Abstract
Several studies have reported that activation of Gq-coupled receptors inhibits PI3K (phosphoinositide 3-kinase) signalling. In the present study, we used purified proteins to demonstrate that Gαq directly inhibits p110α/p85α PI3K in a GTP-dependent manner. Activated Gαq binds to the p110α/p85α PI3K with an apparent affinity that is seven times stronger than that for Gαq·GDP as measured by fluorescence spectroscopy. In contrast, Gαq did not bind to the p110γ PI3K. Fluorescence spectroscopy experiments also showed that Gαq competes with Ras, a PI3K activator, for binding to p110α/p85α. Interestingly, co-precipitation studies using deletion mutants showed that Gαq binds to the p85-binding domain of p110α and not to the Ras-binding domain. Expression of constitutively active GαqQ209L in cells inhibited Ras activation of the PI3K/Akt pathway but had no effect on Ras/Raf/MEK [MAPK (mitogen-activated protein kinase)/ERK (extracellular-signal-regulated kinase) kinase] signalling. These results suggest that activation of Gq-coupled receptors leads to increased binding of Gαq·GTP to some isoforms of PI3K, which might explain why these receptors inhibit this signalling pathway in certain cell types.
Keywords: Akt, fluorescence spectroscopy, Gαq, p85, phosphoinositide 3-kinase, Ras
Abbreviations: coumarin, 7-(dimethylamino)coumarin-4-acetic acid succinimidyl ester; GST, glutathione S-transferase; GTP[S], guanosine 5′-[γ-thio]triphosphate; HA, haemagglutinin; HEK-293 cells, human embryonic kidney 293 cells; IRS-1, insulin receptor substrate-1; MEK, mitogen-activated protein kinase/extracellular-signal-regulated kinase kinase; SH2, Src homology 2; iSH2, inter-SH2; LUV, large unilamellar vesicle; PDGF, platelet-derived growth factor; PI3K, phosphoinositide 3-kinase; PLCβ, phospholipase Cβ; POPC, 1-palmitoyl-2-oleoyl phosphatidylcholine; PS, 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine; RBD, Ras-binding domain
INTRODUCTION
PI3Ks (phosphoinositide 3-kinases) are lipid kinases that regulate diverse physiological processes, including glucose metabolism, cell size regulation, cell survival and cardiac myocyte contractility [1,2]. Of the three classes of PI3K, only the class I enzymes preferentially phosphorylate phosphatidylinositol 4,5-bisphosphate to form phosphatidylinositol 3,4,5-trisphosphate in vivo and exhibit substantial activation in response to growth factor and hormone stimulation. The four class I PI3K isoforms are divided into two subgroups. Class IA PI3Ks are heterodimers consisting of a catalytic subunit (p110α, p110β or p110δ) tightly bound to a regulatory subunit (p85α, p85β, p55γ or variants produced by alternative splicing). The class IB catalytic subunit p110γ does not bind to the p85 or p55 proteins, but instead forms a heterodimer with p101 or p84 regulatory subunits [1,3].
All class IA regulatory subunits (collectively referred to here as p85) contain two SH2 (Src homology 2) domains separated by an iSH2 (inter-SH2) region that binds to the p110 subunit. Binding of the regulatory subunit to p110 inhibits its catalytic activity [4,5]. Receptor tyrosine kinases appear to activate the class IA PI3Ks by using two synergistic mechanisms. First, receptor-mediated phosphorylation of proteins on specific tyrosine residues produces docking sites that bind to the SH2 domains of p85, thus recruiting the enzyme to membranes where its lipid substrates are located and releasing the inhibitory effect of p85 on the catalytic subunit [5–7]. Secondly, many receptor tyrosine kinases activate Ras, which is also located at the membrane, and stimulate its binding to PI3K [8,9]. Initial studies showed that Ras interacts with an RBD (Ras-binding domain) in the p110α and p110β subunits [9,10]. Later studies identified similar domains in p110δ and p110γ [11,12]. Although Ras has been demonstrated to bind to all four class I PI3K catalytic subunits in a GTP-dependent manner, only p110α and p110γ have been shown to be activated by Ras·GTP binding in vitro [10,13]. The p85 subunit of class IA PI3Ks also appears to play an indirect role in controlling the activation of PI3K by Ras [14,15].
G-protein-coupled receptors exert diverse effects on the activity of class I PI3Ks. Activation of these receptors leads to the exchange of GTP for GDP on the heterotrimeric G-protein α subunit, which causes Gα·GTP to dissociate from the Gβγ dimer. Both Gα·GTP and Gβγ can then interact with specific effector proteins to alter cellular functions. p110γ is activated in response to the stimulation of these receptors, and p110β is thought to be activated by both G-protein-coupled and tyrosine kinase receptors [1]. Direct binding of Gβγ to p110γ and p110β activates these enzymes [16,17]. On the other hand, we and others have reported that receptors coupled with the Gαq subfamily of Gα-proteins can inhibit PI3K signalling [18–20]. In subsequent studies, we found that Gαq inhibits PI3K without activating its best-known effector, PLCβ (phospholipase Cβ) [21,22]. Furthermore, we showed that the constitutively active GαqQ209L mutant co-precipitates with the p110α/p85α complex and causes a reduction in its lipid kinase activity. Based on these results, we hypothesized that Gαq binds directly to p110α/p85α to inhibit its activity. In the present study, we used purified recombinant proteins to examine the effect of Gαq on the lipid kinase activity of p110α/p85α in vitro. Next, we measured Gαq binding to p110α/p85α using fluorescence spectroscopy. We found that Gαq binds to PI3K and inhibits its activity in a GTP-dependent manner. Furthermore, in addition to suppressing intrinsic PI3K activity, activated Gαq blocks Ras from binding to p110α/p85α, which might provide an additional mechanism for the inhibitory effect of Gαq on PI3K activation.
MATERIALS AND METHODS
Materials
HA (haemagglutinin) antibody was obtained from Covance (Richmond, CA, U.S.A.). FLAG antibody was from Sigma (St. Louis, MO, U.S.A.). Phospho-Ser217/Ser221 MEK1/2 antibody (MEK is mitogen-activated protein kinase/extracellular-signal-regulated kinase kinase) was obtained from New England Biolabs (Beverly, MA, U.S.A.). Recombinant p110γ protein was purchased from Axxora (San Diego, CA, U.S.A.).
Constructs and recombinant proteins
The cDNA for human H-Ras was obtained by RT (reverse transcriptase)–PCR from HEK-293 (human embryonic kidney 293) cell RNA. It was used as a template to make V12Ras by PCR-based mutagenesis. FLAG–p110α was described previously [21]. FLAG–p110α amino acids 1–116 was made by mutating codon 117 of p110α to a stop codon. p110α amino acids 118–1068 was made by PCR using p110α as a template and the fragment was then subcloned into p3XFLAG-CMV10 (Sigma) to make FLAG–p110α amino acids 118–1068. FLAG–p110γ was constructed by subcloning the p110γ insert of IMAGE clone number 5749986 into p3XFLAG-CMV10. HA–GαqQ209L and GαqQ209L were described previously [21,22]. HA–MEK1 was a gift from N. G. Ahn (University of Colorado, Boulder, CO, U.S.A.). Akt–HA was obtained from R. Roth (Stanford University, Stanford, CA, U.S.A.).
Purification of the p110α/p85α PI3K complex [21] and Gαq [23] from baculovirus-infected Sf9 cells was described previously. Purified recombinant H-Ras (amino acids 1–166) expressed in Escherichia coli was generously provided by Dr N. Nassar (Stony Brook University, Stony Brook, NY, U.S.A.). To make GST (glutathione S-transferase)–p110α fusion proteins, fragments encompassing the various p110α domains were made by PCR and then subcloned into pGEX-5X-3 (Amersham Biosciences, Piscataway, NJ, U.S.A.). GST and the GST–p110α fusion proteins were expressed in E. coli and purified using glutathione–Sepharose 4B (Amersham Biosciences).
Cell culture and lysate preparation
HEK-293 cells and COS7 cells were maintained in Dulbecco's modified Eagle's medium (Sigma) containing 10% (v/v) fetal bovine serum (Sigma) and antibiotics. COS7 cells were transfected using Lipofectamine™ (Invitrogen, Carlsbad, CA, U.S.A.). HEK-293 cells were transfected using TransIT-293 (Mirus, Madison, WI, U.S.A.) following standard methods. The FreeStyle 293 Expression System (Invitrogen) was used to express HA–GαqQ209L. Cells were rinsed with ice-cold PBS 2 days after transfection, and scraped into lysis buffer (50 mM Hepes, pH 7.5, 50 mM NaCl, 5 mM EDTA, 50 mM NaF, 10 mM sodium pyrophosphate, 1 mM sodium orthovanadate, 0.5 mM PMSF and 10 μg/ml aprotinin and leupeptin) with either 1% Triton X-100 or 1% Nonidet P40 plus 0.25% sodium deoxycholate. Homogenates were centrifuged at 15000 g for 15 min at 4 °C, and protein concentrations of the supernatants were determined using a Bradford assay (Bio-Rad, Hercules, CA, U.S.A.).
Kinase assays and Western blots
Akt activity was assayed following a method described previously [18]. PI3K was assayed as described previously [18] except that the PI3K assay buffer contained 40 mM Hepes (pH 7.5), 1 mg/ml fatty-acid-free BSA, 2 mM EGTA and 100 mM NaCl. Western blot signals were visualized using horseradish-peroxidase-linked secondary antibodies (Amersham Biosciences) and chemiluminescence reagents (PerkinElmer Life Sciences, Boston, MA, U.S.A.) [18].
Fluorescence measurements and data analysis
Purified Gαq was stored in a solution containing 20 mM Hepes (pH 7.2), 1 mM EDTA, 3 mM MgCl2, 400 mM NaCl, 0.7% CHAPS and 50 μM GDP (GDP buffer). To activate Gαq, the protein was incubated for 1 h at 30 °C in GTP[S] (guanosine 5′-[γ-thio]triphosphate) buffer (50 mM Hepes, pH 7.2, 100 mM (NH4)2SO4, 150 mM MgSO4, 1 mM EDTA, 0.7% CHAPS and 100 μM GTP[S]) [24]. To activate Ras, the protein was incubated in a buffer containing 20 mM Hepes (pH 7.2), 200 μM (NH4)2SO4, 5 mM EDTA and 5 μM GTP[S] for 1 h on ice. The reaction was stopped by adding 20 mM MgSO4 and the protein was dialysed against buffer A (20 mM Hepes and 160 mM KCl, pH 7.2) plus 1 mM 2-mercaptoethanol for 30 min.
Prior to labelling with 7-(dimethylamino)coumarin-4-acetic acid succinimidyl ester (referred to as coumarin; Molecular Probes, Eugene, OR, U.S.A.), both Gαq and Ras were dialysed overnight against buffer A. The pH of the Gαq and Ras solutions was raised by adding 1 μl of 2 M K2HPO4 (pH 8.5) per 100 μl before labelling with a 4-fold molar excess of coumarin. The reaction mixtures were incubated on ice for 1 h and then free probe was removed by dialysis against buffer A at 4 °C.
LUVs (large unilamellar vesicles), 0.1 μm in diameter, were formed from a 2:1 mixture of POPC (1-palmitoyl-2-oleoyl phosphatidylcholine) and PS (1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine; Avanti Polar Lipids, Alabaster, AL, U.S.A.). Lipids were dissolved in chloroform, dried under vacuum and resuspended in buffer A at room temperature (22 °C). The lipids were then subjected to five cycles of freeze–thawing to form multilamellar vesicles and then extruded through a polycarbonate filter (0.1 μm diameter) for ten cycles to produce LUVs.
Fluorescence measurements were performed on an ISS spectrofluorimeter (ISS, Champaign, IL, U.S.A.) in 3 mm pathlength cuvettes that were filled with 120 μl of sample. All samples contained 80 μM LUVs. Buffer controls used buffer A plus 1 mM dithiothreitol. Coumarin-labelled proteins were excited at 340 nm and fluorescence emission intensity was scanned from 380 to 580 nm. The integrated area was calculated for each point of the titration curve. Binding to either coumarin–Gαq or coumarin–Ras resulted in a maximal 20% increase in fluorescence. The spectra were corrected for loss of fluorescence caused by dilution from addition of buffer alone. These background-corrected spectra were then normalized to the last point on the titration curve to give a final value of 1.0. This process assumes complete binding and allows us to determine apparent dissociation constants (Kd) in nanomolar units when the data are fitted to a bimolecular association curve. Experiments were performed in triplicate and results shown are the means±S.E.M.
RESULTS
Gαq inhibits p110α/p85α PI3K
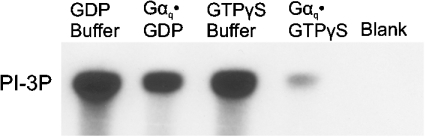
We used recombinant proteins purified from baculovirus-infected insect cells to test if Gαq inhibits the lipid kinase activity of the p110α/p85α PI3K complex in vitro. Addition of Gαq·GDP resulted in a 46±17% (mean±S.E.M., n=4) decrease in PI3K activity as compared with the buffer control, whereas the same amount of Gαq·GTP[S] caused a 92±1% (mean±S.E.M., n=4) decrease in activity (Figure 1). These results show that Gαq directly inhibits the p110α/p85α PI3K complex and activated Gαq is a more potent inhibitor than inactive Gαq.
Figure 1. Inhibition of PI3K by Gαq.
Gαq·GDP and Gαq·GTP[S], along with an equal volume of GDP buffer or GTP[S] buffer as controls, were diluted in PI3K assay buffer and concentrated using a Microcon 30 microconcentrator (Amicon, Beverly, MA, U.S.A.). This was repeated three more times to remove nucleotides and CHAPS, which interferes with the PI3K assay ([26] and results not shown). PI3K assays contained 25 nM purified p110α/p85α complex and either 15 nM Gαq or an equal volume of control buffer. L-α-phosphatidylinositol (Sigma) and [γ-32P]ATP (PerkinElmer LifeSciences) were used as substrates. The autoradiogram shows a typical result from four experiments. Abbreviations: GTPγS, GTP[S]; PI-3P, phosphatidylinositol 3-phosphate.
Gαq binds to p110α/p85α PI3K
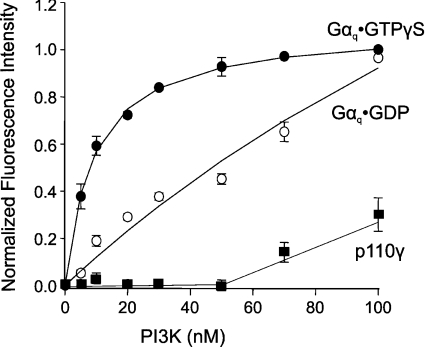
Next, we used a fluorescence spectroscopy technique to measure the Kd between p110α/p85α PI3K and Gαq. Since Gαq is a membrane-bound protein, these measurements were performed in the presence of lipid vesicles consisting of POPC/PS (2:1) to mimic the membrane environment. Gαq was labelled with coumarin and reconstituted into the vesicles, and then PI3K was titrated into the solution. The emission intensity of the coumarin-labelled Gαq, which increases upon binding to other proteins [25], was then measured. Coumarin labelling does not impair Gαq function, since coumarin–Gαq activates PLCβ isoenzymes as efficiently as unlabelled Gαq [25]. Control experiments showed that PI3K also binds to the lipid vesicles with high affinity, with a membrane partition coefficient of approx. 10 μM (results not shown). Since we carried out these studies at lipid concentrations (80 μM) where both proteins are bound, the interactions monitored here occur between proteins confined to vesicle surfaces.
Figure 2 shows that addition of increasing amounts of p110α/p85α PI3K into a solution containing active Gαq·GTP[S] caused an increase in fluorescence that reached a plateau at approx. 100 nM PI3K. The Kd of PI3K binding to Gαq·GTP[S] was calculated to be 11±1 nM. The binding curve using inactive Gαq·GDP yielded a Kd of 77±25 nM. The binding curve for Gαq·GDP appears to consist of two exponential curves, suggesting that two binding species might be present, which may be due to different conformational or oligomeric states. Additional biophysical analysis would be needed to clarify these possibilities. By comparison, Kd values for coumarin-labelled Gαq·GTP[S] binding to three isotypes of PLCβ ranged from 0.3 to 2.8 nM, as determined by fluorescence resonance energy transfer (data in [25] were recalculated to give Kd values at the same lipid concentration used in these studies). The increase in fluorescence was reversed when trypsin was added to the solution (results not shown). In addition, the binding curves shifted to the right with increasing amounts of Gαq·GTP[S], but the Kd values remained the same, indicative of a true protein–protein equilibrium association (results not shown). Gαq·GTP[S] did not bind to the p110γ PI3K under these conditions (Kd>1800 nM; Figure 2), suggesting that Gαq might selectively inhibit only certain PI3K isoforms. These results demonstrate that Gαq binds directly to the p110α/p85α PI3K, and active Gαq binds with a higher affinity than inactive Gαq.
Figure 2. Gαq binds to PI3K.
Purified recombinant p110α/p85α complex was titrated into a solution containing 5 nM coumarin-labelled Gαq·GTP[S] (●) or Gαq·GDP (○) reconstituted into 80 μM POPC/PS (2:1) LUVs. Recombinant p110γ was added to a solution containing 5 nM coumarin-labelled Gαq·GTP[S] (■) in LUVs. Fluorescence measurements and data analysis were performed as described in the Materials and methods section. The results shown are from three independent experiments performed in triplicate.
Gαq interferes with Ras binding to PI3K
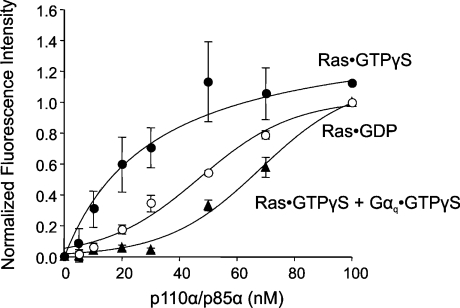
The data in Figure 1 indicate that activated Gαq inhibits intrinsic PI3K activity. We wondered if the interaction with Gαq might also interfere with binding of PI3K to an activator such as Ras. We used fluorescence spectroscopy to first measure the Kd of p110α/p85α complex binding to Ras. Fluorescence intensity measurements were made after adding increasing amounts of PI3K to lipid vesicles preincubated with coumarin-labelled Ras. As expected, PI3K bound to Ras·GTP[S] with a Kd of 31±12 nM, while the Kd with inactive Ras·GDP was estimated to be at least 30 times higher (Figure 3). This correlates well with a value of approx. 150 nM for the interaction between an activated Ras mutant and p110α/p85α measured in solution [10]. When the lipid vesicles were reconstituted with coumarin–Ras in the presence of unlabelled Gαq·GTP[S], specific binding between Ras and PI3K was greatly reduced (Figure 3). This result indicates that Gαq·GTP[S] interferes with Ras binding to p110α/p85α.
Figure 3. Gαq competes with Ras for binding to PI3K.
LUVs were reconstituted with 10 nM coumarin-labelled Ras·GDP (○) or coumarin–Ras·GTP[S] without (●) or with (▲) 10 nM Gαq·GTP[S]. Purified recombinant p110α/p85α was titrated into the solutions and fluorescence measurements were made. The results shown are from three independent experiments performed in triplicate. GTPγS, GTP[S].
We also investigated if Gαq interferes with PI3K binding to tyrosine-phosphorylated proteins that are thought to contribute to receptor-mediated PI3K activation. Using fluorescence spectroscopy, we found that binding between coumarin-labelled tyrosine-phosphorylated IRS-1 (insulin receptor substrate-1) and p110α/p85α was not affected by Gαq·GTP[S] (results not shown).
Gαq binds to the p85-binding domain of p110α
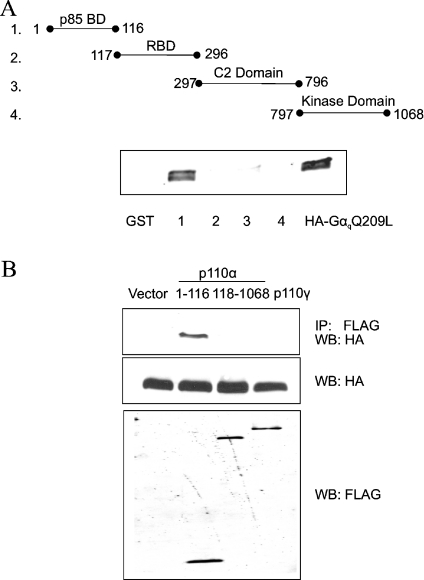
In vitro binding assays using PI3K deletion mutants identified an RBD located in the N-terminal region of p110α and p110β [10] (see Figure 4A). In addition, X-ray crystallography of a Ras·p110γ complex revealed that Ras establishes contacts with the RBD and the catalytic domain [13]. Since Gαq blocks Ras binding to PI3K, we asked whether Gαq also binds to the RBD of p110α. Figure 4(A) illustrates GST fusion proteins encompassing the various domains of p110α that were purified from bacteria and mixed with lysates of mammalian cells expressing HA-tagged GαqQ209L. The GST fusion proteins were pulled down with glutathione–Sepharose, and the presence of HA–GαqQ209L was assessed by Western blotting. Surprisingly, we found that the p85-binding domain of p110α co-precipitated with HA–GαqQ209L, but the RBD did not (Figure 4A).
Figure 4. Gαq binds to the p85-binding domain (p85 BD) of p110α.
(A) Purified GST–p110α fusion proteins encompassing individual domains of p110α (50 ng) were immobilized on glutathione–Sepharose beads and mixed at 4 °C for 1 h with 250 μg of lysate proteins from FreeStyle 293 cells expressing HA–GαqQ209L. Cells were lysed in a buffer containing 1% Nonidet P40 plus 0.25% sodium deoxycholate (see the Materials and methods section). The beads were washed with lysis buffer and bound proteins were analysed on a Western blot probed with HA antibody. Native GST protein immobilized on beads was used as a negative control (first lane). The last lane is a sample of cell lysate expressing HA–GαqQ209L. The experiment was repeated with similar results. (B) COS7 cells were co-transfected with HA–GαqQ209L and either empty vector, FLAG–p110α amino acids 1–116, FLAG–p110α amino acids 118–1068 or FLAG–p110γ. Cell lysates were incubated with FLAG antibody and the immunoprecipitates were analysed on a Western blot probed with HA antibody (top panel). Western-blot analysis of total cell lysate proteins showed appropriate expression of HA–GαqQ209L and the various FLAG-tagged p110 proteins (bottom two panels). IP, immunoprecipitate; WB, Western blot. The experiment was repeated with similar results.
We also tested whether a FLAG-tagged p110α fragment (amino acids 118–1068) that does not contain the p85-binding domain co-immunoprecipitates with HA–GαqQ209L from extracts of COS7 cells expressing both proteins. FLAG–p110α amino acids 118–1068 exhibits substantial lipid kinase activity in an in vitro assay, indicating that the protein is folded correctly (results not shown). Consistent with our findings above using GST fusion proteins, FLAG–p110α amino acids 1–116 co-precipitated with HA–GαqQ209L, but FLAG–p110α amino acids 118–1068 did not (Figure 4B). FLAG-tagged p110γ, which contains an RBD but lacks a p85-binding domain, also did not co-immunoprecipitate with co-transfected HA–GαqQ209L (Figure 4B). We showed earlier that the amount of p85α bound to p110α does not change in the presence of HA–GαqQ209L [21]. Therefore, even though activated Gαq binds to the p85α-binding site on p110α, it does not inhibit PI3K activity by preventing formation of the p110α/p85α complex.
Gαq interferes with Ras/PI3K signalling
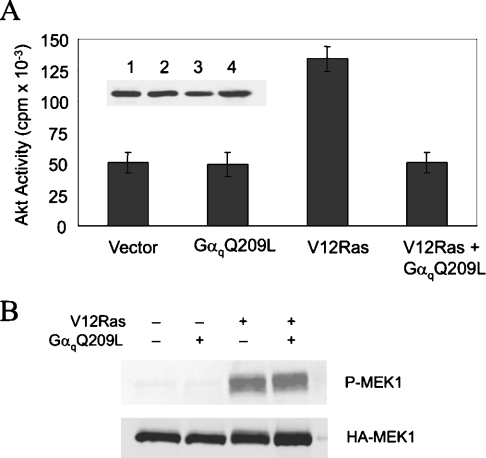
In addition to PI3K, Ras has a number of other direct targets, including the protein kinase Raf. We reasoned that Gαq should specifically inhibit the Ras/PI3K pathway and not the Ras/Raf pathway if Gαq competes with Ras for binding to PI3K. To measure the effect of Gαq on Ras/PI3K signalling, we assayed Akt activity in HEK-293 cells transfected with Akt–HA in the presence or absence of V12Ras and GαqQ209L. The protein kinase Akt is activated by the lipids produced by PI3K and its activity serves as an indirect measure of PI3K activity. Figure 5(A) shows that Akt activity was almost three times higher in cells expressing V12Ras than in vector-transfected control cells, and this increase was completely blocked by GαqQ209L. Bommakanti et al. [19] also observed that activated Gαq exerts a strong inhibitory effect on Ras activation of Akt. Western blotting showed that the amount of Akt–HA in each sample was similar (Figure 5A).
Figure 5. Specific inhibition of the Ras/PI3K/Akt pathway by Gαq.
HEK-293 cells were co-transfected with (A) Akt–HA or (B) HA–MEK1 with or without V12Ras in the presence or absence of GαqQ209L. Cells were incubated in serum-free medium overnight prior to lysate preparation. (A) Akt activity was assayed in HA immunoprecipitates of cell lysates (mean±S.E.M.; n=3). The inset shows a Western blot of cell lysates probed with HA antibody. (B) Activation of MEK1 was assessed on a Western blot probed with phospho-MEK1/2 antibody (upper panel). The blot was reprobed with HA antibody (lower panel).
To assess the effect of Gαq on Ras/Raf signalling, we examined the phosphorylation of MEK1 at Ser217/Ser221. Binding of activated Ras to Raf leads to the phosphorylation and activation of MEK1. HEK-293 cells were transfected with HA–MEK1 in the presence or absence of V12Ras and GαqQ209L. Western blotting with a phospho-specific antibody showed that V12Ras activates MEK1, but GαqQ209L did not block this response (Figure 5B). Reprobing the blot with HA antibody showed that the amount of HA–MEK1 was similar in each lane (Figure 5B). These results indicate that active Gαq does not block all Ras effector pathways.
DISCUSSION
Many reports indicate that activation of Gαq-coupled receptors negatively regulates PI3K signalling. However, these receptors can activate more than one type of Gα subunit, and the Gβγ dimers stimulate some PI3K isoforms. Therefore attributing an effect on PI3K to a particular G-protein subunit after receptor activation can be difficult. In vitro experiments that directly assess the effect of G-protein subunits on p110γ showed that Gβγ dimers activate this enzyme and Gα subunits do not [26]. It was also reported that Gβγ dimers activate p110β but not p110α or p110δ [17,27]. The effect of Gα subunits on class IA PI3Ks has not been investigated up to now using in vitro experiments. Olefsky and co-workers [28] showed that treatment of cultured adipocytes with endothelin-1 increases the amount of endogenous Gαq/11 that co-immunoprecipitates with p110α. We used the GαqQ209L mutant to circumvent the need for receptor activation and showed that this protein also co-precipitates with p110α from extracts of COS7 cells expressing both proteins [21]. Furthermore, we found that GαqQ209L inhibits p110α but not p110β activity in a manner that is independent of PLCβ activation [21,22]. In the present study, we demonstrate using purified proteins that Gαq inhibits the lipid kinase activity of p110α/p85α by binding directly to the enzyme. This result provides a mechanism to explain how Gαq attenuates PI3K signalling in the absence of PLCβ activation.
Evidence obtained using cell systems that are not manipulated to overexpress Gαq or receptors suggests that the inhibitory effect of Gαq on PI3K can have physiological consequences. A number of studies have shown that insulin activation of PI3K is inhibited by agonists that act through Gq-coupled receptors [29–31]. In addition, Exton and co-workers and others [32,33] showed years ago that Gq-coupled receptors inhibit glycogen synthase and counteract the effect of insulin in hepatocytes. It is now known that PI3K plays a key role in regulating glycogen metabolism, and we believe that Gαq inhibition of PI3K may partly explain why activation of these receptors antagonizes insulin responses. In addition, studies on neurons indicate that the L-type voltage-dependent Ca2+ channel is positively regulated by PI3K and negatively regulated by the M1 muscarinic receptor [34,35]. Interestingly, negative regulation by the receptor is mediated by Gαq/11 but does not appear to involve PLCβ [36]. These results are consistent with our recent findings that activation of Gαq in cardiac myocytes of transgenic mice inhibits the L-type Ca2+ channel in a PLCβ-independent manner [37] and that this effect is mediated by PI3K inhibition [38].
We determined that Gαq binds to the p85-binding domain of p110α and does not appear to directly interact with the catalytic domain. It will be of interest to determine if Gαq also binds to the p85-binding domains in p110β and p110δ. It remains unclear how this interaction leads to inhibition of lipid kinase activity. Binding of Gαq to p110α might induce a conformational change in the catalytic site that decreases the kcat (catalytic-centre activity) or the affinity for substrates. Additional studies to elucidate how Gαq inhibits PI3K will require the production of p110α mutants that cannot bind Gαq but that can still form a complex with p85.
The mechanism by which Ras activates PI3K is controversial. Conflicting studies point to either the RBD in p110α or the iSH2 domain in p85α as playing a dominant role in mediating the activating effect of Ras on PI3K [10,15]. The crystal structure of a Ras·p110γ complex shows that Ras makes direct contacts with both the RBD and the C-terminal lobe of the PI3K catalytic domain [13]. Ras binding to p110γ induces conformational changes that affect the putative phosphatidylinositol headgroup binding site and may result in a change of affinity for substrate [13]. Regardless of the exact mechanism for Ras activation, we found that Gαq blocks Ras from binding to PI3K and completely suppresses Ras activation of PI3K signalling. Gαq does not appear to compete directly for Ras binding to p110α, since it does not bind to the RBD. Instead, Gαq binding to PI3K may induce a structural change that disrupts its ability to bind Ras. The fact that GαqQ209L completely blocked Ras activation of PI3K might be due to suppression of intrinsic PI3K activity combined with the additional effect of Gαq in blocking Ras from binding to PI3K.
Gαq binding to PI3K is probably not the only mechanism by which Gq-coupled receptors inhibit PI3K signalling. We showed that stimulation of the α1A adrenergic receptor in Rat-1 cells inhibits PDGF (platelet-derived growth factor)-induced binding of PI3K to the PDGF receptor [39]. This response was due to enhanced dephosphorylation of the PDGF receptor at Tyr751, which forms a docking site for PI3K, by SHP-2 (an SH2-domain-containing protein tyrosine phosphatase). Activation of M3 muscarinic receptors in 1321N1 astrocytoma cells similarly blocked insulin-induced IRS-1 tyrosine phosphorylation and recruitment of PI3K. However, this was apparently a consequence of increased serine phosphorylation of IRS-1, which uncouples it from insulin-induced tyrosine phosphorylation [40]. Finally, activation of Gαq in cardiac myocytes was reported to decrease PI3K signalling due to depletion of intracellular phosphatidylinositol 4,5-bisphosphate, which is a substrate for both PLCβ and PI3K [41]. One difference between the latter two mechanisms cited above and the one reported herein is their dependence on PLCβ activation.
Activation of Gq-coupled receptors can have complex effects on PI3K signalling in cell types that express multiple catalytic and regulatory subunits. Although activated Gαq inhibits p110α, some of the released βγ subunits can activate p110γ and p110β [26]. In addition, different p110/p85 complexes may be differentially affected by Gαq. Future studies that examine the specificity of interactions between Gαq and PI3K may lead to a better understanding of the regulation and cellular function of various PI3K enzyme complexes.
Acknowledgments
This work was supported by the Office of Research and Development, Medical Research Service, Department of Veterans Affairs, and NIH DK62722 (to R.Z.L.) and NIH GM53132 (to S.S.). We thank Y. Yan (Department of Neurobiology and Anatomy, University of Rochester, Rochester, NY, U.S.A.) for making the GST fusion proteins.
References
- 1.Vanhaesebroeck B., Leevers S. J., Ahmadi K., Timms J., Katso R., Driscoll P. C., Woscholski R., Parker P. J., Waterfield M. D. Synthesis and function of 3-phosphorylated inositol lipids. Annu. Rev. Biochem. 2001;70:535–602. doi: 10.1146/annurev.biochem.70.1.535. [DOI] [PubMed] [Google Scholar]
- 2.Oudit G. Y., Sun H., Kerfant B. G., Crackower M. A., Penninger J. M., Backx P. H. The role of phosphoinositide-3 kinase and PTEN in cardiovascular physiology and disease. J. Mol. Cell. Cardiol. 2004;37:449–471. doi: 10.1016/j.yjmcc.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 3.Suire S., Coadwell J., Ferguson G. J., Davidson K., Hawkins P., Stephens L. p84, a new Gbetagamma-activated regulatory subunit of the type IB phosphoinositide 3-kinase p110gamma. Curr. Biol. 2005;15:566–570. doi: 10.1016/j.cub.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 4.Kodaki T., Woscholski R., Hallberg B., Rodriguez-Viciana P., Downward J., Parker P. J. The activation of phosphatidylinositol 3-kinase by Ras. Curr. Biol. 1994;4:798–806. doi: 10.1016/s0960-9822(00)00177-9. [DOI] [PubMed] [Google Scholar]
- 5.Yu J., Zhang Y., McIlroy J., Rordorf-Nikolic T., Orr G. A., Backer J. M. Regulation of the p85/p110 phosphatidylinositol 3′-kinase: stabilization and inhibition of the p110alpha catalytic subunit by the p85 regulatory subunit. Mol. Cell. Biol. 1998;18:1379–1387. doi: 10.1128/mcb.18.3.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Backer J. M., Myers M. G., Jr, Shoelson S. E., Chin D. J., Sun X. J., Miralpeix M., Hu P., Margolis B., Skolnik E. Y., Schlessinger J., et al. Phosphatidylinositol 3′-kinase is activated by association with IRS-1 during insulin stimulation. EMBO J. 1992;11:3469–3479. doi: 10.1002/j.1460-2075.1992.tb05426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carpenter C. L., Auger K. R., Chanudhuri M., Yoakim M., Schaffhausen B., Shoelson S., Cantley L. C. Phosphoinositide 3-kinase is activated by phosphopeptides that bind to the SH2 domains of the 85-kDa subunit. J. Biol. Chem. 1993;268:9478–9483. [PubMed] [Google Scholar]
- 8.Sjolander A., Yamamoto K., Huber B. E., Lapetina E. G. Association of p21ras with phosphatidylinositol 3-kinase. Proc. Natl. Acad. Sci. U.S.A. 1991;88:7908–7912. doi: 10.1073/pnas.88.18.7908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodriguez-Viciana P., Warne P. H., Dhand R., Vanhaesebroeck B., Gout I., Fry M. J., Waterfield M. D., Downward J. Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature (London) 1994;370:527–532. doi: 10.1038/370527a0. [DOI] [PubMed] [Google Scholar]
- 10.Rodriguez-Viciana P., Warne P. H., Vanhaesebroeck B., Waterfield M. D., Downward J. Activation of phosphoinositide 3-kinase by interaction with Ras and by point mutation. EMBO J. 1996;15:2442–2451. [PMC free article] [PubMed] [Google Scholar]
- 11.Vanhaesebroeck B., Welham M. J., Kotani K., Stein R., Warne P. H., Zvelebil M. J., Higashi K., Volinia S., Downward J., Waterfield M. D. P110delta, a novel phosphoinositide 3-kinase in leukocytes. Proc. Natl. Acad. Sci. U.S.A. 1997;94:4330–4335. doi: 10.1073/pnas.94.9.4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walker E. H., Perisic O., Ried C., Stephens L., Williams R. L. Structural insights into phosphoinositide 3-kinase catalysis and signalling. Nature (London) 1999;402:313–320. doi: 10.1038/46319. [DOI] [PubMed] [Google Scholar]
- 13.Pacold M. E., Suire S., Perisic O., Lara-Gonzalez S., Davis C. T., Walker E. H., Hawkins P. T., Stephens L., Eccleston J. F., Williams R. L. Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase gamma. Cell (Cambridge, Mass.) 2000;103:931–943. doi: 10.1016/s0092-8674(00)00196-3. [DOI] [PubMed] [Google Scholar]
- 14.Jimenez C., Hernandez C., Pimentel B., Carrera A. C. The p85 regulatory subunit controls sequential activation of phosphoinositide 3-kinase by Tyr kinases and Ras. J. Biol. Chem. 2002;277:41556–41562. doi: 10.1074/jbc.M205893200. [DOI] [PubMed] [Google Scholar]
- 15.Chan T. O., Rodeck U., Chan A. M., Kimmelman A. C., Rittenhouse S. E., Panayotou G., Tsichlis P. N. Small GTPases and tyrosine kinases coregulate a molecular switch in the phosphoinositide 3-kinase regulatory subunit. Cancer Cell. 2002;1:181–191. doi: 10.1016/s1535-6108(02)00033-8. [DOI] [PubMed] [Google Scholar]
- 16.Stoyanov B., Volinia S., Hanck T., Rubio I., Loubtchenkov M., Malek D., Stoyanova S., Vanhaesebroeck B., Dhand R., Nurnberg B., et al. Cloning and characterization of a G protein-activated human phosphoinositide-3 kinase. Science. 1995;269:690–693. doi: 10.1126/science.7624799. [DOI] [PubMed] [Google Scholar]
- 17.Kurosu H., Maehama T., Okada T., Yamamoto T., Hoshino S., Fukui Y., Ui M., Hazeki O., Katada T. Heterodimeric phosphoinositide 3-kinase consisting of p85 and p110beta is synergistically activated by the betagamma subunits of G proteins and phosphotyrosyl peptide. J. Biol. Chem. 1997;272:24252–24256. doi: 10.1074/jbc.272.39.24252. [DOI] [PubMed] [Google Scholar]
- 18.Ballou L. M., Cross M. E., Huang S., McReynolds E. M., Zhang B. X., Lin R. Z. Differential regulation of the phosphatidylinositol 3-kinase/Akt and p70 S6 kinase pathways by the alpha(1A)-adrenergic receptor in rat-1 fibroblasts. J. Biol. Chem. 2000;275:4803–4809. doi: 10.1074/jbc.275.7.4803. [DOI] [PubMed] [Google Scholar]
- 19.Bommakanti R. K., Vinayak S., Simonds W. F. Dual regulation of Akt/protein kinase B by heterotrimeric G protein subunits. J. Biol. Chem. 2000;275:38870–38876. doi: 10.1074/jbc.M007403200. [DOI] [PubMed] [Google Scholar]
- 20.Liu F., Yang P., Baez M., Ni B. Neurotensin negatively modulates Akt activity in neurotensin receptor-1-transfected AV12 cells. J. Cell. Biochem. 2004;92:603–611. doi: 10.1002/jcb.20098. [DOI] [PubMed] [Google Scholar]
- 21.Ballou L. M., Lin H. Y., Fan G., Jiang Y. P., Lin R. Z. Activated G alpha q inhibits p110 alpha phosphatidylinositol 3-kinase and Akt. J. Biol. Chem. 2003;278:23472–23479. doi: 10.1074/jbc.M212232200. [DOI] [PubMed] [Google Scholar]
- 22.Fan G., Ballou L. M., Lin R. Z. Phospholipase C-independent activation of glycogen synthase kinase-3-beta and C-terminal Src kinase by Galpha(q) J. Biol. Chem. 2003;278:52432–52436. doi: 10.1074/jbc.M310982200. [DOI] [PubMed] [Google Scholar]
- 23.Kozasa T., Gilman A. G. Purification of recombinant G proteins from Sf9 cells by hexahistidine tagging of associated subunits. Characterization of alpha 12 and inhibition of adenylyl cyclase by alpha z. J. Biol. Chem. 1995;270:1734–1741. doi: 10.1074/jbc.270.4.1734. [DOI] [PubMed] [Google Scholar]
- 24.Chidiac P., Markin V. S., Ross E. M. Kinetic control of guanine nucleotide binding to soluble Galpha(q) Biochem. Pharmacol. 1999;58:39–48. doi: 10.1016/s0006-2952(99)00080-5. [DOI] [PubMed] [Google Scholar]
- 25.Runnels L. W., Scarlata S. F. Determination of the affinities between heterotrimeric G protein subunits and their phospholipase C-beta effectors. Biochemistry. 1999;38:1488–1496. doi: 10.1021/bi9821519. [DOI] [PubMed] [Google Scholar]
- 26.Kerchner K. R., Clay R. L., McCleery G., Watson N., McIntire W. E., Myung C. S., Garrison J. C. Differential sensitivity of phosphatidylinositol 3-kinase p110gamma to isoforms of G protein betagamma dimers. J. Biol. Chem. 2004;279:44554–44562. doi: 10.1074/jbc.M406071200. [DOI] [PubMed] [Google Scholar]
- 27.Maier U., Babich A., Nurnberg B. Roles of non-catalytic subunits in gbetagamma-induced activation of class I phosphoinositide 3-kinase isoforms beta and gamma. J. Biol. Chem. 1999;274:29311–29317. doi: 10.1074/jbc.274.41.29311. [DOI] [PubMed] [Google Scholar]
- 28.Imamura T., Ishibashi K., Dalle S., Ugi S., Olefsky J. M. Endothelin-1-induced GLUT4 translocation is mediated via Galpha(q/11) protein and phosphatidylinositol 3-kinase in 3T3-L1 adipocytes. J. Biol. Chem. 1999;274:33691–33695. doi: 10.1074/jbc.274.47.33691. [DOI] [PubMed] [Google Scholar]
- 29.Velloso L. A., Folli F., Sun X. J., White M. F., Saad M. J., Kahn C. R. Cross-talk between the insulin and angiotensin signaling systems. Proc. Natl. Acad. Sci. U.S.A. 1996;93:12490–12495. doi: 10.1073/pnas.93.22.12490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Folli F., Kahn C. R., Hansen H., Bouchie J. L., Feener E. P. Angiotensin II inhibits insulin signaling in aortic smooth muscle cells at multiple levels. A potential role for serine phosphorylation in insulin/angiotensin II crosstalk. J. Clin. Invest. 1997;100:2158–2169. doi: 10.1172/JCI119752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Batty I. H., Downes C. P. Thrombin receptors modulate insulin-stimulated phosphatidylinositol 3,4,5-trisphosphate accumulation in 1321N1 astrocytoma cells. Biochem. J. 1996;317:347–351. doi: 10.1042/bj3170347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hutson N. J., Brumley F. T., Assimacopoulos F. D., Harper S. C., Exton J. H. Studies on the alpha-adrenergic activation of hepatic glucose output. I. Studies on the alpha-adrenergic activation of phosphorylase and gluconeogenesis and inactivation of glycogen synthase in isolated rat liver parenchymal cells. J. Biol. Chem. 1976;251:5200–5208. [PubMed] [Google Scholar]
- 33.Thomas A. P., Martin-Requero A., Williamson J. R. Interactions between insulin and alpha 1-adrenergic agents in the regulation of glycogen metabolism in isolated hepatocytes. J. Biol. Chem. 1985;260:5963–5973. [PubMed] [Google Scholar]
- 34.Blair L. A., Marshall J. IGF-1 modulates N and L calcium channels in a PI 3-kinase-dependent manner. Neuron. 1997;19:421–429. doi: 10.1016/s0896-6273(00)80950-2. [DOI] [PubMed] [Google Scholar]
- 35.Shapiro M. S., Loose M. D., Hamilton S. E., Nathanson N. M., Gomeza J., Wess J., Hille B. Assignment of muscarinic receptor subtypes mediating G-protein modulation of Ca(2+) channels by using knockout mice. Proc. Natl. Acad. Sci. U.S.A. 1999;96:10899–10904. doi: 10.1073/pnas.96.19.10899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bannister R. A., Melliti K., Adams B. A. Reconstituted slow muscarinic inhibition of neuronal (Ca(v)1.2c) L-type Ca2+ channels. Biophys. J. 2002;83:3256–3267. doi: 10.1016/S0006-3495(02)75327-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fan G., Jiang Y. P., Lu Z., Martin D. W., Kelly D. J., Zuckerman J. M., Ballou L. M., Cohen I. S., Lin R. Z. A transgenic mouse model of heart failure using inducible Galpha(q) J. Biol. Chem. 2005;280:40337–40346. doi: 10.1074/jbc.M506810200. [DOI] [PubMed] [Google Scholar]
- 38.Lu Z., Jiang Y. P., Ballou L. M., Cohen I. S., Lin R. Z. Galpha(q) inhibits cardiac L-type Ca2+ channels through phosphatidylinositol 3-kinase. J. Biol. Chem. 2005;280:40347–40354. doi: 10.1074/jbc.M508441200. [DOI] [PubMed] [Google Scholar]
- 39.Lin H. Y., Ballou L. M., Lin R. Z. Stimulation of the alpha1A adrenergic receptor inhibits PDGF-induced PDGF beta receptor Tyr751 phosphorylation and PI 3-kinase activation. FEBS Lett. 2003;540:106–110. doi: 10.1016/s0014-5793(03)00233-3. [DOI] [PubMed] [Google Scholar]
- 40.Batty I. H., Fleming I. N., Downes C. P. Muscarinic-receptor-mediated inhibition of insulin-like growth factor-1 receptor-stimulated phosphoinositide 3-kinase signalling in 1321N1 astrocytoma cells. Biochem. J. 2004;379:641–651. doi: 10.1042/BJ20031700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Howes A. L., Arthur J. F., Zhang T., Miyamoto S., Adams J. W., Dorn I. G., Woodcock E. A., Brown J. H. Akt-mediated cardiomyocyte survival pathways are compromised by G alpha q-induced phosphoinositide 4,5-bisphosphate depletion. J. Biol. Chem. 2003;278:40343–40351. doi: 10.1074/jbc.M305964200. [DOI] [PubMed] [Google Scholar]