Abstract
The Arabidopsis-mei2-Like (AML) genes comprise a five-member gene family related to the mei2 gene, which is a master regulator of meiosis in Schizosaccharomyces pombe and encodes an RNA binding protein. We have analyzed the AML genes to assess their role in plant meiosis and development. All five AML genes were expressed in both vegetative and reproductive tissues. Analysis of AML1-AML5 expression at the cellular level indicated a closely similar expression pattern. In the inflorescence, expression was concentrated in the shoot apical meristem, young buds, and reproductive organ primordia. Within the reproductive organs, strong expression was observed in meiocytes and developing gametes. Functional analysis using RNA interference (RNAi) and combinations of insertion alleles revealed a role for the AML genes in meiosis, with RNAi lines and specific multiple mutant combinations displaying sterility and a range of defects in meiotic chromosome behavior. Defects in seedling growth were also observed at low penetrance. These results indicate that the AML genes play a role in meiosis as well as in vegetative growth and reveal conservation in the genetic mechanisms controlling meiosis in yeast and plants.
INTRODUCTION
The plant life cycle alternates between a diploid sporophyte and a haploid gametophyte. Meiosis in plants represents the transition from the sporophyte to the gametophyte generation. In higher plants, meiosis takes place in specialized cells, the sporocytes, which are formed in the anthers and ovules. The molecular analysis of events leading up to and including meiosis and spore formation in plants has advanced considerably in recent years, based largely upon studies in Arabidopsis thaliana and maize (Zea mays) (reviewed in Yang and Sundaresan, 2000; Bhatt et al., 2001). Many of the genes encoding basic structural components of the meiotic machinery that is common to all eukaryotes, such as that required for chromosome organization and segregation, show conservation with genes in yeast and other organisms. Others are unique to plants and do not have obvious counterparts in other species (Caryl et al., 2003).
The sporocyte develops from an archesporial cell either directly or after further division. The SPOROCYTELESS (SPL)/NOZZLE gene of Arabidopsis is required for specification of the sporocyte (Schiefthaler et al., 1999; Yang et al., 1999) and encodes a nuclear protein related to MADS box transcription factors. Within the anthers, SPL expression is concentrated in the sporogenous cells, which form sporocytes. SPL is positively regulated by the floral organ identity gene AGAMOUS, which specifies reproductive organs and encodes a MADS box family transcription factor (Ito et al., 2004). The evidence supports a model wherein sporocyte specification is part of a process of organogenesis in which specific cell types are formed within reproductive organs through a mechanism involving a hierarchy of transcriptional controls headed by AGAMOUS. Meiotic division of the sporocyte gives rise to four haploid spores. The mechanisms that control meiosis in flowering plants, in particular early events involving premeiotic DNA synthesis and entry into the meiotic program, are not well understood.
The switch from mitosis to meiosis in yeast (both Schizosaccharomyces pombe and Saccharomyces cerevisiae) has been well studied and involves a combination of nutritional and developmental controls (reviewed in Yamamoto, 1996; Honigberg and Purnapatre, 2003). In S. pombe, Mei2p is considered to be a master regulator of meiosis and is required for premeiotic DNA synthesis as well as entry into meiosis (Watanabe and Yamamoto, 1994). The mei2 gene encodes an RNA binding protein with three RNA recognition motifs (RRMs), of which the C-terminal RRM3 is critical for function (Watanabe et al., 1997). The control of meiosis by Mei2p is determined by its phosphorylation status (Watanabe et al., 1997). When haploid cells are placed in sporulation medium, they accumulate mei2 transcript, but Mei2p remains inactive due to phosphorylation at two sites, S438 and T527, by the pat1 kinase. After fusion between two haploid cells of opposite mating type, Mei2p accumulates in the unphosphorylated form that promotes premeiotic DNA synthesis as well as entry into meiosis I. At the start of meiosis 1, Mei2p is localized to the nucleus, where it is observed in the form of a dot. The nuclear localization of Mei2p as a dot is promoted by meiRNA, a small noncoding RNA molecule, by a mechanism wherein meiRNA is thought to entrap Mei2p within the nucleus (Yamashita et al., 1998; Sato et al., 2001). The dot corresponds to the location of the sme2 gene, which encodes meiRNA (Shimada et al., 2003). Mei2p also interacts with Mei2-interacting protein 1 (Mip1p), a protein with WD-40 and HEAT domains that are involved in protein–protein interactions (Shinozaki-Yabana et al., 2000). Mip1p is essential for spore viability and cell growth in addition to being required for sexual development and meiosis and is a founding member of the Raptor (for regulatory-associated protein of TOR) family of proteins that associates with the TOR kinase, a major regulator of translation and cell growth found in all eukaryotes (reviewed in Kim and Sabatini, 2004). TOR and Raptor are considered to act as part of a complex, TORC1, that regulates multiple aspects of cell growth and physiology in response to nutrient status. Raptor is considered to act as a scaffold for recruitment of substrates that are phosphorylated by TOR.
The presence of mei2-like genes in plants was first revealed by the identification and characterization of Arabidopsis-mei2-Like1 (AML1). AML1 was isolated in a screen for Arabidopsis cDNAs that could allow meiosis to proceed in a mam2 map3 mutant of S. pombe that is deficient in the mating receptors and hence in the receptor signaling that is required for initiation of meiosis (Hirayama et al., 1997). Although AML1 could rescue the mating receptor signaling requirement for meiosis, it could not complement a mei2 mutant. This suggests that the rescue of the meiotic defect by AML1 is not by replacement of endogenous mei2 function but by some other mechanism that perhaps interferes with negative control of mei2. AML1 was also shown to be expressed in a number of tissues: leaves, roots, flowers, and siliques. The TERMINAL EAR1 (TE1) gene of maize was the first plant mei2-like gene to be functionally characterized, and the analysis indicated that it acts to control phyllotaxy and leaf initiation in the meristem by negatively regulating the number and position of the sites of leaf initiation (Veit et al., 1998). A bioinformatic study has shown that mei2-like genes are widespread in the plant kingdom, where they constitute a diversified group comprising distinct clades, and are also found in alveolates (Anderson et al., 2004; Jeffares et al., 2004). In Arabidopsis, there are nine mei2-related genes distributed over three clades. The five genes, AML1-AML5, which are most similar to mei2, cluster together and show a broad expression pattern encompassing vegetative and reproductive tissues. The AML1 protein has recently been shown to bind At Raptor1B in a yeast two-hybrid assay, a finding that implicates the AML family of proteins in TOR-dependent signaling (Anderson and Hanson, 2005).
The only plant phenotype described for AML genes has been early flowering (Anderson and Hanson, 2005), and it is not known whether any of the plant mei2-related genes play a role in meiosis. TE1 is expressed in the shoot and root meristems and not in the meiocytes, and the te1 mutant phenotype does not include any meiotic defects (Veit et al., 1998; Jeffares et al., 2004). In this study, we have performed a functional analysis of AML1-AML5. We present evidence that the AML genes play a role in vegetative meristem activity. We also show that the AML genes are strongly expressed in meiocytes and play a role in meiosis. Our findings suggest conservation in the control of meiosis between S. pombe and plants.
RESULTS
The AML Genes Are Widely Expressed in Vegetative and Reproductive Tissues, with Strong Expression in Meristems and Meiocytes
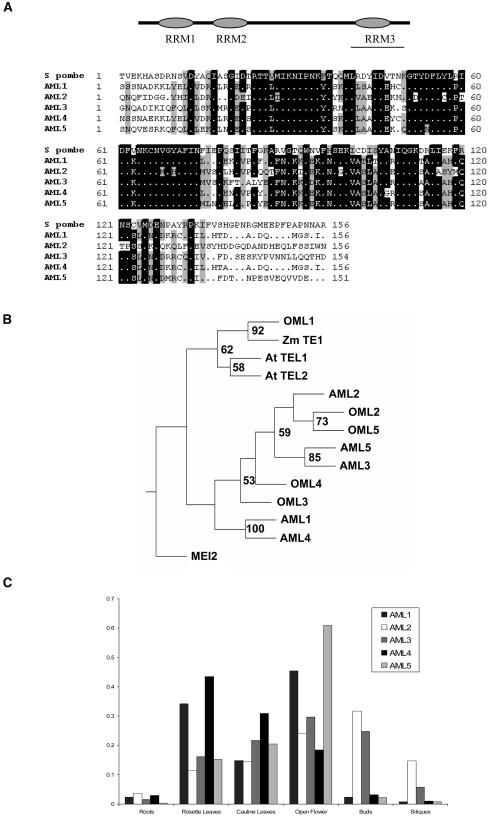
The predicted AML proteins show greatest similarity to Mei2p in the C-terminal portion within a region that encompasses RRM3 (Figure 1A). A phylogenetic analysis of the mei2-related proteins AML1-AML5, At TEL1, At TEL2, OML1-OML5, and Zm TE1 from Arabidopsis, rice (Oryza sativa), and maize along with Mei2p is depicted in Figure 1B. AML2, AML3, and AML5 are clustered in one clade that includes OML2 and OML5, whereas AML1 and AML4, which are closely related, cluster together with OML3 and OML4. At TEL1 and At TEL2 cluster with OML1 and Zm TE1 in a distinct clade. These relationships are in general agreement with those reported by Anderson et al. (2005). Semiquantitative RT-PCR analyses were performed to examine expression of the AML genes in different tissues. Expression of AML1-AML5 was detected in buds, open flowers, green siliques, leaves, and roots, indicating a broad expression pattern of these genes (Figure 1C). To compare these findings with other expression patterns, we queried the Genevestigator microarray expression database https://www.genevestigator.ethz.ch (Zimmermann et al., 2004).
Figure 1.
AML Genes Are Expressed in Vegetative and Reproductive Tissues in Arabidopsis.
(A) AML genes encode RNA binding proteins with three RRMs. Bar indicates the region of greatest similarity. ClustalX alignment of AML1-AML5 and S. pombe mei2 proteins for RRM3.
(B) Phylogenetic tree of mei2-related genes from Arabidosis, rice, maize, and S. pombe generated using Phylip. Bootstrap values >50 are indicated.
(C) RT-PCR analysis of relative expression of AML1-AML5 in different tissues normalized with GAPC.
The analysis indicated that all five AML genes are expressed in all organs examined, covering both vegetative and reproductive development. The normalized expression values for the different genes ranged from 823 ± 18 to 1976 ± 66 in adult leaves and from 1911 ± 82 to 2136 ± 162 in the inflorescence. This contrasts, for example, with AP1, whose expression is known to be confined to reproductive tissues (Gustafson-Brown et al., 1994) and for which the normalized expression value for the inflorescence was 20-fold higher than for adult leaves (1879 versus 98).
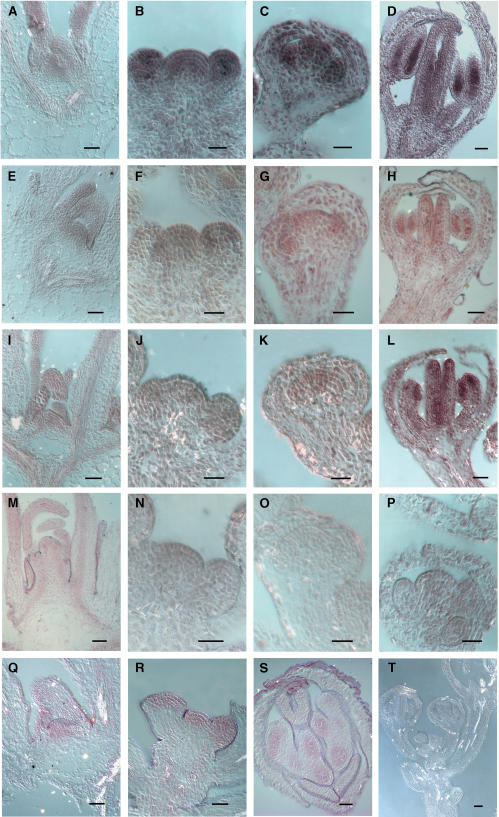
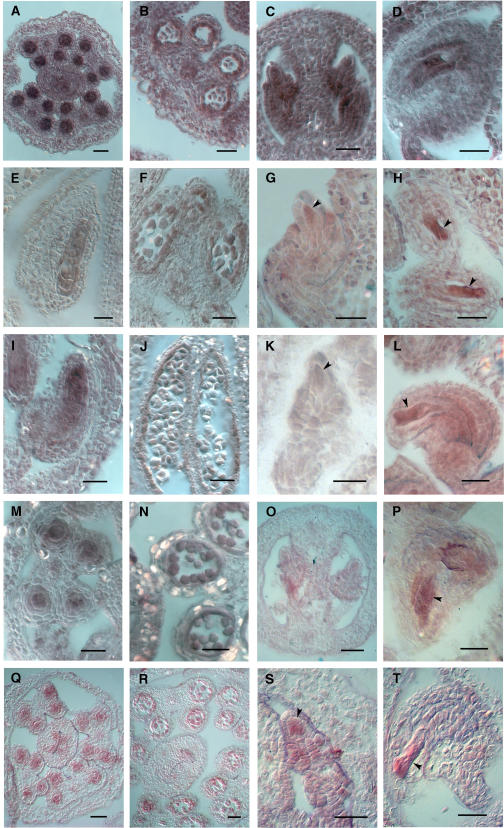
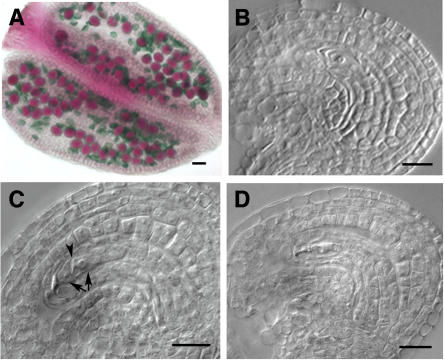
To obtain specific expression patterns, we examined expression of AML1-AML5 in aerial vegetative and reproductive tissues, using RNA in situ hybridization. The results indicated an overall expression pattern for all five genes that was very similar (Figures 2 and 3). In each case, a basal level of expression was present in most tissues. More concentrated expression was seen in the vegetative shoot apical meristem and the inflorescence meristem (Figure 2) as well as in the axillary buds (data not shown). For all the genes, a strong signal was present in the vegetative apical meristem covering all cell layers and in the emerging leaves (Figure 2A). All the AML genes except AML3 showed strong expression in the inflorescence meristem and floral buds (Figure 2B). In stage 6 flowers, expression was concentrated in the developing anther and pistil primordia and to a lesser extent in the sepals (Figure 2C). In flowers at stage 9, increased expression was observed for all AML genes in developing carpels along the walls of the placenta and ovule primordia (Figure 2D) and in male meiocytes, which showed a strong signal (Figure 3A). After meiosis, reduced expression was observed in developing tetrads (Figure 3B). Ovules showed expression in all cells, including the female meiocyte (Figure 3C), and later in the embryo sac, which showed strong expression (Figure 3D). Therefore, a detailed examination of the expression patterns for AML1-AML5 demonstrated that all genes are expressed at multiple stages of vegetative and reproductive development, including meiosis and gametogenesis, and overlap in their expression patterns. The strong meiotic expression was particularly striking and prompted us to look for a possible function during meiosis as is the case for the related gene mei2 of S. pombe.
Figure 2.
RNA in Situ Expression Analysis of AML1-AML5 in Apical Meristems and Reproductive Organs.
AML5 ([A] to [D]); AML1 ([E] to [H]); AML2 ([I] to [L]); AML3 ([M] to [P]); AML4 ([Q] to [S]); AML1 sense (T). Bars = 10 μm.
(A), (E), (I), (M), and (Q) Vegetative SAM.
(B), (F), (J), (N), and (R) Inflorescence SAM.
(C), (G), (K), and (O) Floral stage 5 and 6 buds showing increased signal in stamen and pistil primordia.
(D), (H), (L), and (S) Concentrated signal is detected in anther locules and inner margins of the pistil in stage 8 and 9 buds.
(P) Floral stage 7.
Figure 3.
RNA in Situ Expression Analysis of AML1-AML5 in Meiocytes and Gametophytes.
AML5 ([A] to [D]); AML1 ([E] to [H]); AML2 ([I] to [L]); AML3 ([M] to [P]); AML4 ([Q] to [T]). Bars = 25 μm.
(A), (E), (I), (M), and (Q) Transverse ([A], [M], and [Q]) and longitudinal ([E] and [I]) sections of anthers showing strong expression in pollen mother cells.
(B), (F), (J), (N), and (R) Expression in developing pollen.
(C), (G), (K), (O), and (S) Expression in young ovules, including the megaspore mother cell (arrowhead).
(D), (H), (L), (P), and (T) Mature ovules showing strong expression in the embryo sac (arrowhead).
RNA Interference with AML5 Causes Sterility and Defects in Gametogenesis
Given the close similarity in expression pattern between all five AML genes, the possibility of functional redundancy between members of the AML family appeared likely. This was in fact borne out in subsequent experiments (see below). We initially adopted an RNA interference (RNAi) approach to address the function of AML genes in meiosis. A 572-bp region of the AML5 cDNA (GenBank entry NM_179396; coordinates 2112 to 2683) encoding the C-terminal part of the protein was selected for RNAi experiments. This contains the region most conserved in all of the AML genes and includes RRM3. To estimate similarity and specificity, we searched the Arabidopsis genome using BLASTN for short nearly exact matches to the 572-bp region. Close similarity was detected only to each of the other four AML genes at E values between 1e−44 and 1e−11. The next level of similarity was at E = 0.034. Hence, the RNAi strategy was designed to specifically target the five AML genes.
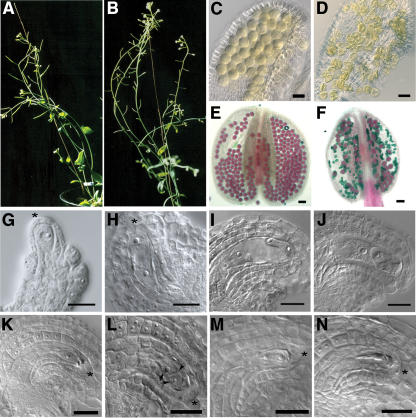
The 572-bp region was cloned in both sense and antisense orientations in the vector pKANNIBAL (Wesley et al., 2001). Lines carrying the AML5 RNAi construct expressed under control of the 35S promoter were generated and examined for phenotypes. The main phenotypes we observed in T1 plants were sterility coupled with increased branching, suggesting a loss of apical dominance (Figure 4). Out of 70 T1 plants, 11 displayed >90% sterility, whereas none of the control transformants (0/80) showed sterility. To investigate the basis of sterility, we examined embryo sac and pollen development in T1 and T2 plants from lines exhibiting strong sterility (>90%). In ovules, the phenotype ranged from arrest of embryo sac development at early stages to degeneration of the embryo sac. At low frequency (∼5% of ovules), we also observed two enlarged cells along with degenerating material from the nonfunctional megaspores (Figure 4L). The two cells could represent the products of an abnormal division of the functional megaspore, or they could possibly represent two surviving megaspores. Although the frequency of occurrence of such two-celled structures was low, they were observed in several independent RNAi plants as well as in antisense transformant plants (J. Kaur, unpublished data) and in plants carrying triple mutant combinations of aml alleles (described below). We did not observe such a phenotype in wild-type or control transformants. Defects in male gametophyte development were also observed. The pollen was shrunken and showed reduced viability as revealed by Alexander staining. In addition to defects in embryo sac and pollen development, we also observed defects in seedling development in several T2 and T3 generation RNAi lines (described below).
Figure 4.
AML5-RNAi Lines Display Sterility and Defective Gametogenesis.
(A) Wild-type fertile plant with elongated siliques.
(B) AML5-RNAi line showing strong sterility. The siliques have failed to elongate.
(C) and (D) Optical sections of cleared pollen from the wild type (C) and a strong AML5-RNAi line (D). Majority of the pollen in the RNAi anther are shriveled.
(E) and (F) Alexander staining of anthers showing viable pollen (purple) in the wild-type anthers (E) compared with a majority of nonviable pollen (green) in the AML5-RNAi line (F).
(G) to (N) Different stages of female gametophyte development in the wild type ([G] to [K]) and AML5-RNAi ([L] to [N]).
(G) Ovule stage 2-1, wherein the megaspore mother cell has differentiated.
(H) and (I) Two-nuclear ([H]; ovule stage 3-2) and four-nuclear ([I]; ovule stage 3-4) embryo sac.
(J) Ovule stage 3-6 showing a mature embryo sac.
(K) Stage 3-6 ovule showing an embryo sac arrested at the uninucleate stage.
(L) Two enlarged cells (arrows) separated by a cell wall (arrowhead) in place of a mature embryo sac.
(M) Arrested two-nuclear embryo sac.
(N) Degenerating embryo sac based on the presence of optically dense material.
Asterisks mark the apical epidermis below which the megaspore mother cell differentiates. Bars = 25 μm.
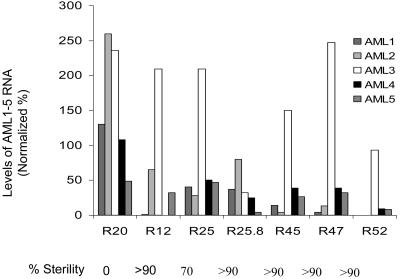
To test if the defects in gametogenesis correlated with a reduction in the mRNA levels of the AML genes, we quantitated RNA levels for each of the five AML genes in the inflorescence using semiquantitative RT-PCR (Figure 5). Lines showing strong sterility, such as R12, R45, and R47, exhibited a twofold or greater reduction in level for at least three of the AML genes, whereas line R25, which showed moderate sterility, exhibited less reduction in RNA levels. RNAi line R20 that did not show sterility failed to show such a reduction. Therefore, the phenotype correlates with reduction in RNA levels. The analysis suggested that AML1 and AML4 are more important, as their expression was substantially reduced in all the strong RNAi lines. A subset of RNAi lines exhibiting sterility was also crossed to the wild type, and in each case, the phenotype was found to be heritable and dominant in F1 (data not shown), consistent with it being due to RNAi.
Figure 5.
AML1-AML5 Transcript Levels Are Reduced in AML5-RNAi Lines Showing Strong Sterility.
Quantitation of AML1-AML5 transcript levels in five independent T1 lines and one T2 line (R25.8) showing strong sterility. The levels of each transcript are represented as percentages normalized against the levels of AML1-AML5 gene expression observed in control plants. In each line showing strong sterility, at least three members of the AML family show a more than twofold reduction in the transcript level. The control values represent the mean for two plants: C3 and C21, representing two control vector transformants. R20 is an RNAi line that did not show any sterility.
Combinations of aml Insertion Alleles Cause Defects in Gametogenesis and Vegetative Growth
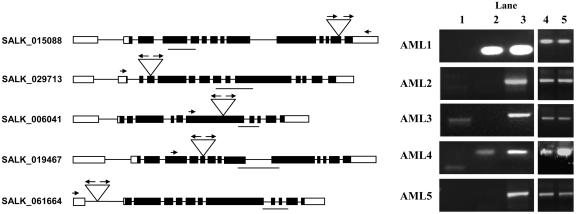
We obtained one insertion line for each of the five AML genes from the SALK T-DNA collection (Alonso et al., 2003). The presence of the T-DNA insertion in each of the lines was confirmed by PCR using T-DNA border primers in combination with gene-specific primers on both sides of the insertion for each of the genes so as to also establish the presence and orientation of T-DNA border sequences at both ends of the insertion (Figure 6; data not shown). The aml1, aml2, and aml4 insertions are in exons, whereas the aml3 and aml5 insertions are in introns. We used gene-specific primers within the coding region and downstream from the point of insertion to examine transcript levels by RT-PCR. For aml2, aml3, and aml5, no product was detected, indicating that they are probably null alleles. For aml1, we failed to detect a transcript using a T-DNA primer and a gene-specific primer downstream of the insertion, although we could detect a transcript using primers that were upstream of the insertion. These results suggest that the transcript is truncated. The aml1 insertion site is 12 bases after the predicted end of RRM3 and, hence, in a region that is likely to be important for function. In the case of aml4, which carries an insertion in exon 6, we detected the presence of transcripts downstream of the insertion site. However, we could also detect the presence in transcripts of T-DNA, which interrupted the coding region. Our analysis therefore indicated that for each of the insertion lines, the insertion is likely to cause a severe reduction in function of the respective AML gene.
Figure 6.
Characterization of AML Expression in Lines Carrying T-DNA Insertions in the AML1-AML5 Genes.
Line diagram showing the site of T-DNA insertion in each of the AML genes together with RT-PCR analysis of AML1-AML5 expression in the insertion mutants. Exons are indicated as black boxes and the untranslated regions as white boxes. Inverted triangles represent the site of T-DNA insertion in each line. The arrows represent the LB1 primers, and the short arrows represent the gene-specific primers used. The line below the gene represents the cDNA sequence amplified using gene-specific primers. Lane 1: PCR done using left border primer (LB1) in combination with a gene-specific primer. For AML1, the gene-specific primer is downstream of the site of insertion, whereas for all others, the gene-specific primer is upstream of the site of insertion. For AML1, AML2, and AML5, no amplification is seen. For AML3 and AML4, the gene and T-DNA left border junction can be identified, indicating the presence of T-DNA in the processed transcript. Lane 2: A pair of gene-specific primers was used to detect the presence of the transcripts in the insertion mutants. In aml2, aml3, and aml5 insertion mutants, the transcripts downstream of the site of insertion could not be detected. Lane 3: The primer pairs used in lane 2 were used to amplify corresponding transcripts from wild-type plants. Lanes 4 and 5: GAPC controls for mutant and wild type, respectively.
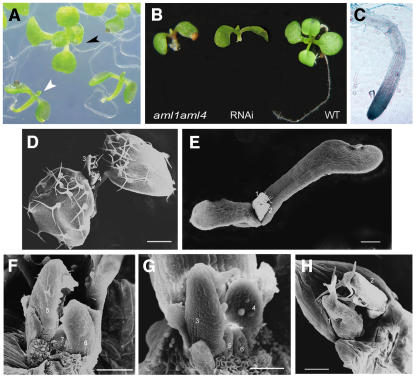
We identified plants homozygous for the insertion in each case. None of the single mutant lines displayed a visible phenotype; therefore, double mutants were generated in all possible combinations between aml1, aml2, aml3, aml4, and aml5. In the aml1 aml4 double mutant, we observed a partially penetrant phenotype at the seedling stage, wherein approximately one-quarter (20/86) of the seedlings were arrested at cotyledon expansion and failed to grow new leaves (Figure 7). A similar phenotype was observed in the aml1 aml2 aml4 triple mutant (obtained by crossing aml1 aml2 and aml1 aml4) as well as in T2 and T3 generation RNAi lines, and some of these seedlings also showed defects in root development (Figure 7B). Scanning electron microscopy of arrested seedlings (Figures 7D to 7H) showed that the shoot apical meristem did initiate leaf primordia; however, these were much slower in growth and expansion when compared with the wild type, suggesting a defect in meristem activity. The AML1 and AML4 genes show the highest level of sequence similarity (75% amino acid sequence identity) among the five AML genes and are hence likely to share a greater degree of functional redundancy. No phenotype was observed for any of the other double mutant combinations.
Figure 7.
Seedling Arrest in aml Mutant Combinations and RNAi Lines.
(A) aml1 aml4 double mutant showing seedling arrest: wild-type seedling (black arrowhead) and arrested seedling (white arrowhead).
(B) Defective root growth.
(C) ProAML2:β-glucuronidase expression in root tips.
(D) to (H) Scanning electron microscopy of 10-d-old seedlings: the wild type ([D] and [F]), aml1 aml2 aml4 ([E] and [G]), and AML5 RNAi (H).
(D) Low magnification; cotyledons have been removed. Leaves 1 and 2 have expanded. Primordia of leaves 3 and 4 are visible.
(E) Whole seedling. Leaves fail to expand.
(F) High magnification; cotyledons and leaves 1, 2, and 4 have been removed. Primordia of leaves 5 and 6 are visible.
(G) Primordia of leaves 3 and 4 are indicated.
(H) Cotyledons have been removed. Primordia of leaves 1 and 2 are indicated.
Bars = 100 μm in (D) and (E) and 30 μm in (F) to (H).
Triple mutant combinations were generated by crossing together homozygous double mutants. Sterility and defects in male and female gametogenesis were observed in 8/8 of the heterozygous aml1 aml2/+ aml4/+ F1 plants. Homozygous triple mutant plants were identified and analyzed in the F2 generation. The 5/5 aml1 aml2 aml4 triple mutant plants that were identified showed sterility and gametogenesis defects (Figure 8) but not aml1 aml2 aml3 and aml2 aml3 aml4 combinations. The sterility was ∼30 to 60% and correlated with defects in female gametogenesis (Tables 1 and 2). Male gametogenesis was defective as indicated by the presence of ∼30 to 40% shrunken pollen. The female phenotypes representing arrest at early stages or degenerating embryo sacs were similar to those observed for the RNAi lines.
Figure 8.
Defective Gametogenesis in aml1 aml2 aml4 Triple Mutants.
(A) Alexander staining of anther showing viable (purple) and nonviable (green) pollen.
(B) Stage 3-6 ovule with embryo sac arrested at the functional megaspore stage.
(C) Two cells (arrows) present along with degenerating megaspores in place of a mature embryo sac. Arrowhead marks the cell wall.
(D) Degenerating embryo sac.
Bars = 20 μm.
Table 1.
Reduced Fertility in aml1 aml2 aml4 Mutant Combinations
| Genotype | Sterility |
|---|---|
| aml1 aml2/+ aml4/+ | 28 ± 10 |
| aml1 aml2 aml4/+ | 34 ± 10 |
| aml1 aml2/+ aml4 | 47 ± 10 |
| aml1 aml2 aml4 | 61 ± 6 |
Table 2.
Defective Female Gametogenesis
| Ovule Stages
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 3-4
|
3-6
|
|||||||||
| Genotype | 1n | 2n | 4n | mesa | degb | 1n | 2n | 4n | mes | deg |
| Wild type | 1 | 5 | 46 | 0 | 0 | 0 | 2 | 3 | 48 | 0 |
| RNAi | 40 | 6 | 1 | 0 | 12 | 27 | 5 | 1 | 36 | 123 |
| aml1 aml2 aml4 | 23 | 4 | 28 | 0 | 10 | 209 | 9 | 14 | 572 | 190 |
Mature embryo sac.
Degenerating embryo sac.
AML Genes Play a Role in Meiosis
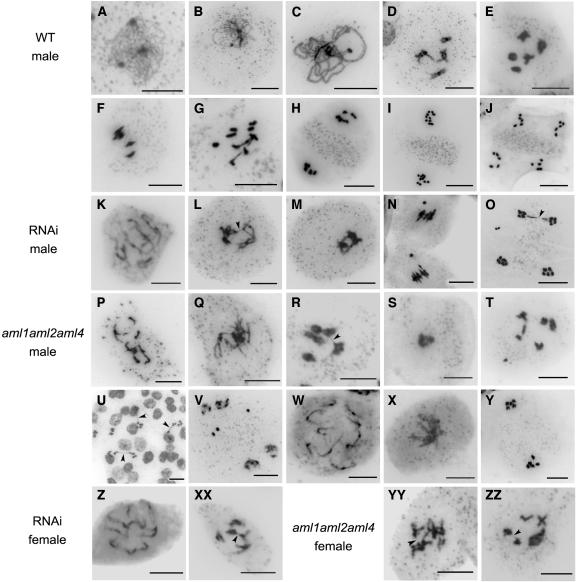
The defects in male and female gametophyte development described above could arise from a defective meiosis. We therefore examined chromosome spreads from various stages of meiosis in meioctyes from T1 and T2 generation RNAi plants and the aml1 aml2 aml4 triple mutant combination. In observations on several T1 and T2 RNAi lines, an overall 115/457 (25%) of the male meioses showed abnormalities (Table 3), whereas for the wild type, 0/500 meioses were abnormal. The abnormalities included pairing defects ranging from partial desynapsis to the formation of univalents, fragmentation and appearance of acentric pieces, and clumping of chromosomes (Figure 9). Chromosome bridges were observed resulting from exchange between what appeared to be nonhomologous chromosomes (Figure 9L). Similar defects were also observed in female meiosis (4/11 at diplotene) (Figure 9XX). We observed multiple instances of the appearance of an acentric fragment prior to anaphase I, in what was an otherwise normal-looking meiocyte (Figure 9N). At prometaphase I to metaphase I, there was clumping of chromosomes (Figure 9M). Defects were also observed during meiosis in the triple mutant combination of aml1 aml2 aml4 and aml1 aml2/+ aml4/+ plants. The 181/738 meioses covering diplotene to tetrad stages in aml1 aml2 aml4 plants were abnormal. Abnormalities included desynapsis and the formation of univalents (Figures 9P and 9W), chromosome bridges (Figures 9R and 9U), presence of an acentric fragment (Figure 9T), and clumping (Figures 9S and 9X). In addition, 5/25 prometaphase II meiocytes lacked an organelle band at the center (Figure 9Y), whereas all (26/26) wild-type prometaphase II stages examined showed an organelle band. The range of meiotic abnormalities is therefore similar between the RNAi lines and the triple mutant allelic combinations examined here. Together, these data provide evidence for a role for the AML genes in meiosis in plants.
Table 3.
Meiotic Chromosome Organization Defects
| RNAi
|
aml1 aml2 aml4
|
Wild Type
|
||||
|---|---|---|---|---|---|---|
| Meiosis Stage | Normal Abnormal | Normal Abnormal | Normal Abnormal | |||
| Diplotene | 133 | 52 (28) | 207 | 77 (27) | 114 | 0 |
| Diakinesis–metaphase I | 32 | 12 (24) | 116 | 19 (14) | 42 | 0 |
| Anaphase I | 16 | 12 (43) | 24 | 51 (68) | 23 | 0 |
| Telophase I–II | 40 | 7 (15) | 92 | 12 (11) | 219 | 0 |
| Tetrad | 121 | 32 (21) | 118 | 22 (16) | 102 | 0 |
| Total | 342 | 115 (25) | 557 | 181 (25) | 500 | 0 |
Figure 9.
Defective Meiotic Chromosome Organization in AML5-RNAi and aml1 aml2 aml4 Plants.
(A) Leptotene
(B) Zygotene.
(C) Pachytene.
(D) Late diplotene.
(E) Diakinesis.
(F) Metaphase I.
(G) Anaphase I.
(H) Metaphase II showing organelle band.
(I) Anaphase II.
(J) Telophase II.
(K) Partial desynapsis resulting in a mix of univalents and bivalents at diplotene.
(L) Chromosome bridge (arrowhead) between nonhomologous chromosomes at diakinesis.
(M) Clumping of chromosomes.
(N) Acentric chromosome in addition to five condensed bivalents at metaphase I.
(O) Chromosome bridge (arrowhead) and laggard chromosome at telophase II.
(P) Univalents and chromosome fragments corresponding to pachytene stage.
(Q) Disorganized diplotene stage.
(R) Chromosome bridge (arrowhead) at diakinesis.
(S) Clumped chromosomes at metaphase I.
(T) Two univalents and acentric chromosome at diakinesis.
(U) Anaphase I bridges (arrowheads).
(V) Laggard chromosomes at telophase II.
(W) Ten univalents.
(X) Clumped chromosomes at prometaphase I.
(Y) Telophase I showing absence of organelle band.
(Z) Ten univalents.
(XX) Chromosome bridge (arrowhead).
(YY) and (ZZ) Chromosome bridges (arrowheads) at diakinesis.
Bars = 10 μm.
Evidence for a Gametophytic Function for AML Genes
The postmeiotic defects observed during gametophyte development could arise from a defective meiosis or additionally from a gametophytic requirement for AML function after meiosis. Evidence in support of the latter came from the finding that transmission of the aml1 aml4 double mutant haplotype is defective in a parental background of aml1 aml4/+, where there was no evidence for sterility. Genotyping of all seeds (51 total) from a single silique (100% seed set and germination efficiency) obtained from an aml1 aml4/+ plant indicated recovery of the aml4, aml4/+, and +/+ genotypes at frequencies (5/51, 17/51, and 29/51, respectively) that were significantly different from those predicted by randomness [χ2 = 28.3; P(χ2 > 13.8) = 0.001 for 2 df]. A likely explanation is that the aml1 aml4 pollen haplotype shows reduced fitness and is outcompeted by the aml1 AML4 haplotype during pollination.
DISCUSSION
The control of meiosis is a key step in the transition from the sporophytic to the gametophytic phase of the plant life cycle. The mei2 gene of S. pombe is a positive regulator of meiosis and encodes an RNA binding protein required for premeiotic DNA synthesis and entry into meiosis I. Here, we have addressed the issue of whether the AML genes AML1-AML5 also play a role in plant meiosis. Examination of the expression pattern for AML1-AML5 indicated strong expression in meiocytes. Expression was also observed in the shoot apical meristem during vegetative and reproductive development and in the developing reproductive organs. These observations suggested that the members of this gene family may be required at multiple stages of plant development. The expression patterns for the five genes were closely related and showed substantial overlap, suggesting likely functional redundancy within the gene family. Expression of the AML genes in meristems has also been reported by Anderson et al. (2004). To determine the function of the AML genes, we first employed an RNAi-based approach using a conserved region that could specifically target all members of the AML gene family for inhibition of expression. In addition, we generated multiple mutant combinations of AML insertion alleles. The strong expression of AML1-AML5 observed in the meiocytes, as well as the associated meiotic defects (both in the male and female) observed in RNAi lines and in the aml1 aml2 aml4 triple mutant combination, provided evidence that the AML genes play an important role in meiosis. The absence of a phenotype in single mutants indicated redundancy of function within the AML gene family.
AML1 and AML4, which are most similar to each other and to mei2, appear to contribute to the role in meiosis. The phenotypes observed were due to a range of abnormalities in chromosome organization during meiotic prophase and later stages. These included desynapsis, formation of interchromosomal bridges, chromosome fragmentation, defects in chromosome segregation, and cytoplasmic defects as revealed by the absence of an organelle band at the end of meiosis I. The chromosomal phenotypes first become apparent at postpachytene stages. Other Arabidopsis genes and mutants affecting meiosis for which defects are mainly seen after pachytene are dsy1 (Ross et al., 1997), dsy10 (Cai and Makaroff, 2001), mei1 (He et al., 1996; Grelon et al., 2003), mmd1/duet (Reddy et al., 2003; Yang et al., 2003), At CAPE1, At CAPE2 (Siddiqui et al., 2003), and At CDC45 (Stevens et al., 2004). Several of these encode proteins that are directly implicated in chromatin organization and include proteins that are considered to act in mitosis as well as meiosis. At CDC45 is likely to play a role in the initiation of DNA replication in both mitosis and meiosis, and it has been suggested that the meiotic phenotype of At CDC45 RNAi lines is due to requirement of a higher level of At CDC45 for meiosis than for mitosis. This possibility is also supported in the case of AML genes by the strong expression observed in meiocytes. Even though the AML mutant phenotypes do not include a block in premeiotic DNA synthesis as is the case for mei2, they may still play a role in premeiotic DNA synthesis. At CDC45, based upon information on CDC45 in yeast and mammalian system, is thought to play a role in DNA replication, and the At CDC45 RNAi meiotic phenotypes are primarily postpachytene rather than earlier and likely arise from structural defects during replication that manifest later in meiosis. It is possible that AML proteins could directly affect chromatin organization as part of a chromatin remodeling complex. There is accumulating evidence for the direct involvement of RNA binding proteins in chromatin organization and remodeling: for example, through coordination of pre-mRNA splicing with chromatin remodeling (Dellaire et al., 2002), the finding that components of the DNA methylation system are also RNA binding proteins (Jeffery and Nakielny, 2004), and the presence of components of the RNAi machinery in complexes that play a role in assembly of heterochromatin (Verdel et al., 2004). Examples from plant systems include the FCA gene of Arabidopsis, which is a regulator of flowering and encodes an RNA binding protein (Macknight et al., 1997) that interacts with At SWI3B, a homolog of the yeast Swi/Snf complex (Sarnowski et al., 2002), and RS2-Interacting KH protein, which is part of a complex that is implicated in epigenetic silencing of KNOX genes (Phelps-Durr et al., 2005). Alternatively, AML proteins could act indirectly through posttranscriptional control of factors that play a role in meiosis and chromatin organization. In metazoans, RNA binding proteins play a central role in germline development and the control of meiosis and act at the level of translational regulation (reviewed in Haag, 2001; Crittenden et al., 2003).
There are two ways in which Mei2p has been shown to be involved in chromatin organization. The first is directly, through the formation of a nuclear dot in association with the sme2 locus, which encodes meiRNA (Shimada et al., 2003). The second, which may be indirect, is reflected in the requirement of mei2-dependent signaling for chromatin remodeling at the ade2-M26 recombination hotspot (Mizuno et al., 2001). M26 chromatin remodeling is considered to be mediated by binding of the ATF/CREB family transcription factor Atf1-Pcr1 to the CRE element–related sequence created by the M26 mutation (Wahls and Smith, 1994). The function of Atf1-Pcr1 is regulated through phosphorylation by Spc1/Sty1 in response to nitrogen starvation (Shiozaki and Russell, 1996; Wilkinson et al., 1996; Kon et al., 1998). Binding of transcription factors is thought to increase accessibility of the DNA to the recombination machinery (Nicolas, 1998). The defects in meiotic chromosome organization in aml mutants and RNAi lines may similarly reflect a requirement for AML-dependent signaling in chromatin organization during meiosis possibly by controlling the activity of factors that bind DNA and control access to chromatin.
The finding that the AML genes play a role in meiosis in Arabidopsis suggests a level of conservation in the control of meiosis between plants and S. pombe. Apart from sterility, we also observed vegetative phenotypes both in RNAi lines and in aml1 aml4 double mutants as well as in aml1 aml2 aml4 triple mutants at low penetrance. The vegetative phenotypes were slow growth leading to seedling arrest and defects in root growth. It therefore appears that AML genes also play a role in vegetative development. The AML genes show strong expression throughout the meristem as well as in the emerging leaves, which would be consistent with a role in growth and cell division in the meristem and young organs. In the case of Mei2p, no function has been ascribed during vegetative development, and the small amount of Mei2p that is present during vegetative growth is considered to be in an inactive form (Watanabe et al., 1997). We therefore speculate that the ancestral function of the AML genes was in meiosis and that the vegetative function evolved subsequently.
Recent studies have provided information on the action of the TOR signaling pathway in Arabidopsis (Menand et al., 2002; Anderson et al., 2005; Deprost et al., 2005). The At TOR gene is essential for embryogenesis and endosperm development and is expressed in the embryo and meristematic regions (Menand et al., 2002). The At Raptor1/1B and At Raptor2/1A genes have been proposed to play a role in postembryonic growth (Anderson et al., 2005). The At Raptor1/1B At Raptor2/1A double mutant shows seedling arrest after germination due to defective meristem activity, and the At Raptor1/1B single mutant shows slow growth, delayed flowering, reduced apical dominance, and reduced fertility (Anderson et al., 2005). Some of these phenotypes, such as slow growth, seedling arrest, defective root growth, and reduced fertility, are similar to those found in aml1 aml4 double mutants, aml1 aml2 aml4 triple mutants, and RNAi lines and would be consistent with the AML and At Raptor genes acting in the same pathway. The At TOR gene, whose inactivation leads to embryo lethality, is likely to play a central role in the pathway. The finding that the AML genes play a role in meiosis implicates the involvement of the TOR signaling pathway in plant meiosis. Taken together, these studies provide evidence for the action of the TOR pathway during growth and organogenesis at multiple stages of plant development. The TOR pathway has been implicated in the control of meiosis and sexual development in both S. cerevisiae (Zheng and Schreiber, 1997) and S. pombe (Kawai et al., 2001; Weisman and Choder, 2001). TOR has also been shown to regulate nucleolar structure through association of the Rpd3 histone deacetylase with rDNA chromatin (Tsang et al., 2003). In Arabidopsis, the absence of a gametophytic effect of loss of TOR would suggest that TOR function in the gametophyte is not essential (Menand et al., 2002). On the other hand, we find that AML function does have a gametophytic component and is also required for growth of organ primordia in the vegetative meristem. There is substantial overlap in expression pattern between At TOR (Menand et al., 2002) and the AML genes (Anderson et al., 2005; this study), with strong expression being observed in proliferating tissues, such as the embryo and shoot apical meristems. Hence, the AML genes also appear to be active in tissues where At TOR expression is high. In S. pombe, the evidence suggests that Mei2p action is associated with conditions under which TOR activity is inhibited. Therefore, there may be differences in the mechanism of action between mei2 and AMLs. AML1 has been shown to bind At Raptor1B (Anderson and Hanson, 2005); however, whether there is a functional relationship between At TOR and AML remains to be determined.
We note that there are differences between our findings regarding the aml mutant phenotypes and those of Anderson and Hanson (2005), who did not observe such phenotypes. The only phenotype they observed for aml mutants was that they were early flowering. We have not examined flowering time in detail in the aml mutants, though we do find preliminary evidence for early flowering in aml multiple mutant lines (J. Sebastian, unpublished data). In addition, we find that aml RNAi lines and the aml1 aml2 aml4 triple mutant show sterility that can be traced back to defects in meiosis. The difference between our findings and those of Anderson and Hanson (2005) could be due to differences in the genetic background or strength of the aml alleles, particularly aml1, which appears to play an important role and for which the allele used in this study may be stronger than the one used by Anderson and Hanson (2005).
In conclusion, we have provided evidence in favor of the involvement of the AML genes in meiosis as well as in vegetative development. Our findings suggest that the control of meiosis in plants involves the integration of nutrition-dependent signaling pathways within a developmental context as has been shown to be the case for yeast.
METHODS
Plant Material and Growth Conditions
The Columbia ecotype of Arabidopsis thaliana was used for generating transgenic lines expressing the AML5 RNAi construct. Lines carrying a T-DNA insertion in each of the AML1-AML5 genes were obtained from the ABRC (SALK_015088, SALK_029713, SALK_006041, SALK_019467, and SALK_061664). Plants were grown at 21°C under a 16-h-light and 8-h-dark cycle as described previously (Siddiqi et al., 2000).
Phylogenetic Analysis
Sequences were aligned using ClustalW with default parameters (Higgins et al., 1994). The Phylip 3.6a3 package (Felsenstein, 1989; http://bioweb.pasteur.fr) was used for construction of the phylogenetic tree using 100 replicates with neighbor joining as the set criterion and Mei2 as the outgroup in the consensus tree.
Generation of AML5 RNAi Lines
A 572-bp fragment corresponding to the C-terminal region of AML5 was amplified using primer pair AML2F1 (5′-GACTCGAGTCTAGAGTCCGGGATTCTCGGACTACC-3′) and AML2R1 (5′-TAGAATTCGGATCCTGCATGCATCACTTAACTCTG-3′), which engineered XhoI-XbaI and EcoRI-BamHI enzyme sites at the 5′ and 3′ ends, respectively. The 572-bp fragment was cloned in the sense orientation as a XhoI-EcoRI fragment and in the antisense orientation as a BamHI-XbaI fragment in the pKANNIBAL vector (Wesley et al., 2001). This AML5 RNAi construct was further subcloned into the binary vector pART27 and transformed into Agrobacterium tumefaciens strain AGL1 for in planta transformations. The pKANNIBAL vector without the insert was used as a control. In planta transformation was performed as described by Bechtold and Pelletier (1998). The AML5-RNAi transformants were selected on Murashige and Skoog plates containing 2% sucrose and kanamycin at 50 μg/mL and transferred to soil. The sterility was scored as the mean percentage reduction of seeds in approximately five siliques, assuming a wild-type seed set of 50 per silique. Plants scored as >90% sterile gave less than three seeds per silique. Wild-type plants set 52 ± 6 seeds per silique.
Expression Analysis by Semiquantitative RT-PCR
Plant tissue was frozen in liquid nitrogen and stored at −80°C. Total RNA was isolated from the inflorescence using Trizol (Invitrogen) according to the manufacturer's protocol and treated with RNase-free DNase1 (RQ1; Promega) before proceeding for reverse transcription. Equal amounts of RNA (∼1 μg) from different samples was reverse transcribed using MMLV RT (Gibco) according to the manufacturer's instructions. cDNA product was diluted 10 to 50 times, and 2 μL of these dilutions was used to determine the linear range for AML1-AML5. GAPC expression was measured using GAPC1 and GAPC2 primers after determining the linear amplification range and used for normalization. PCR was performed after equalizing the reverse transcription products for each sample with respect to GAPC. Primers AML1up and AML1sac were used for detecting expression of AML1, 140jUp and 140jD for AML2, Chr4F and Chr4R for AML3, AML5F and AML5R2 for AML4, and 337c and 40BD for AML5. Sequence information for all the primers used in this study is given in Supplemental Table 1 online. RT-PCR products were separated on a 1% agarose gel, blotted onto Hybond N+, and hybridized with the respective α32P-labeled probes for AML1-AML5 and GAPC. Hybridization signals were detected using a Fuji FLA-3000 phosphor imager and quantified using Image Gauge software.
Microscopy
Developmental analysis of whole-mount anthers and ovules was done after fixing and clearing the inflorescence in methyl benzoate as described previously (Siddiqi et al., 2000). The slides were observed on a Zeiss Axioplan 2 imaging microscope under differential interference contrast optics using a ×40 oil immersion objective. Pollen viability was examined using the method of Alexander staining (Alexander, 1969). Images were captured on an Axioplan CCD camera using Axiovision (version 3.1) and processed using Adobe Photoshop 6.0.
Meiotic chromosome spreads of male meiocytes were prepared and analyzed according to the protocol of Ross et al. (1996), with minor modifications as described in Agashe et al. (2002). Chromosomes were stained with 4′,6-diamidino-2-phenylindole (1 μg/mL) and observed on a Zeiss Axioplan 2 imaging microscope using a 365-nm excitation and 420-nm long-pass emission filter and a ×100 oil objective. The images were captured on an Axioplan CCD camera using Axiovision (version 3.2) and processed using Adobe Photoshop 6.0.
For scanning electron microscopy analysis, the plant material was fixed in formaldehyde-acetic acid-alcohol containing 1% Triton X-100 at 4°C overnight and dehydrated through an acetone series followed by critical point drying in liquid CO2. The dried samples were mounted on stubs and sputter coated prior to examination using a Leica 240 scanning electron microscope.
Analysis of T-DNA Mutant Lines and Multiple Mutant Combinations
Lines carrying T-DNA insertions in AML1-AML5 were obtained from the ABRC. In each case, the presence of the T-DNA insert was confirmed using a left border outwardly directed primer, LB1, in combination with a gene-specific primer flanking the site of insertion. Both junction fragments were amplified for aml1 to aml5 using gene-specific primers on each side of the insertion in combination with a T-DNA left border primer. In the case of aml1, the two junction fragments were amplified using LB1-K22R and K22F-InvF (a left border inwardly directed primer), respectively. For aml2, the junction fragments were amplified using the gene-specific primers F7F1-LB1 and F7R1 in combination with internal left border primers Tail4 and Tail3 for the other end. This structure is consistent with a head-to-head insertion of the T-DNA. The aml3, aml4, and aml5 insertions were also head-to-head insertions, and both the junction fragments were identified using the same internal left border primers in combination with the gene-specific primers F15J5F1 and F15J5R1 for aml3, T28F1 and T28R1 for aml4, and F15D2F1 and F15D2R1 for aml5. For genotyping of plants, primers flanking the insertion were used to identify the wild-type alleles, and the mutant alleles were identified using LB1 in combination with K22R1, F7F1, F15J5F1, T28F1, and F15D2F1 for aml1 to aml5, respectively.
For generating double mutant combinations, homozygous insertion lines were crossed, and the F2 population was genotyped to identify plants carrying both the mutations in homozygous condition. For generating triple mutants, homozygous double mutants were used as parents. The two parental lines carried one common mutation in the homozygous condition. Homozygous triple mutants were identified and analyzed in the F2 generation.
In Situ Hybridizations
In situ hybridizations were performed as described previously (Siddiqi et al., 2000). Antisense RNAs specific to each of the AML genes were used as probes along with sense controls. The regions used for strand-specific cDNA probes were as follows: AML1 (NM_125589), 1583 to 2120; AML2 (NM_201945), 1080 to 1853; AML3 (NM_117922), 923 to 1761; AML4 (NM_120811), 1590 to 2623; AML5 (AY091327), 946 to 1612. Specificity of the probe was validated in cross-hybridization experiments using radioactively labeled probe under conditions of stringency that were the same as those used for in situ hybridizations. For each gene, the probe used showed >12-fold specificity over hybridization to the related genes. Floral stages are according to Smyth et al. (1990).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers X07180 (Mei2p), At5g61960 (AML1), At2g42890 (AML2), At4g18120 (AML3), At5g07290 (AML4), At1g29400 (AML5), At3g26120 (At TEL1), At1g67770 (At TEL2), BAB92568 (OML1), BAD12869 (OML2), AAW56930 (OML3), BAD46727 (OML4), BAD28947 (OML5), and AF047852 (Zm TE1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. ClustalW Multiple Sequence Alignment.
Supplemental Table 1. List of Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank Peter Waterhouse for providing pKANNIBAL and Ashok Kumar and R. Kumaresan for help with scanning electron microscopy analysis. We also thank Animesh Ray, Jyotsna Dhawan, and members of our laboratory for comments and suggestions as well as two anonymous reviewers for changes that improved the manuscript. In addition, we thank Mehar Sultana for synthesis of oligonucleotides. This work was supported by a grant from the Department of Biotechnology, Government of India, and by the Council for Scientific and Industrial Research. J.K. and J.S. were supported by fellowships from the Council for Scientific and Industrial Research. We also thank the ABRC for supply of DNA clones and seed material.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Imran Siddiqi (imran@ccmb.res.in).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.105.039156.
References
- Agashe, B., Prasad, C.K., and Siddiqi, I. (2002). Identification and analysis of DYAD: A gene required for meiotic chromosome organisation and female meiotic progression in Arabidopsis. Development 129 3935–3943. [DOI] [PubMed] [Google Scholar]
- Alexander, M.P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44 117–122. [DOI] [PubMed] [Google Scholar]
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. [DOI] [PubMed] [Google Scholar]
- Anderson, G.H., Alvarez, N.D., Gilman, C., Jeffares, D.C., Trainor, V.C., Hanson, M.R., and Veit, B. (2004). Diversification of genes encoding mei2-like RNA binding proteins in plants. Plant Mol. Biol. 54 653–670. [DOI] [PubMed] [Google Scholar]
- Anderson, G.H., and Hanson, M.R. (2005). The Arabidopsis Mei2 homologue AML1 binds AtRaptor1B, the plant homologue of a major regulator of eukaryotic cell growth. BMC Plant Biol. 5 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, G.H., Veit, B., and Hanson, M.R. (2005). The Arabidopsis AtRaptor genes are essential for post-embryonic plant growth. BMC Biol. 3 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold, N., and Pelletier, G. (1998). In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 82 259–266. [DOI] [PubMed] [Google Scholar]
- Bhatt, A.M., Canales, C., and Dickinson, H.G. (2001). Plant meiosis: The means to 1N. Trends Plant Sci. 6 114–121. [DOI] [PubMed] [Google Scholar]
- Cai, X., and Makaroff, C.A. (2001). The dsy10 mutation of Arabidopsis results in desynapsis and a general breakdown in meiosis. Sex. Plant Reprod. 14 63–67. [Google Scholar]
- Caryl, A.P., Jones, G.H., and Franklin, F.C. (2003). Dissecting plant meiosis using Arabidopsis thaliana mutants. J. Exp. Bot. 54 25–38. [DOI] [PubMed] [Google Scholar]
- Crittenden, S.L., Eckmann, C.R., Wang, L., Bernstein, D.S., Wickens, M., and Kimble, J. (2003). Regulation of the mitosis/meiosis decision in the Caenorhabditis elegans germline. Philos. Trans. R. Soc. Lond. B Biol. Sci. 358 1359–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaire, G., Makarov, E.M., Cowger, J.J., Longman, D., Sutherland, H.G., Luhrmann, R., Torchia, J., and Bickmore, W.A. (2002). Mammalian PRP4 kinase copurifies and interacts with components of both the U5 snRNP and N-CoR deacetylase complexes. Mol. Cell. Biol. 22 5141–5156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deprost, D., Truong, H.N., Robaglia, C., and Meyer, C. (2005). An Arabidopsis homolog of RAPTOR/KOG1 is essential for early embryo development. Biochem. Biophys. Res. Commun. 326 844–850. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J. (1989). PHYLIP–Phylogeny inference package (version 3.2). Cladistics 5 164–166. [Google Scholar]
- Grelon, M., Gendrot, G., Vezon, D., and Pelletier, G. (2003). The Arabidopsis MEI1 gene encodes a protein with five BRCT domains that is involved in meiosis-specific DNA repair events independent of SPO11-induced DSBs. Plant J. 35 465–475. [DOI] [PubMed] [Google Scholar]
- Gustafson-Brown, C., Savidge, B., and Yanofsky, M.F. (1994). Regulation of the Arabidopsis floral homeotic gene APETALA1. Cell 76 131–143. [DOI] [PubMed] [Google Scholar]
- Haag, E.S. (2001). Rolling back to BOULE. Proc. Natl. Acad. Sci. USA 98 6983–6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, C., Tirlapur, U., Cresti, M., Peja, M., Crone, D.E., Mascarenhas, J.P., and He, C.P. (1996). An Arabidopsis mutant showing aberrations in male meiosis. Sex. Plant Reprod. 9 54–57. [Google Scholar]
- Higgins, D., Thompson, J., Gibson, T., Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties, and weight matrix choice. Nucleic Acids Res. 22 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama, T., Ishida, C., Kuromori, T., Obata, S., Shimoda, C., Yamamoto, M., Shinozaki, K., and Ohto, C. (1997). Functional cloning of a cDNA encoding Mei2-like protein from Arabidopsis thaliana using a fission yeast pheromone receptor deficient mutant. FEBS Lett. 413 16–20. [DOI] [PubMed] [Google Scholar]
- Honigberg, S.M., and Purnapatre, K. (2003). Signal pathway integration in the switch from the mitotic cell cycle to meiosis in yeast. J. Cell Sci. 116 2137–2147. [DOI] [PubMed] [Google Scholar]
- Ito, T., Wellmer, F., Yu, H., Das, P., Ito, N., Alves-Ferreira, M., Riechmann, J.L., and Meyerowitz, E.M. (2004). The homeotic protein AGAMOUS controls microsporogenesis by regulation of SPOROCYTELESS. Nature 430 356–360. [DOI] [PubMed] [Google Scholar]
- Jeffares, D.C., Phillips, M.J., Moore, S., and Veit, B. (2004). A description of the Mei2-like protein family; structure, phylogenetic distribution and biological context. Dev. Genes Evol. 214 149–158. [DOI] [PubMed] [Google Scholar]
- Jeffery, L., and Nakielny, S. (2004). Components of the DNA methylation system of chromatin control are RNA-binding proteins. J. Biol. Chem. 279 49479–49487. [DOI] [PubMed] [Google Scholar]
- Kawai, M., Nakashima, A., Ueno, M., Ushimaru, T., Aiba, K., Doi, H., and Uritani, M. (2001). Fission yeast tor1 functions in response to various stresses including nitrogen starvation, high osmolarity, and high temperature. Curr. Genet. 39 166–174. [DOI] [PubMed] [Google Scholar]
- Kim, D.H., and Sabatini, D.M. (2004). Raptor and mTOR: Subunits of a nutrient-sensitive complex. Curr. Top. Microbiol. Immunol. 279 259–270. [DOI] [PubMed] [Google Scholar]
- Kon, N., Schroeder, S.C., Krawchuk, M.D., and Wahls, W.P. (1998). Regulation of the Mts1-Mts2-dependent ade6–M26 meiotic recombination hot spot and developmental decisions by the Spc1 mitogen-activated protein kinase of fission yeast. Mol. Cell. Biol. 18 7575–7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macknight, R., Bancroft, I., Page, T., Lister, C., Schmidt, R., Love, K., Westphal, L., Murphy, G., Sherson, S., Cobbett, C., and Dean, C. (1997). FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell 89 737–745. [DOI] [PubMed] [Google Scholar]
- Menand, B., Desnos, T., Nussaume, L., Berger, F., Bouchez, D., Meyer, C., and Robaglia, C. (2002). Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proc. Natl. Acad. Sci. USA 99 6422–6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno, K., et al. (2001). Counteracting regulation of chromatin remodeling at a fission yeast cAMP response element-related recombination hotspot by stress-activated protein kinase, cAMP-dependent kinase and meiosis regulators. Genetics 159 1467–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas, A. (1998). Relationship between transcription and initiation of meiotic recombination: Toward chromatin accessibility. Proc. Natl. Acad. Sci. USA 95 87–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelps-Durr, T.L., Thomas, J., Vahab, P., and Timmermans, M.C. (2005). Maize rough sheath2 and its Arabidopsis orthologue ASYMMETRIC LEAVES1 interact with HIRA, a predicted histone chaperone, to maintain knox gene silencing and determinacy during organogenesis. Plant Cell 17 2886–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, T.V., Kaur, J., Agashe, B., Sundaresan, V., and Siddiqi, I. (2003). The DUET gene is necessary for chromosome organization and progression during male meiosis in Arabidopsis and encodes a PHD finger protein. Development 130 5975–5987. [DOI] [PubMed] [Google Scholar]
- Ross, K.J., Fransz, P., Armstrong, S.J., Vizir, I., Mulligan, B., Franklin, F.C., and Jones, G.H. (1997). Cytological characterization of four meiotic mutants of Arabidopsis isolated from T-DNA-transformed lines. Chromosome Res. 5 551–559. [DOI] [PubMed] [Google Scholar]
- Ross, K.J., Fransz, P., and Jones, G.H. (1996). A light microscopic atlas of meiosis in Arabidopsis thaliana. Chromosome Res. 4 507–516. [DOI] [PubMed] [Google Scholar]
- Sarnowski, T.J., Swiezewski, S., Pawlikowska, K., Kaczanowski, S., and Jerzmanowski, A. (2002). AtSWI3B, an Arabidopsis homolog of SWI3, a core subunit of yeast Swi/Snf chromatin remodeling complex, interacts with FCA, a regulator of flowering time. Nucleic Acids Res. 30 3412–3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, M., Shinozaki-Yabana, S., Yamashita, A., Watanabe, Y., and Yamamoto, M. (2001). The fission yeast meiotic regulator Mei2p undergoes nucleocytoplasmic shuttling. FEBS Lett. 499 251–255. [DOI] [PubMed] [Google Scholar]
- Schiefthaler, U., Balasubramanian, S., Sieber, P., Chevalier, D., Wisman, E., and Schneitz, K. (1999). Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96 11664–11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada, T., Yamashita, A., and Yamamoto, M. (2003). The fission yeast meiotic regulator Mei2p forms a dot structure in the horse-tail nucleus in association with the sme2 locus on chromosome II. Mol. Biol. Cell 14 2461–2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki-Yabana, S., Watanabe, Y., and Yamamoto, M. (2000). Novel WD-repeat protein Mip1p facilitates function of the meiotic regulator Mei2p in fission yeast. Mol. Cell. Biol. 20 1234–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiozaki, K., and Russell, P. (1996). Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes Dev. 10 2276–2288. [DOI] [PubMed] [Google Scholar]
- Siddiqi, I., Ganesh, G., Grossniklaus, U., and Subbiah, V. (2000). The dyad gene is required for progression through female meiosis in Arabidopsis. Development 127 197–207. [DOI] [PubMed] [Google Scholar]
- Siddiqui, N.U., Stronghill, P.E., Dengler, R.E., Hasenkampf, C.A., and Riggs, C.D. (2003). Mutations in Arabidopsis condensin genes disrupt embryogenesis, meristem organization and segregation of homologous chromosomes during meiosis. Development 130 3283–3295. [DOI] [PubMed] [Google Scholar]
- Smyth, D.R., Bowman, J.L., and Meyerowitz, E.M. (1990). Early flower development in Arabidopsis. Plant Cell 2 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, R., Grelon, M., Vezon, D., Oh, J., Meyer, P., Perennes, C., Domenichini, S., and Bergounioux, C. (2004). A CDC45 homolog in Arabidopsis is essential for meiosis, as shown by RNA interference-induced gene silencing. Plant Cell 16 99–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang, C.K., Bertram, P.G., Ai, W., Drenan, R., and Zheng, X.F. (2003). Chromatin-mediated regulation of nucleolar structure and RNA Pol I localization by TOR. EMBO J. 22 6045–6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veit, B., Briggs, S.P., Schmidt, R.J., Yanofsky, M.F., and Hake, S. (1998). Regulation of leaf initiation by the terminal ear 1 gene of maize. Nature 393 166–168. [DOI] [PubMed] [Google Scholar]
- Verdel, A., Jia, S., Gerber, S., Sugiyama, T., Gygi, S., Grewal, S.I., and Moazed, D. (2004). RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303 672–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahls, W.P., and Smith, G.R. (1994). A heteromeric protein that binds to a meiotic homologous recombination hot spot: Correlation of binding and hot spot activity. Genes Dev. 8 1693–1702. [DOI] [PubMed] [Google Scholar]
- Watanabe, Y., Shinozaki-Yabana, S., Chikashige, Y., Hiraoka, Y., and Yamamoto, M. (1997). Phosphorylation of RNA-binding protein controls cell cycle switch from mitotic to meiotic in fission yeast. Nature 386 187–190. [DOI] [PubMed] [Google Scholar]
- Watanabe, Y., and Yamamoto, M. (1994). S. pombe mei2+ encodes an RNA-binding protein essential for premeiotic DNA synthesis and meiosis I, which cooperates with a novel RNA species meiRNA. Cell 78 487–498. [DOI] [PubMed] [Google Scholar]
- Weisman, R., and Choder, M. (2001). The fission yeast TOR homolog, tor1+, is required for the response to starvation and other stresses via a conserved serine. J. Biol. Chem. 276 7027–7032. [DOI] [PubMed] [Google Scholar]
- Wesley, S.V., et al. (2001). Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27 581–590. [DOI] [PubMed] [Google Scholar]
- Wilkinson, M.G., Samuels, M., Takeda, T., Toone, W.M., Shieh, J.C., Toda, T., Millar, J.B., and Jones, N. (1996). The Atf1 transcription factor is a target for the Sty1 stress-activated MAP kinase pathway in fission yeast. Genes Dev. 10 2289–2301. [DOI] [PubMed] [Google Scholar]
- Yamamoto, M. (1996). Regulation of meiosis in fission yeast. Cell Struct. Funct. 21 431–436. [DOI] [PubMed] [Google Scholar]
- Yamashita, A., Watanabe, Y., Nukina, N., and Yamamoto, M. (1998). RNA-assisted nuclear transport of the meiotic regulator Mei2p in fission yeast. Cell 95 115–123. [DOI] [PubMed] [Google Scholar]
- Yang, W.C., and Sundaresan, V. (2000). Genetics of gametophyte biogenesis in Arabidopsis. Curr. Opin. Plant Biol. 3 53–57. [DOI] [PubMed] [Google Scholar]
- Yang, W.C., Ye, D., Xu, J., and Sundaresan, V. (1999). The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev. 13 2108–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, X., Makaroff, C.A., and Ma, H. (2003). The Arabidopsis MALE MEIOCYTE DEATH1 gene encodes a PHD-finger protein that is required for male meiosis. Plant Cell 15 1281–1295. [PMC free article] [PubMed] [Google Scholar]
- Zheng, X.F., and Schreiber, S.L. (1997). Target of rapamycin proteins and their kinase activities are required for meiosis. Proc. Natl. Acad. Sci. USA 94 3070–3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, P., Hirsch-Hoffmann, M., Hennig, L., and Gruissem, W. (2004). GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 136 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.