Abstract
The calcium/calmodulin-dependent protein phosphatase calcineurin stimulates cardiac hypertrophy in response to numerous stimuli. Calcineurin activity is suppressed by association with modulatory calcineurin-interacting protein (MCIP)1/DSCR1, which is up-regulated by calcineurin signaling and has been proposed to function in a negative feedback loop to modulate calcineurin activity. To investigate the involvement of MCIP1 in cardiac hypertrophy in vivo, we generated MCIP1 null mice and subjected them to a variety of stress stimuli that induce cardiac hypertrophy. In the absence of stress, MCIP1−/− animals exhibited no overt phenotype. However, the lack of MCIP1 exacerbated the hypertrophic response to activated calcineurin expressed from a muscle-specific transgene, consistent with a role of MCIP1 as a negative regulator of calcineurin signaling. Paradoxically, however, cardiac hypertrophy in response to pressure overload or chronic adrenergic stimulation was blunted in MCIP1−/− mice. These findings suggest that MCIP1 can facilitate or suppress cardiac calcineurin signaling depending on the nature of the hypertrophic stimulus. These opposing roles of MCIP have important implications for therapeutic strategies to regulate cardiac hypertrophy through modulation of calcineurin–MCIP activity.
The heart responds to physiological and pathological stimuli by hypertrophic growth. Although physiological hypertrophy is beneficial and enhances cardiac function, pathological hypertrophy is associated with maladaptive changes in cardiac contractility and calcium handling, cardiac fibrosis, and sudden death from arrhythmias. Thus, deciphering the details of the signaling pathways that convey hypertrophic signals is an important problem with therapeutic relevance.
Numerous signal transduction pathways have been implicated in cardiac hypertrophy and heart failure, many of which are mediated by intracellular calcium signaling (reviewed in ref. 1). The calcium/calmodulin-dependent protein phosphatase calcineurin is an especially effective inducer of cardiac growth and has been shown to be necessary and sufficient for hypertrophy in response to physiological and pathological stimuli (2). Inhibition of calcineurin activity with cyclosporin A or FK-506 can block cardiac hypertrophy in response to pressure overload, β-adrenergic stimulation, and sarcomere dysfunction (reviewed in ref. 3). Knockout mice lacking the calcineurin A (CnA) β catalytic subunit also show a diminished hypertrophic response to these stimuli (4). Members of the nuclear factor of activated T cells (NFAT) family of transcription factors serve as mediators of calcineurin signaling (5). Consistent with the notion that NFAT mediates cardiac hypertrophy, forced expression of an active form of NFATc4 is sufficient to induce hypertrophy (2), and NFATc3 knockout mice have a diminished hypertrophic response (6).
Several protein inhibitors of calcineurin have been identified including AKAP79, Cabin/Cain, and the calcineurin B homology protein (CHP) (7–10). Modulatory calcineurin-interacting protein (MCIP)1, first cloned as the product of the Down syndrome critical region 1 (DSCR1) gene on chromosome 21 (11), interacts with the catalytic A subunit of calcineurin and inhibits its phosphatase activity (12). Although we initially termed the protein myocyte-enriched calcineurin-interacting protein 1, its expression is not limited to muscle and its biological function is unlikely limited to muscle tissues. Therefore, we suggest that “modulatory” calcineurin-interacting protein may be a more appropriate term. Two other members of the MCIP gene family, MCIP2 and MCIP3, are not located on chromosome 21 (13, 14). MCIP1 and MCIP2 are expressed at the highest levels in striated muscles and brain, whereas MCIP3 is expressed at a low level in a broad range of tissues. MCIP orthologs have also been identified in other eukaryotes as well as in yeast (15) and the pathogenic fungus Cryptococcus neoformans (16).
The function of MCIP proteins as suppressors of calcineurin activity has been defined largely from studies of MCIP overexpression and support the notion that MCIP proteins antagonize the pro-hypertrophic actions of calcineurin. For example, cardiac overexpression of MCIP1 in transgenic mice can diminish hypertrophy in response to overexpression of a constitutively active form of calcineurin, as well as to chronic isoproterenol administration, pressure overload, and exercise (17, 18). MCIP1 gene expression is also up-regulated in response to calcineurin signaling, suggesting that MCIP1 functions in a feedback inhibition loop to suppress calcineurin activity (19). No loss-of-function MCIP phenotypes have been reported yet in mammalian cells, leaving open the possibility that MCIP may play additional unanticipated roles in the calcineurin pathway or other signaling pathways.
To assess the role of MCIP1 as a mediator of calcineurin signaling during cardiac hypertrophy, we disrupted the mouse MCIP1 gene. MCIP1−/− animals were viable and fertile and exhibited no overt abnormalities. However, when crossed to a transgenic line expressing activated calcineurin in the heart, MCIP1−/− mice showed an exacerbated hypertrophic response accompanied by severe fibrosis. In contrast, MCIP1−/− animals developed less hypertrophy than control littermates when subjected to aortic banding or chronic adrenergic stimulation. These findings suggest that MCIP1 plays dual roles in hypertrophic signaling by potentiating or suppressing hypertrophy depending on the stimulus.
Materials and Methods
Generation and Analysis of MCIP1−/− Mice.
MCIP1 genomic clones were isolated from a murine bacterial artificial chromosome library (Research Genetics, Birmingham, AL). The MCIP1 targeting vector was designed to remove exons 5 and 6. The left arm of the targeting construct contained ≈2.5 kb of the intron between exons 4 and 5 including the splice acceptor site of exon 5. A β-galactosidase reporter gene was fused to the splice acceptor site of exon 5. An NcoI site was engineered at the beginning of the β-galactosidase reporter to assist in genotyping. The right arm of the targeting construct contained ≈4.5 kb of a region of the MCIP1 genomic locus consisting of the intron between exons 6 and 7 and most of exon 7. The targeting vector was linearized and electroporated into mouse embryonic stem cells of strain 129 origin. Properly targeted embryonic stem cell clones were identified by Southern blot analysis and injected into blastocysts isolated from C57BL6/J mice. Chimeras obtained from the blastocyst injections were bred with C57BL6 mice to obtain offspring with the targeted allele. All experiments presented were performed in the mixed C57BL6/129 background. The muscle creatine kinase (MCK)-constitutively active CnA (CnA*) mice have been described (20). For β-galactosidase staining, embryos from timed matings or adult tissues were fixed in PBS-buffered formalin and subsequently stained overnight with 5-bromo-4-chloro-3-indolyl β-D-galactoside. Northern blot analysis was performed by using total RNA isolated from the hearts of 6-week-old mice using Trizol reagent (Invitrogen).
Histology.
Hearts were fixed in PBS-buffered formalin, embedded in paraffin, and sectioned at 5 μm for histological examination. Sections were stained with Masson's trichrome and photographed under normal bright-field microscopic conditions.
Transverse Aortic Constriction (TAC).
The protocol for constricting the transverse aorta of mice is an established procedure for producing left ventricular (LV) hypertrophy. Adult mice 10–12 weeks of age were anesthetized with ketamine (87 mg/kg) and xylazine (13 mg/kg) administered i.p. After blunt dissection through the intercostal muscles, the aorta was identified and freed by additional blunt dissection. A 7.0 silk suture was placed around the great vessel and tied around a blunt needle of 27 gauge. The blunt needle then was removed rapidly, causing a tight constriction. The surgical incision was closed in two layers with an interrupted suture pattern. Seven days after surgery, mice were killed, and their hearts were removed rapidly for assessment of chamber weight and snap-frozen for RNA isolation. Noninvasive transthoracic echocardiograms were performed according to the methods described (21).
Isoproterenol Infusion.
Adult male mice 10–12 weeks of age were anesthetized by i.p. administration of Avertin. A miniosmotic pump (Alzet, Palo Alto, CA) containing isoproterenol or saline vehicle was inserted s.c. in the back of the animal. This pump delivered 28 μg/hour per 25 kg of body weight (BW) of isoproterenol in a 0.9% saline solution or saline solution alone. Mice were killed 7 days later for assessment of cardiac hypertrophy.
Calcineurin Activity Assay.
Cardiac tissue was minced finely and homogenized in lysis buffer (50 mM Tris, pH 7.4/5 mM ascorbic acid/protease inhibitors, Roche, Indianapolis) followed by further disruption by sonication. Lysates then were cleared by centrifugation. Free phosphate was removed by passing the lysate through a Micro Bio 6 desalting column (BioRad). Calcineurin activity was measured by using the synthetic phosphorylated RII peptide (Biomol, Plymouth Meeting, PA). The amount of free phosphate released was measured with molybdate dye solution (Promega). Calcineurin-specific phosphatase activity was measured as the amount inhibited by cyclosporin A. Western blots to detect calcineurin protein were performed with the above lysates by using a calcineurin A antibody (Transduction Laboratories, Lexington, KY).
Northern Blot Analysis.
Total RNA was isolated from cardiac ventricular tissue by using Trizol (Invitrogen). Four micrograms of total RNA was spotted onto dot blots, and 15 μg of total RNA was loaded in each gel lane. Gene-specific probes were hybridized by using QuikHyb (Ambion, Austin, TX). Measurements of bound probe were made by using a Storm PhosphorImager (Molecular Dynamics).
Statistical Analysis.
Statistical significance was determined by the two-tailed Student's t test.
Results
Generation of MCIP1−/− Mice.
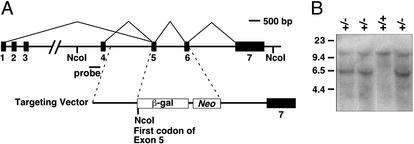
The mouse MCIP1 gene contains seven exons including four potential alternative first exons (Fig. 1A). Exons 1 and 4 contain separate promoter regions and seem to be the primary first exons used (22). We replaced exons 5 and 6, which correspond to the second and third exons of the MCIP1 mRNA expressed in the heart, with a β-galactosidase reporter fused to the splice acceptor site of exon 5 (Fig. 1A). Proper targeting and processing of the mutant mRNA creates a transcript with either exon 1 or 4 fused in frame to β-galactosidase. Because exons 1 and 4 each encode 29 amino acids of the MCIP1 protein, the targeted gene is expected to generate a fusion protein containing only the most amino-terminal portion of the MCIP1 protein, which does not interact with or inhibit calcineurin (data not shown).
Figure 1.
Targeting of the mouse MCIP1 gene. (A) Schematic representation of the MCIP1 targeting construct in which the β-galactosidase (β-gal) reporter and neomycin (Neo) gene replace exons 5 and 6 of the MCIP1 locus. The splice acceptor site of exon 5 is fused in frame with the β-galactosidase-coding region to allow for proper processing and expression of the targeted allele. NcoI sites are shown. (B) A representative Southern blot with genomic DNA digested with NcoI detecting both the wild-type (12 kb) and targeted (6 kb) allele by using the probe as shown in A.
Embryonic stem cells heterozygous for the targeted MCIP1 allele were injected into C57BL6 blastocysts to generate chimeric mice, which transmitted the mutant allele through the germ line. Southern blot analysis of offspring from chimeric mice confirmed proper targeting of the MCIP1 locus (Fig. 1B). Breeding of heterozygous mutant mice yielded MCIP1−/− mice at the expected Mendelian frequency (Table 1). Homozygous mutants were viable and fertile and showed no obvious abnormalities except that their heart weight (HW)/BW ratios were slightly reduced relative to those of wild-type controls (P < 0.05; Table 1). Given the ability of MCIP1 to suppress the cardiac growth-promoting activity of calcineurin, this finding seemed paradoxical (see below). Because MCIP1 is also expressed in the brain and skeletal muscle and at lower levels in other tissues (11), we performed detailed histopathological analyses of tissues and organs of mutant mice. No abnormalities other than the slight reduction in HW were observed.
Table 1.
Genotype and HW/BW ratios of offspring from MCIP1+/− intercrosses
| MCIP1+/+ | MCIP1+/− | MCIP1−/− | |
|---|---|---|---|
| Mendelian ratio | |||
| n | 66 | 126 | 58 |
| % | 26.4 | 50.4 | 23.2 |
| HW/BW ratio | |||
| n | 7 | 4 | 8 |
| BW, g | 22.64 ± 0.61 | 21.22 ± 1.64 | 20.65 ± 0.92 |
| HW/BW, mg/g | 5.35 ± 0.18 | 4.90 ± 0.14 | 4.80 ± 0.07* |
Ratio of genotypes represented at time of weaning is shown. HW/BW measurements were performed in female mice 12–15 weeks of age. HW/BW is expressed as mg/g, and all data are presented as mean ± SEM.
P < 0.05, MCIP1+/+ vs. MCIP1−/−.
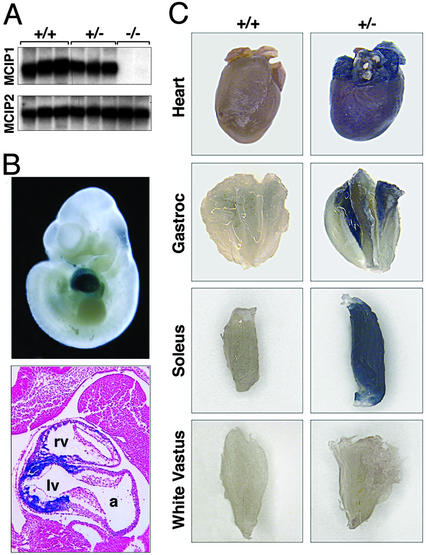
Northern blot analysis revealed no detectable MCIP1 mRNA in hearts of homozygous mutant animals (Fig. 2A). Western blot analysis also showed no detectable MCIP1 protein in the hearts or brains of mutant mice (data not shown and E. Bush, personal communication). There was no difference in expression of MCIP2 mRNA in hearts from wild-type and MCIP1−/− mice (Fig. 2A), ruling out the possibility that it might be up-regulated to compensate for the absence of MCIP1.
Figure 2.
MCIP1 and LacZ expression in MCIP1−/− mice. (A) Northern blot analysis was performed by using total RNA isolated from hearts of wild-type (+/+), heterozygous (+/−), or MCIP1 null (−/−) mice. Probes for the MCIP1 and MCIP2 genes were used. (B) Whole mount (Upper) and histological section (Lower) of a lacZ-stained MCIP1+/− embryo at day 10. Only ventricular myocardium staining is detected at this time point. a, atrium; lv, left ventricle; rv, right ventricle. (C) The heart and gastrocnemius, soleus, and white vastus muscle groups were isolated from either wild-type (+/+) or MCIP1 heterozygous (+/−) adult mice and stained for β-galactosidase activity.
The β-galactosidase reporter integrated into the MCIP1 locus was expressed in the same pattern as the endogenous MCIP1 gene during embryonic development. At embryonic day 10, β-galactosidase activity was detectable only in the developing heart (Fig. 2B), which is consistent with earlier published results showing MCIP1 expression primarily in the developing heart at embryonic days 9.5 and 10.5 (23). Examination of histological sections revealed that this staining was localized to the ventricular myocardium. We also examined reporter gene activity in various postnatal tissues of adult MCIP1+/− animals. β-Galactosidase activity was present throughout the adult heart including the atria (Fig. 2C). Staining was also detected in skeletal muscle containing a high proportion of slow-twitch-type fibers including the soleus and was absent in primarily fast-twitch muscle groups such as white vastus. β-Galactosidase activity was also present in the adult brain, particularly in the hippocampus and dentate gyrus (data not shown).
Calcineurin Activity Is Decreased in MCIP1−/− Mice.
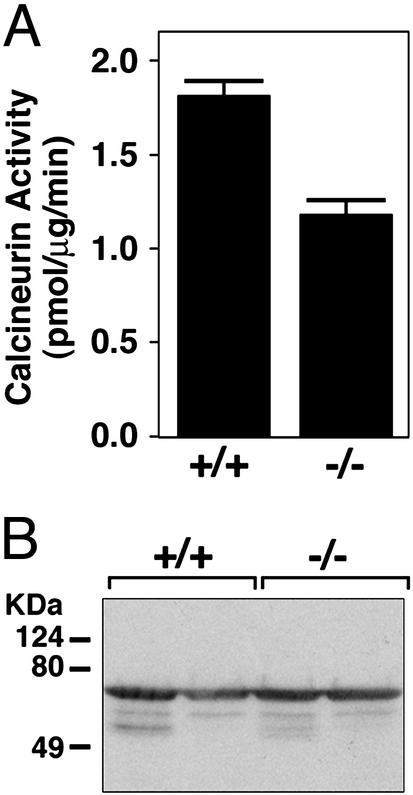
Based on its up-regulation in response to calcineurin signaling, MCIP1 has been hypothesized to participate in a negative feedback loop to suppress calcineurin activity (19). Surprisingly, we observed a decrease of ≈45% in the level of calcineurin activity in cardiac extracts from MCIP1 null mice (Fig. 3A) that was not due to a decrease in calcineurin protein levels (Fig. 3B). These findings suggested that MCIP1 might be required for maximal activity of calcineurin. It should be pointed out that this calcineurin assay detects maximal potential calcineurin activity in the presence of optimal amounts of calcium and calmodulin. If there were differences in calcineurin activity in wild-type and mutant hearts in vivo due to, for example, differences in subcellular localization or association of the enzyme with positive or negative cofactors, it is unlikely such differences would be detected in this assay.
Figure 3.
Calcineurin activity in MCIP1 mutant mice. (A) Calcineurin activity was measured from cardiac lysates prepared from wild-type (+/+) or MCIP1 mutant (−/−) mice. The values shown are mean of eight hearts ± SEM. (B) A representative Western blot is shown to detect CnA levels using extracts prepared from individual hearts for the calcineurin activity assay described for A.
Exacerbated Response of MCIP1−/− Mice to Calcineurin Overexpression.
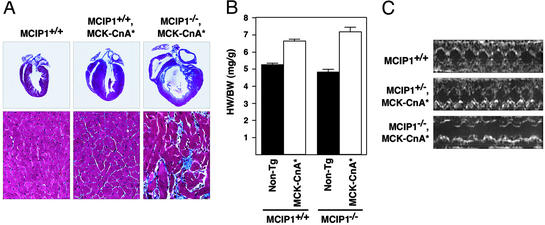
Transgenic mice expressing an activated form of calcineurin (CnA*) in the heart under control of the α myosin heavy chain (αMHC) promoter develop marked cardiac hypertrophy leading to failure and death (2). We bred mice bearing an αMHC-CnA* transgene into the MCIP1−/− background to determine whether cardiac hypertrophy would be influenced by the absence of MCIP1. However, few MCIP1−/− animals harboring the CnA* transgene were obtained, preventing a careful examination of the phenotype. Therefore, we also crossed the MCIP1−/− mice to transgenic mice expressing CnA* under the control of the MCK promoter (MCK-CnA*) (20) that develop cardiac hypertrophy, albeit to a lesser extent than the αMHC-CnA* animals (ref. 20 and Fig. 4A). The hypertrophic response to calcineurin overexpression in this model was exacerbated in the MCIP1−/− background (Fig. 4A). Mutant mice harboring the MCK-CnA* transgene also developed severe cardiac fibrosis, with large areas of myocyte dropout surrounding the fibrotic zones (Fig. 4A). Terminal deoxynucleotidyltransferase-mediated dUTP end-labeling staining of heart sections revealed no increase in myocyte apoptosis in these hearts (data not shown). The increase of HW/BW ratios caused by the CnA* transgene was larger in the MCIP1−/− animals than in wild-type littermates (49% vs. 26% increase; Fig. 4B), although this may be an underestimate due to the amount of myocyte dropout. Transthoracic echocardiography was performed to determine whether the lack of MCIP1 exacerbated the functional deficit resulting from calcineurin activation. M-mode echocardiographic analysis revealed dilation and decreased fractional shortening in the MCIP1−/−, MCK-CnA* background compared with the MCIP1+/−, MCK-CnA* background (Fig. 4C). The MCK-CnA* transgene is expressed in skeletal muscle as well as cardiac myocytes and has been shown to increase the percentage of type I fibers (20). The skeletal muscle fiber type composition of MCIP−/−, MCK-CnA* remains to be determined.
Figure 4.
Response of MCIP1−/− mice to calcineurin overexpression. (A) Hearts from 8-week-old wild-type and MCK-CnA* transgenic mice with the indicated MCIP1 genotypes were sectioned and stained with Masson's trichrome. High magnification of the sections are also shown. Large areas of fibrosis (stained blue) as well as myofibrillar disarray are evident in the MCIP1 null mouse with the MCK-CnA* transgene. (B) HW/BW ratios of nontransgenic (filled bars) and MCK-CnA* transgenic (open bars) mice of the indicated MCIP1 genotype were measured at 8 weeks of age. Data are represented as mean ± SEM. (C) M-mode echocardiographic tracings of unanesthetized mice of the indicated genotype are shown.
Diminished Hypertrophic Response of MCIP1−/− Mice to TAC.
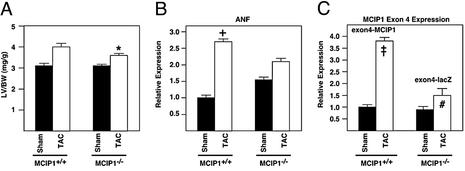
We next examined the hypertrophic response of the MCIP1−/− animals in a pressure-overload model of TAC. Unexpectedly, the MCIP1−/− animals exhibited a smaller degree of hypertrophy than wild-type controls (16% vs. 29% increase in LV weight/BW; Fig. 5A). Echocardiographic measurements confirmed this finding (Table 2). Increases in LV and septal wall thickness in TAC mice were also smaller in the MCIP1−/− animals compared with wild-type controls. Chamber diameter was similar in the wild-type and MCIP1−/− animals, indicating that no dilation occurred.
Figure 5.
Response of MCIP1−/− mice to TAC. (A) LV weight/BW ratios were measured in sham-operated (filled bars) and TAC (open bars) wild-type and MCIP1 null mice. The MCIP1 null animals developed less pressure-overload-induced hypertrophy than wild-type controls (*, P = 0.05, LV weight/BW of TAC MCIP1+/+ vs. TAC MCIP1−/−). The levels of transcripts for ANF (B) and MCIP1 exon 4 variant (C) were measured by dot blot analysis. The levels of MCIP1 exon 4 variant shown in C represent the endogenous exon 4-MCIP1 transcript (MCIP1+/+) or the exon 4-lacZ fusion transcript (MCIP1−/−). Increases in ANF and the calcineurin-responsive MCIP1 exon 4 variant were diminished in the MCIP1 mutant mice compared with wild-type controls. ANF and MCIP1 levels were normalized against GAPDH expression levels. All data are represented as mean ± SEM. †, P < 0.05 wild-type TAC vs. wild-type sham; ‡, P < 0.01 wild-type TAC vs. wild-type sham; #, P < 0.01 MCIP1−/− TAC vs. wild-type TAC.
Table 2.
Echocardiographic assessment of the LV chamber
| MCIP1+/+ | MCIP1+/+ | MCIP1−/− | MCIP1−/− | |
|---|---|---|---|---|
| Procedure | Sham | TAC | Sham | TAC |
| No. of animals | 4 | 4 | 5 | 5 |
| LVPWd, mm | 0.58 ± 0.03 | 0.79 ± 0.02 | 0.55 ± 0.02 | 0.62 ± 0.06 |
| IVSd, mm | 0.58 ± 0.03 | 0.81 ± 0.04 | 0.62 ± 0.03 | 0.70 ± 0.06 |
| LVIDd, mm | 3.65 ± 0.17 | 3.30 ± 0.13 | 3.26 ± 0.07 | 3.09 ± 0.15 |
| LVPWs, mm | 1.35 ± 0.09 | 1.60 ± 0.05 | 1.43 ± 0.13 | 1.39 ± 0.06 |
| IVSs, mm | 1.51 ± 0.01 | 1.81 ± 0.12 | 1.38 ± 0.08 | 1.44 ± 0.09 |
| LVIDs, mm | 1.50 ± 0.11 | 1.24 ± 0.16 | 1.19 ± 0.08 | 1.25 ± 0.12 |
| LVM, mg | 66.1 ± 7.8 | 85.6 ± 6.6 | 54.9 ± 4.3 | 60.9 ± 11.6 |
| FS, % | 59.1 ± 1.4 | 62.7 ± 3.4 | 63.5 ± 2.5 | 59.7 ± 2.4 |
Measurements using transthoracic M-mode echocardiography were made from sham and banded MCIP1+/+ and MCIP1−/− mice 7 days postoperative TAC procedure. LVPWd and LVPWs, end-diastolic and end-systolic posterior wall thickness, respectively; IVSd and IVSs, end-diastolic and end-systolic intraventricular septal thickness, respectively; LVIDd and LVIDs, end-diastolic and end-systolic LV internal dimensions, respectively; LVM, echocardiagraphic-derived LV mass; FS, fractional shortening (LVIDd − LVIDs/LVIDd). Data are represented as mean ± SEM.
Expression of atrial natriuretic factor (ANF), a fetal cardiac gene and marker of hypertrophy, correlated with the relative changes in LV weight/BW of wild-type and null mice (Fig. 5B). Namely, the increase in ANF expression in banded animals was greater in the wild-type mice compared with the MCIP1−/− animals (2.8- vs. 1.3-fold). Interestingly, ANF expression was slightly higher in sham MCIP1−/− animals as compared with wild-type sham controls.
Because MCIP1 expression is activated by calcineurin and expression of the calcineurin-responsive exon 4 isoform of MCIP1 is induced in response to pressure overload (17), we measured induction of MCIP1 gene expression as a measure of calcineurin activation in hearts after TAC. It is possible to measure the level of expression of MCIP1 isoforms initiated from exons 1 or 4, because the targeted allele contains both exons, which are spliced properly to the β-galactosidase reporter. Induction of the MCIP1 exon 4 isoform was greater in wild-type than homozygous null animals in response to aortic banding (3.8- vs. 1.5-fold; Fig. 5C). However, we cannot rule out the possibility that the targeted MCIP1 locus is somehow unresponsive for reasons not related to calcineurin activation or that the stability of the exon 4-LacZ fusion transcript does not allow for an accurate measurement of promoter activity. Expression of the MCIP2 gene did not change in banded animals in either the wild-type or MCIP1−/− group (data not shown).
Diminished Hypertrophic Response of MCIP1−/− Mice to Chronic Adrenergic Stimulation.
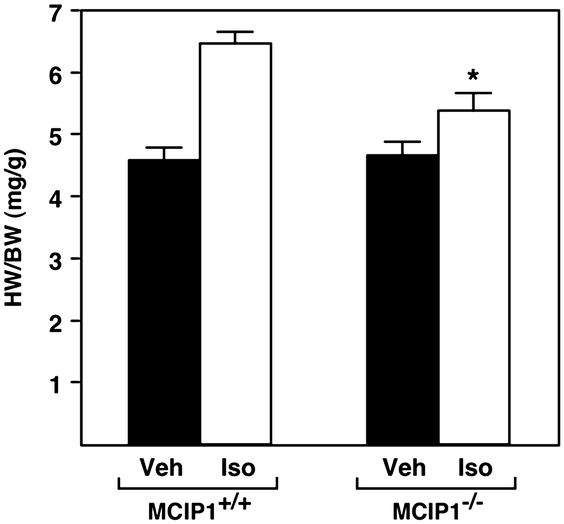
Chronic delivery of the β-adrenergic agonist, isoproterenol, through an osmotic minipump also results in hypertrophic growth. As shown in Fig. 6, MCIP1−/− mice showed a diminished hypertrophic response to isoproterenol compared with wild-type controls (16% vs. 41% increase in LV weight/BW, P < 0.05).
Figure 6.
Response of MCIP1 mutant mice to chronic adrenergic stimulation. HW/BW ratios were measured in wild-type or MCIP1 mutant mice treated for 7 days with isoproterenol infusion (Iso, open bars) or vehicle alone (Veh, filled bars). The hypertrophic response of the MCIP1 mutant mice compared with wild-type controls was significantly lower (*, P < 0.05, HW/BW of isoproterenol-treated MCIP1−/− mice vs. isoproterenol-treated MCIP1+/+ mice).
Discussion
Cardiac hypertrophy is a complex process involving multiple interconnected signaling pathways. It has become increasingly clear that calcineurin plays a critical role in this process. The ability of MCIP1 to inhibit calcineurin activity and of calcineurin to stimulate MCIP1 expression led to the hypothesis that MCIP1 functions in an inhibitory feedback loop to suppress calcineurin activity (19). Consistent with this hypothesis, MCIP1−/− mice showed an exaggerated hypertrophic response to constitutively active calcineurin expressed from a muscle-specific promoter. However, contrary to expectations, MCIP1−/− mice were partially resistant to other hypertrophic stimuli including aortic banding and isoproterenol infusion.
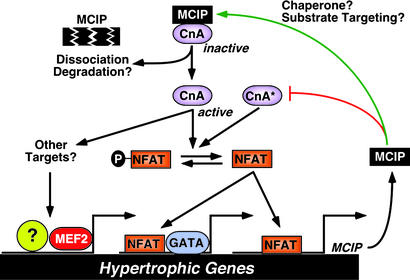
How can the paradoxical responses of MCIP1 mutant mice to these different hypertrophic stimuli be explained? We favor the interpretation that MCIP1 plays dual roles in the regulation of calcineurin activity. These roles depend on both the nature and strength of the hypertrophic stimulus and on the level of expression of MCIP1 (Fig. 7). High concentrations of MCIP1 inhibit calcineurin and attenuate hypertrophy resulting from diverse stimuli. Under conditions in which calcineurin activity is high, as in the case of MCK-CnA* transgenic mice, MCIP1 acts to suppress calcineurin signaling, but more subtle permissive effects of MCIP1 on calcineurin activity are lost. Thus, in this setting the hypertrophic response is enhanced by the lack of MCIP1. More striking than the increased hypertrophic response of the MCIP1−/−, MCK-CnA* hearts is the gross histologic abnormalities. Although there is an exaggerated hypertrophic response, the degree of hypertrophy is not as great as seen in other genetic models of cardiac hypertrophy such as the αMHC-CnA* transgenic animal (2). The marked fibrosis and myocyte dropout evident in the MCIP1−/−, MCK-CnA* hearts may indicate a severe dysregulation of calcineurin activity. Echocardiographic analysis of MCIP1−/−, MCK-CnA* animals confirmed decreased cardiac function. In contrast, in the presence of physiological levels of calcineurin signaling, as under conditions of pressure overload or chronic β-adrenergic stimulation, MCIP1 seems to be required for the full hypertrophic response. We speculate that the diminished responsiveness of MCIP1 mutant mice to the latter stimuli reflects a permissive role of MCIP1 in calcineurin activation for two reasons. First, activation of the calcineurin-responsive MCIP1 promoter upstream of exon 4 was impaired in mutant mice after aortic banding. Second, calcineurin activity was reduced in cardiac lysates from MCIP1−/− mice. Although we have attributed the antithetical cardiac responses of MCIP1 mutant mice to opposing roles of MCIP1 in the control of calcineurin activity, it is also possible that MCIP1 has additional targets other than calcineurin that differentially affect hypertrophic signaling. It should also be pointed out that the incomplete effect of MCIP1 deletion on calcineurin responsiveness may be due to functional redundancy with MCIP2 and MCIP3.
Figure 7.
A model for the dual roles of MCIP1 in cardiac growth. MCIP1 performs an initial permissive function for calcineurin activity, possibly serving as a chaperone-like protein or to properly localize calcineurin to target substrates. However, activation of calcineurin requires release from MCIP1 inhibition through an as-yet-unknown mechanism that may include MCIP1 degradation or dissociation from calcineurin. An activated calcineurin molecule (CnA*) is predicted to bypass the permissive function of MCIP1 and to be hyperactivated in the absence of MCIP1. The hypertrophic effect of calcineurin can be mediated through dephosphorylation of NFAT and its subsequent translocation to the nucleus, where it interacts with specific binding partners such as GATA to activate the cardiac hypertrophic growth program. Calcineurin also has other targets that influence cardiac growth, such as MEF2.
The notion that MCIP1 may play dual roles in the control of calcineurin activity is supported by loss-of-function phenotypes of MCIP orthologs in other organisms. The yeast MCIP ortholog Rcn1p was identified as a high copy suppressor of calcineurin (15). However, rcn1 mutants exhibit impaired calcineurin signaling. Targeted disruption of the MCIP1 ortholog, calcineurin-binding protein 1 (CBP1), in the pathogenic fungus C. neoformans, also results in partial loss of calcineurin function, again suggesting that MCIP proteins contribute to calcineurin activity (16).
Precisely how MCIP1 might potentiate calcineurin function remains to be determined. In yeast, it seems that calcineurin protein levels are partially stabilized by Rcn1p (15), although this does not seem sufficient to account for the full effect of Rcn1p on calcineurin signaling. Although we detected no differences in calcineurin protein levels in the MCIP1−/− mice, we have observed an increase in MCIP1 protein stability in the presence of calcineurin (data not shown). In this regard, the protein phosphatase 1 (PP1) regulatory subunit, inhibitor 2, has been proposed to assist in proper folding of PP1, and its association with PP1 inhibits phosphatase activity (24, 25). Although MCIP1 seems to potentiate calcineurin activity under certain conditions, calcineurin bound by MCIP1 is inactive. Presumably, there must be mechanisms to relieve inhibition by MCIP1, which may occur by changes in MCIP1 phosphorylation, stability, or by other means. Interestingly, we have shown previously that MCIP1 can be phosphorylated at the conserved SP repeat of the protein in vitro by glycogen synthase kinase 3β, a negative regulator of cardiac hypertrophy (26, 27). In addition, mutation of these serines in the C. neoformans ortholog, calcineurin-binding protein 1, leads to decreased stability of the protein (16). MCIP1 may also be required to properly localize calcineurin to its cellular substrates. Overexpression of MCIP1 such as in the αMHC-MCIP1 transgenic model would be expected to shift the equilibrium toward the inactive, MCIP1-bound state (18). The constitutively active form of calcineurin expressed in the MCK-CnA* transgenic mouse may function independently of a permissive influence of MCIP1. Alternatively, or in addition, the constitutively active form of calcineurin, which lacks the carboxyl-terminal regulatory region, could be independent of a permissive function due to altered subcellular localization or improper substrate targeting such that calcineurin signaling by this form of the protein would be dysregulated by the lack of MCIP1. Irrespective of the molecular basis for the reduction in calcineurin function in MCIP1 mutant mice, it is clear that the exact stoichiometry between calcineurin and MCIP1 is critical in maintaining proper calcineurin activity levels in the cell.
These phenotypes of MCIP1 mutant mice underscore the complexity of signaling pathways involved in cardiac hypertrophy. Further investigation of the regulatory interactions between calcineurin and MCIP proteins in the heart and other organs, and of the functions of MCIP2 and MCIP3, undoubtedly will lead to a greater understanding of calcineurin function.
Acknowledgments
We thank A. Tizenor for graphics assistance, J. Page for editorial assistance, Y. Cheah for blastocyst injections, and Dr. J. Richardson and J. Shelton for histological sections. This work was supported by the National Institutes of Health, the Donald W. Reynolds Foundation, and the Muscular Dystrophy Association (to E.N.O.).
Abbreviations
- CnA
calcineurin A subunit
- CnA*
constitutively active CnA
- NFAT
nuclear factor of activated T cells
- MCIP
modulatory calcineurin-interacting protein
- MCK
muscle creatine kinase
- TAC
transverse aortic constriction
- LV
left ventricular
- BW
body weight
- HW
heart weight
- αMHC
α myosin heavy chain
- ANF
atrial natriuretic factor
References
- 1.Frey N, Olson E N. Annu Rev Physiol. 2003;65:45–80. doi: 10.1146/annurev.physiol.65.092101.142243. [DOI] [PubMed] [Google Scholar]
- 2.Molkentin J D, Lu J R, Antos C L, Markham B, Richardson J, Robbins J, Grant S R, Olson E N. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leinwand L A. Proc Natl Acad Sci USA. 2001;98:2947–2949. doi: 10.1073/pnas.051033698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bueno O F, Wilkins B J, Tymitz K M, Glascock B J, Kimball T F, Lorenz J N, Molkentin J D. Proc Natl Acad Sci USA. 2002;99:4586–4591. doi: 10.1073/pnas.072647999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crabtree G R, Olson E N. Cell. 2002;109,Suppl.:S67–S79. doi: 10.1016/s0092-8674(02)00699-2. [DOI] [PubMed] [Google Scholar]
- 6.Wilkins B J, De Windt L J, Bueno O F, Braz J C, Glascock B J, Kimball T F, Molkentin J D. Mol Cell Biol. 2002;22:7603–7613. doi: 10.1128/MCB.22.21.7603-7613.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kashishian A, Howard M, Loh C, Gallatin W M, Hoekstra M F, Lai Y. J Biol Chem. 1998;273:27412–27419. doi: 10.1074/jbc.273.42.27412. [DOI] [PubMed] [Google Scholar]
- 8.Lai M M, Burnett P E, Wolosker H, Blackshaw S, Snyder S H. J Biol Chem. 1998;273:18325–18331. doi: 10.1074/jbc.273.29.18325. [DOI] [PubMed] [Google Scholar]
- 9.Lin X, Sikkink R A, Rusnak F, Barber D L. J Biol Chem. 1999;274:36125–36131. doi: 10.1074/jbc.274.51.36125. [DOI] [PubMed] [Google Scholar]
- 10.Sun L, Youn H D, Loh C, Stolow M, He W, Liu J O. Immunity. 1998;8:703–711. doi: 10.1016/s1074-7613(00)80575-0. [DOI] [PubMed] [Google Scholar]
- 11.Fuentes J J, Pritchard M A, Planas A M, Bosch A, Ferrer I, Estivill X. Hum Mol Genet. 1995;4:1935–1944. doi: 10.1093/hmg/4.10.1935. [DOI] [PubMed] [Google Scholar]
- 12.Rothermel B, Vega R B, Yang J, Wu H, Bassel-Duby R, Williams R S. J Biol Chem. 2000;275:8719–8725. doi: 10.1074/jbc.275.12.8719. [DOI] [PubMed] [Google Scholar]
- 13.Miyazaki T, Kanou Y, Murata Y, Ohmori S, Niwa T, Maeda K, Yamamura H, Seo H. J Biol Chem. 1996;271:14567–14571. doi: 10.1074/jbc.271.24.14567. [DOI] [PubMed] [Google Scholar]
- 14.Strippoli P, Lenzi L, Petrini M, Carinci P, Zannotti M. Genomics. 2000;64:252–263. doi: 10.1006/geno.2000.6127. [DOI] [PubMed] [Google Scholar]
- 15.Kingsbury T J, Cunningham K W. Genes Dev. 2000;14:1595–1604. [PMC free article] [PubMed] [Google Scholar]
- 16.Gorlach J, Fox D S, Cutler N S, Cox G M, Perfect J R, Heitman J. EMBO J. 2000;19:3618–3629. doi: 10.1093/emboj/19.14.3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hill J A, Rothermel B, Yoo K D, Cabuay B, Demetroulis E, Weiss R M, Kutschke W, Bassel-Duby R, Williams R S. J Biol Chem. 2002;277:10251–10255. doi: 10.1074/jbc.M110722200. [DOI] [PubMed] [Google Scholar]
- 18.Rothermel B A, McKinsey T A, Vega R B, Nicol R L, Mammen P, Yang J, Antos C L, Shelton J M, Bassel-Duby R, Olson E N, Williams R S. Proc Natl Acad Sci USA. 2001;98:3328–3333. doi: 10.1073/pnas.041614798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang J, Rothermel B, Vega R B, Frey N, McKinsey T A, Olson E N, Bassel-Duby R, Williams R S. Circ Res. 2000;87:E61–E68. doi: 10.1161/01.res.87.12.e61. [DOI] [PubMed] [Google Scholar]
- 20.Naya F J, Mercer B, Shelton J, Richardson J A, Williams R S, Olson E N. J Biol Chem. 2000;275:4545–4548. doi: 10.1074/jbc.275.7.4545. [DOI] [PubMed] [Google Scholar]
- 21.Rogers J H, Tamirisa P, Kovacs A, Weinheimer C, Courtois M, Blumer K J, Kelly D P, Muslin A J. J Clin Invest. 1999;104:567–576. doi: 10.1172/JCI6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuentes J J, Pritchard M A, Estivill X. Genomics. 1997;44:358–361. doi: 10.1006/geno.1997.4866. [DOI] [PubMed] [Google Scholar]
- 23.Casas C, Martinez S, Pritchard M A, Fuentes J J, Nadal M, Guimera J, Arbones M, Florez J, Soriano E, Estivill X, Alcantara S. Mech Dev. 2001;101:289–292. doi: 10.1016/s0925-4773(00)00583-9. [DOI] [PubMed] [Google Scholar]
- 24.MacKintosh C, Garton A J, McDonnell A, Barford D, Cohen P T, Tonks N K, Cohen P. FEBS Lett. 1996;397:235–238. doi: 10.1016/s0014-5793(96)01175-1. [DOI] [PubMed] [Google Scholar]
- 25.Alessi D R, Street A J, Cohen P, Cohen P T. Eur J Biochem. 1993;213:1055–1066. doi: 10.1111/j.1432-1033.1993.tb17853.x. [DOI] [PubMed] [Google Scholar]
- 26.Antos C L, McKinsey T A, Frey N, Kutschke W, McAnally J, Shelton J M, Richardson J A, Hill J A, Olson E N. Proc Natl Acad Sci USA. 2002;99:907–912. doi: 10.1073/pnas.231619298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vega R B, Yang J, Rothermel B A, Bassel-Duby R, Williams R S. J Biol Chem. 2002;277:30401–30407. doi: 10.1074/jbc.M200123200. [DOI] [PubMed] [Google Scholar]