Abstract
Detailed knowledge of the composition and structure of the spliceosome and its assembly intermediates is a prerequisite for understanding the complex process of pre-mRNA splicing. To this end, we have developed a tobramycin affinity-selection method that is generally applicable for the purification of native RNP complexes. By using this method, we have isolated human prespliceosomes that are ideally suited for both biochemical and structural studies. MS identified >70 prespliceosome-associated proteins, including nearly all known U1 and U2 snRNP proteins, and expected non-snRNP splicing factors. In addition, the DEAD-box protein p68, RNA helicase A, and a number of proteins that appear to perform multiple functions in the cell, such as YB-1 and TLS, were detected. Several previously uncharacterized proteins of unknown function were also identified, suggesting that they play a role in splicing and potentially act during prespliceosome assembly. These data provide insight into the complexity of the splicing machinery at an early stage of its assembly.
Spliceosomes, the complex enzymes responsible for pre-mRNA splicing, consist of the U1, U2, U5, and U4/U6 small nuclear ribonucleoproteins (snRNP), plus numerous non-snRNP proteins. During the stepwise assembly of spliceosomes, several distinct complexes, which form in a defined order (i.e., E complex, followed by A, B, and C), can be distinguished biochemically in vitro (reviewed in refs. 1 and 2). Before association of snRNPs and splicing factors, the pre-mRNA is bound by heterogenous nuclear ribonucleoprotein (hnRNP) proteins, forming the H complex. Spliceosome assembly is initiated by the ATP-independent interaction of the U1 snRNP with the 5′ splice site which, in part, involves base pairing between the U1 snRNA and the pre-mRNA. This initial complex, termed E (early), also contains the U2 snRNP, which at this stage is loosely associated with the pre-mRNA (3). In a subsequent ATP-dependent step, the U2 snRNA base-pairs with the branch site, leading to the stable association of the U2 snRNP and formation of the A complex (also called the prespliceosome). Contacts between snRNA/proteins and the pre-mRNA that are established during these early stages of spliceosome assembly play crucial roles in the recognition and selection of splice sites. Thus, regulation of alternatively spliced pre-mRNAs is often achieved by modulating the association of spliceosomal components during E and A complex formation. In a subsequent step, the U4/U6.U5 tri-snRNP binds, leading to the formation of spliceosomal B complex. After a major conformational change and the first transesterification reaction of splicing, complex C is generated. After the second transesterification step, the mRNA and excised intron are released, and the spliceosome dissociates.
To date, >200 spliceosomal proteins have been identified in mammals by MS (4–6). However, in these studies, a mixture of purified splicing complexes was analyzed, preventing the assignment of a particular protein to a specific splicing complex (4). More recently, affinity selection with amylose beads of splicing complexes formed on pre-mRNA prebound with the maltose-binding protein has been used to isolate highly purified, native E and C complexes (3, 7). However, to date, the protein composition of only the spliceosomal C complex has been determined in detail by MS (7). Thus, comprehensive information about the composition of splicing complexes formed before the catalytic steps of splicing is currently lacking. Similarly, approaches to isolate native splicing complexes that are suitable for both biochemical and structural studies are quite limited.
Here, we have developed a tobramycin affinity-selection method and used it to purify preparative amounts of human prespliceosomes under native conditions. MS analyses revealed that prespliceosomes are highly complex, with >70 associated proteins. These studies provide detailed information about the composition of prespliceosomes and suggest a role for the newly identified prespliceosomal proteins during the early stages of spliceosome assembly.
Methods
Plasmids, Oligonucleotides, and RNA Preparation.
Oligonucleotides used in this study were M13f (GTAAAACGACGGCCAGT), HPV23 (GATCCGGCTTAGTATAGCGAGGTTTAGCTACACTCGTGCTGAGCCGGATCCGCATG), and HPV24 (CGGATCCGGCTCAGCACGAGTGTAGCTAAACCTCGCTATACTAAGCCG). The J6f1 RNA aptamer (8) noncoding and coding sequences, respectively, are underlined. The EcoRI-BamH1 insert of pMINX (9) was first cloned into the pGEM-3Zf(+), yielding pGEM-MINX. The HPV23 and HPV24 oligonucleotides were annealed and cloned into BamH1/SphI linearized pGEM-MINX to yield pGEM MINX-T5. PCR with the HPV24 and M13f primers generated the transcription template for the aptamer-containing pre-mRNA. To obtain control pre-mRNA lacking the aptamer, the PCR product was cleaved with BamHI. m7G-capped transcripts were synthesized with T7 RNA polymerase (MegaScript, Ambion, Austin, TX). For quantitation, [α-32P]UTP (3000 Ci/mmol; 1 Ci = 37 GBq) was added to 0.23 μM.
Solid-Phase Splicing Reaction and Spliceosome Purification.
N-hydroxysuccinimide-activated Sepharose 4 Fast Flow was derivatized with 5 mM tobramycin as described (10). All further procedures were performed at 4°C, unless otherwise stated. For affinity selection, 4× binding buffer (4× BP)/80 mM Tris⋅HCl, pH 9.1 at 4°C/4 mM CaCl2/4 mM MgCl2/2 mM DTT was freshly prepared. The tobramycin matrix was blocked with 400 μl of blocking buffer (1× BP/300 mM KCl/0.1 mg/ml tRNA/0.5 mg/ml BSA/0.01% Nonidet P-40) per 15-μl aliquot by head over tail (HOT) rotation overnight. The matrix was collected, and 400 μl of a mixture containing ≈60 pmol of tobramycin-tagged pre-mRNA and 40 μg of tRNA in binding buffer (1× BP/145 mM KCl), were added to 15 μl of matrix and incubated for 1.5 h. The matrix was then washed three times (750 μl each) with binding buffer containing 0.1% Nonidet P-40. A pre-mRNA concentration of 22–25 nM was found to be optimal for A complex formation (not shown). For a standard splicing assay, four 15-μl aliquots of matrix bound with pre-mRNA were prepared. Splicing mix (1.5 ml) was added per 15-μl matrix and incubated at the indicated time and temperature with constant HOT agitation. The splicing mix contained 35% HeLa cell nuclear extract (11) in buffer D [20 mM Hepes-KOH, pH 7.9/100 mM KCl/1.5 mM MgCl2/0.2 mM EDTA, pH 8.0/0.5 mM DTT/0.5 mM PMSF/10% (vol/vol) glycerol] and was supplemented with 25 mM KCl/3 mM MgCl2/2 mM ATP/20 mM creatine phosphate. After splicing, reactions were chilled on ice, and the matrix was collected by centrifugation. Matrix aliquots were pooled and washed three times with 750 μl of washing buffer (1× BP/75 mM KCl/0.1% Nonidet P-40). Complexes were eluted by incubating with 250 μl of elution buffer (1× BP/5 mM tobramycin/145 mM KCl/2 mM MgCl2) with HOT rotation for 10 min. Typically, ≈75% of the bound complexes could be eluted. RNA and protein were recovered and analyzed on 8.3 M urea/10% polyacrylamide gels (RNA) or by SDS/10–13% PAGE (proteins), or for splicing, on 8.3 M urea-15% polyacrylamide gels.
Glycerol Gradient Centrifugation.
Tobramycin eluate (220 μl) was loaded onto a 3.8-ml linear 10–30% glycerol gradient in 20 mM Hepes-KOH, pH 7.9/75 mM KCl/1.5 mM MgCl2/0.5 mM DTT. Gradients were centrifuged for 107 min at 60,000 rpm in a Sorvall TH660 rotor and harvested manually in 175-μl fractions from the top. RNA and protein were extracted from 100 μl of every odd fraction and analyzed as above.
Psoralen Crosslinking and Northern Blotting.
Crosslinking was initiated by adding psoralen (4′-aminomethyl-4,5′,8-trimethylpsoralen hydrochloride) in DMSO (2 mg/ml) to 75 μl of the even gradient fractions to a final concentration of 40 μg/ml and incubating for 10 min on ice. Samples were irradiated with 365 nm light for 30 min at 4°C. RNA was recovered (12) and resolved on a 5% polyacrylamide/8.3 M urea gel and analyzed by Northern blotting essentially as described (13). DNA probes were generated by random priming (Prime-it II, Promega) using pMINX DNA (pre-mRNA) or PCR-amplified U1 or U2 snRNA coding sequences and [α-32P]dATP (6,000 Ci/mmol).
MS.
Proteins separated by SDS/PAGE were analyzed by matrix-assisted laser desorption/ionization MS (MALDI-MS) or liquid chromatography-coupled tandem MS [LC-MSMS as described in ref. 14; for details, see Supporting Methods, which is published as supporting information on the PNAS web site, www.pnas.org) and identified in the National Center for Biotechnology Information nonredundant database by using MASCOT (Matrix Science, London) as a search engine.
Results
Tobramycin Affinity Selection of Native Spliceosomal Complexes.
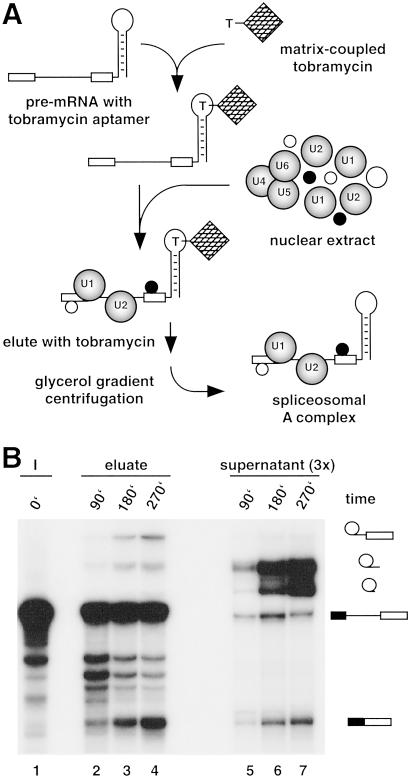
To affinity-purify spliceosomes under native conditions, we introduced a 40 nt RNA aptamer at the 3′ end of the adenovirus-derived MINX pre-mRNA. This aptamer binds with high affinity (5 nM) the aminoglycoside antibiotic tobramycin under physiological conditions (8). Indeed, the aptamer-tagged MINX pre-mRNA bound efficiently to tobramycin-derivatized Sepharose beads when the binding reaction was performed at pH 9.1 in buffer containing 145 mM KCl and 1 mM Mg2+. Under these conditions, 60–70% of aptamer-tagged pre-mRNA, but only ≈2% of untagged pre-mRNA, bound. More importantly, 80% of the bound pre-mRNA could be eluted by incubating with 5 mM tobramycin. The aptamer-tagged MINX pre-mRNA formed spliceosomal complexes and was spliced in vitro with efficiencies and kinetics similar to that of untagged MINX pre-mRNA (not shown). However, in the presence of nuclear extract, <1% of the tagged pre-mRNA bound to the tobramycin matrix, suggesting that the RNA aptamer is no longer accessible or the conditions used for splicing are suboptimal for binding. Solid-phase splicing was thus performed by first immobilizing the tagged pre-mRNA on the tobramycin matrix and subsequently incubating with nuclear extract under splicing conditions (Fig. 1A). The matrix was then washed extensively, and bound material was eluted with tobramycin. RNA was isolated from the eluate or reaction supernatant and analyzed by denaturing PAGE (Fig. 1B). Splicing intermediates/products were first observed in those complexes eluted from the matrix after 90 min and increased markedly after 180 and 270 min, when mRNA product became readily detectable (Fig. 1B, lanes 2–4). Thus, splicing proceeds with significantly reduced kinetics as compared with the nonimmobilized, tagged MINX pre-mRNA, which yielded splicing products already after 15 min (not shown). Most of the pre-mRNA remained bound to the matrix even after extended incubation times, as evidenced by the presence of only a very low level (maximally 7%) of pre-mRNA and splicing intermediates in the supernatant of the solid-phase splicing reaction (Fig. 1B, lanes 5–7). However, consistent with it's release from spliceosomes after the second step of splicing, elevated amounts of excised intron were detected. These results demonstrate that despite reduced reaction kinetics, the matrix-bound, aptamer-tagged pre-mRNA is spliced efficiently and, therefore, functional spliceosomal E, A, B, and C complexes must be assembled as well.
Fig 1.
(A) Purification strategy. (B) Matrix-bound, aptamer-tagged pre-mRNA is efficiently spliced. Solid-phase splicing was performed for the indicated times at 30°C. RNA was recovered after elution (lanes 2–4) or from the reaction supernatant (lanes 5–7; note that three times more supernatant than eluate was assayed), analyzed by denaturing PAGE, and visualized by autoradiography. Lane 1, input pre-mRNA. The positions of the pre-mRNA and splicing intermediates/products are indicated on the right.
Purification of Spliceosomal Complex A.
To isolate prespliceosomes, we performed solid-phase splicing for only 45 min to minimize formation of B/C complexes. The RNA and protein content of the tobramycin eluate was then analyzed by denaturing PAGE or SDS/PAGE, respectively. As a control for nonspecific interactions, an affinity selection was also performed in an identical manner, but without pre-mRNA. As shown in Fig. 2A, in the absence of pre-mRNA, relatively little snRNA was detected in the eluate (lane 2). By contrast, the eluate of the aptamer-tagged pre-mRNA (lane 1) contained similar amounts of pre-mRNA, U1 and U2 snRNA, and only a low level of U4, U5, and U6 snRNA. This result indicated that predominantly splicing complex A (and/or potentially E) had formed after 45 min and was subsequently eluted with tobramycin. Correspondingly, the eluate from the affinity selection performed with pre-mRNA exhibited a complex but distinct protein pattern, whereas only a few proteins migrating in the 55- to 65-kDa range were observed without pre-mRNA (Fig. 2A, compare lanes 3 and 4).
Fig 2.
(A) RNA and protein content of splicing complexes eluted with tobramycin. Solid-phase splicing was performed for 45 min at 30°C in the presence (lanes 1 and 3) or absence (lanes 2 and 4) of pre-mRNA, and the complexes were subsequently eluted with tobramycin. RNA and protein were recovered, analyzed by PAGE, and visualized by silver staining. (B) Glycerol gradient analysis of affinity-purified spliceosomal complexes. Naked pre-mRNA (•) or complexes formed after splicing for 15 min at 4°C (○) or for 45 min at 30°C (▴) were subjected to 10–30% glycerol gradient centrifugation, and the percent of 32P-labeled pre-mRNA in each fraction was determined by Cherenkov counting. (C) RNA profile of the glycerol gradient fractionated, 45-min splicing complexes. RNA was recovered from odd-numbered fractions and analyzed as in A. PRE, pre-mRNA; *, contaminating RNAs.
To characterize the affinity-purified splicing complexes in more detail, the eluate was subjected to glycerol gradient centrifugation (Fig. 2B), and the sedimentation behavior of complexes containing 32P-labeled pre-mRNA was compared with that of naked pre-mRNA or affinity-selected H complexes (i.e., complexes formed after a 15-min incubation with nuclear extract at 4°C). The vast majority of complexes in the 45-min eluate peaked in fraction 11, with a sedimentation value of ≈22S; minor peaks sedimenting at ≈28–42S were also observed. In contrast, naked pre-mRNA and H complexes sedimented at ≈12 and ≈13S, respectively. Thus, the 45-min eluate seems to consist of a relatively homogeneous population of splicing complexes which exhibit an S value significantly higher than the H complex, which is consistent with the idea that predominantly complex A or E had been isolated. Indeed, analysis of the RNA composition of the gradient fractions from the 45-min eluate revealed equimolar amounts of the pre-mRNA and U1 and U2 snRNAs in fractions 9–13 (Fig. 2C). These fractions were nearly free of U4, U5, and U6 snRNAs, which, for the most part, were found in either slower or faster migrating complexes; the latter, which contain equimolar amounts of the pre-mRNA and all of the major spliceosomal snRNAs (lanes 17–23), likely correspond to spliceosomal complex B. Thus, glycerol gradient centrifugation allows the efficient separation of H and B complexes from the affinity-selected A/E complexes.
Identification of E and A Complexes via RNA–RNA Crosslinking.
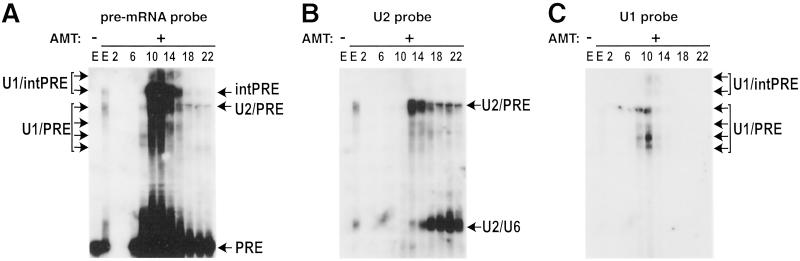
E and A complexes both contain U1 and U2 snRNPs, but a U2/pre-mRNA base pairing interaction is first observed in the A complex (3). Thus, RNA–RNA crosslinking (12) was performed to identify precisely complexes migrating in the 22S region of the gradient. Gradient fractions were UV-irradiated in the presence of psoralen, and the RNA was analyzed by Northern blotting, sequentially probing for the pre-mRNA, U1 and U2 snRNAs (Fig. 3 A–C). Multiple U1 snRNA-containing crosslinks, which peaked in fraction 10, were observed in fractions 8–14 (Fig. 3C). Each of these bands was also detected with a probe specific for the pre-mRNA, confirming that they represent crosslinks between the splicing substrate and U1 (Fig. 3A). In contrast, a U2/pre-mRNA crosslink was first detected in fraction 12, and its intensity gradually diminished from fraction 14 to 22 (Fig. 3B). In an independent experiment, U2/pre-mRNA crosslinks were also observed in fraction 11, but not in the preceding fractions (not shown). Thus, fractions 11–14 contain predominantly the spliceosomal A complex, whereas fractions 8–10 likely contain mainly E complex. A major U2/U6 crosslink (verified by probing for U6; not shown), which is indicative of complex B, peaked in fractions 16–22. However, only small amounts were detected below fraction 14, confirming that fractions 11–13 (the A complex peak) are essentially free of complex B.
Fig 3.
Characterization of RNA–RNA interactions in gradient-fractionated spliceosomal complexes by psoralen crosslinking. Affinity-selected complexes were fractionated on a 10–30% glycerol gradient, and crosslinking was performed with even-numbered gradient fractions (as indicated above each lane) or ≈2% of unfractionated eluate (E lanes). In the left-most E lane, the eluate was UV-irradiated in the absence of psoralen to control for nonspecific crosslinking. RNA was transferred to a nylon membrane that was hybridized sequentially with 32P-labeled probes specific for pre-mRNA (A) U2 snRNA (B) and U1 snRNA (C). The identity of the crosslinked species is indicated on the right. AMT, psoralen; PRE, pre-mRNA; intPRE, internally crosslinked pre-mRNA.
Protein Composition of Prespliceosomes.
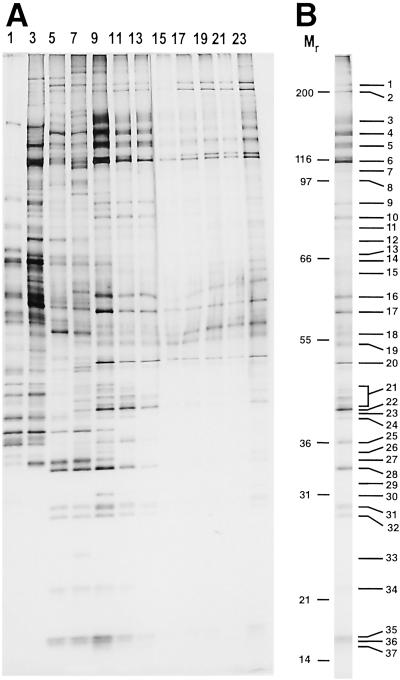
The protein composition of the various gradient fractions was analyzed by SDS/PAGE (Fig. 4A). A differential distribution of proteins across the gradient was observed, with high molecular weight U2 and U5 proteins peaking in fractions 9–13 and 17–23, respectively. In addition, a distinct protein pattern was found in the uppermost fractions, which essentially lack pre-mRNA, suggesting that some proteins (or snRNPs; see also Fig. 2C for RNA composition) have dissociated during elution and/or gradient centrifugation. Proteins present in fractions 11–13 (the A complex peak) were pooled, fractionated by SDS/PAGE, and analyzed by MALDI-MS or, additionally, by LC-MSMS.
Fig 4.
Protein content of the glycerol gradient fractionated, 45-min splicing complexes (A), or of pooled gradient fractions 11–13 (B). Protein was recovered from odd-numbered or pooled fractions and analyzed as in Fig. 2.
More than 70 proteins were identified, including almost all proteins known to be present in 12S U1 and 17S U2 snRNPs (Fig. 4B and Table 1). In contrast, no U4/U6 and only high molecular weight U5 and/or tri-snRNP proteins were detected, confirming the absence of large amounts of 25S U4/U6.U5 tri-snRNPs. Consistent with their known roles during E/A complex formation, most members of the SR protein family and the essential factors U2AF65 and U2AF35 were identified. A large number of hnRNP proteins and both m7G-cap-binding proteins (CBP20 and CBP80) were also identified. In addition to the 17S U2 snRNP-associated proteins hPrp5 and hPrp43, two more members of the DExH/D-box family of RNA unwindases/RNPases, namely, RNA helicase A (DDX9) and p68, were detected. Several proteins implicated in pre-mRNA splicing that appear to perform multiple roles in the cell, such as Y-box-binding protein 1 (YB-1), TLS/FUS, and NFAR-2 (15–19), were identified, as well as the mRNA export factors Aly and HuR (20–22). Interestingly, affinity-purified prespliceosomes also contained a number of proteins not previously known to be involved in splicing (Table 1).
Table 1.
Prespliceosome-associated proteins
| Protein | BN | SC | AC | Feature |
|---|---|---|---|---|
| U1 snRNP | ||||
| U1-70K | 15 | +++ | P08621 | RRM, SR |
| U1A | 28 | +++ | P09012 | 2 RRM |
| U1C | 34 | + | NP_003084 | C2H2-ZF |
| 17S U2 snRNP | ||||
| SF3b155 | 3 | +++ | AF054284 | HEAT |
| SF3b145 | 4 | +++ | NP_006833 | SAP |
| SF3b130 | 5 | +++ | NP_036558 | CPSF A |
| SF3b49 | 20 | ++ | NP_005841 | 2 RRM |
| p14 | 37 | +++ | AAK94041 | RRM |
| SF3b14b | 37 | + | Q9UH06 | |
| SF3b10 | n.s. | ++ | NP_112577 | |
| SF3a120 | 6 | +++ | NP_005868 | 2 SURP, UBQ |
| SF3a66 | 16 | +++ | NP_009096 | C2H2-ZF |
| SF3a60 | 17 | +++ | NP_006793 | C2H2-ZF, SAP |
| U2A′ | 30 | +++ | P09661 | LRR |
| U2B" | 31 | +++ | P08579 | 2 RRM |
| 17S U2-associated proteins | ||||
| hPrp5 | 4 | + | XP_044949 | SR, DEAD |
| SR140 | 4 | + | BAA20790 | RRM, SURP, SR |
| CHERP | 5 | ++ | NP_006378 | SURP, SR, G-patch |
| hPrp43 | 9 | +++ | O43143 | DEAH |
| SPF45 | 19 | +++ | NP_006441 | G-patch, RRM |
| SPF31 | 26 | ++ | NP_055095 | DnaJ |
| SPF30 | 27 | ++ | AAC64086 | Tudor |
| U5 snRNP | ||||
| hPrp8 | 1 | + | AAC61776 | |
| 200K (Brr2) | 2 | + | O75643 | DEIH, DxxH |
| 116K (Snu114) | 6 | + | NP_004238 | GTP-EF2 |
| 102K (Prp6) | 7 | ++ | AAF66128 | TPR |
| 100K (Prp28) | 7,8 | + | NP_004809 | DEAD |
| tri-snRNP | ||||
| 110K (Snu66) | 8 | + | AAK49523 | SR |
| Sm proteins | ||||
| SmB/B′ | 31/32 | +++ | NP_003082 | Sm |
| SmD3 | 35 | +++ | NP_004166 | Sm |
| SmD2 | 36 | +++ | NP_004588 | Sm |
| SmD1 | 37 | +++ | NP_008869 | Sm |
| SmE | n.s. | ++ | NP_003085 | Sm |
| SmF | n.s. | ++ | NP_003086 | Sm |
| SmG | n.s. | ++ | NP_003087 | Sm |
| hnRNP proteins | ||||
| U | 6 | +++ | NP_114032 | SAP, SPRY |
| R | 17 | ++ | NP_005817 | 3 RRM, RGG |
| K | 15 | ++ | NP_112552 | 3 KH, (RGG) |
| G | 21 | ++ | NP_002130 | RRM |
| A3 | 21 | ++ | P51991 | 2 RRM |
| C1/C2 | 22 | +++ | NP_004491 | RRM |
| A2/B1 | 23 | +++ | NP_112533 | 2 RRM |
| A1 | 21/25 | +++ | NP_002127 | 2 RRM |
| SR proteins | ||||
| SRp55 | 18 | +++ | NP_006266 | 2 RRM, SR |
| SRp40 | 23 | + | Q13243 | 2 RRM, SR |
| 9G8 | 25/26 | ++ | NP_006267 | RRM, SR |
| ASF/SF2 | 27 | +++ | Q07955 | 2 RRM, SR |
| SC35 | 26 | ++ | A42701 | RRM, SR |
| SRp30 | 27 | + | NP_003760 | 2 RRM, SR |
| SRp20 | 33 | +++ | P23152 | RRM, SR |
| Splicing/transport factors | ||||
| CBP80 | 10 | +++ | A54748 | MIF4G |
| CBP20 | 35 | ++ | P52298 | RRM |
| U2AF65 | 15 | ++ | NP_009210 | SR, 3 RRM |
| U2AF35 | 37 | + | NP_006749 | C3H1-ZF, RRM, SR |
| DDX9 | 3 | + | Q08211 | 2 DSRM, DEAH |
| p68 | 14 | +++ | NP_004387 | DEAH |
| NFAR-2 | 8 | ++ | AAK07425 | 2 DSRM |
| TLS/FUS | 13 | +++ | P35637 | RRM, RANBP-ZF |
| YB-1 | 20 | +++ | P16991 | CSB |
| F23858_1 | 11 | +++ | T02299 | 2 SURP, G-patch |
| hPrp19 | 18 | + | NP_055317 | 7 WD40 |
| ALY | 27 | ++ | NP_005773 | RRM |
| HuR | 25 | ++ | NP_001410 | 3 RRM |
| No previous connection to splicing | ||||
| FLJ10839 | 3 | +++ | XP_167600 | (CSB), SAP |
| RBM 5 | 6 | ++ | NP_005769 | 2 RRM, RanBP-ZF, C2H2-ZF, G-patch |
| ASR2B | 7 | ++ | AAK21006 | RRM |
| E1B-AP5 | 8 | ++ | NP_008971 | SAP, SPRY, (RGG) |
| CC1.3 | 12 | ++ | NP_004893 | 3 RRM, RS |
| FLJ21007 | 12 | ++ | NP_110421 | UBA, Tudor |
| CD2BP2 | 19 | ++ | NP_006101 | GYF |
| DnaJ hom. | 6 | + | NP_055602 | DnaJ |
| SMC1 | 4 | + | BAA11495 | P-loop, DA-box |
| SMC2 | 5 | + | XP_050024 | P-loop, DA-box |
One peptide of U2AF35 was found in a band containing D1–D3, p14, and SF3b14b (Band 37). n.s., not shown in Fig. 4B. Human proteins are listed, with the names of selected yeast homologs shown in brackets. BN, band numbering in Fig. 4B. SC (score): a high (+++), middle (++), or low (+) score was assigned based on the percent sequence coverage determined by MALDI-MS and/or the number of peptides identified by LC-MSMS. Note that the intensity of the gel bands was not taken into account. AC, accession number as found in the protein sequence database of the ENTREZ databases at the National Center for Biotechnology Information. Feature, listed features are derived from Prosite Scan (http://hits.isb-sib.ch/cgi-bin/PFSCAN) and Pfam (www.sanger.ac.uk/Software/Pfam/index.shtml).
Discussion
By using a tobramycin affinity-selection procedure, followed by glycerol gradient centrifugation, it was possible to purify human prespliceosomes (i.e., A complex) under native conditions. Several lines of evidence support the conclusion that the complexes characterized by MS represent prespliceosomes as opposed to B/C complexes: (i) both the tobramycin eluate and MS-analyzed gradient fractions contained nearly equimolar amounts of pre-mRNA, U1 and U2 snRNA, and only minimal amounts of U5 or U4/U6 snRNA; (ii) most proteins identified by MS were U1 and U2 snRNP proteins or known prespliceosomal-splicing factors; and (iii) RNA–RNA crosslinks characteristic for B/C complex were not observed. Although several high molecular weight tri-snRNP proteins were present in low amounts (as judged by silver staining of the protein gel), no splicing factors known to associate first during or after B complex formation were detected by MS (Table 1). Tri-snRNP proteins could be present because of a low level of contaminating B/C complex (or a dissociation product thereof) or they may be bona fide prespliceosomal components. Indeed, recent studies suggest that the U4/U6.U5 tri-snRNP complex interacts with the pre-mRNA already at the early stages of splicing complex formation (23).
E and A complexes both contain U1 and U2 snRNPs, and, thus, it is difficult to distinguish between them based on snRNA and snRNP protein composition alone. However, two observations support the conclusion that the complexes analyzed by MS are predominantly A complexes: (i) the E complex protein SF1/mBBP was not detected, and (ii) psoralen crosslinking identified U1/pre-mRNA and U2/pre-mRNA base-pairing interactions in the MS-analyzed fractions. As the U2 snRNP first base-pairs with the pre-mRNA in the A complex (3), these data provide the best evidence that A complexes, as opposed to E complexes, were analyzed. However, because of the intrinsically low efficiency of psoralen crosslinking, we cannot rule out that gradient fractions 11–13 contain E complexes in addition to A. Gradient-fractionated, affinity-purified splicing complexes did not withstand native PAGE, and thus, this method could not be used to address this question.
Most proteins identified in our affinity-purified prespliceosomes are known to be involved in pre-mRNA splicing, a fact which attests to the high purity of these complexes. Furthermore, nearly all proteins expected to be present in mammalian prespliceosomes were found, suggesting that the inventory of prespliceosomal proteins presented in Table 1 is relatively complete. Interestingly, most of the newly identified 17S U2 snRNP proteins, such as hPrp5, hPrp43, SPF45, SF3b14, and SF3b10, were present in purified prespliceosomes, consistent with the idea that they potentially facilitate the association of U2 with the pre-mRNA (24). Based on the combined total mass of the identified proteins, it is unlikely that all of them are found in every prespliceosome. Indeed, some proteins are clearly present in substoichiometric amounts, indicating that they are present in a subpopulation of the affinity-purified complexes. As multiple members of the SR and hnRNP protein families were detected, it is also likely that the complexes we have analyzed may consist of a mixture of prespliceosomes containing different subsets of these proteins.
In addition to SR, hnRNP, and snRNP proteins, ≈20 additional polypeptides were identified. Each of the newly identified prespliceosomal proteins, by virtue of their copurification with these complexes, potentially plays a role in splicing. Several multifunctional proteins, including YB-1 and TLS/FUS were among those identified, consistent with the idea that they are general splicing factors and may act at an early stage of the splicing process. Indeed, numerous studies have implicated TLS/FUS in pre-mRNA splicing (17, 25–27), and YB-1 is known to play a role in alternative splicing (15). In addition to hPrp5 and hPrp43, the DExH-box proteins, p68 and RNA helicase A (DDX9) were also identified, suggesting that they may contribute to the dynamics of spliceosome formation. Correspondingly, recent evidence suggests that p68 destabilizes the U1/5′ splice-site interaction during spliceosome assembly (28).
MS revealed several proteins cofractionating with splicing complex A whose precise function is not known. Three of them (CC1.3, F23858_1, and RBM 5) contain domains (i.e., SURP, SR) that are highly characteristic of pre-mRNA splicing factors, making them good candidates for previously uncharacterized, bona fide prespliceosomal proteins. Additional experiments are needed, however, to clarify whether any are indeed involved in pre-mRNA splicing. Although the majority of proteins identified by MS are most likely bona fide splicing factors, some (e.g., hnRNP proteins) may bind flanking regions of the pre-mRNA or internal intron sequences that are not involved in the splicing process. Future studies with truncated MINX pre-mRNAs should reveal if these and potentially other proteins bind the substrate, but are not directly involved in splicing. At present, it is also not clear which of the newly identified prespliceosomal proteins are core components (i.e., are generally present in prespliceosomes) and which may specifically associate with the MINX pre-mRNA used in our studies.
Our data provide the first detailed inventory of the components of the spliceosome at the early stages of its assembly. The characterization of complexes formed at later stages should allow a clear cut determination of the order and dynamics of the association of proteins with the spliceosome during its assembly, catalytic activity, and subsequent disassembly. Similarly, the affinity purification method described here could potentially be used to identify substrate-specific splicing factors, such as those involved in alternative splicing events. Finally, the ability to isolate prespliceosomes, as well as potentially other spliceosomal complexes under native conditions, is a pivotal step toward the elucidation of their molecular architecture. Ultrastructural investigations, e.g., electron microscopy, performed with native spliceosomal complexes at defined stages of the splicing reaction, may ultimately reveal the structural dynamics of the pre-mRNA-splicing machinery.
Supplementary Material
Acknowledgments
We thank Irene Öchsner and Monika Raabe for excellent technical assistance. This work was supported by Deutsche Forschungsgemeinschaft Grant Lu 294/12-1 and Bundesministerium für Bildung und Forschung (Germany) Grant 031U215B (to R.L.).
Abbreviations
snRNP, small nuclear ribonucleoprotein
hnRNP, heterogenous nuclear ribonucleoprotein
SR, serine/arginine rich
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Reed R. & Palandijan, L. (1997) in Eukaryotic mRNA Processing, ed. Krainer, A. R. (IRL, Oxford), pp. 103–129.
- 2.Burge C. B., Tuschl, T. & Sharp, P. A. (1999) in The RNA World, eds. Gesteland, R. F., Cech, T. R. & Atkins, J. F. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 525–560.
- 3.Das R., Zhou, Z. & Reed, R. (2000) Mol. Cell 5 779-787. [DOI] [PubMed] [Google Scholar]
- 4.Neubauer G., King, A., Rappsilber, J., Calvio, C., Watson, M., Ajuh, P., Sleeman, J., Lamond, A. & Mann, M. (1998) Nat. Genet. 20 46-50. [DOI] [PubMed] [Google Scholar]
- 5.Rappsilber J., Ryder, U., Lamond, A. I. & Mann, M. (2002) Genome Res. 12 1231-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou Z., Licklider, L. J., Gygi, S. P. & Reed, R. (2002) Nature 419 182-185. [DOI] [PubMed] [Google Scholar]
- 7.Jurica M. S., Licklider, L. J., Gygi, S. R., Grigorieff, N. & Moore, M. J. (2002) RNA 8 426-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hamasaki K., Killian, J., Cho, J. & Rando, R. R. (1998) Biochemistry 37 656-663. [DOI] [PubMed] [Google Scholar]
- 9.Zillmann M., Zapp, M. L. & Berget, S. M. (1988) Mol. Cell. Biol. 8 814-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Y. & Rando, R. R. (1995) Chem. Biol. 2 281-290. [DOI] [PubMed] [Google Scholar]
- 11.Dignam J. D., Lebovitz, R. M. & Roeder, R. G. (1983) Nucleic Acids Res. 11 1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wassarman D. A. & Steitz, J. A. (1992) Science 257 1918-1925. [DOI] [PubMed] [Google Scholar]
- 13.Fabrizio P., McPheeters, D. S. & Abelson, J. (1989) Genes Dev. 3 2137-2150. [DOI] [PubMed] [Google Scholar]
- 14.Shevchenko A., Wilm, M., Vorm, O. & Mann, M. (1996) Anal. Chem. 68 850-858. [DOI] [PubMed] [Google Scholar]
- 15.Stickeler E., Fraser, S. D., Honig, A., Chen, A. L., Berget, S. M. & Cooper, T. A. (2001) EMBO J. 20 3821-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lerga A., Hallier, M., Delva, L., Orvain, C., Gallais, I., Marie, J. & Moreau-Gachelin, F. (2001) J. Biol. Chem. 276 6807-6816. [DOI] [PubMed] [Google Scholar]
- 17.Hallier M., Lerga, A., Barnache, S., Tavitian, A. & Moreau-Gachelin, F. (1998) J. Biol. Chem. 273 4838-4842. [DOI] [PubMed] [Google Scholar]
- 18.Saunders L. R., Jurecic, V. & Barber, G. N. (2001) Genomics 71 256-259. [DOI] [PubMed] [Google Scholar]
- 19.Saunders L. R., Perkins, D. J., Balachandran, S., Michaels, R., Ford, R., Mayeda, A. & Barber, G. N. (2001) J. Biol. Chem. 276 32300-32312. [DOI] [PubMed] [Google Scholar]
- 20.Zhou Z., Luo, M. J., Straesser, K., Katahira, J., Hurt, E. & Reed, R. (2000) Nature 407 401-405. [DOI] [PubMed] [Google Scholar]
- 21.Strasser K. & Hurt, E. (2001) Nature 413 648-652. [DOI] [PubMed] [Google Scholar]
- 22.Gallouzi I. E. & Steitz, J. A. (2001) Science 294 1895-1901. [DOI] [PubMed] [Google Scholar]
- 23.Maroney P. A., Romfo, C. M. & Nilsen, T. W. (2000) Mol. Cell 6 317-328. [DOI] [PubMed] [Google Scholar]
- 24.Will C. L., Urlaub, H., Achsel, T., Gentzel, M., Wilm, M. & Lührmann, R. (2002) EMBO J. 21 4978-4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hackl W. & Lührmann, R. (1996) J. Mol. Biol. 264 843-851. [DOI] [PubMed] [Google Scholar]
- 26.Wu S. & Green, M. R. (1997) EMBO J. 16 4421-4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang L., Embree, L. J., Tsai, S. & Hickstein, D. D. (1998) J. Biol. Chem. 273 27761-27764. [DOI] [PubMed] [Google Scholar]
- 28.Liu Z. R. (2002) Mol. Cell. Biol. 22 5443-5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.