Abstract
ADP-ribosylation factors (ARFs) are critical in vesicular trafficking. Brefeldin A-inhibited guanine nucleotide-exchange protein (BIG)1 and BIG2 activate ARFs by accelerating replacement of bound GDP with GTP. Additional and differing functions of these ≈200-kDa proteins are now being recognized, as are their independent intracellular movements. Here, we describe the localization in COS7 cells by immunofluorescence microscopy of BIG2, but not BIG1, with structures that have characteristics of recycling endosomes during transferrin (Tfn) uptake and Tfn receptor (TfnR) recycling. Cell content of BIG2 and Rab11, but not TfnR, BIG1, Rab4, or Exo70, was increased after 60 min of Tfn uptake. BIG2, but not BIG1, appeared in density-gradient fractions containing TfnR, Rab11, and Exo70 after 60 min of Tfn uptake. Treatment of cells with BIG2 small interfering RNA (siRNA), but not BIG1 or control siRNAs, decreased BIG2 protein >90% without affecting BIG1, ARF, or actin content, whereas TfnR was significantly increased as was its accumulation in perinuclear recycling endosomes. Tfn release appeared unaffected by BIG1 siRNA but was significantly slowed from cells treated with BIG2 siRNA alone or plus BIG1 siRNA. We suggest that BIG2 has an important role in Tfn uptake and TfnR recycling, perhaps through its demonstrated interaction with Exo70 and the exocyst complex.
Keywords: ADP-ribosylation factor, transferrin receptor, Exo70, Rab11
Formation of vesicles for specific translocation of protein and lipid molecules among eukaryotic cell organelles is initiated by binding of ADP-ribosylation factors (ARFs) or SAR family GTPase proteins to a donor membrane with localized alteration of its curvature or budding (1, 2). The mature vesicle later fuses with another target membrane to deliver its cargo. ARFs cycle between GDP-bound inactive and GTP-bound active forms. Activation requires a guanine nucleotide-exchange factor (GEF) to accelerate release of bound nucleotide from inactive ARF-GDP and permit GTP binding (3–5). All known ARF GEFs, which differ widely in molecular size and structure, contain a ≈200-aa Sec7 domain that catalyzes replacement of ARF-bound GDP with GTP (6, 7). A major functional distinction that correlates with Sec7 domain structure is the sensitivity of ARF activation to inhibition by brefeldin A (BFA). BFA-inhibited guanine nucleotide-exchange protein (BIG)1 (≈200 kDa) and BIG2 (≈190 kDa) are mammalian examples of the relatively large BFA-inhibited GEF molecules. They were initially purified together in a ≈670-kDa complex from bovine brain cytosol (8).
Endogenous BIG1 and BIG2 were immunoprecipitated together from HepG2 cells by antibodies specific for either one and colocalized microscopically, at least partially, with Golgi markers (9). Shinotsuka et al. (10, 11) were among the first to clearly distinguish BIG1 and BIG2 functions. Overexpression of a BIG2 mutant that is unable to activate ARF caused redistribution of adaptor protein 1 (AP-1) and GGA1 with tubulation of trans-Golgi network (TGN) membranes (10). Shinotsuka et al. (10, 11) concluded that BIG2 was necessary for the formation of the AP-1-clathrin adaptor complex in cells but not the coat protein complex I. Other studies implicated BIG2 in the structural integrity of recycling endosomes (REs) (12). Much of the still incomplete understanding of multiple endocytic and recycling vesicular compartments with their connecting pathways (13) is based on studies of transferrin (Tfn) and Tfn receptor (TfnR). Identification of specific molecular components, as well as information about trafficking among early endosomes (EEs), REs, and the TGN, established a role for REs in the return of internalized TfnR to the cell surface (14–17).
The interaction of an N-terminal fragment of human BIG2 in a yeast two-hybrid screen with the RIα subunit of protein kinase A and identification of three short sequences in it that bind selectively one or more of the four regulatory R subunits have been reported (18). Another product of the same yeast two-hybrid screen was an alternatively spliced Exo70 clone. Endogenous BIG2 and Exo70, a component of the exocyst complex required for exocytosis, were present in preparations of microtubules purified from human cells and with γ-tubulin in centrosome fractions from the same cells (19). We report here findings that indicate a function for endogenous BIG2 in TfnR recycling from REs to the plasma membrane in COS7 cells, perhaps involving Exo70.
Results
Endocytosis of Tfn and Localization of BIG1 and BIG2 in COS7 Cells.
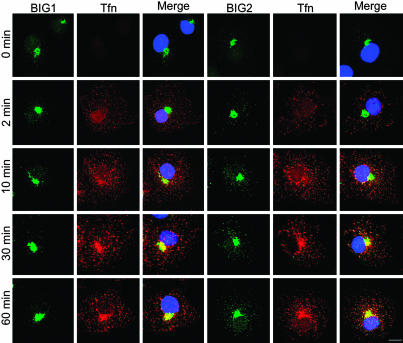
To evaluate the relationship of BIG1 and BIG2 to endosomal structures, the proteins were observed microscopically during the uptake of Alexa Fluor 568-tagged Tfn. Before addition of Tfn and after 2 min of uptake, when most of the Tfn would still be in EEs, it did not overlap with BIG1 or BIG2, which remained largely concentrated near cell nuclei, perhaps with Golgi structures (Fig. 1). After 10 min however, when some of the Tfn was expected to be in REs, BIG2 partially coincided with Tfn in small punctate collections around the initial BIG2 concentration, whereas no change in BIG1 was seen. After 30 and 60 min of uptake, when Tfn was expected to have reached Rab4/Rab11-positive REs (16), its colocalization in punctate concentrations with BIG2, but not BIG1, was widespread (Fig. 1); colocalization of BIG2 and EEA1 was not detected (data not shown). With increasing time of Tfn uptake, BIG2-stained structures increased in size, number, and distribution, consistent with BIG2 accumulation in REs.
Fig. 1.
Localization of endogenous BIG1 and BIG2 in COS7 cells by immunofluorescence microscopy during Tfn uptake. Cells were incubated without serum for 3 h and then with Alexa Fluor 568-tagged Tfn (red) at 37°C for 2, 10, 30, or 60 min before fixation and staining with anti-BIG 1 or anti-BIG2 antibodies, followed by Alexa Fluor 488-conjugated anti-rabbit IgG. The experiment was repeated three times. (Scale bar, 16 μm.)
Separation by Density Gradient Centrifugation.
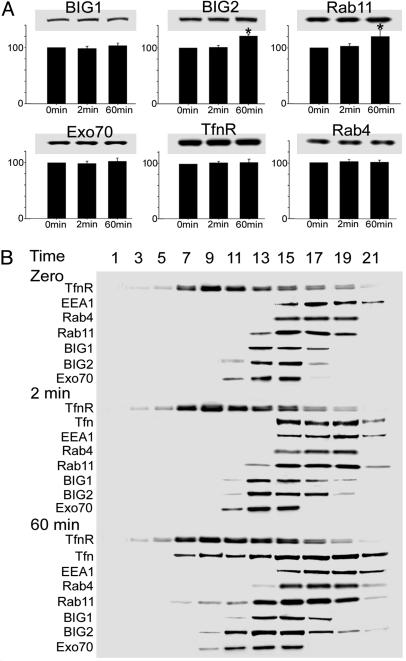
To evaluate the effects of Tfn uptake on the distribution of BIG1 and BIG2 and several proteins associated with specific intracellular structures, postnuclear supernatant fractions from cells before and after 2 or 60 min of incubation with Tfn were subjected to centrifugation in iodixanol density gradients. Western blotting of total proteins before gradient fractionation revealed that amounts of BIG2 and Rab11(but not BIG1, Rab4, Exo70, or TfnR) were significantly greater (19% and 17%, respectively) after 60 min of Tfn uptake than they were at zero time or 2 min (Fig. 2A).
Fig. 2.
Separation of EEs and REs by Optiprep gradient centrifugation. (A) Cells grown to 70% confluence on 100-mm culture plates were incubated for 3 h without serum and then with Alexa Fluor 488-tagged Tfn at 37°C for 2 or 60 min before rapid chilling, washing, and homogenization. Samples of proteins (10 μg) in postnuclear supernatant before gradient fractionation were separated in 4–12% NuPAGE Bis-Tris gel, followed by immunoblotting and densitometric quantification of BIG1, BIG2, Rab11, Rab4, Exo70, and TfnR. Data are expressed relative to those of the same protein in cells at zero time = 100 and are reported as means ± SD of values from three experiments, with blots from a representative experiment. ∗, P < 0.05. (B) Homogenates of cells prepared as described in A were centrifuged for 5 min at 1,000 × g. Samples (1 mg/ml) of supernatants were applied to 5–20% iodixanol gradients (10 ml), which were centrifuged. Samples of the indicated fractions (collected from the bottom) were analyzed by immunoblotting with antibodies against the indicated proteins. The experiment was repeated three times.
In cells fractionated after 2 min of Tfn uptake, Tfn was confined to fractions 15–21, which also contained all of Rab4 and EEA1, plus a large fraction of Rab11, as they did before Tfn addition (Fig. 2B). Most of the BIG1 and BIG2 were also present in those fractions, but their distributions quantitatively did not parallel those of the other proteins. After 60 min of uptake, Tfn was seen additionally in fractions 7–13, as was Rab11. At no time did distribution of Rab11 in the gradient parallel that of Tfn or TfnR (Fig. 2B). Relative amounts of both BIG2 and Exo70 in fractions 9–11, which may include a population of REs, were clearly greater at 60 min than at zero or 2 min (Fig. 2B). Distributions of BIG1, Rab4, and EEA1 were apparently unchanged with time, whereas TfnR was at all times most abundant in fractions 7–15, consistent with the presence of REs in these fractions (14).
Efficacy and Specificity of BIG1 and BIG2 Small Interfering RNA (siRNA).
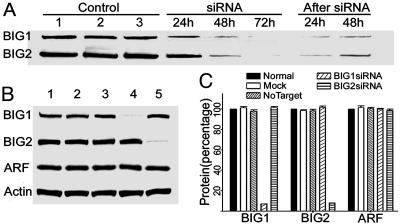
To assess in a different way the involvement of BIG2 (and BIG1) in Tfn/TfnR recycling, we used siRNA to decrease amounts of endogenous BIG1 and/or BIG2 in COS7 cells. Amounts of endogenous BIG1 and BIG2 were clearly decreased after 24 h of incubation with BIG1 or BIG2 siRNA, respectively, and were even lower after 48 h (Fig. 3A). Quantification of immunoreactive proteins by densitometry 72 h after addition of BIG1 or BIG2 siRNA (Fig. 3 B and C) showed that endogenous BIG1 and BIG2 were decreased by 93% (7 ± 0.04% vs. mock) and 92% (8 ± 0.05% vs. mock), respectively.
Fig. 3.
Effects of BIG1 and BIG2 siRNA and controls on protein levels. (A) Cells (1.5 × 105 cells per well) in six-well plates were incubated without or with siRNA as indicated. After 72 h of siRNA treatment, some cells were washed and incubated in complete DMEM for 24 or 48 h. Proteins (35 μg) in samples of cell lysates [sonified in 1 ml of buffer (20 mM Tris·HCl, pH 8.0/1 mM EDTA/1 mM NaN3 2 mM MgCl2/2 mM DTT/250 mM sucrose)] were separated in 4–12% NuPAGE Bis-Tris gel, followed by immunoblotting with antibodies against BIG1 or BIG2. Control lanes: 1, untreated; 2, mock-transfected (vehicle only); 3, nontarget siRNA. siRNA lanes: BIG1 or BIG2 treatment for 24, 48, or 72 h. After siRNA lanes: 24 or 48 h after removal of siRNA following 72-h treatment. (B) As in A, cells were incubated for 72 h with or without the indicated siRNA, and proteins were immunoblotted with antibodies against BIG1, BIG2, ARF, or actin. Lanes: 1, untreated; 2, vehicle only; 3, non-target siRNA; 4, BIG1 siRNA; 5, BIG2 siRNA. Data were similar in more than two other experiments. (C) Data are means ± SD of values from three experiments like that in B. Amounts of the indicated proteins were quantified by densitometry and expressed relative to that of the same protein in untreated normal cells = 100.
When incubation was continued after removal of siRNA at 72 h, increased BIG1 or BIG2 was detectable after 24 h and more so after 48 h (Fig. 3A). Treatment of cells with BIG1 or BIG2 siRNA for 72 h did not affect amounts of the other BIG protein or of ARF or actin (Fig. 3 B and C). Likewise amounts of these proteins were unaffected by the vehicle alone (mock) or siRNA negative control (nontargeting) (Fig. 3 B and C).
Effects of BIG1 and BIG2 siRNA on TfnR.
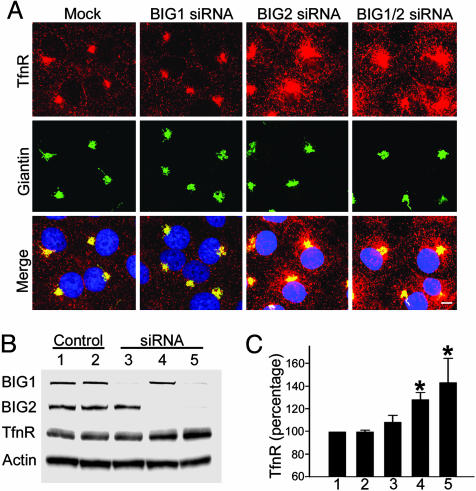
In polarized and nonpolarized cells, TfnR and Tfn are believed to remain together and, after release of iron from Tfn, to return together to the cell surface from the peripheral endosomes, either directly or indirectly via pericentriolar REs (13, 14). BIG1 siRNA did not alter the amount or distribution of TfnR seen on confocal microscopy (Fig. 4A), but BIG2 siRNA alone or with BIG1 siRNA clearly increased TfnR in pericentriolar REs very close to structures containing giantin, a marker for cis- and median-Golgi, which were not discernably affected by the siRNAs (Fig. 4A), or by Tfn uptake (data not shown). Quantification of TfnR on Western blots by densitometry confirmed that it was 30% and 40% higher in cells incubated with BIG2 siRNA or BIG2 plus BIG1 siRNA, respectively, and the smaller increment with BIG1 siRNA alone was not statistically significant (Fig. 4 B and C).
Fig. 4.
Effects of BIG1 and BIG2 siRNA on subcellular distribution of TfnR. (A) Cells (2 × 104 cells per well) in four-well culture slides were incubated for 3 h without serum after 72 h in complete medium with or without the indicated siRNA before confocal immunofluorescence microscopy with antibodies against TfnR and giantin (a cis- and medial-Golgi marker). (Scale bar, 8 μm.) (B) Proteins (10 μg) from postnuclear supernatant of cells from five 100-mm plates incubated for 72 h with or without the indicated siRNAs were separated in 4–12% NuPAGE Bis-Tris gel, followed by immunoblotting with antibodies against BIG1, BIG2, TfnR, and actin. Lanes: 1, mock-transfected; 2, non-target siRNA; 3, BIG1 siRNA; 4, BIG2 siRNA; 5, BIG1 and BIG2 siRNA. (C) Data are means ± SD of values from three experiments like that in B. TfnR was quantified by densitometry of immunoblots and expressed relative to that of the same protein in mock-transfected cells. ∗, P < 0.01 vs. mock.
Effect of BIG1 and BIG2 siRNA on Tfn Uptake and Intracellular Trafficking.
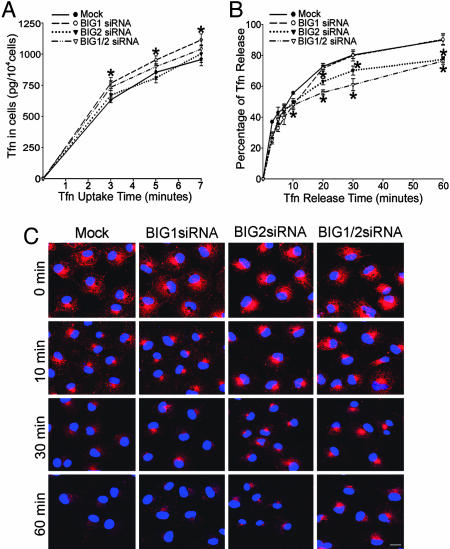
Tfn bound to the TfnR is believed to move sequentially through Rab4-positive EEs and, at least in part, through REs that contain Rab11 before being returned to the cell surface and released. To try to quantify initial rates of Tfn uptake, we measured Tfn content of cells in the first 7 min of uptake (Fig. 5A). Accumulation of Tfn in cells was not constant during this time, presumably because TfnR is cycling even without external ligand, and Tfn accumulation is always the difference between uptake and release (Fig. 5A). Although TfnR was significantly increased in cells incubated with BIG2 siRNA (Fig. 4C), confocal microscopy failed to reveal the effects of BIG1 or BIG2 siRNA alone or together on the amounts or localization of TfnR. These cells did accumulate significantly more Tfn after 3, 5, and 7 min of uptake than did cells incubated with vehicle or BIG2 siRNA alone, both of which had no effect (Fig. 5A).
Fig. 5.
Effects of BIG1 and BIG2 siRNA on Tfn uptake and release. (A) After incubation for 72 h with vehicle (mock) or the indicated siRNA, cells (1 × 104 cells per well) in 24-well plates were incubated at 37°C overnight in complete DMEM and then without serum for 3 h before addition of Alexa Fluor 488-tagged Tfn. At the indicated time thereafter, cells were washed extensively and lysed. Alexa Fluor 488-tagged Tfn in lysates was quantified by ELISA. Data are means ± SD of values from three experiments. ∗, P < 0.01 vs. mock. (B) Cells incubated with Alexa Fluor 488-tagged Tfn for 60 min as described in A were washed extensively, stripped of surface-bound Tfn, and incubated in DMEM containing unlabeled Tfn. At the indicated time, medium and cells were collected separately, Alexa Fluor 488-tagged Tfn in samples of cell lysate and medium was quantified by ELISA, and the amount in medium was expressed as the percentage of the total in medium plus cell lysate. Data are means ± SD of values from three experiments. ∗, P < 0.01 vs. mock. (C) Cells (2 × 104 cells per well) in four-well culture slides were incubated at 37°C for 72 h without or with siRNA as indicated, for 3 h without serum, and then with Alexa Fluor 568-tagged Tfn for 60 min. After being washed extensively, cells were incubated in DMEM containing unlabeled Tfn at 37°C for the indicated time before microscopy. (Scale bar, 20 μm.)
To assess Tfn release from REs, cells that had taken up Alexa Fluor 488-tagged Tfn for 60 min were washed and incubated in medium containing unlabeled Tfn; amounts of 488-tagged Tfn in medium and cells were quantified at intervals thereafter, as shown in Fig. 5B. There were no significant differences in release among mock-transfected (vehicle alone) and siRNA-transfected cells before 10 min. At later times, however, Tfn release was significantly slower from cells transfected with BIG2 or BIG2 plus BIG1 siRNA than from cells transfected with vehicle alone or BIG1 siRNA (Fig. 5B).
Microscopically (Fig. 5C), Alexa Fluor 568-tagged Tfn appeared to be largely absent from peripheral sites in most cells within 10 min, albeit a bit more persistent, perhaps, in cells transfected with BIG2 siRNA, with or without BIG1 siRNA. This finding would be consistent with the initial, faster release from all cells of ≈50% of intracellular Tfn representing largely the clearance of Tfn from EEs. After 30 and 60 min, cells transfected with BIG2, with or without BIG1, siRNA appeared to contain more Tfn than mock- or BIG1 siRNA-transfected cells (Fig. 5C).
Discussion
After endocytosis, the TfnR and Tfn are transported via clathrin-coated vesicles to peripheral EEs (20–22). From there, Tfn-TfnR directly (t1/2 = 5 min) or indirectly (t1/2 = 15–30 min) via REs are returned to the cell surface (20, 23). In our experiments, no colocalization of BIG2 (or BIG1) with Tfn was observed by confocal immunofluorescence microscopy after 2 min of internalization. With increasing time of Tfn uptake, more and larger punctate concentrations of BIG2 (but not BIG1) peripheral to its initial site did coincide with those of Tfn. Colocalization of overexpressed hemagglutinin-tagged BIG2 with TfnR was described in the “peripheral punctate structures” of HeLa cells (12). Our failure to detect TfnR and BIG2 together in gradient fractions of cells not taking up Tfn may reflect differences in the sensitivity of detection with anti-BIG2 and anti-hemagglutinin antibodies and/or differences in intracellular distribution of very different amounts of endogenous and overexpressed BIG2. In fractions of cell membranes separated on Optiprep gradients after 60 min of Tfn uptake, we found more BIG2 in fractions containing TfnR and Rab11 (a marker of REs) than at zero time or after 2 min of uptake. In our experiments, amounts of both BIG2 and Rab11 were slightly, but significantly, greater after 60 min of Tfn uptake than they were before, but the source of the increase remains to be determined.
Our group and others had reported that BIG2 was localized mainly in TGN (8–11). Shinotsuka et al. (11) reported that overexpressed hemagglutinin–BIG2 appeared to function in trafficking between the TGN and endosomes by regulating membrane association of AP-1 and GGA via ARF activation. AP-1 adaptor complexes comprise a ubiquitous AP-1A complex and an epithelial cell-specific AP-1B complex. AP-1A was implicated in sorting events between the TGN and endosomes (24), whereas AP-1B was associated with REs functioning in the polarized transport of membrane proteins to the basolateral surface of epithelial cells (25–27). It was, therefore, not surprising to find endogenous BIG2 in REs, where overexpressed BIG2 had been described (12). REs form a network of vesicles and tubules, which is often situated close to the TGN, where it appeared by confocal microscopy in the COS7 cells. There is increasing evidence that AP-1B has a sorting function in the perinuclear region, possibly in the TGN or an endosomal compartment (28, 29). Ang et al. (17) suggested that REs in polarized cells may serve not only in transport from TGN but also as a locus of sorting for both endocytic and secretory pathways. Learning from what site(s) BIG2 is translocated to REs will help us to understand better the structural and functional relationships between TGN and perinuclear REs.
The presence of BIG2 in REs is consistent with a role in regulating cargo and/or membrane traffic through this compartment. To obtain new clues to the potential functions of BIG1 and BIG2 in Tfn recycling, we used siRNAs of confirmed efficacy and specificity to decrease endogenous BIG1 or BIG2 or both. Immunofluorescence microscopy of cells 72 h after addition of BIG2 siRNA alone or with BIG1 siRNA revealed widespread, markedly increased accumulation of TfnR, including in perinuclear REs. Quantification of Tfn uptake and release with time revealed no significant effect of BIG2 siRNA on uptake in the first 10 min; the slower release of Tfn from those cells suggested delayed or impaired transport from REs to the cell surface, consistent with the accumulation of TfnR seen in such cells in the absence of Tfn uptake. Release of Tfn from cells transfected with BIG1 in addition to BIG2 siRNA was not significantly slower. All of our observations are consistent with the conclusion that BIG2 siRNA interfered only with the transit of Tfn and TfnR from REs toward the cell surface and not with that from more peripheral endocytic structures.
Sheen et al. (30) reported that BFA, which inhibits ARF activation by BIG1 and BIG2, perturbed the localization of E-cadherin and β-catenin in cultured cells by preventing their transport from Golgi membranes. Overexpression of dominant-negative BIG2, which presumably did not affect BIG1, had similar effects (30). We described the interaction in a yeast two-hybrid system of the N-terminal region of the BIG2 molecule with Exo70 (19), a protein of the exocyst complex required for exocytosis. Exo70 was reported to associate with Tfn and TfnR-positive REs, where it was recruited by overexpression of AP-1B (26). We found some Exo70 in density-gradient fractions of membranes that contained BIG2, TfnR, and Rab11. The same four proteins were seen in different fractions after 60 min of Tfn uptake, consonant with a role for BIG2 in TfnR trafficking from endoplasmic reticulum, Golgi, or REs in the perinuclear region toward the cell surface via interaction with Exo70 in the exocyst complex (14). Exocyst function has been implicated in the delivery of both neurotransmitter and hormone receptors to the cell surface (31).
Among the relatively few proteins now known to function in TfnR recycling, two Rab proteins have been convincingly implicated. Overexpression of presumably GDP-bound mutant Rab11 and GTP-bound mutant Rab5 had different effects on the pathways and kinetics of TfnR recycling (15, 32). Similarly, interference with the function of syntaxin 13 (33) or RME-1 (34) only partially altered TfnR recycling, which was also inhibited by BFA (35). None of these actions, however, seemed to affect 100% of the internalized TfnR, leading to the conclusion that not all of the multiple recycling pathways and/or molecular mechanisms depend on BFA-sensitive proteins (36). All of our evidence is consistent with a role for BIG2 in the return of a significant fraction of TfnR from REs to the cell surface to serve in Tfn uptake.
It was only several years after BIG1 and BIG2 were purified together in a macromolecular complex (8) that some of their diverse and independent activities were first reported. Although these ≈200-kDa proteins, overall, are 74% identical in amino acid sequences, 90% identical in Sec7 domains (37), and almost 90% in more than one-half of the molecule (18), there are clear localized sequence differences associated with specific functions, e.g., three AKAP sequences in BIG2, only one of which is present in BIG1 (18). Mechanisms that bring about BIG1 accumulation in the cell nucleus are still unclear. BIG2 has never been found in nuclei, but it contains sequence identical to the nuclear localization signal recently identified in BIG1 (C. Citterio, H. D. Jones, G.P.-R., A. Islam, J.M., and M.V., unpublished data). In our gradient fractionation studies of cells taking up Tfn, most of the BIG1 was recovered in the same two fractions before and after addition of Tfn, whereas, after only 2 min of Tfn uptake, the distribution of BIG2 was beginning to change (Fig. 2). In addition, incubation of cells with BIG1 siRNA for 72 h, which decreased BIG1 by >90%, had no effect on amounts of TfnR, which were significantly increased in cells treated similarly with BIG2 siRNA.
A role for iron in exacerbating neurodegenerative processes has been widely discussed. In Alzheimer’s, Parkinson’s, and Huntington’s diseases, significant accumulations of iron in affected regions of the brain suggested its deleterious effects (38). BIG2, which is relatively abundant in brain, was first purified from bovine brain cytosol (8), and gross defects in embryonic human brain development were associated with mutations of BIG2 (30). We had wondered whether those might be related to the reported interaction of BIG2 with Exo70 and a potential role in exocyst function (19). Now, we might ask whether such developmental abnormalities could result from a perturbation of iron metabolism, which is so critical in all cells, in the embryonic brain.
Materials and Methods
Antibodies and Reagents.
Rabbit anti-BIG1 and anti-BIG2 antibodies against synthetic peptides CSQPPEQELGINKQ (italic C added to facilitate coupling to keyhole limpet hemocyanin) and RLKHSQAQSK, respectively (9), were affinity-purified with the peptides used for immunization coupled to a SulfoLink kit (Pierce) according to the manufacturer’s instructions. Mouse monoclonal antibody against ARF (catalog no. MA3-060) was purchased from Affinity BioReagents (Neshanic Station, NJ); mouse monoclonal antibody against TfnR (catalog no. 13-6800) and rabbit anti-Rab11 antibodies (catalog no. 71-5300) were purchased from Zymed Laboratories; mouse anti-Tfn monoclonal antibody (catalog no. ab10208) was from Abcam (Cambridge, MA); rabbit anti-giantin antibody (catalog no. PRB-114C) was from Covance (Richmond, CA); Alexa Fluor 488- or 594-conjugated secondary antibodies, Alexa Fluor 568- or 488 -tagged Tfn, anti-Alexa Fluor 488 rabbit IgG fraction (catalog no. A-11094), and goat anti-mouse IgG–horseradish peroxidase were from Molecular Probes; o-phenylenediamine was from Sigma; EDTA-free protease inhibitor was from Roche; and all of the reagents for siRNA were from Dharmacon Research (Layfayette, CO).
Immunofluorescence Microscopy.
COS7 cells (used because they are more favorable than HepG2 cells for immunofluorescence microscopy) were grown at 37°C in 5% CO2/95% and DMEM with 10% FBS (GIBCO), 100 units/ml penicillin G, and 100 μg/ml streptomycin. For experiments, 2 × 104 cells were incubated in four-well culture slides for 24 h and then for 3 h in serum-free medium containing 0.2% BSA to deplete endogenous Tfn. After incubation with Alexa Fluor 568-tagged Tfn (25 μg/ml) for the time indicated in the figures, cells were fixed with 3% paraformaldehyde in PBS for 12 min at room temperature, washed with PBS, and incubated in blocking buffer (10% normal goat serum and 0.1% saponin in PBS) for 30 min. Cells were incubated with anti-BIG1 or -BIG2 antibodies (diluted with blocking buffer, respectively, 1:1,000 and 1:500) for 1 h at room temperature. After being washed with PBS, cells were incubated for 1 h with Alexa Fluor 488-labeled anti-rabbit IgG (diluted 1:1,000 in blocking buffer) and washed with PBS. Coverslips were mounted in Prolong Gold anti-fade reagent with DAPI (Molecular Probes) and inspected with a confocal microscope (Leica model TCS4D/DMIRBE).
Separation of EEs and REs on Optiprep Iodixanol Gradient.
COS7 cells grown to 70% confluence on 100-mm culture plates were incubated without serum for 3 h at 37°C, then without or with Alexa Fluor 488-tagged Tfn for the time indicated in the figures before washing and scraping in 50 mM Hepes (pH 7.2) containing 0.25 M sucrose, 78 mM KCl, 8.37 mM CaCl2, 10 mM EGTA, and 4 mM MgCl2 (1 ml per plate), followed by homogenization (20 strokes in a Dounce homogenizer), and centrifugation for 5 min at 1,000 × g. Samples (1 ml) of supernatants were applied to 5–20% iodixanol gradients (10 ml), which were centrifuged for 16 h at 27,000 rpm on a SW41 rotor before collection from the bottom of 500-μl fractions. Trichloroacetic acid-precipitated proteins from each fraction were dispersed in 50 μl of NuPAGE LDS sample buffer, proteins in samples (25 μl) were separated in NuPAGE gels, and blots were incubated with antibodies against the indicated proteins. Signals were detected by using LAS-3000 (Fujifilm), and the intensities of the blots were quantified by densitometry using multi gauge software (Fujifilm).
siRNA Treatment and Tfn Uptake and Release.
siRNAs against human BIG1 (5′-TGAATCACCTCAACTTAGA-3′) and BIG2 (5′-CAACTACGACTGTGAT TTA-3′) were designed and synthesized as Option A4 by Dharmacon Research. Dharmacon siCONTROL nontargeting siRNA was used as a negative control. COS7 cells were incubated for 24 h in four-well culture slides for immunofluorescence microscopy or in 100-mm plates for Western blotting, followed by incubation for 72 h in DharmaFECT2 siRNA transfection reagent containing 100 pmol of siRNA duplexes or other additions as indicated in the figures according to the manufacturer’s instructions. The cells were then washed and incubated for 3 h in serum-free medium containing 1% BSA.
For Tfn uptake, cells were incubated with Alexa Fluor 488-tagged Tfn (25 μg/ml) for the time indicated in the figures, placed on ice, quickly washed four times with ice-cold PBS, and lysed in 0.5 ml of lysis buffer (1% Triton X-100/0.1% SDS/0.2% BSA/50 mM NaCl/1 mM EDTA/10 mM Tris, pH 7.4/1:1,000 protease inhibitors). For Tfn recycling, cells were incubated with Alexa Fluor 488-tagged Tfn at 37°C for 60 min, placed on ice, washed four times with ice-cold PBS containing 0.1% BSA, stripped of surface-bound Tfn with stripping buffer (25 mM citric acid/24.5 mM sodium citrate/280 mM sucrose, pH 4.6), and incubated with serum-free DMEM containing 1% BSA and 2.5 mg/ml unlabeled Tfn for the times indicated in the figures. Medium was collected, and cells were lysed in 0.5 ml of lysis buffer. Alexa Fluor 488-tagged Tfn was quantified by ELISA.
Quantification of Tfn in Cells and Medium by ELISA.
ELISA plates were prepared by using Immulon 2HB microplates (Fisher) incubated overnight at 4°C with 10 μg/ml rabbit anti-488 IgG in 0.1 M sodium bicarbonate, pH 9.3. After being washed four times in PBST (PBS containing 0.05% Tween 20), plates were incubated overnight at 4°C with PBS containing 1% BSA and washed three times with PBST. Sample (50 μl) and PBS (50 μl) were then added to each well, followed by incubation for 2 h at room temperature. After five washes with PBST, 100 μl of 50 ng/ml mouse anti-Tfn in PBS with 0.1% BSA were added to each well, and 60 min later, the plates were washed five times with PBST before adding 100 μl of 50 ng/ml goat anti-mouse IgG–horseradish peroxidase in PBS with 0.1% BSA to each well. After 60 min and five washes with PBST, 100 μl of substrate solution (o-phenylenedianine dihydrochloride, 30 mg tablet dissolved in 15 ml of 20 mM sodium citrate buffer, pH 5.0, plus 100 μl of 3% H2O2). Addition of 100 μl of 1 M H2SO4 to each well 20 min later terminated the reactions. Absorbance at 490 nm was quantified by using an ELISA plate reader (Bio-Rad).
Acknowledgments
We thank Dr. Christian Combs (National Heart, Lung, and Blood Institute Confocal Microscopy Core Facility) and Dr. Zu-Xi Yu (National Heart, Lung, and Blood Institute Pathology Core Facility) for invaluable help and Dr. Vincent C. Manganiello for important discussions and manuscript review. This research was supported by the Intramural Research Program of the National Heart, Lung, and Blood Institute, National Institutes of Health.
Abbreviations
- ARF
ADP-ribosylation factor
- BFA
brefeldin A
- BIG
BFA-inhibited guanine nucleotide-exchange protein
- EE
early endosome
- RE
recycling endosome
- Tfn
transferrin
- TfnR
transferrin receptor
- AP-1
adaptor protein 1
- siRNA
small interfering RNA
- TGN
trans-Golgi network.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Rothman J. E., Wieland F. T. Science. 1996;272:227–234. doi: 10.1126/science.272.5259.227. [DOI] [PubMed] [Google Scholar]
- 2.Springer S., Spang A., Schekman R. Cell. 1999;97:145–148. doi: 10.1016/s0092-8674(00)80722-9. [DOI] [PubMed] [Google Scholar]
- 3.Moss J., Vaughan M. J. Biol. Chem. 1998;273:21431–21434. doi: 10.1074/jbc.273.34.21431. [DOI] [PubMed] [Google Scholar]
- 4.Jackson C. L., Casanova J. E. Trends Cell Biol. 2000;10:60–67. doi: 10.1016/s0962-8924(99)01699-2. [DOI] [PubMed] [Google Scholar]
- 5.Cox R., Mason-Gamer R. J., Jackson C. L., Segev N. Mol. Biol. Cell. 2004;15:1487–1505. doi: 10.1091/mbc.E03-06-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peyroche A., Paris S., Jackson C. L. Nature. 1996;384:479. doi: 10.1038/384479a0. [DOI] [PubMed] [Google Scholar]
- 7.Sata M., Donaldson J. G., Moss J., Vaughan M. Proc. Natl. Acad. Sci. USA. 1998;95:4204–4208. doi: 10.1073/pnas.95.8.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morinaga N., Tsai S.-C., Moss J., Vaughan M. Proc. Natl. Acad. Sci. USA. 1996;93:12856–12860. doi: 10.1073/pnas.93.23.12856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamaji R., Adamik R., Takeda K., Togawa A., Pacheco-Rodriguez G., Ferrans V. J., Moss J., Vaughan M. Proc. Natl. Acad. Sci. USA. 2000;97:2567–2572. doi: 10.1073/pnas.97.6.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shinotsuka C., Waguri S., Wakasugi M., Uchiyama Y., Nakayama K. Biochem. Biophys. Res. Commun. 2002;294:254–260. doi: 10.1016/S0006-291X(02)00456-4. [DOI] [PubMed] [Google Scholar]
- 11.Shinotsuka C., Yoshida Y., Kawamoto K., Takatsu H., Nakayama K. J. Biol. Chem. 2002;277:9468–9473. doi: 10.1074/jbc.M112427200. [DOI] [PubMed] [Google Scholar]
- 12.Shin H. W., Morinaga N., Noda M., Nakayama K. Mol. Biol. Cell. 2004;15:5283–5294. doi: 10.1091/mbc.E04-05-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maxfield F. R., McGraw T. E. Nat. Rev. Mol. Cell Biol. 2004;5:121–132. doi: 10.1038/nrm1315. [DOI] [PubMed] [Google Scholar]
- 14.Sheff D. R., Daro E. A., Hull M., Mellman I. J. Cell Biol. 1999;145:123–139. doi: 10.1083/jcb.145.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilcke M., Johannes L., Galli T., Mayau V., Goud B., Salamero J. J. Cell Biol. 2000;151:1207–1220. doi: 10.1083/jcb.151.6.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sheff D., Pelletier L., O’Connell C. B., Warren G., Mellman I. J. Cell Biol. 2002;156:797–804. doi: 10.1083/jcb.20111048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ang A. L., Taguchi T., Francis S., Fölsch H., Murrells L. J., Pypaert M., Warren G., Mellman I. J. Cell Biol. 2004;167:531–543. doi: 10.1083/jcb.200408165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H., Adamik R., Pacheco-Rodriguez G., Moss J., Vaughan M. Proc. Natl. Acad. Sci. USA. 2003;100:1627–1632. doi: 10.1073/pnas.0337678100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu K. F., Shen X., Li H., Pacheco-Rodriguez G., Moss J., Vaughan M. Proc. Natl. Acad. Sci. USA. 2005;102:2784–2789. doi: 10.1073/pnas.0409871102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gruenberg J., Maxfield F. R. Curr. Opin. Cell Biol. 1995;7:552–563. doi: 10.1016/0955-0674(95)80013-1. [DOI] [PubMed] [Google Scholar]
- 21.Mellman I. Annu. Rev. Cell Dev. Biol. 1996;12:575–625. doi: 10.1146/annurev.cellbio.12.1.575. [DOI] [PubMed] [Google Scholar]
- 22.Clague M. J. Biochem. J. 1998;336:271–282. doi: 10.1042/bj3360271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mayor S., Presley J. F., Maxfield F. R. J. Cell Biol. 1993;121:1257–1269. doi: 10.1083/jcb.121.6.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doray B., Ghosh P., Griffith J., Geuze H. J., Kornfeld S. Science. 2002;297:1700–1703. doi: 10.1126/science.1075327. [DOI] [PubMed] [Google Scholar]
- 25.Fölsch H., Pypaert M., Schu P., Mellman I. J. Cell Biol. 2001;152:595–606. doi: 10.1083/jcb.152.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fölsch H., Pypaert M., Maday S., Pelletier L., Mellman I. J. Cell Biol. 2003;163:351–362. doi: 10.1083/jcb.200309020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Taguchi T., Pypaert M., Warren G. Traffic (Oxford, U.K.) 2003;4:344–352. doi: 10.1034/j.1600-0854.2003.00091.x. [DOI] [PubMed] [Google Scholar]
- 28.Gan Y., McGraw T. E., Rodriguez-Boulan E. Nat. Cell Biol. 2002;4:605–609. doi: 10.1038/ncb827. [DOI] [PubMed] [Google Scholar]
- 29.Traub L. M., Apodaca G. Nat. Cell Biol. 2003;5:1045–1047. doi: 10.1038/ncb1203-1045. [DOI] [PubMed] [Google Scholar]
- 30.Sheen V. L., Ganesh V. S., Topcu M., Sebire G., Bodell A., Hill R. S., Grant P.E., Shugart Y. Y., Imitola J., Khoury S. J., et al. Nat. Genet. 2004;36:69–76. doi: 10.1038/ng1276. [DOI] [PubMed] [Google Scholar]
- 31.Hoogenraad C. C., Sheng M. Nat. Cell Biol. 2003;6:493–495. doi: 10.1038/ncb0603-493. [DOI] [PubMed] [Google Scholar]
- 32.Ullrich O., Reinsch S., Urbe S., Zerial M., Parton R. G. J. Cell Biol. 1996;135:913–924. doi: 10.1083/jcb.135.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prekeris R., Klumperman J., Chen Y. A., Scheller R. H. J. Cell Biol. 1998;143:957–971. doi: 10.1083/jcb.143.4.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin S. X., Grant B., Hirsh D., Maxfield F. R. Nat. Cell Biol. 2001;3:567–572. doi: 10.1038/35078543. [DOI] [PubMed] [Google Scholar]
- 35.Stoorvogel W., Oorschot V., Geuze H. J. J. Cell Biol. 1996;132:21–33. doi: 10.1083/jcb.132.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Dam E. M., Stoorvogel W. Mol. Biol. Cell. 2002;13:169–182. doi: 10.1091/mbc.01-07-0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Togawa A., Morinaga N., Ogasawara M., Moss J., Vaughan M. J. Biol. Chem. 1999;274:12308–12315. doi: 10.1074/jbc.274.18.12308. [DOI] [PubMed] [Google Scholar]
- 38.Dexter D. T., Wells F. R., Lees A. J., Agid F., Agid Y., Jenner P., Marsden C. D. J. Neurochem. 1989;52:1830–1836. doi: 10.1111/j.1471-4159.1989.tb07264.x. [DOI] [PubMed] [Google Scholar]