Abstract
Four phosphoproteins, of 34, 43, 50, and 84 kDa, with common amino termini are synthesized via alternative splicing from the UL112-113 region of the human cytomegalovirus genome. Although genetic studies provided evidence that both the UL112 and UL113 loci in the viral genome are required for efficient viral replication, whether the four proteins play specific roles or cooperate in replication is not understood. Here we present evidence, using in vitro and in vivo coimmunoprecipitation assays, that the four UL112-113 proteins both self-interact and interact with each other. A mapping study of the 84-kDa protein showed that the N-terminal region encompassing amino acids 1 to 125, which is shared in all UL112-113 proteins and highly conserved among betaherpesviruses, is required for both self-interaction and nuclear localization as foci. Further localization studies revealed that, unlike the 43-, 50-, and 84-kDa proteins, which were distributed as nuclear punctate forms, the 34-kDa form was located predominantly in the cytoplasm. However, when all four proteins were coexpressed simultaneously, all of the UL112-113 proteins were efficiently localized to the promyelocytic leukemia oncogenic domains. We also found that the ability of the UL112-113 proteins to relocate UL44 (the viral polymerase processivity factor) to prereplication foci relied on self-interaction and reached maximal levels when the four proteins were coexpressed. Therefore, our data suggest that interactions occurring among UL112-113 proteins via their shared N-terminal regions are important to both their intranuclear targeting and the recruitment of UL44 to subnuclear sites for viral replication.
Human cytomegalovirus (HCMV), a member of the betaherpesvirus subfamily, is a ubiquitous pathogen that causes congenital disease and disease in immunocompromised individuals (16). Like many of the other alpha- and gammaherpesviruses, HCMV carries a conserved set of six replication core proteins, namely, the DNA polymerase (UL54) and its associated polymerase processivity factor (UL44), the single-stranded DNA binding protein (UL57), and the heterotrimer consisting of the DNA helicase (UL105), primase (UL70), and primase-associated factor (UL102) subunits (3, 4). Transient oriLyt-dependent in vitro replication assays have shown that in addition to these six core proteins, a total of five additional functional proteins (UL36-38, TRS1/IRS1, IE1/IE2, UL84, and UL112-113) are required for efficient DNA replication (14, 15, 21). Among these, the UL36-38 region, TRS1/IRS1, IE1/IE2, and UL112-113 have all been initially ascribed roles in the transactivation of replication genes (9), although the key functions of UL36 and UL37 have recently been proven to be in the inhibition of apoptosis (6, 20). In cotransfection assays, only UL84 was essential for the promotion of oriLyt-dependent DNA replication, whereas UL112-113 was determined to be needed for efficient replication only when the core replication proteins, UL84, and IE2 were expressed (19).
The UL112-113 region encodes four nuclear phosphoproteins via alternative splicing. These phosphoproteins have molecular masses of 34, 43, 50, and 84 kDa, and all contain common amino termini consisting of 252 amino acids (23, 24). The accumulation of four UL112-113 proteins in virus-infected cells appears to be regulated in a time-dependent manner. The 43-kDa protein is expressed first and most abundantly and remains at a relatively constant level throughout infection, whereas the levels of the other proteins increase significantly during the late stage of infection (24). Two important experiments provided genetic evidence for the key role of the UL112-113 proteins in DNA replication. Firstly, the expression of antisense RNA specific to the UL112-113 proteins was shown to significantly suppress both the expression of UL112-113 proteins and the replication of viral DNA (25). Secondly, deletions in the UL112 or UL113 region of the bacterial artificial chromosome-based HCMV genome revealed significantly impaired viral growth (5, 26).
Recent studies have revealed that the UL112-113 proteins are targeted to the peripheries of the subnuclear structures referred to as the promyelocytic leukemia (PML) oncogenic domains (PODs) or nuclear domain 10 (ND10) during the early stages of viral infection in cells but accumulate in the viral DNA replication compartments during the later stages of infection (2, 17, 25). Interestingly, when coexpressed with six core proteins in transfected cells, the UL112-113 proteins were shown to be responsible for the relocalization of the UL44 protein (p52, the viral polymerase processivity factor) to POD-associated sites, indicating that the UL112-113 proteins perform a vital function in the recruitment of the viral replication core proteins to the periphery of the PODs, in which the formation of viral replication compartments is initiated (2, 17).
Despite the genetic evidence suggesting that both the UL112 and UL113 loci in the viral genome are required for efficient viral replication, it remains to be determined whether the four splice variants encoded by the UL112-113 region actually play a specific role or cooperate during replication. For this study, we expressed the four UL112-113 proteins either separately or simultaneously. We then investigated whether they interact with each other and have cooperative effects with regard to their intracellular localization and their ability to relocate UL44 to prereplication foci.
MATERIALS AND METHODS
Cell culture and virus infection.
HF, Vero, and 293T cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. The HCMV(Towne) virus stocks used in this study were prepared as previously described (12). In order to initiate infection, subconfluent HF cells were infected with HCMV at a multiplicity of infection (MOI) of 2.0 PFU per cell. The input supernatant virus was then adsorbed for 1.5 h at 37°C, and the inoculum was replaced with fresh warm medium at time zero.
Plasmid construction.
The pΔ60SV, pΔ6SV, and pΔ45SV plasmids, which contain AD169 UL112-113 cDNAs encoding 43-, 50-, and 84-kDa proteins, respectively (23), were kindly provided by Deborah H. Spector (University of California, San Diego, Calif.). The cDNA encoding the 34-kDa protein was amplified by PCR (top primer, 5′ GAAGATCTATGGATCTCCCTACTACCGTC 3′; bottom primer, 5′ GAAGATCTTCACGAGTCCACTCGGAAAGC 3′) from pRTS26, which harbors the AD169 UL112-113 genomic gene (19), and cloned into the pENTR vector (Invitrogen). The cDNAs for the 43-, 50-, and 84-kDa proteins were amplified by PCR (top primer, 5′ GAAGATCTATGGATCTCCCTACTACCGTC 3′; bottom primer, 5′ GAAGATCTTTAATCGTCGAAAAACGCCGCGAT 3) from the pΔ60SV, pΔ6SV, and pΔ45SV plasmids, respectively, and cloned into the pENTR vector. Expression plasmids encoding 5′ hemagglutinin (HA)- or Flag-tagged 34-, 43-, 50-, and 84-kDa proteins were then generated in a background of pSG5 (7), using Gateway technology (Invitrogen).
Two large truncation mutants were produced in the background of the 84-kDa protein. The 84(ΔExon-1) mutant, harboring a deletion of the N-terminal 252 amino acids (exon 1), was amplified by PCR (top primer, 5′ GAAGATCTATGGATCTCCCTACTACCGTC 3′; bottom primer, 5′ GAAGATCTTTAATCGTCGAAAAACGCCGCGAT 3′) and placed into pENTR. The 84(ΔC) mutant has a stop codon (UGA) at amino acid position 355, and the mutation was generated by PCR (top primer, 5′ CTACCCATTGAGCGCTGAGCGGTGGTTTCGTCG 3′; bottom primer, 5′ CGACGAAACCACCGCTCAGCGCTCAATGGGTAG 3′) according to the Stratagene QuickChange site-directed mutagenesis protocol. In addition, a series of small deletion mutants, each harboring a 25-amino-acid deletion within exon 1, were also generated by PCR in the same 84-kDa protein background. Expression plasmids encoding 5′ HA- or Flag-tagged mutant variants were generated in pSG5 as described above. In order to construct template plasmids for in vitro transcription/translation reactions, we cloned the wild-type or mutant cDNAs into the pSPUTK (without a tag) (Stratagene) or pCS3-MT (with a six-Myc tag) plasmid (18), using Gateway technology. pCS3-MT-driven plasmids were also employed to express proteins in the cultured cells via transfection.
The pJHA309 plasmid, which expresses 5′ Flag-tagged UL112-113 proteins together from the genomic gene, has been previously described (2). To disrupt the reported nuclear localization signal (NLS) (13) in the pJHA309 background, we replaced the Lys-Arg-Gln-Lys residues, which correspond to positions 261 to 264 of the 43-, 50-, and 84-kDa proteins, with His-Met residues using PCR mutagenesis, resulting in pJHA331. The Δ1-25 and ΔC mutations in the 84-kDa protein background were also introduced into a plasmid which harbors the HA-tagged genomic gene (pRYK21), and this resulted in the formation of pMY69 encoding HA-UL112-113(Δ1-25) and pRYK60 encoding HA-UL112-113(ΔC).
The pSP38 plasmid harboring the HCMV replication origin DNA and plasmids which express six core replication proteins (UL54, UL44, UL57, UL105, UL70, and UL102) and auxiliary proteins (UL36-38, IE1/IE2, and UL84) have been previously described (19) and were provided by Gary S. Hayward (Johns Hopkins University School of Medicine, Baltimore, MD).
Transient DNA transfection.
With HF cells, DNA transfection was conducted using Lipofectamine 2000 reagents (Invitrogen). 293T cells were transfected via the N,N-bis-(2-hydroxyethyl)-2-aminoethanesulfonic acid-buffered saline (BBS) version of the calcium phosphate method, as described previously (12). With Vero cells, the DNA mixtures were introduced into the cells using FuGene 6 reagents (Roche).
Antibodies and indirect immunofluorescence assay (IFA).
A rabbit antipeptide polyclonal antibody (PAb) specific to the 84-kDa form of the UL112-113 protein was described previously (2). Mouse monoclonal antibody (MAb) M23, which recognizes the shared exon 1 of all four UL112-113 proteins (10), was kindly provided by Kanji Hirai (Tokyo Medical and Dental University, Tokyo, Japan). The anti-HA rat MAb 3F10, either conjugated with peroxidase or labeled with fluorescein, and the anti-Myc mouse MAb 9E10 were purchased from Roche. The anti-Flag mouse MAb M2 was obtained from Sigma. The anti-UL44 (p52) mouse MAb was obtained from Advanced Biotechnologies, Inc. The rabbit anti-PML PAb, referred to as PML(C), has been previously described (1).
For IFA, the cells were fixed with 1% paraformaldehyde and permeabilized with 0.2% Triton X-100. All of the subsequent procedures performed have been previously described (12). All slides were examined and photographed with a Zeiss Axiophot microscope.
In vitro transcription/translation and cross-linking assays.
The UL112-113 proteins were synthesized in vitro by using the TNT Quick Coupled transcription/translation system (Promega). For cross-linking assays, 5-μl in vitro-translated protein samples were diluted with 95 μl of 10 mM potassium phosphate buffer (pH 8.0). The mixtures were then incubated in 0.0001% glutaraldehyde (Sigma) for 30 min at room temperature. Fifteen microliters of each sample was then boiled for 5 min and separated by sodium dodecyl sulfate-8% polyacrylamide gel electrophoresis (SDS-8% PAGE). The gels were dried, and the protein bands were detected by autoradiography.
Coimmunoprecipitation assays.
DNA-transfected cells were harvested and sonicated in coimmunoprecipitation buffer (50 mM Tris-Cl, pH 7.4, 50 mM NaF, 5 mM sodium phosphate, and 0.1% Triton X-100) supplemented with protease inhibitor cocktail (Sigma). Cell lysates or proteins synthesized by coupled in vitro transcription/translation reactions were then incubated with the appropriate Ab. After 2 h of incubation at 4°C, 30 μl of a 50% slurry of protein A- and G-Sepharose (Amersham) was added and adsorbed for 16 h at 4°C. The mixture was pelleted and washed six times with coimmunoprecipitation buffer. The beads were then resuspended and boiled for 5 min in loading buffer. The samples were analyzed by SDS-PAGE and immunoblotting when necessary.
Metabolic labeling and immunoprecipitation.
HF cells (1 × 107 cells) in 150-mm tissue culture dishes were infected with HCMV(Towne) at an MOI of 2.0. For [35S]methionine-cysteine labeling, the cells were washed with phosphate-buffered saline at 48 h and incubated for 30 min with methionine- and cysteine-free Dulbecco's modified Eagle's medium supplemented with 5% dialyzed fetal bovine serum. The medium was then replaced with fresh medium containing 300 μCi of [35S]methionine-cysteine (14.3 mCi/ml) (Promix; Amersham). After 4 h of incubation at 37°C, the cells were harvested and lysed with RIPA buffer (1% NP-40, 1% sodium deoxycholate, 0.1% SDS, 150 mM NaCl, 10 mM Tris-HCl [pH 7.4]) for 30 min on ice. The cell lysates were then utilized in immunoprecipitation experiments.
Immunoblot analysis.
DNA-transfected cells were washed with phosphate-buffered saline, and the total extracts were prepared by incubation of the cells with RIPA buffer. The clarified cell extracts were separated in an SDS-8% polyacrylamide gel, followed by performance of the standard procedure using an enhanced chemiluminescence system (Amersham).
RESULTS
The UL112-113 proteins both self-interact and interact with each other via the common N-terminal region in vitro.
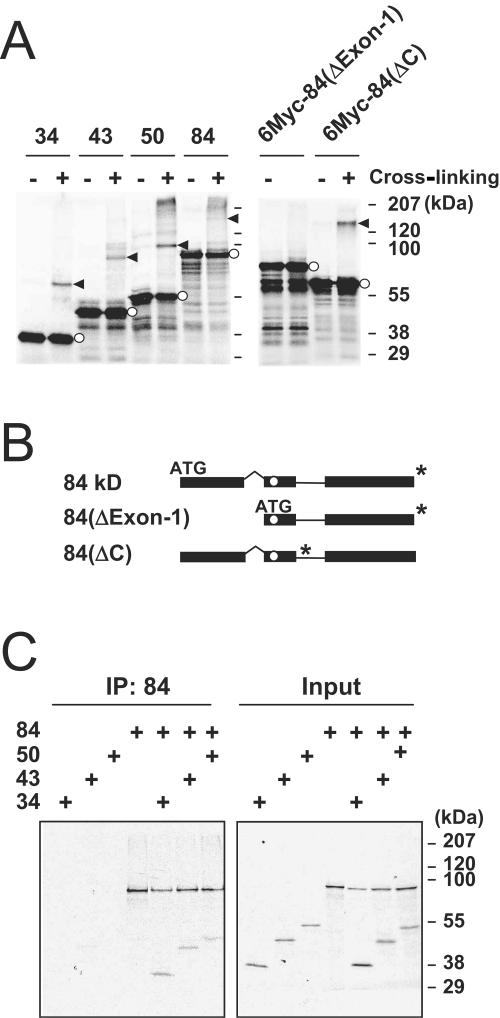
To investigate interactions among the four proteins encoded by the UL112-113 region of the HCMV genome, we generated expression plasmids encoding the 34-, 43-, 50-, and 84-kDa proteins with N-terminal Flag tags (Fig. 1A). The expression of these proteins was verified by immunoblot analysis of the cell extracts prepared from transfected Vero cells (Fig. 1B). Notably, the cDNA for the 84-kDa protein also expressed a small amount of the 43-kDa protein. We also generated 35S-labeled 34-, 43-, 50-, and 84-kDa proteins via coupled in vitro transcription and translation. When the individually translated proteins were incubated with glutaraldehyde, cross-linked dimeric variants were detected for all of the 34-, 43-, 50-, and 84-kDa proteins (Fig. 2A). Similar cross-linking assays were conducted with two mutant variants of the 84-kDa protein. The ΔExon-1 mutant, which is devoid of the 252 amino acids at the N terminus, and the ΔC mutant, which lacks the C-terminal region encompassing amino acids 348 to 684, were both synthesized with N-terminal six-Myc tags (Fig. 2B). Interestingly, the ΔExon-1 mutant failed to form dimers, whereas the ΔC mutant continued to generate dimers (Fig. 2A). This clearly suggests that the UL112-113 proteins may self-interact via their common 252 N-terminal amino acids.
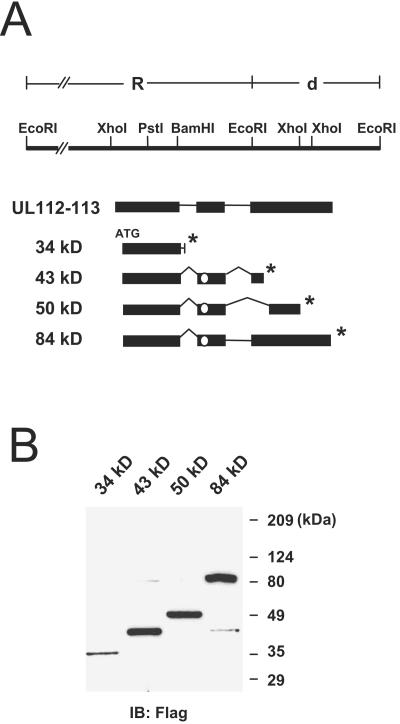
FIG. 1.
Construction of expression plasmids encoding the different forms of UL112-113 proteins. (A) The R-d region of the EcoRI restriction profile of HCMV strain AD169 is shown. From the UL112-113 locus within this R-d region, four splicing variants, of 34, 43, 50, and 84 kDa, are synthesized. The positions of the exons and introns of the RNAs are summarized. The ATG start codon and the in-frame stop codon for each cDNA (*) are indicated. The position of the reported NLS within exon 2 is also indicated with open circles. (B) Expression of the UL112-113 proteins in transfected cells. Vero cells were transfected with plasmids encoding Flag-tagged individual 34-, 43-, 50-, and 84-kDa proteins. At 48 h, the total cell extracts were prepared and subjected to SDS-10% PAGE, followed by immunoblot analysis with anti-Flag MAb M2. Protein size markers are also indicated.
FIG. 2.
Self-interaction and interaction among the UL112-113 proteins in vitro. (A) Cross-linking of individual UL112-113 proteins. The 35S-labeled 34-, 43-, 50-, and 84-kDa proteins and the six-Myc-tagged 84(ΔExon-1) and 84(ΔC) forms were synthesized individually by in vitro transcription/translation reactions. Equal amounts of proteins were incubated in both the presence and absence of glutaraldehyde and then subjected to SDS-8% PAGE, followed by autoradiography. Bands representing dimers are shown by arrowheads, and the positions of monomers are denoted by open circles. (B) Structures of 84(ΔExon-1) and 84(ΔC) mutant forms. The ATG start codon and the in-frame stop codon (*) for each cDNA are indicated. The position of the reported NLS is also indicated within exon 2 with open circles. (C) Immunoprecipitation of cotranslated UL112-113 proteins. The 35S-labeled 34-, 43-, 50-, and 84-kDa proteins were individually synthesized or cotranslated with the 84-kDa protein via in vitro transcription/translation reactions. The products then were immunoprecipitated (IP) with anti-84-kDa-protein Ab and subjected to SDS-8% PAGE, followed by autoradiography (left). The input translation products are also shown (right).
We then attempted to determine whether the different UL112-113 proteins were capable of interacting with each other. The 34-, 43-, 50-, and 84-kDa proteins were cotranslated with the 84-kDa protein. When the 35S-labeled proteins were immunoprecipitated with a rabbit PAb which recognizes only the 84-kDa form of the UL112-113 proteins, we observed coprecipitation of the 34-, 43-, and 50-kDa proteins (Fig. 2C). This also suggests that the 84-kDa UL112-113 protein may interact with other 34-, 43-, and 50-kDa UL112-113 proteins.
Interactions among four UL112-113 proteins in vivo.
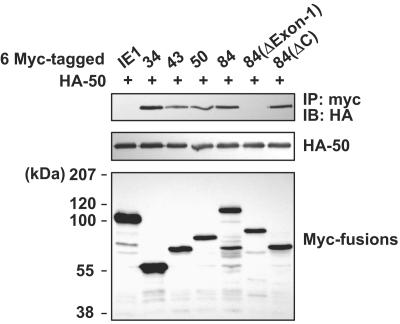
The interactions occurring among the different UL112-113 proteins were investigated further by using coimmunoprecipitation assays with DNA-cotransfected cells (Fig. 3). When the Myc-tagged 34-, 43-, 50-, and 84-kDa proteins were coexpressed simultaneously with the HA-tagged 50-kDa protein in 293T cells, immunoprecipitation of the cell extracts with anti-Myc Ab resulted in coprecipitation of the HA-tagged 50-kDa protein. When the myc-84(ΔExon-1) and myc-84(ΔC) mutants were employed, the ΔExon-1 mutant was not coprecipitated with the HA-tagged 50-kDa protein, although the ΔC mutant continued to be coprecipitated with it. These results showed that four different UL112-113 proteins could interact with one another via their common N-terminal 252-amino-acid regions (exon 1). In a control experiment, the IE1 protein was determined not to interact with the HA-tagged 50-kDa protein. To exclude the possibility that the lack of interaction of the ΔExon-1 mutant with the 50-kDa protein was attributable to its cytoplasmic retention, we assessed the cellular localization of the ΔExon-1 mutant by IFA. We found that although some ΔExon-1 mutants were distributed in the cytoplasm, most of the ΔExon-1 mutants continued to exhibit nuclear localization (data not shown).
FIG. 3.
Interactions among UL112-113 proteins in cotransfected cells. 293T cells were cotransfected with the HA-tagged 50-kDa protein and six-Myc-tagged intact or mutant UL112-113 proteins. The Myc-tagged IE1 protein was also included as a negative control. The cell lysates were prepared and immunoprecipitated with anti-Myc Ab, followed by SDS-PAGE and immunoblotting with anti-HA Ab (top). The expression level of each of the proteins was confirmed by immunoblotting of the total cell extracts with anti-HA Ab (middle) and anti-Myc Ab (bottom).
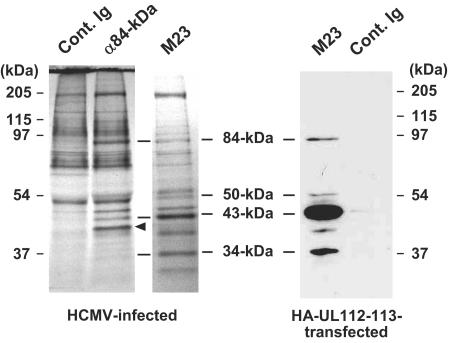
To confirm whether or not these interactions between the different UL112-113 proteins occur in virus-infected cells, the proteins of HCMV-infected HF cells were labeled in vivo with [35S]Met-Cys, followed by immunoprecipitation with an 84-kDa form-specific Ab. In order to identify the UL112-113 proteins, we also conducted immunoprecipitation with the mouse MAb M23, which was reported to detect all four of the UL112-113 proteins together (10). We determined that immunoprecipitation with an anti-84-kDa-protein Ab coprecipitated the 43-kDa protein and also probably the 34-kDa protein (Fig. 4). The 50-kDa UL112-113 protein, which might be coprecipitated with the 84-kDa protein, could not be identified. This was attributed to interference of the immunoglobulin G (IgG) heavy chains. This result generally appears to support the idea that the four UL112-113 proteins are able to interact with one another and may manifest as a complex in virus-infected cells. Note that a protein with a molecular mass of about 40 kDa could be coprecipitated efficiently with the anti-84-kDa-protein Ab but not with the M23 Ab (Fig. 4, arrowhead).
FIG. 4.
Coimmunoprecipitation of UL112-113 proteins in HCMV-infected cells. HF cells were infected with HCMV(Towne) at an MOI of 2.0. At 48 h, the cell proteins were labeled in vivo with [35S]methionine-cysteine for 4 h at 37°C. The cell lysates were prepared, and immunoprecipitation was carried out with control rabbit IgG or rabbit anti-84-kDa-protein PAb. Immunoprecipitation was also conducted with mouse MAb M23. The precipitates were subjected to SDS-8% PAGE, followed by autoradiography. The positions in the gel corresponding to the 34-, 43-, 50-, and 84-kDa UL112-113 proteins are indicated, along with protein size markers. A protein band which appears to be specifically immunoprecipitated with the anti-84-kDa-protein Ab but not with M23 is indicated with an arrowhead. As a control for the specificity of the M23 Ab and the positions of the UL112-113 proteins by SDS-PAGE, 293T cells were transfected with a plasmid encoding HA-UL112-113 proteins. The cell lysates were immunoprecipitated with the M23 Ab or the control mouse IgG, followed by immunoblotting with anti-HA Ab.
The highly conserved N-terminal 125-amino-acid region of the 84-kDa protein is required for both self-interaction and nuclear localization as foci.
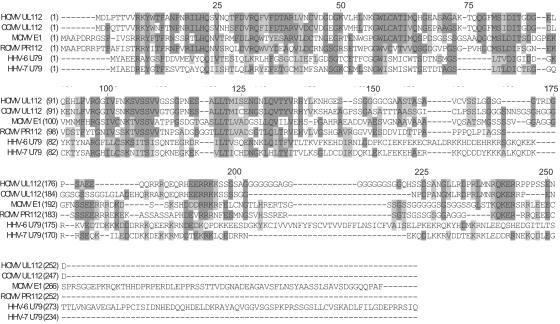
The sequence of the N-terminal 252 amino acids (exon 1) encoded by HCMV UL112 appears to be well conserved among betaherpesviruses, including chimpanzee CMV, rat CMV, mouse CMV, human herpesvirus 6 (HHV-6), and HHV-7 (Fig. 5). Interestingly, the N-terminal region of exon 1 exhibits a higher degree of homology than does the C-terminal portion.
FIG. 5.
Alignment of the amino acid sequences of the protein domains encoded by HCMV UL112 and its homologues encoded by other betaherpesviruses. The aligned amino acid sequences are those encoded by HCMV UL112, chimpanzee CMV (CCMV) UL112, mouse CMV (MCMV) E1, rat CMV (RCMV) PR112, HHV-6 U79, and HHV-7 U79. The consensus residues derived from primarily conserved residues at given positions are indicated by dark shading, whereas similar residues at given positions are indicated by light shading. Numbers along the top of the aligned sequences indicate the amino acid positions of the exon 1 domain, which is encoded by the HCMV UL112 region.
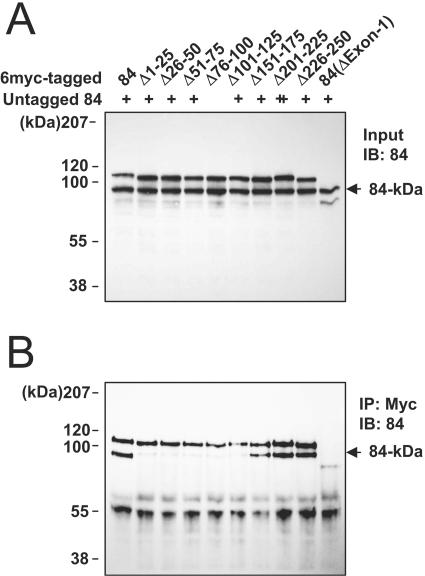
To map domains within exon 1 required for self-interaction, we constructed a series of mutant 84-kDa variants, all containing a 25-amino-acid deletion within the exon 1 region. The untagged 84-kDa protein and the Myc-tagged wild-type or deletion mutant proteins were then cotranslated in vitro and were immunoprecipitated with anti-Myc Ab, with a subsequent immunoblot analysis with an anti-84-kDa-protein Ab (Fig. 6). The results indicated that like the ΔExon-1 mutant, the small deletion mutants Δ1-25, Δ26-50, Δ51-75, Δ76-100, and Δ101-125 did not bind to the intact 84-kDa protein, whereas the mutants Δ151-175, Δ201-225, and Δ226-250 continued to bind to the intact 84-kDa protein as efficiently as was observed for the wild-type protein. Our results show that the self-interaction of the 84-kDa protein requires the highly conserved 125 amino acids located in the N-terminal region.
FIG. 6.
Self-interactions of mutant 84-kDa proteins. (A) Input of in vitro cotranslation products. Both the untagged 84-kDa protein and the six-Myc-tagged wild-type or mutant 84-kDa proteins were synthesized by in vitro transcription/translation reactions. The mutant forms used were as follows: Δ1-25, Δ26-50, Δ50-75, Δ76-100, Δ101-125, Δ126-150, Δ176-200, Δ201-225, Δ226-250, and ΔExon-1. Equal amounts of translated proteins were separated by SDS-8% PAGE, followed by immunoblot analysis with the anti-84-kDa-protein Ab. The position of the untagged 84-kDa protein is indicated with an arrow. (B) Immunoprecipitation of cotranslated proteins. The cotranslated proteins were immunoprecipitated with anti-Myc Ab 9E10 and subjected to SDS-8% PAGE, followed by immunoblot analysis with the anti-84-kDa-protein Ab. The position of the coimmunoprecipitated 84-kDa form is shown with an arrow.
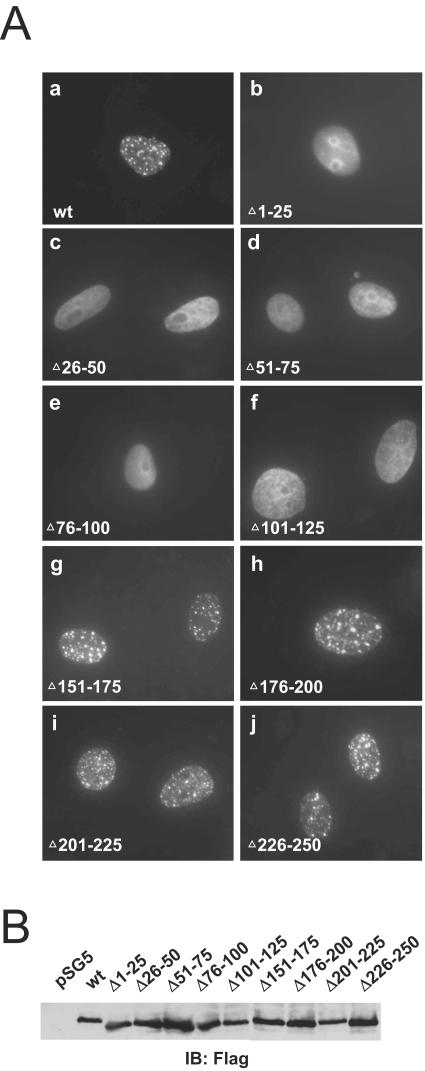
The UL112-113 proteins were shown to be distributed in nuclear punctate forms (2). To determine whether self-interaction of the 84-kDa protein is required for its localization in nuclear foci, we expressed the above mutant proteins in Vero cells and characterized their localization patterns by IFA. Our results indicated that the mutant proteins containing 25-amino-acid deletions within the highly conserved N-terminal portion of exon 1, from positions 1 to 125 (Δ1-25, Δ26-50, Δ51-75, Δ76-100, and Δ101-125), were localized as uniform nuclear diffuse forms, whereas the mutant proteins harboring deletions at positions 126 to 250 (Δ151-175, Δ176-200, Δ201-225, and Δ226-250) yielded the typical localization pattern of the intact 84-kDa protein (Fig. 7A). The expression levels of the wild-type and mutant proteins in transfected Vero cells proved to be comparable to the results of immunoblotting analysis (Fig. 7B). This demonstrates that the highly conserved N-terminal region, located from positions 1 to 125 of exon 1 and essential for self-interaction, is also required for the formation of nuclear foci from the 84-kDa protein.
FIG. 7.
Localization of mutant 84-kDa proteins. (A) Vero cells were transfected with plasmids encoding wild-type or mutant forms of N-terminally Flag-tagged 84-kDa protein, as follows: a, wild-type; b, Δ1-25; c, Δ26-50; d, Δ50-75; e, Δ76-100; f, Δ101-125; g, Δ151-165; h, Δ176-200; i, Δ201-225; and j, Δ226-250. After 48 h, the cells were fixed with paraformaldehyde, followed by IFA with anti-Flag Ab M2 and fluorescein isothiocyanate (FITC)-labeled anti-mouse IgG. (B) Expression levels of wild-type and mutant 84-kDa proteins. Vero cells were transfected with the plasmids described above. At 48 h, the total cell extracts were prepared and subjected to SDS-8% PAGE, followed by immunoblot analysis with anti-Flag Ab M2.
Differential localization of four UL112-113 proteins.
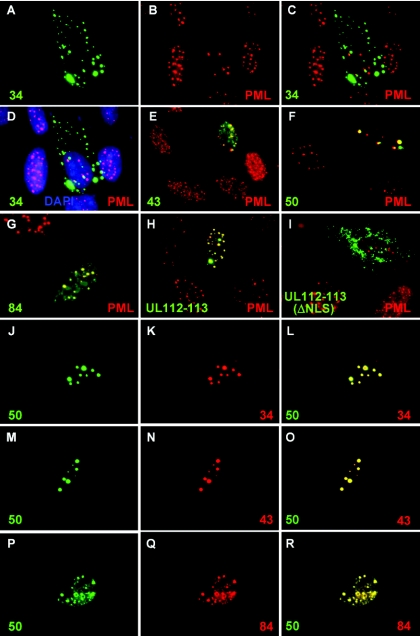
The localization pattern of each of the UL112-113 proteins was further examined in transfected HF cells by IFA. When the Flag-tagged UL112-113 proteins were individually expressed in HF cells, the 34-kDa protein was determined to be predominantly distributed in the cytoplasm, although some aggregates were also located in the nucleus. Double labeling with PML indicated that the nuclear forms of the 34-kDa protein were not associated with the PODs (Fig. 8A to D). The 43-kDa protein was distributed in nuclear punctate forms with some diffuse nuclear background staining, whereas the 50-kDa protein was found to be localized only as nuclear punctate forms (Fig. 8E and F). The nuclear punctate forms of these proteins partially overlapped with the PODs. The localization pattern of the 84-kDa protein was similar to that of the 43-kDa protein, but its nuclear foci appeared to overlap the PODs more efficiently than did those of the 43- and 50-kDa proteins in transfected cells (Fig. 8G). Considering that a small amount of the 43-kDa protein was also expressed from the cDNA for the 84-kDa protein (Fig. 1 and 3), this efficient POD targeting may also be explained by the coexpression of both the 84-kDa and 43-kDa proteins.
FIG. 8.
Colocalization interactions among UL112-113 proteins. (A to I) HF cells were transfected with plasmids encoding the Flag-tagged 34-kDa (A to D), 43-kDa (E), 50-kDa (F), or 84-kDa (G) UL112-113 protein or plasmids containing the UL112-113 genomic gene encoding all four Flag-tagged proteins together (H) or their ΔNLS versions (I). After 48 h, the cells were fixed with paraformaldehyde, followed by double-label IFA with anti-Flag M2 (green) and anti-PML (red) Abs. FITC-labeled anti-mouse IgG and rhodamine/RedX-coupled anti-rabbit IgG were used for visualization. To stain the cell nucleus, mounting solution containing 4′,6′-diamidino-2-phenylindole (DAPI; Vector Laboratories, Inc., Burlingame, Calif.) was used. For the 34-kDa protein, two side-by-side panels of single-labeled FITC (A, for 34-kDa) and rhodamine/RedX (B, for PML) IFA images and two-color merge (C, for 34-kDa and PML) and three-color merge (D, for 34-kDa, PML, and DAPI) images are shown. (J to R) HF cells were cotransfected with plasmids encoding the HA-50-kDa and Flag-34-kDa proteins (J to L), the HA-50-kDa and Flag-43-kDa proteins (M to O), or the HA-50-kDa and Flag-84-kDa proteins (P to R). After 48 h, the cells were fixed with paraformaldehyde, followed by double-label IFA with anti-HA (green) and anti-Flag (red). Two side-by-side panels of single-labeled IFA images and a third panel of merged images are also shown.
An expression plasmid containing the UL112-113 genomic gene expressing an N-terminal Flag tag was previously described and shown to encode four UL112-113 proteins simultaneously (2). When the cells received this plasmid, cell staining with anti-Flag Ab revealed that the UL112-113 proteins yielded a localization pattern reminiscent of that of the 84-kDa protein (Fig. 8H). Interestingly, no cytoplasmic form of the 34-kDa protein could be detected in these cells. This suggests that the simultaneous presence of four splice variants might have some influence on the localization patterns of individual UL112-113 proteins, thereby resulting in efficient targeting to the PODs.
A study involving the 43-kDa protein allowed for the mapping of an NLS to the region encompassing amino acid positions 245 to 272 and also revealed that the KRQK residues located from positions 261 to 264 are vitally important for the functioning of the NLS (13). Apparently, the failure of nuclear targeting by the 34-kDa protein in this study can be attributed to the absence of this reported NLS. When we deleted these KRQK residues within the background of the genomic gene expression plasmid, the mutant ΔNLS UL112-113 proteins were localized as cytoplasmic punctate forms (Fig. 8I), thereby verifying that these amino acids also perform an NLS function in both the 50- and 84-kDa proteins as well as the 43-kDa protein.
Colocalization interactions among the UL112-113 proteins.
Our results suggest that the simultaneous coexpression of the different UL112-113 proteins might affect the localization patterns of individual proteins. We further evaluated this possibility by using a series of cotransfection assays. When the HA-tagged 50-kDa protein and the Flag-tagged 34-kDa protein were expressed simultaneously in HF cells, both of the proteins were colocalized in a nuclear punctate pattern typical of the 50-kDa protein, thereby suggesting that the majority of the cytoplasmic forms of the 34-kDa protein had been relocalized to the nucleus in the presence of the 50-kDa protein (Fig. 8J to L). In a similar fashion, when the 50-kDa and 43-kDa proteins were coexpressed simultaneously, they were colocalized in a pattern reminiscent of that of the 50-kDa protein (Fig. 8M to O). Interestingly, when the 50-kDa and 84-kDa proteins were coexpressed simultaneously, both proteins were colocalized in a distribution pattern reminiscent of that of the 84-kDa protein (Fig. 8P to R). These findings indicate that there may be some cooperative behavior inherent to the localization of different UL112-113 proteins and that either the expression of the 84-kDa protein or the coexpression of both the 84-kDa and 43-kDa proteins may regulate, to some degree, the localization patterns of these other UL112-113 proteins.
Effects of self-interaction and coexpression of the four UL112-113 proteins on the recruitment of UL44 protein to prereplication foci.
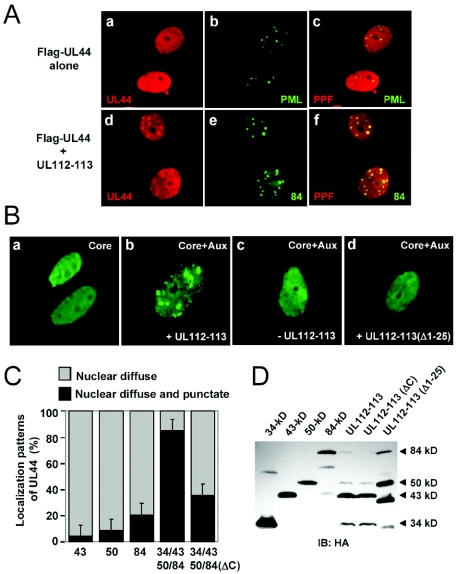
When the six core replication complex components (UL54, UL44, UL57, UL105, UL70, and UL102) were coexpressed along with the UL112-113 proteins, the UL44 protein (the polymerase processivity factor) was shown to be recruited to the POD-associated sites in DNA cotransfection assays (2). For this study, we investigated whether the UL112-113 proteins are able to do so when coexpressed along with the UL44 protein only. The Flag-tagged UL44 protein was localized in a uniform nuclear diffuse pattern in transfected Vero cells (Fig. 9A, panels a to c). When Flag-UL44 was cotransfected with the plasmid harboring the UL112-113 genomic gene, which encodes all four of the UL112-113 proteins simultaneously, the distribution pattern of Flag-UL44 changed to include typical punctate forms which were colocalized with the 84-kDa protein (Fig. 9A, panels d to f). This result demonstrates that the UL112-113 proteins are able to recruit UL44 into the POD-associated sites, even in the absence of other core replication complex components.
FIG. 9.
Recruitment of the UL44 protein to prereplication foci by UL112-113 proteins. (A) Localization patterns of the UL44 protein in cells cotransfected with UL112-113 proteins. Vero cells were transfected with a plasmid encoding Flag-UL44 alone (a to c) or cotransfected with a plasmid encoding Flag-UL44 and a plasmid containing the UL112-113 genomic gene (d to f). After 48 h, the cells were fixed with paraformaldehyde, followed by double-label IFA with anti-Flag (red) and either anti-PML or anti-84-kDa-protein Ab (green). Two side-by-side panels of single-labeled IFA images and a third panel of merged images are shown. (B) Role of self-interaction of UL112-113 proteins in the formation of prereplication foci. Vero cells were cotransfected with plasmids containing the replication origin (pSP38) and plasmids expressing six core replication proteins (UL54, UL44, UL57, UL105, UL70, and UL102) (a), six core proteins plus four auxiliary proteins (UL36-38, IE1/IE2, UL84, and UL112-113) (b), six core proteins plus three auxiliary proteins without UL112-113 (c), and six core proteins plus four auxiliary proteins with the mutant UL112-113(Δ1-25) protein (d). The cells were fixed at 72 h, and IFA was carried out with an anti-UL44 mouse MAb. (C) Quantitation of the relocalization of UL44 by UL112-113 proteins. Vero cells were transfected with plasmids encoding the individual 43-, 50-, and 84-kDa proteins or a plasmid containing the UL112-113 genomic gene or its ΔC mutant version. The percentages of cotransfected cells exhibiting only the nuclear diffuse pattern of the UL44 protein and displaying both the nuclear diffuse and nuclear punctate patterns of the UL44 protein, which represents the relocalization of UL44 to the POD-associated sites by the UL112-113 proteins, were counted. The experiments were repeated three times, and the average numbers are shown. (D) Expression levels of UL112-113 proteins used in these experiments. Vero cells were transfected with the indicated plasmids encoding the HA-tagged UL112-113 proteins. At 48 h, the total cell lysates were prepared and subjected to SDS-8% PAGE, followed by immunoblotting with anti-HA Ab.
We next investigated the importance of UL112-113 self-interaction with regard to the recruitment of UL44 to the nuclear foci. Vero cells were cotransfected with plasmids containing the HCMV replication origin and plasmids encoding six replication core proteins. When we conducted IFA with anti-UL44 Ab, we observed localization of UL44 as a nuclear diffuse form (Fig. 9B, panel a). When four auxiliary proteins (such as UL36-38, IE1/IE2, UL84, and UL112-113) were added, we observed efficient generation of UL44-containing nuclear foci which resembled the viral prereplication foci in HCMV-infected cells (2) (Fig. 9B, panel b). However, those UL44 foci were not produced at all in cases in which the UL112-113 plasmid was omitted from the transfection mix (Fig. 9B, panel c) or the expression plasmid for the UL112-113 genomic gene was replaced with one containing a deletion of the N-terminal 25 amino acids, UL112-113(Δ1-25) (Fig. 9B, panel d). Considering that the Δ1-25 mutant produces self-interaction-deficient mutant variants of the UL112-113 proteins, our findings demonstrate that the self-interaction exhibited by UL112-113 is a prerequisite for the recruitment of the UL44 protein to the prereplication foci.
Finally, we investigated whether simultaneous coexpression of all four UL112-113 proteins exerts a cooperative effect on their capacities to form UL44 foci (Fig. 9C). When the UL44 protein was coexpressed with the 43-, 50-, or 84-kDa protein, the formation of UL44 foci was detected in 4, 9, and 21% of the UL44-positive cotransfected cells, respectively. When the four UL112-113 proteins were simultaneously coexpressed from the genomic gene, the efficiency with which the UL44 foci were formed increased drastically, to 88%, thereby suggesting that the expression of all four UL112-113 proteins facilitates the recruitment of UL44. We also introduced the ΔC mutation of the 84(ΔC) mutant form (Fig. 2B) into the background of the genomic gene. This mutant UL112-113(ΔC) gene was shown to encode the 34-, 43-, and 50-kDa proteins normally, but not the intact 84-kDa protein (Fig. 9D). When the UL112-113(ΔC) gene was employed instead of the intact UL112-113 gene, the formation of UL44 foci was reduced to 36%. The expression levels of the UL112-113 proteins in these experiments are shown in Fig. 9D. This immunoblot analysis verified both the expression of the 25-amino-acid-deleted UL112-113 proteins from the UL112-113(Δ1-25) gene and the lack of expression of the intact 84-kDa form of the UL112-113(ΔC) gene. Taken together, our results clearly demonstrate that the simultaneous coexpression of the four UL112-113 proteins results in a cooperative effect on their activity of recruiting UL44 into the prereplication foci.
DISCUSSION
In the present study, we have clearly demonstrated that the 34-, 43-, 50-, and 84-kDa proteins encoded by the HCMV UL112-113 region are able to self-interact and interact with each other via the shared N-terminal exon 1 region. In addition, we have also provided evidence that there is a strong correlation between the activities of the UL112-113 proteins of self-interaction, formation of nuclear foci, and recruitment of UL44 to the viral prereplication foci. It is likely that the self-interaction of UL112-113 proteins constitutes a prerequisite for their ability to form nuclear foci, which is itself a prerequisite for the recruitment of UL44 to the viral prereplication sites. It has been reported that the 34-, 43-, and 50-kDa proteins have DNA-binding activity (10). It is also quite conceivable that self-interaction is a prerequisite for the DNA-binding activity of these proteins. Considering genetic evidence that both the UL112 and UL113 loci are required for efficient viral replication in cultured HF cells, it appears that these correlating activities of UL112-113 proteins may play important roles in initiating the efficient DNA replication process in HF cells.
The reasons underlying the synthesis of 34-, 43-, 50-, and 84-kDa proteins via alternative splicing from the UL112-113 region have long remained an enigma. The 43-kDa protein has been reported to be the first and most abundantly expressed of these proteins, whereas the expression of the others increases significantly during the late stages of infection (24). This temporal regulation of protein accumulation may reflect some distinct role for each of the UL112-113 proteins during the progression of viral infection. We showed previously that the UL112-113 proteins initially are targeted to the POD-associated sites as early as 6 h after infection and that they are colocalized with the IE2 protein (2). Taking into consideration that the IE2 domains are also sites in which immediate-early transcription occurs (8), it is likely that the UL112-113 proteins have some role in the regulation of viral gene transcription, at least within the very early stages after the initiation of infection. In this regard, it is notable that the UL112-113 proteins enhanced the activation of the viral polymerase (UL54) promoter by the major immediate-early proteins (11, 13). Furthermore, the 43-kDa protein has been demonstrated to enhance the IE2-mediated activation of the promoters of several early genes involved in DNA replication (9). Recently, mouse CMV M112/113 proteins, the homologues of the HCMV UL112-113 proteins, have been demonstrated to bind to IE3 and to interfere with IE3-mediated repressive effects on the major immediate-early promoter (22). However, all of these transactivation assays using target reporter genes were carried out in transiently DNA-transfected cells. Therefore, whether or not the UL112-113 proteins indeed act as regulators of viral gene expression in virus-infected cells needs to be addressed.
During the later stages of infection, the UL112-113 proteins have been demonstrated to accumulate in prereplication foci and in the viral DNA replication compartments. The UL112-113 proteins have been proposed to play a role in orchestrating the formation of these virus-specific subnuclear structures involved in DNA replication (2, 17). In the present study, we have provided some lines of evidence suggesting that the 84-kDa protein may play unique roles in both the efficient targeting of all of the UL112-113 proteins to the POD-associated sites and the recruitment of UL44 to prereplication foci. Coexpression of the 84-kDa protein affected the localization patterns of the other UL112-113 proteins, resulting in more efficient targeting of all of the UL112-113 proteins to the PODs. Importantly, when the 84-kDa form-specific region was not expressed in the mutant genomic UL112-113(ΔC) gene, we noted a significant impairment of the relocalization of UL44 to the prereplication foci. This strongly suggests that the simultaneous coexpression of the four intact UL112-113 proteins, especially the expression of the intact 84-kDa protein, may constitute a prerequisite for the efficient initiation of viral DNA replication. In this regard, it is also notable that a protein of approximately 40 kDa could be coimmunoprecipitated efficiently from virus-infected cell lysates using the anti-84-kDa-protein Ab, but this did not occur when Ab M23 was used (Fig. 4). Identification and study regarding the specific function served by this interaction would be very helpful in understanding the specific role of the 84-kDa protein in viral replication.
Previous studies demonstrated that the oriLyt plasmid could undergo transient DNA replication in the presence of the core replication proteins, UL84, and IE2 when they were expressed using constitutive promoters. Furthermore, the UL112- or UL113-deleted mutant virus could still replicate to a very low titer. Therefore, it may be argued that UL44 is recruited to prereplication foci and replication compartments even in the absence of the UL112-113 proteins. Whether other viral proteins are able to help in the association of UL44 with prereplication foci in UL112-113-deleted mutant virus-infected cells needs to be further investigated. However, the data presented in this study suggest that UL112-113 may play an important role in the relocalization of UL44 to prereplication and replication sites, given that the other known interacting partner of UL44 is the relatively low abundance Pol catalytic protein (UL54). Consistent with this notion, we also found that UL112-113 interacted with UL44 in cotransfection assays (data not shown), suggesting that UL112-113 may directly recruit UL44 to prereplication foci.
In summary, we have shown here for the first time that the enigmatic UL112-113 proteins self-interact and interact with each other via their common N-terminal regions, which are encoded by the UL112 locus. The observed self-interaction was responsible for the distributions of these proteins as nuclear foci. In addition, the 84-kDa form-specific region encoded by the UL113 locus was also required for the efficient targeting of other UL112-113 proteins and the UL44 protein to the viral prereplication sites. The detailed action mechanisms of specific forms of UL112-113 proteins need to be further investigated to understand their exact roles in viral DNA replication.
Acknowledgments
We thank Deborah H. Spector (University of California, San Diego, Calif.) and Kanji Hirai (Tokyo Medical and Dental University, Tokyo, Japan) for providing us with the UL112-113 cDNAs and M23 Ab, respectively. We also thank Gary S. Hayward (Johns Hopkins University School of Medicine, Baltimore, Md.) for the anti-84-kDa protein Ab and the expression plasmids for replication proteins.
This work was supported by a Korea Research Foundation grant (KRF-2004-041-C00287) to J.-H.A.
REFERENCES
- 1.Ahn, J. H., and G. S. Hayward. 2000. Disruption of PML-associated nuclear bodies by IE1 correlates with efficient early stages of viral gene expression and DNA replication in human cytomegalovirus infection. Virology 274:39-55. [DOI] [PubMed] [Google Scholar]
- 2.Ahn, J. H., W. J. Jang, and G. S. Hayward. 1999. The human cytomegalovirus IE2 and UL112-113 proteins accumulate in viral DNA replication compartments that initiate from the periphery of promyelocytic leukemia protein-associated nuclear bodies (PODs or ND10). J. Virol. 73:10458-10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison III, T. Kouzarides, J. A. Martignetti, et al. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 4.Davison, A. J., A. Dolan, P. Akter, C. Addison, D. J. Dargan, D. J. Alcendor, D. J. McGeoch, and G. S. Hayward. 2003. The human cytomegalovirus genome revisited: comparison with the chimpanzee cytomegalovirus genome. J. Gen. Virol. 84:17-28. [DOI] [PubMed] [Google Scholar]
- 5.Dunn, W., C. Chou, H. Li, R. Hai, D. Patterson, V. Stolc, H. Zhu, and F. Liu. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. USA 100:14223-14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldmacher, V. S., L. M. Bartle, A. Skaletskaya, C. A. Dionne, N. L. Kedersha, C. A. Vater, J. W. Han, R. J. Lutz, S. Watanabe, E. D. Cahir McFarland, E. D. Kieff, E. S. Mocarski, and T. Chittenden. 1999. A cytomegalovirus-encoded mitochondria-localized inhibitor of apoptosis structurally unrelated to Bcl-2. Proc. Natl. Acad. Sci. USA 96:12536-12541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Green, S., P. Isseman, and E. Sheer. 1988. A versatile in vivo and in vitro eukaryotic expression vector for protein engineering. Nucleic Acids Res. 16:369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishov, A. M., R. M. Stenberg, and G. G. Maul. 1997. Human cytomegalovirus immediate early interaction with host nuclear structures: definition of an immediate transcript environment. J. Cell Biol. 138:5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iskenderian, A. C., L. Huang, A. Reilly, R. M. Stenberg, and D. G. Anders. 1996. Four of eleven loci required for transient complementation of human cytomegalovirus DNA replication cooperate to activate expression of replication genes. J. Virol. 70:383-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iwayama, S., T. Yamamoto, T. Furuya, R. Kobayashi, K. Ikuta, and K. Hirai. 1994. Intracellular localization and DNA-binding activity of a class of viral early phosphoproteins in human fibroblasts infected with human cytomegalovirus (Towne strain). J. Gen. Virol. 75:3309-3318. [DOI] [PubMed] [Google Scholar]
- 11.Kerry, J. A., M. A. Priddy, T. Y. Jervey, C. P. Kohler, T. L. Staley, C. D. Vanson, T. R. Jones, A. C. Iskenderian, D. G. Anders, and R. M. Stenberg. 1996. Multiple regulatory events influence human cytomegalovirus DNA polymerase (UL54) expression during viral infection. J. Virol. 70:373-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee, H. R., D. J. Kim, J. M. Lee, C. Y. Choi, B. Y. Ahn, G. S. Hayward, and J. H. Ahn. 2004. Ability of the human cytomegalovirus IE1 protein to modulate sumoylation of PML correlates with its functional activities in transcriptional regulation and infectivity in cultured fibroblast cells. J. Virol. 78:6527-6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li, J., T. Yamamoto, K. Ohtsubo, M. Shirakata, and K. Hirai. 1999. Major product pp43 of human cytomegalovirus U(L)112-113 gene is a transcriptional coactivator with two functionally distinct domains. Virology 260:89-97. [DOI] [PubMed] [Google Scholar]
- 14.Pari, G. S., and D. G. Anders. 1993. Eleven loci encoding trans-acting factors are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA replication. J. Virol. 67:6979-6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pari, G. S., M. A. Kacica, and D. G. Anders. 1993. Open reading frames UL44, IRS1/TRS1, and UL36-38 are required for transient complementation of human cytomegalovirus oriLyt-dependent DNA synthesis. J. Virol. 67:2575-2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pass, R. F. 2001. Cytomegalovirus, vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 17.Penfold, M. E., and E. S. Mocarski. 1997. Formation of cytomegalovirus DNA replication compartments defined by localization of viral proteins and DNA synthesis. Virology 239:46-61. [DOI] [PubMed] [Google Scholar]
- 18.Roth, M. B., A. M. Zahler, and J. A. Stolk. 1991. A conserved family of nuclear phosphoproteins localized to sites of polymerase II transcription. J. Cell Biol. 115:587-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarisky, R. T., and G. S. Hayward. 1996. Evidence that the UL84 gene product of human cytomegalovirus is essential for promoting oriLyt-dependent DNA replication and formation of replication compartments in cotransfection assays. J. Virol. 70:7398-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skaletskaya, A., L. M. Bartle, T. Chittenden, A. L. McCormick, E. S. Mocarski, and V. S. Goldmacher. 2001. A cytomegalovirus-encoded inhibitor of apoptosis that suppresses caspase-8 activation. Proc. Natl. Acad. Sci. USA 98:7829-7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith, J. A., and G. S. Pari. 1995. Expression of human cytomegalovirus UL36 and UL37 genes is required for viral DNA replication. J. Virol. 69:1925-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang, Q., L. Li, and G. G. Maul. 2005. Mouse cytomegalovirus early M112/113 proteins control the repressive effect of IE3 on the major immediate-early promoter. J. Virol. 79:257-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wright, D. A., and D. H. Spector. 1989. Posttranscriptional regulation of a class of human cytomegalovirus phosphoproteins encoded by an early transcription unit. J. Virol. 63:3117-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wright, D. A., S. I. Staprans, and D. H. Spector. 1988. Four phosphoproteins with common amino termini are encoded by human cytomegalovirus AD169. J. Virol. 62:331-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yamamoto, T., S. Suzuki, K. Radsak, and K. Hirai. 1998. The UL112/113 gene products of human cytomegalovirus which colocalize with viral DNA in infected cell nuclei are related to efficient viral DNA replication. Virus Res. 56:107-114. [DOI] [PubMed] [Google Scholar]
- 26.Yu, D., M. C. Silva, and T. Shenk. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. USA 100:12396-12401. [DOI] [PMC free article] [PubMed] [Google Scholar]