Abstract
Adenosine deaminases that act on RNA (ADARs) catalyze the site-specific conversion of adenosine to inosine in primary mRNA transcripts, thereby affecting coding potential of mature mRNAs. Structural determinants that define the adenosine moieties for specific ADARs-mediated deaminations are currently unknown. We report the solution structure of the central region of the human R/G stem–loop pre-mRNA, a natural ADAR2 substrate encoding the subunit B of the glutamate receptor (R/G site). The structure reveals that the GCU(A/C)A pentaloop that is conserved in mammals and birds adopts a novel fold. The fold is stabilized by a complex interplay of hydrogen bonds and stacking interactions. We propose that this new pentaloop structure is an important determinant of the R/G site recognition by ADAR2.
Keywords: adenosine deaminase, NMR, RNA editing, RNA structure
INTRODUCTION
Adenosine deaminases that act on RNA (ADARs) convert adenosines to inosines embedded in double-stranded RNA (dsRNA; Polson et al. 1991; Bass et al. 1997; Emeson and Singh 2000; Gerber and Keller 2001; Bass 2002; Schaub and Keller 2002). The enzymatic activity requires the C-terminal adenosine deaminase domain to catalyze the hydrolytic deamination of adenosine, whereas RNA substrate recognition is mediated by the N-terminal double-stranded RNA-binding motifs (dsRBMs; Stefl et al. 2005). Depending on the RNA substrate structure, ADARs can deaminate as many as 50% of the adenosine residues within a perfect dsRNA (in a nonselective fashion) or as little as a single, specific adenosine moiety (Emeson and Singh 2000; Bass 2002). The majority of nonselective editing occurs in untranslated regions (UTRs) and introns where large regular duplexes are formed (Morse and Bass 1999; Rueter et al. 1999; Morse et al. 2002; Athanasiadis et al. 2004; Levanon et al. 2004). Such modifications can modulate gene silencing triggered by intramolecular structures in mRNA (Tonkin and Bass 2003), lead to the nuclear retention of RNA transcripts (Kumar and Carmichael 1997; Zhang and Carmichael 2001), or participate in an antiviral response (Wong et al. 1991) by extensive modification of viral RNAs. Conversely, selective editing has been shown to take place largely within codons, so that multiple RNA and protein isoforms can be created from a single genomic locus. For example, ADARs have been shown to produce functionally important isoforms for several proteins involved in synaptic neurotransmission, including ligand and voltage-gated ion channels and G-protein coupled receptors (Sommer et al. 1991; Egebjerg and Heinemann 1993; Kohler et al. 1993; Lomeli et al. 1994; Burns et al. 1997; Hoopengardner et al. 2003).
ADAR2 (a member of the mammalian ADAR family) specifically modifies the glutamate receptor subunit B (GluR-B) pre-mRNA at the R/G site (referred as R/G stem–loop), where the genomic AGA encoding an arginine is modified after editing into IGA encoding a glycine (the ribosome interprets I as G). This amino acid change affects the biophysical properties of the ion channel, the R/G edited protein recovering faster from desensitization (Lomeli et al. 1994). The R/G site is located to a 71-nt intramolecular stem–loop in its pre-mRNA consisting of a stem with a few mismatches and a pentaloop (Fig. 1A ▶). The secondary structure of the R/G stem–loop is strongly conserved in mammals and birds with the GCUMA pentaloop consensus (where M is A/C) (Aruscavage and Bass 2000). To date, there is still no structural insight into the process of A-to-I editing.
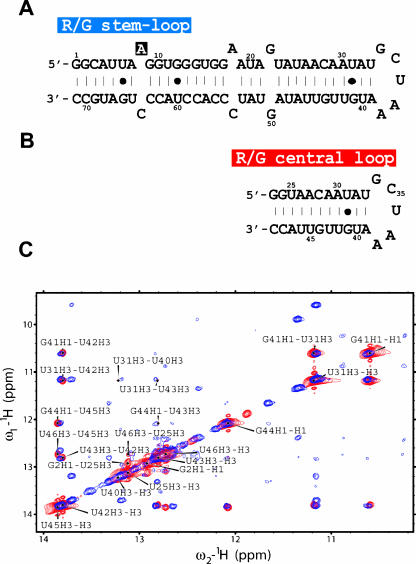
FIGURE 1.
Sequence and folding of the R/G stem–loop and central loop. (A) R/G stem–loop, a 71-nt intramolecular stem–loop that includes the 67 nt of the human GluR-B R/G pre-mRNA sequence and is closed by two G·C base pairs to improve yield of in vitro transcription. The R/G stem–loop pre-mRNA involves base pairing of intron and exon; the intron/exon junction occurs at the +2 position (G10) relative to the R/G editing site (A8). (B) R/G central loop, a 27-nt intramolecular stem–loop that includes the 23 nt of the central part of the human GluR-B R/G pre-mRNA closed by two G·C base pairs. (C) 2D [1H-1H] NOESY spectra (in 90% H2O/10% D2O) of a 71-nt R/G stem–loop RNA recorded at 900 MHz spectrometer (in blue) and 27-nt R/G central loop recorded at 600 MHz spectrometer (in red). The assignments are labeled by the one-letter code of nucleotides accompanied by a sequence number and a proton name.
RESULTS AND DISCUSSION
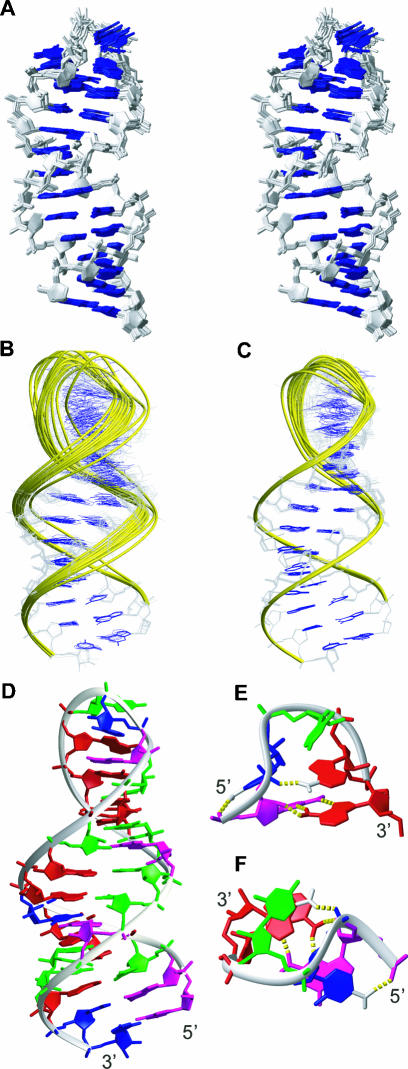
We report the solution structure of the central region of the human R/G stem–loop, embedding the pentaloop GCUAA (referred as R/G central loop; Fig. 1B ▶). Prior to structure determination, we assessed the folding of the R/G central loop and the full-length R/G stem–loop (Fig. 1 ▶) by comparing their [1H,1H]-TOCSY and [1H,1H]-NOESY spectra (Fig. 1C ▶). These data confirmed that the central fragment (nt 25–47; Fig. 1 ▶) maintains the same conformation adopted within the context of the full-length R/G stem–loop. The structure of the R/G central loop was determined using 602 conformationally restrictive NOE-derived distance restraints, 128 torsion angle restraints, and 85 restraints derived from residual dipolar couplings. The R/G central loop adopts a stable stem–loop structure with a well-defined stem, adopting a standard A-form double-helical architecture, and with a structured GCUAA pentaloop, stabilized by a complex interplay of hydrogen bonding and stacking interactions (Fig. 2 ▶). The resulting ensemble of structures is well defined, as demonstrated by the superimposition of the final 20 lowest energy conformers (Fig. 2A ▶). Inclusion of residual dipolar coupling restraints that contain long-range angular information improved the global precision of the resulting NMR ensemble (Fig. 2B,C ▶). A full summary of structural statistics is given in Table 1 ▶. The GCUAA pentaloop adopts a novel fold in which G34 and A38 form a sheared G·A mismatch, C35 is flipped out toward the major groove, and the central U36 stacks over the following A37 in the minor groove (Fig. 2E,F ▶). The flipped out C35 is stabilized by a hydrogen bond between an amino proton and a phosphate oxygen of the preceding residue G34. Similarly, A37 is stabilized by a hydrogen bond between an amino proton and a phosphate oxygen of C35. This latter hydrogen bond explains the A/C conservation at this position in mammals and birds, as the alternative C base (present in rat and mouse species) also has an amino group at a similar position that is likely to be hydrogen bonded to a phosphate oxygen of C35 as well (Fig. 3 ▶).
FIGURE 2.
Structure of the R/G central loop. (A) Stereo view of the NMR ensemble of the R/G central loop, the 13 final lowest energy structures are superimposed over all residues, sugar-phosphate backbones (in gray sticks), and RNA bases (in blue sticks) are shown. (B) NMR ensemble calculated without RDCs (13 lowest energy structures) superimposed over the three terminal base pairs. Overall RMSD is 0.84 ± 0.23 Å. (C) NMR ensemble calculated with RDCs (13 lowest energy structures) superimposed over the three terminal base pairs. Overall RMSD is 0.72 ± 0.20 Å. (D) The lowest energy structure of the R/G central loop represented as a ribbon-and-stick model. Nucleotides are colored red (A), green (U), magenta (G), and blue (C). Only heavy atoms are shown. (E,F) Close-up views of the GCUAA pentaloop from the minor groove and from the top, respectively. Hydrogen bonds are shown by yellow dashed sticks and the hydrogens involved are displayed.
TABLE 1.
NMR experimental restraints and structure statistics
| NMR restraintsa | ||
| Total restraints | 840 (31 per residue) | |
| NOEs | 602 | |
| Torsion anglesb | 128 | |
| H-bonds | 25 | |
| RDCs | 85 | |
| No. RDCs | RDCs | |
| Structure statisticsc | ||
| NOE violations | ||
| Number (>0.2 Å) | 2.61 ± 1.19 | 3.69 ± 1.32 |
| Maximum violations (Å) | 0.25 | 0.28 |
| Torsion violations | ||
| Number (>5°) | 0 | 1.0 ± 1.29 |
| Maximum violations (°) | 4.8 | 9.8 |
| RCDs violations | ||
| Number (>2 Hz) | 7.38 ± 4.13 | |
| Maximum violations (Hz) | 4.3 | |
| R.M.S.D. from the mean coordinates (Å)c | ||
| Heavy atoms | 0.84 ± 0.23 | 0.72 ± 0.20 |
| R.M.S.D. from ideal geometryc | ||
| Bonds lengths (Å) | 0.011 ± 0.0002 | 0.012 ± 0.0002 |
| Bonds angles (°) | 2.789 ± 0.0309 | 2.941 ± 0.0320 |
aFinal force constants used for square-well penalty functions of individual types of restraints were 32 kcal·mol−1·Å−2 for NOEs and Watson–Crick hydrogen bonds, 200 kcal·mol−1·rad−2 for torsion angles and 0.3 kcal·mol−1·Hz−2 for residual dipolar couplings (RDCs). The force constants were ramped up during the simulated annealing protocol as described (Padrta et al. 2002). Error bounds of ±2 Hz were used for all residual dipolar coupling restraints.
bThe A-form duplex backbone torsion angles derived from high-resolution crystal structures were used for the double-helical part as described in Materials and Methods: α (270°–330°), β (150°–210°), γ (30°–90°), δ (50°–110°), ɛ (180°–240°), and ζ (260°–320°).
cThe statistics (average ± standard deviations) calculated for the bundle of the 13 lowest energy conformers.
FIGURE 3.
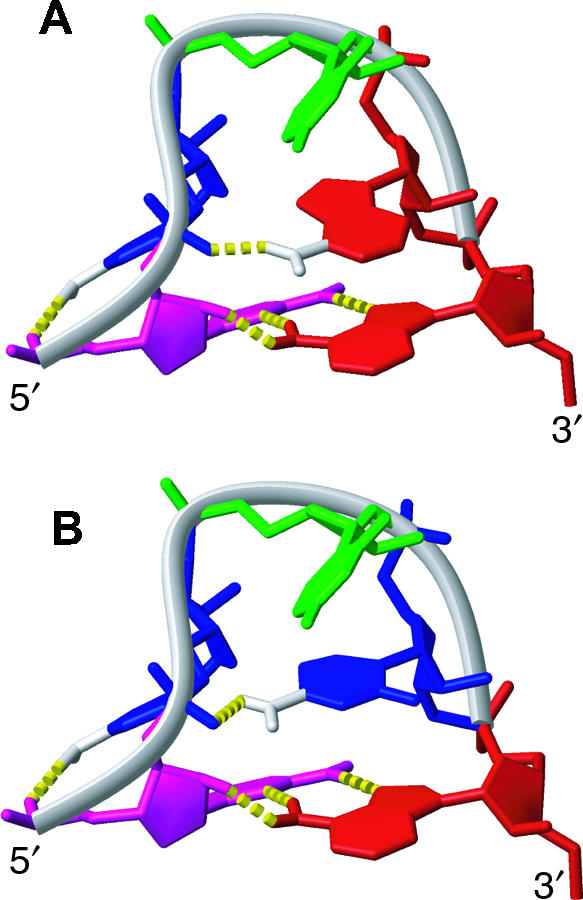
Comparison of GCUAA and GCUCA pentaloops, demonstrating the conservation of the GCUMA fold. (A) NMR structure of GCUAA pentaloop. Nucleotides are colored red (A), green (U), magenta (G), and blue (C). (B) Model of GCUCA pentaloop structure created based on the GCUAA pentaloop structure. The A of GCUAA was replaced by the C of GCUCA while keeping the conformation of the χ torsion angle.
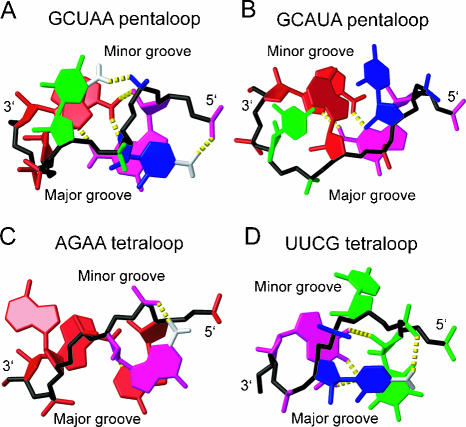
Surprisingly, the structure of the GCUAA pentaloop significantly differs from the structures of GCAUA (Huppler et al. 2002; Fig. 4B ▶), GAAGA (Legault et al. 1998), and GAAAA (Scharpf et al. 2000), which all belong to the GNR(N)A pentaloop family (where N is any nucleotide, R is a purine, and (N) denotes any bulged nucleotide). The only similar feature between the GCUAA pentaloop structure and the GNR(N)A family is the G·A sheared mismatch formed between the 5′-G and 3′-A. In contrast to the GNR(N)A fold that is reminiscent of the most frequently occurring GNRA-type fold (Jucker et al. 1996), except for the additional bulged (N) nucleotide, the GCUAA pentaloop displays rather structural features reminiscent of the UUCG (Allain and Varani 1995; Fig. 4D ▶) and AGNN (Lebars et al. 2001; Wu et al. 2001; Fig. 4C ▶) tetraloop folds. Specifically, the C of the UUCG and the G of AGNN tetraloops are, similar to the C of the GCUAA pentaloop, flipped out toward the major groove, stacked over the first base of the loop, and hydrogen bonded to the closest phosphate-oxygen through its amino group (Fig. 4 ▶). The GCUAA pentaloop and UUCG and AGNN tetraloops also have a very similar “S-shape” conformation of the sugar-phosphate backbone (Fig. 4 ▶, in black).
FIGURE 4.
Comparison between the structures of the GCUAA (A), GCAUA (Huppler et al. 2002) (B), AGAA (Lebars et al. 2001; Wu et al. 2001) (C), and UUCG (Allain and Varani 1995) loops (D). Nucleotides are colored red (A), green (U), magenta (G), and blue (C). Sugar-phosphate backbones are shown in black. Hydrogen bonds are shown as yellow dashed sticks. Amino groups involved in H bonding are depicted; those involved in contacting phosphate oxygen are in white. Otherwise only heavy atoms are shown.
The structure of the GCUAA pentaloop explains well the phylogenetic conservation of GCUMA (where M is A/C; Aruscavage and Bass 2000). The first and last positions are invariantly G and A, respectively, to allow formation of the G·A sheared mismatch. Furthermore, the second position is invariantly a C and the fourth position an A or a C, nucleotides conserved to stabilize the conformation of the pentaloop via their amino-phosphate hydrogen bonds. Finally the central nucleotide is a U, probably to prevent the pentaloop from adopting the GNR(N)A-fold.
The structural similarity between the AGNN and UUCG tetraloops and the GCUAA pentaloop is particularly interesting considering that both the AGNN and the UUCG tetraloops are specifically recognized by the dsRBMs of Rnt1p (Wu et al. 2004) and Staufen (Ramos et al. 2000), respectively. In both dsRBM–RNA complexes, the α-helix 1 of the dsRBMs recognizes the specific shapes of the minor groove created by the conserved AGNN and UUCG tetraloop folds (Ramos et al. 2000; Wu et al. 2004). Analogously, we recently observed that the GCUCA pentaloop, rat analog of the human GCUAA pentaloop of the R/G stem–loop pre-mRNA, is specifically recognized by the first dsRBM of rat ADAR2 (R. Stefl and F.H.-T. Allain, unpubl.). In addition, pre-mRNA encoding the R/G site of subunit C of the glutamate receptor that is also specifically edited by ADAR2 has a UCCR tetraloop (Aruscavage and Bass 2000) that belongs to UNCG-fold. Altogether, similar architectures of the reported GCUCA pentaloop and AGNN and UUCG tetraloops indicates that these loops might play a more important role in the recognition processes mediated by dsRBM-containing proteins than was previously expected.
MATERIALS AND METHODS
RNA sample preparations
Unlabeled, uniformly 13C,15N-labeled RNA, and base-type-specifically 13C,15N-AC/GU-labeled RNA oligonucleotides were prepared by in vitro transcription using T7 polymerase and DNA synthetic primers (Price et al. 1998) and purified either on a denaturing polyacrylamide gel or using HPLC under denaturing conditions. RNA oligonucleotides were annealed at low salt, pH 6.0, and dilute conditions by heating to 95°C and snap cooling on ice to ensure homogenous formation of the monomeric stem–loop conformation. The homogenous formation of the monomeric stem–loop conformation was confirmed by gel filtration coupled with in-line laser light scattering measurement (Wyatt Technology) and native PAGE analysis.
NMR spectroscopy
All the NMR experiments were conducted at 303 K on Bruker Avance-900 and DRX-750, 600, and 500 MHz spectrometers. Exchangeable proton resonances were assigned using two-dimensional (2D) NOESY spectrum (mixing time of 200 msec) of the RNA in 90% H2O/10% D2O at 278 K. Nonexchangeable proton resonances were assigned using 2D NOESY, 2D TOCSY, [1H-13C] HSQC, 3D [1H-13C-1H] HCCH TOCSY, and 3D [1H-13C-1H] NOESY-HSQC spectra in 99.99% D2O. NOE restraints of nonexchangeable protons were obtained from 2D NOESY experiments with mixing times of 30, 90, 150, and 250 msec, and 3D NOESY-HSQC experiment with 100 msec mixing time. The NOEs were semi-quantitatively classified based on their intensities in the 2D and 3D NOESY spectra as follows: very strong (1.8–2.4 Å), strong (1.8–3.0 Å) medium (1.8–4.0 Å), weak (1.8–5.0 Å), and very weak (1.8–6.0 Å). NOEs involving exchangeable protons were observed in 2D NOESY in 90% H2O/10% D2O and all classified as very weak (1.8–6.0 Å). Hydrogen-bonding distance restraints were used only for the Watson–Crick base pairs, when the imino-proton was observed experimentally. The imino protons of U33 and G34 were not observed in the 2D NOESY, probably due to the exchange with solvent. For G34, it is expected since G34 H1 is not hydrogen bonded. However, the absence of U33 H3 is surprising since our structure shows that U33 is involved in a Watson–Crick base pair with A39. It is possible that the presence of the G34–A38 non-Watson–Crick base pair above U33 made the U33–A39 base pair less stable and therefore U33 H3 more prone to exchange with the solvent. Residual dipolar couplings (RDCs) were measured from 1JCH differences in t2-coupled HSQCs in the presence or absence of a C12E9/hexanol mixture (Ruckert and Otting 2000). All spectra were processed using XWINNMR (Bruker) and analyzed using Sparky (T.D. Goddard and D.G. Kneller, SPARKY 3, University of California, San Francisco).
Structure calculations
Structures were calculated using AMBER 7.0 software (University of California, San Francisco) with the Cornell et al. (1995) force field, with the generalized-Born solvation model (Bashford and Case 2000), and with a refinement protocol as described (Padrta et al. 2002). Square-well penalty functions with force constants as indicated in Table 1 ▶ were used for all experimental restraints. Refinements were performed in two steps. In the first step, refinement with NOEs and torsion angles was done starting from completely randomized structures. In the second step, the RDCs were employed in addition to NOE and torsion angle restraints. Initial estimates of the alignment tensors were obtained from the preliminary structures resulting from the first refinement step, as described (Tsui et al. 2000). To impose better convergence of the ensembles some artificial torsion angle restraints were used for α (270°–330°), β (150°–210°), γ (30°–90°), δ (50°–110°), ɛ (180°–240°), and ζ (260°–320°); these torsion angle restraints cover the A-form RNA double-helical range derived from high-resolution crystal structures of RNA (NDB ID: AR0010 [R = 1.16 Å], AR0013 [R = 1.2 Å], AR0020 [R = 1.20 Å], AR0021 [R = 1.20 Å], ARB002 [R = 0.8 Å], ARB003 [R = 1.1 Å], ARB004 [R = 0.9 Å], ARB005 [R = 0.8 Å], ARF0108 [R = 1.8 Å], and ARH064 [R = 1.8 Å]). These torsion angle restraints do not exert any force on the final structures. To prevent high violations of local geometries while accommodating the RDC restraints, additional angle restraints to maintain proper local geometries were employed (Tsui et al. 2000). The family of best structures (13 out of 30 calculated) was selected on the basis of lowest energy criteria. Molecular graphics was generated using MOLMOL (Koradi et al. 1996). The hydrogen bonds found in the pentaloop were not observed experimentally but were deduced based on distances and angles between the proton, the donor, and the acceptor in the ensemble of structures. Atomic coordinates and NMR restraints have been deposited into the Protein Data Bank under accession code (1YSV). The chemical shifts have been deposited into the BioMagResBank Database (accession code BMRB-6485).
Acknowledgments
This work was supported by the Swiss National Science Foundation and the Roche Research Fund for Biology at the ETH Zurich (F.H.T.A.) and the European Molecular Biology Organization and the Human Frontier Science Program postdoctoral fellowships (R.S.). F.H.T.A. is an EMBO Young Investigator.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.7276805.
REFERENCES
- Allain, F.H. and Varani, G. 1995. Structure of the P1 helix from group I self-splicing introns. J. Mol. Biol. 250: 333–353. [DOI] [PubMed] [Google Scholar]
- Aruscavage, P.J. and Bass, B.L. 2000. A phylogenetic analysis reveals an unusual sequence conservation within introns involved in RNA editing. RNA 6: 257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiadis, A., Rich, A., and Maas, S. 2004. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS 2: e391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashford, D. and Case, D.A. 2000. Generalized Born models of macromolecular solvation effects. Annu. Rev. Phys. Chem. 51: 129–152. [DOI] [PubMed] [Google Scholar]
- Bass, B.L. 2002. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 71: 817–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass, B.L., Nishikura, K., Keller, W., Seeburg, P.H., Emeson, R.B., O’Connell, M.A., Samuel, C.E., and Herbert, A. 1997. A standardized nomenclature for adenosine deaminases that act on RNA. RNA 3: 947–949. [PMC free article] [PubMed] [Google Scholar]
- Burns, C.M., Chu, H., Rueter, S.M., Hutchinson, L.K., Canton, H., Sanders-Bush, E., and Emeson, R.B. 1997. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature 387: 303–308. [DOI] [PubMed] [Google Scholar]
- Cornell, W.D., Cieplak, P., Bayly, C.I., Gould, I.R., Merz, K.M., Ferguson, D.M., Spellmeyer, D.C., Fox, T., Caldwell, J.W., and Kollman, P.A. 1995. A 2nd generation force-field for the simulation of proteins, nucleic-acids, and organic-molecules. J. Am. Chem. Soc. 117: 5179–5197. [Google Scholar]
- Egebjerg, J. and Heinemann, S.F. 1993. Ca2+ permeability of unedited and edited versions of the kainate selective glutamate receptor GluR6. Proc. Natl. Acad. Sci. 90: 755–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emeson, R.B. and Singh, M. 2000. Adenosine to inosine RNA editing: Substrates and consequences. In RNA editing: Frontiers in molecular biology (ed. B.L. Bass), pp. 109–138. Oxford University Press, London.
- Gerber, A.P. and Keller, W. 2001. RNA editing by base deamination: Enzymes, more targets, new mysteries. Trends Biochem. Sci. 26: 376–384. [DOI] [PubMed] [Google Scholar]
- Hoopengardner, B., Bhalla, T., Staber, C., and Reenan, R. 2003. Nervous system targets of RNA editing identified by comparative genomics. Science 301: 832–836. [DOI] [PubMed] [Google Scholar]
- Huppler, A., Nikstad, L.J., Allmann, A.M., Brow, D.A., and Butcher, S.E. 2002. Metal binding and base ionization in the U6 RNA intramolecular stem-loop structure. Nat. Struct. Biol. 9: 431–435. [DOI] [PubMed] [Google Scholar]
- Jucker, F.M., Heus, H.A., Yip, P.F., Moors, E.H., and Pardi, A. 1996. A network of heterogeneous hydrogen bonds in GNRA tetraloops. J. Mol. Biol. 264: 968–980. [DOI] [PubMed] [Google Scholar]
- Kohler, M., Burnashev, N., Sakmann, B., and Seeburg, P.H. 1993. Determinants of Ca2+ permeability in both TM1 and TM2 of high affinity kainate receptor channels: Diversity by RNA editing. Neuron 10: 491–500. [DOI] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., and Wuthrich, K. 1996. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 14: 29–32, 51–55. [DOI] [PubMed] [Google Scholar]
- Kumar, M. and Carmichael, G.G. 1997. Nuclear antisense RNA induces extensive adenosine modifications and nuclear retention of target transcripts. Proc. Natl. Acad. Sci. 94: 3542–3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebars, I., Lamontagne, B., Yoshizawa, S., Aboul-Elela, S., and Fourmy, D. 2001. Solution structure of conserved AGNN tetraloops: Insights into Rnt1p RNA processing. EMBO J. 20: 7250–7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legault, P., Li, J., Mogridge, J., Kay, L.E., and Greenblatt, J. 1998. NMR structure of the bacteriophage λ N peptide/boxB RNA complex: Recognition of a GNRA fold by an arginine-rich motif. Cell 93: 289–299. [DOI] [PubMed] [Google Scholar]
- Levanon, E.Y., Eisenberg, E., Yelin, R., Nemzer, S., Hallegger, M., Shemesh, R., Fligelman, Z.Y., Shoshan, A., Pollock, S.R., Sztybel, D., et al. 2004. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 22: 1001–1005. [DOI] [PubMed] [Google Scholar]
- Lomeli, H., Mosbacher, J., Melcher, T., Hoger, T., Geiger, J.R., Kuner, T., Monyer, H., Higuchi, M., Bach, A., and Seeburg, P.H. 1994. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science 266: 1709–1713. [DOI] [PubMed] [Google Scholar]
- Morse, D.P. and Bass, B.L. 1999. Long RNA hairpins that contain inosine are present in Caenorhabditis elegans poly(A)+ RNA. Proc. Natl. Acad. Sci. 96: 6048–6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse, D.P., Aruscavage, P.J., and Bass, B.L. 2002. RNA hairpins in noncoding regions of human brain and Caenorhabditis elegans mRNA are edited by adenosine deaminases that act on RNA. Proc. Natl. Acad. Sci. 99: 7906–7911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padrta, P., Stefl, R., Kralik, L., Zidek, L., and Sklenar, V. 2002. Refinement of d(GCGAAGC) hairpin structure using one- and two-bond residual dipolar couplings. J. Biomol. NMR 24: 1–14. [DOI] [PubMed] [Google Scholar]
- Polson, A.G., Crain, P.F., Pomerantz, S.C., McCloskey, J.A., and Bass, B.L. 1991. The mechanism of adenosine to inosine conversion by the double-stranded RNA unwinding/modifying activity: A high-performance liquid chromatography–mass spectrometry analysis. Biochemistry 30: 11507–11514. [DOI] [PubMed] [Google Scholar]
- Price, S., Oubridge, C., Varani, G., and Nagai, K. 1998. Preparation of RNA–protein complexes for X-ray crystallography and NMR. In RNA–protein interaction: Practical approach (ed. C. Smith), pp. 37–74. Oxford University Press, Oxford, UK.
- Ramos, A., Grunert, S., Adams, J., Micklem, D.R., Proctor, M.R., Freund, S., Bycroft, M., St Johnston, D., and Varani, G. 2000. RNA recognition by a Staufen double-stranded RNA-binding domain. EMBO J. 19: 997–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruckert, M. and Otting, G. 2000. Alignment of biological macromolecules in novel nonionic liquid crystalline media for NMR experiments. J. Am. Chem. Soc. 122: 7793–7797. [Google Scholar]
- Rueter, S.M., Dawson, T.R., and Emeson, R.B. 1999. Regulation of alternative splicing by RNA editing. Nature 399: 75–80. [DOI] [PubMed] [Google Scholar]
- Scharpf, M., Sticht, H., Schweimer, K., Boehm, M., Hoffmann, S., and Rosch, P. 2000. Antitermination in bacteriophage λ. The structure of the N36 peptide–boxB RNA complex. Eur. J. Biochem. 267: 2397–2408. [DOI] [PubMed] [Google Scholar]
- Schaub, M. and Keller, W. 2002. RNA editing by adenosine deaminases generates RNA and protein diversity. Biochimie 84: 791–803. [DOI] [PubMed] [Google Scholar]
- Sommer, B., Kohler, M., Sprengel, R., and Seeburg, P.H. 1991. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell 67: 11–19. [DOI] [PubMed] [Google Scholar]
- Stefl, R., Skrisovska, L., and Allain, F.H. 2005. RNA sequence- and shape-dependent recognition by proteins in the ribonucleoprotein particle. EMBO Rep. 6: 33–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonkin, L.A. and Bass, B.L. 2003. Mutations in RNAi rescue aberrant chemotaxis of ADAR mutants. Science 302: 1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui, V., Zhu, L., Huang, T.H., Wright, P.E., and Case, D.A. 2000. Assessment of zinc finger orientations by residual dipolar coupling constants. J. Biomol. NMR 16: 9–21. [DOI] [PubMed] [Google Scholar]
- Wong, T.C., Ayata, M., Ueda, S., and Hirano, A. 1991. Role of biased hypermutation in evolution of subacute sclerosing panencephalitis virus from progenitor acute measles virus. J. Virol. 65: 2191–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, H., Yang, P.K., Butcher, S.E., Kang, S., Chanfreau, G., and Feigon, J. 2001. A novel family of RNA tetraloop structure forms the recognition site for Saccharomyces cerevisiae RNase III. EMBO J. 20: 7240–7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, H., Henras, A., Chanfreau, G., and Feigon, J. 2004. Structural basis for recognition of the AGNN tetraloop RNA fold by the double-stranded RNA-binding domain of Rnt1p RNase III. Proc. Natl. Acad. Sci. 101: 8307–8312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. and Carmichael, G.G. 2001. The fate of dsRNA in the nucleus: A p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell 106: 465–475. [DOI] [PubMed] [Google Scholar]