London) 289:366-373. [DOI] [PubMed] [Google Scholar]
- 54.Wong, S. K., W. Li, M. J. Moore, H. Choe, and M. Farzan. 2004. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 279:3197-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiao, X., S. Chakraborti, A. S. Dimitrov, K. Gramatikoff, and D. S. Dimitrov. 2003. The SARS-CoV S glycoprotein: expression and functional characterization. Biochem. Biophys. Res. Commun. 312:1159-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu, Y., Y. Liu, Z. Lou, L. Qin, X. Li, Z. Bai, H. Pang, P. Tien, G. F. Gao, and Z. Rao. 2004. Structural basis for coronavirus-mediated membrane fusion. Crystal structure of mouse hepatitis virus spike protein fusion core. J. Biol. Chem. 279:30514-30522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu, Y., Z. Lou, Y. Liu, H. Pang, P. Tien, G. F. Gao, and Z. Rao. 2004. Crystal structure of severe acute respiratory syndrome coronavirus spike protein fusion core. J. Biol. Chem. 279:49414-49419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu, Y., J. Zhu, Y. Liu, Z. Lou, F. Yuan, Y. Liu, D. K. Cole, L. Ni, N. Su, L. Qin, X. Li, Z. Bai, J. I. Bell, H. Pang, P. Tien, G. F. Gao, and Z. Rao. 2004. Characterization of the heptad repeat regions, HR1 and HR2, and design of a fusion core structure model of the spike protein from severe acute respiratory syndrome (SARS) coronavirus. Biochemistry 43:14064-14071. [DOI] [PubMed] [Google Scholar]
- 59.Yang, Z. Y., Y. Huang, L. Ganesh, K. Leung, W. P. Kong, O. Schwartz, K. Subbarao, and G. J. Nabel. 2004. pH-dependent entry of severe acute respiratory syndrome coronavirus is mediated by the spike glycoprotein and enhanced by dendritic cell transfer through DC-SIGN. J. Virol. 78:5642-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yuan, K., L. Yi, J. Chen, X. Qu, T. Qing, X. Rao, P. Jiang, J. Hu, Z. Xiong, Y. Nie, X. Shi, W. Wang, C. Ling, X. Yin, K. Fan, L. Lai, M. Ding, and H. Deng. 2004. Suppression of SARS-CoV entry by peptides corresponding to heptad regions on spike glycoprotein. Biochem. Biophys. Res. Commun. 319:746-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhu, J., G. Xiao, Y. Xu, F. Yuan, C. Zheng, Y. Liu, H. Yan, D. K. Cole, J. I. Bell, Z. Rao, P. Tien, and G. F. Gao. 2004. Following the rule: formation of the 6-helix bundle of the fusion core from severe acute respiratory syndrome coronavirus spike protein and identification of potent peptide inhibitors. Biochem. Biophys. Res. Commun. 319:283-288. [DOI] [PMC free article] [PubMed] [Google Scholar]