Abstract
Human herpesvirus 8-encoded interleukin-6 (vIL-6) signals through the gp130 signal transducer but is not dependent on the IL-6 receptor α subunit (IL-6R, gp80) that is required for signaling by endogenous IL-6 proteins; however, IL-6R can enhance vIL-6 activity and can enable signaling through a gp130 variant, gp130.PM5, that is itself unable to support vIL-6 signaling. These findings suggest that the vIL-6-gp130 interactions are qualitatively different from those of human IL-6 (hIL-6) and that vIL-6 signaling may be more promiscuous than that of hIL-6 but that IL-6R may play a role in vIL-6 signaling in vivo. To examine the receptor binding requirements of vIL-6, we have undertaken mutational analyses of regions of gp130 and IL-6R potentially involved in interactions with ligand or in functional complex formation and used these variants in functional, ligand-binding, and receptor dimerization assays. The data presented identify positions within two interstrand loops of the gp130 cytokine-receptor homology domain that are important for vIL-6 signaling and vIL-6-induced receptor dimerization and show that vIL-6, like hIL-6, can form complexes with IL-6R and gp130 but that the roles of putative cytokine-binding residues of IL-6R in ligand-induced functional complex formation are qualitatively different in the case of vIL-6 and hIL-6.
The discovery of a homologue of interleukin-6 (IL-6) encoded by human herpesvirus 8 (HHV-8) was significant for two reasons: first, because an IL-6 gene had not previously been identified in a virus, and second, because HHV-8 was known to be associated with Kaposi's sarcoma, a disease in which endogenous IL-6 may play a role (6, 20, 23, 28). Subsequently, it was shown that HHV-8 was also associated with multicentric Castleman's disease, in which IL-6 has also been implicated as an important factor in disease development (2, 29, 30, 34, 39), and with primary effusion lymphoma (5, 26). In the case of primary effusion lymphoma, both human and viral IL-6 (hIL-6 and vIL-6, respectively) act as mitogenic factors (1, 9, 15).
The mechanism by which vIL-6 induces intracellular signal transduction is generally the same as that utilized by the cellular IL-6 proteins in that both viral and cellular proteins bind to the gp130 signal transducer to effect dimerization, phosphorylation of gp130 cytoplasmic tyrosine residues through the recruitment of Janus kinases (Jaks), and the consequent recruitment and activation through tyrosine phosphorylation of SH2 domain-containing STAT transcription factors (10, 14, 25, 27, 35, 36). The mitogen-activated protein kinase pathway is also activated via gp130 by both hIL-6 and vIL-6 (12, 31, 37). However, unlike endogenous IL-6 proteins, vIL-6 can signal through gp130 in the absence of the α receptor subunit, IL-6R (gp80), although there is evidence that IL-6R can enhance vIL-6 signaling (4, 12, 22, 28, 37). That vIL-6 can form functional complexes with IL-6R and gp130 has been inferred also from the finding that a gp130 variant that has been altered in the cytokine-binding E-F loop of the cytokine-receptor homology region (CHR) can support signaling by vIL-6 only in the presence of IL-6R (37). Nevertheless, the ability of vIL-6 to signal through gp130 alone suggests that the viral cytokine may be able to signal more promiscuously than its cellular counterpart in vivo, with possible implications for its role in viral biology and pathogenesis.
IL-6R and gp130 contain homologous domains that are found in other receptor subunits that associate with gp130 to mediate signal transduction. These domains include immunoglobulin-like and cytokine-receptor homology (comprising two protein domains) regions, both of which are involved in hIL-6 binding in the case of gp130, with the CHR region of IL-6R also interacting with hIL-6 (11, 13, 16, 17, 19, 24, 38). Three regions of hIL-6 are known to interact with receptor: site I is involved in binding to IL-6R, while sites II and III interact with gp130 CHR and immunoglobulin domain residues, respectively. In the case of vIL-6, alteration of a triplet of amino acids within the CHR E-F loop, a component of the hIL-6 interacting interface, was found to abrogate IL-6R-independent signaling, suggesting that one or more the altered residues are important for ligand recognition (37). However, whether this is the case and whether other regions of the CHR are required for interactions with vIL-6, via the equivalent of hIL-6 site II, has not been resolved, and the nature of the interactions of vIL-6 with IL-6R has not previously been investigated.
We present here data that identify residues in gp130 that are involved in vIL-6-mediated functional dimerization and heterodimerization with IL-6R and residues of IL-6R that are necessary for vIL-6-induced association with gp130. Our results suggest that the E-F and C-D loops of gp130 CHR and regions of IL-6R equivalent to gp130 CHR B2-C2 are involved in receptor recognition by vIL-6.
MATERIALS AND METHODS
Cell culture and transfections.
HEK293-T cells were grown in Dulbecco modified Eagle medium supplemented with 5% fetal calf serum. Hep3B cells (used in some of the functional assays) were grown in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum. Cells were passaged 12 to 24 h before transfection to produce monolayers of 40 to 60% confluency in 25-cm2 flasks or six-well plates (for functional assays) or 100-mm dishes (for generation of secreted proteins for receptor-ligand binding assays). Transfections were carried out by calcium phosphate-DNA coprecipitation with HEPES-buffered saline; cells or media were harvested 48 h posttransfection. For functional analyses of receptor variants, pEFBOS-based expression constructions (0.5 to 1 μg) were used together with 1 to 3 μg of pSG5-based vIL-6 or hIL-6 expression vector (37) or pSG5 (negative control) and 0.5 to 1 μg of the pα2MCAT reporter plasmid. Where necessary, total amounts of transfected DNA were made up to a standard amount by using pEF-BOS (receptor expression vector) DNA. For the generation of proteins for use in ligand-receptor and receptor-receptor binding assays, 2 to 10 μg of expression vector was used for transfection.
Mutagenesis.
Mutagenesis was conducted on M13-gp130 and M13-IL-6R uracil-containing single-stranded DNA templates by using the Kunkel method (18), essentially as described previously (37). Coding sequences containing introduced mutations were excised from M13 replicative-form DNA by restriction digestion for cloning into pEF-BOS or were amplified by PCR with primers containing introduced restriction enzyme sites, and the appropriately digested PCR products were cloned into pSG5 or immunoglobulin G (IgG) Fc-containing pSG5 (pSG5.X.Fc) eukaryotic expression vectors (see below).
Plasmids and oligonucleotides.
The pSG5-based eukaryotic expression plasmids for vIL-6 and hIL-6 comprise the respective coding sequences cloned into the BamHI site of pSG5 (37). The human IL-6R and human gp130 expression vectors pEF-BOS-hIL-6R and pEF-BOS-hgp130 were provided by M. Narazaki and T. Kishimoto; they comprise the receptor coding sequences cloned between the XbaI (IL-6R) or SacI and BamHI sites of the pEF-BOS eukaryotic expression vector (21). The pα2MCAT reporter plasmid comprises α2-macroglobulin promoter sequences cloned upstream of the chloramphenicol acetyltransferase (CAT) gene (33). The generation of the signaling-defective gp130 variant, gp130.PM5, has been described previously (37). Other altered gp130 and IL-6R coding sequences were generated similarly by using the Kunkel mutagenesis procedure (18). Sequences encoding the receptor proteins were derived by digestion with XbaI (IL-6R) or SacI and BamHI (gp130) of M13-receptor replicative-form DNA for cloning into the same sites in pEF-BOS. Sequences encoding the extracellular regions of the proteins were PCR amplified from M13 vectors and cloned into a polylinker-modified pSG5 vector, pSG5.X, between the HindIII and BamHI (IL-6R) and SmaI and BamHI (gp130) sites. Sequences specifying the extracellular regions of gp130 and IL-6R were cloned between the HindIII and BamHI (sIL-6R) and EcoRV and BamHI (sgp130) sites of a similar vector, pSG5.X.Fc, containing human IgG Fc coding sequences downstream of the polylinker, to generate contiguous sIL-6R-Fc and sgp130-Fc open reading frames for the expression of these fusion proteins by transfected cells. Expression vectors for Flag-tagged, soluble versions of gp130 variants PM5, PM16, PM17, PM29, PM31, and PM33 were made by cloning of SmaI-BamHI-digested PCR-amplified sgp130 sequences between the SmaI and BamHI sites of a Flag epitope-containing pSG5-based vector, pSG5.S.Flag. The primers used for cloning sIL-6R and sgp130 were the universal sequencing primer (GTAAAACGACGGCCAGT; 5′, IL-6R; HindIII site in the M13 polylinker), IL-6R.P8 (acggatcCTTGCACTGGGAGGCT; 3′, sIL-6R; lowercase indicates noncomplementary sequences containing introduced restriction site), gp130.P1 (acacccgggGATGTTGACGTTGCAGA; 5′, gp130), and gp130.P5 (acggatccGGCTTCAATTTCTCCTT; 3′, sgp130).
Western blotting.
vIL-6, sgp130, and sIL-6R proteins or Fc- or Flag-fused derivatives obtained from transfected cell media were size fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electrophoretically transferred to nitrocellulose membranes. These were blocked in TBS-T (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.05% Tween 20) containing 5% nonfat milk, prior to the addition of primary antibody (0.1 to 0.5 μg/ml). Antibodies used for the detection of gp130, IL-6R, and Flag epitope were obtained from PharMingen (San Diego, Calif.; catalog no. 33831A), R&D Systems (Minneapolis, Minn.; catalog no. AB-227-NA), and Sigma (St. Louis, Mo.; catalog no. F3165), respectively; the vIL-6 peptide antiserum used for the detection of vIL-6 has been described previously (37). After a washing in TBS-T containing 5% nonfat milk, horseradish peroxidase-conjugated anti-goat (hIL-6R), anti-mouse (gp130 and Flag), or anti-rabbit (vIL-6) secondary antibody (Santa Cruz Biotechnology, Santa Cruz, Calif.; catalog numbers sc-2020, sc-2064, and sc-2004, respectively) diluted 1:2,000 was used to detect filter-bound primary antibody. Horseradish peroxidase on TBS-washed filters was visualized by a chemiluminescence assay.
Ligand-receptor and receptor-receptor binding assays.
Assays of vIL-6 binding to sgp130-Fc, or altered derivatives, and receptor-receptor dimerization (gp130-gp130 and gp130-IL-6R) assays were carried out with the component proteins derived from growth media of HEK293-T cells transfected with the appropriate expression vectors; sgp130 and sIL-6R were also obtained as purified recombinant proteins from a commercial source (R&D Systems; catalog numbers 228-GP and 227-SR). For assays involving exogenous mixing of separate components, 400-μl aliquots of ligand and receptor samples (or pSG5 controls) were incubated with protein A-Sepharose (Pharmacia Biotech; catalog no. 17-0974) at 4°C overnight, and the matrix and bound material was then subjected to repeated centrifugation and washing in phosphate-buffered saline prior to Western analysis. For coexpressed (cotransfected) components, 1-ml aliquots of media were treated similarly. Precipitated, washed samples were heat-denatured, applied to sodium dodecyl sulfate-polyacrylamide gels, and transferred to nitrocellulose for Western analysis to detect vIL-6, sgp130 (native, or Fc or Flag fusions), or sIL-6R (Fc fusion).
RESULTS
Alanine-scanning mutagenesis of putative cytokine-binding loops of gp130.
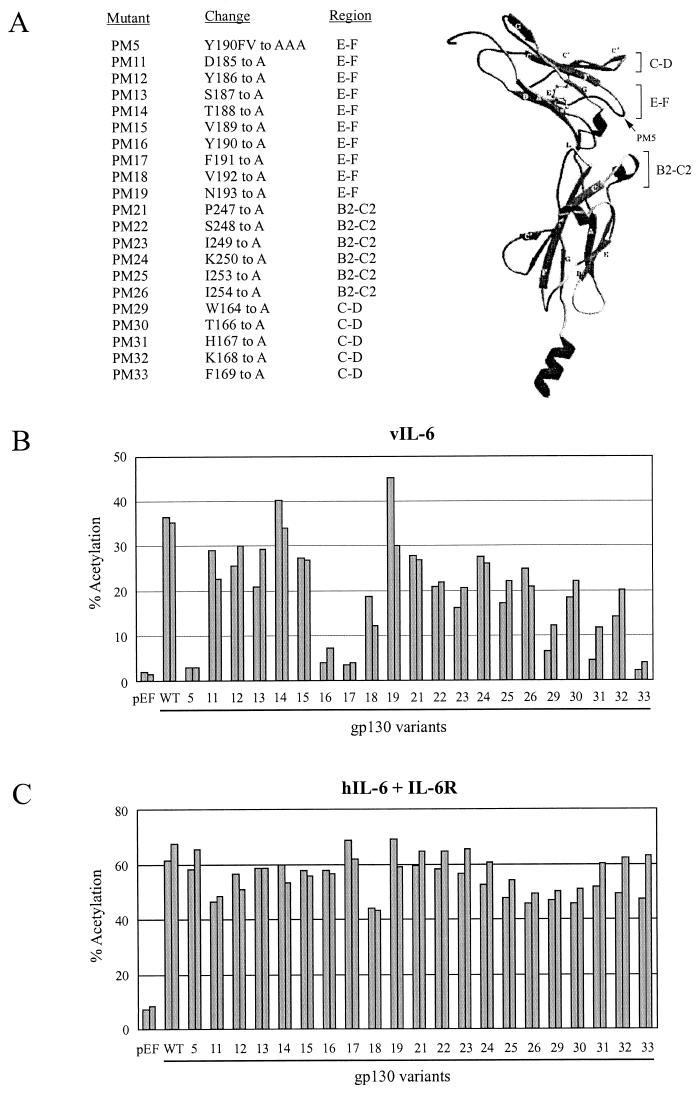
The structure of the CHR region of gp130 has been determined by X-ray crystallography (3). These data generally confirmed the structures predicted from the primary structure and from the known structure of the related receptor for human growth factor (8). Salient features of gp130 structure with respect to cytokine binding are the two previously predicted cytokine binding loops, E-F and B2-C2 (3, 13, 19), and another loop region, C-D, that potentially is available for interaction with ligand (Fig. 1A). We reported previously that a gp130 variant, gp130.PM5, in which CHR E-F loop residues Y190FV192 were changed to alanines, was abrogated in its ability to support vIL-6 signaling in the absence of cotransfected IL-6R (37). The B2-C2 region variant gp130.PM7 (S251V252 to AA), however, showed wild-type levels of activity.
FIG. 1.
Functional analyses of gp130 CHR variants. (A) Positions of alanine-scanning substitutions in the C-D, E-F, and B2-C2 loops of gp130. (B) Activities of the gp130 variants in comparison to wild-type gp130 in vIL-6 signaling assays. Expression vectors for each of the gp130 proteins, or empty vector (pEF-BOS), were cotransfected with vIL-6 and the STAT-responsive CAT target pα2MCAT (33). Cell extracts were harvested for determinations of CAT activities by standard methods. The results of duplicate experiments are shown. (C) Activities of the gp130 variants relative to wild-type gp130 in hIL-6 signaling assays in which an IL-6R expression vector was cotransfected with ligand and gp130 expression plasmids and pα2MCAT. WT, wild type. (The ribbon diagram of sgp130 in panel A was reproduced from Fig. 1A of reference 3 with the permission of Yvonne Jones and Oxford University Press.)
To determine which of the PM5 residues were important for activity and whether other residues within the E-F loop and in the B2-C2 and C-D regions may be involved in vIL-6 binding, we undertook alanine-scanning mutagenesis of these regions to derive a series of gp130 variant expression constructions for use in transient-transfection assays (see Materials and Methods). The gp130 variants generated are listed in Fig. 1A. Each of the pEF-BOS-cloned altered gp130 coding sequences (or pEF-BOS, negative control) was cotransfected with the vIL-6 expression vector pSVvIL-6 (28) and the STAT-CAT reporter pα2MCAT (33) into HEK293-T cells. CAT activities in cell extracts were subsequently determined. The results of these experiments are shown in Fig. 1B. While several of the constructions showed reduced activities relative to the wild-type gp130 expression vector, mutations PM16, PM17, PM29, PM31, and PM33 (in addition to PM5) showed greatly reduced activities (<25% compared to the wild type). Similar assays performed with expression vectors for hIL-6 and IL-6R (required for hIL-6 signaling) cotransfected with the each of the gp130 expression plasmids and pα2MCAT revealed essentially wild-type activities of each of the gp130 variants (Fig. 1C).
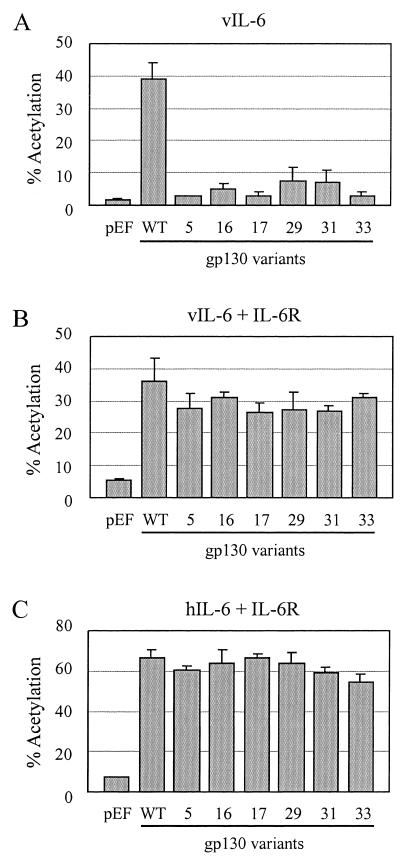
Additional experiments were performed with the gp130 variants showing reduced vIL-6 signaling to determine whether IL-6R could “rescue” the altered gp130 proteins to allow efficient vIL-6 signal transduction, as seen for hIL-6. Parallel, multiply repeated experiments were performed in the presence or absence of IL-6R for vIL-6 and in the presence of IL-6R for hIL-6. The data from these experiments are presented in Fig. 2. The results revealed positive effects of IL-6R coexpression on signaling by each of the gp130 variants in response to vIL-6, a finding analogous to the situation for IL-6R-dependent hIL-6 signal transduction by these altered gp130 proteins.
FIG. 2.
Effect of IL-6R coexpression with gp130 variants on vIL-6 signaling. Data derived from triplicate cotransfection experiments for vIL-6 signaling through gp130 and functionally altered gp130 variants either in the presence or absence of overexpressed IL-6R are shown, together with corresponding data for hIL-6 signal transduction through the same gp130 proteins in the presence IL-6R. WT, wild type.
The data shown in Fig. 1 and 2 identify individual residues within the E-F and C-D loops that are important for IL-6R-independent signaling through gp130, with these possibly representing vIL-6-gp130 binding interfaces, and demonstrate that, as for previously reported results for gp130.PM5 (37), IL-6R can enable increased signaling by vIL-6 through the corresponding functionally defective gp130 variants. This suggests that IL-6R can stabilize vIL-6 interactions with gp130 to enable the formation of functional signaling complexes.
Binding of vIL-6 to functionally defective gp130 variants.
We reasoned that the inability of gp130 variants PM16, PM17, PM29, PM31, and PM33 to support high-level signaling by vIL-6 could be due to reduced affinity of vIL-6-gp130 interactions or to reduced or altered formation of higher order vIL-6-gp130 signaling complexes. To test the former hypothesis, we cloned sequences specifying the soluble components (lacking the transmembrane domain and cytoplasmic region) of gp130 and the respective PM variants into a pSG5-based human IgG-Fc-containing vector to generate eukaryotic expression plasmids that could be used to express the respective gp130-Fc fusion proteins in transfected cells. These constructions, pSVvIL-6, or pSG5 (negative control), were transfected separately into HEK293-T cells to obtain the component proteins for use in protein A-Sepharose-mediated coprecipitation binding assays (see Materials and Methods). Coprecipitated vIL-6 was identified by Western analysis by using vIL-6-specific antiserum (37). These experiments were performed in the absence or presence of recombinant sIL-6R (obtained commercially). An alternative procedure that involved cotransfection of each of the gp130-Fc expression vectors with pSVvIL-6 into HEK293-T-cell cultures was also used to control for the possibility that coexpression of the ligand and receptor proteins may be important for their interaction.
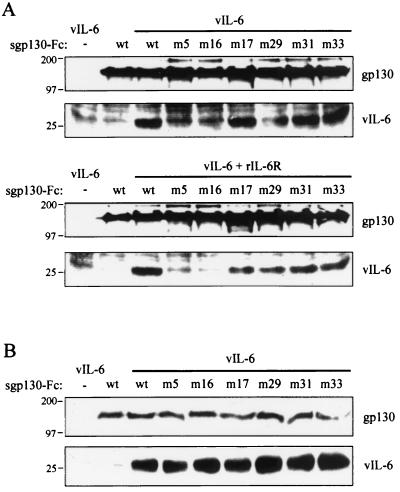
Representative results from experiments in which the protein components were expressed independently and then mixed exogenously are shown in Fig. 3A. Wild-type sgp130-Fc was able to interact with vIL-6 in this assay, with vIL-6 protein being coprecipitated with the protein A-bound Fc fusion protein, but not by protein A-Sepharose alone. The gp130 variants PM17, PM29, PM31, and PM33 were also able to coprecipitate vIL-6, whereas variants PM5 (Y190FV to AAA) and PM16 (Y190 to A) were unable to bind vIL-6, either with or without added sIL-6R. The data in the absence of sIL-6R for gp130.PM5 and gp130.PM16 are consistent with the inability of these altered proteins to support vIL-6 signaling and with recently published vIL-6:gp130 crystallographic data demonstrating that gp130 Y190 is positioned appropriately for interactions with vIL-6 (7).
FIG. 3.
Binding of IL-6 by gp130 variants as determined by coprecipitation assays. (A) Soluble Fc fusion proteins of wild-type and specifically altered gp130 were obtained from the media of HEK293-T cells transfected with expression vectors for each of the proteins. Separate cultures were transfected with pSV-vIL-6 to generate vIL-6-containing media. Samples of each of the sgp130-Fc media were mixed with samples of vIL-6 medium and incubated with protein A-Sepharose, and then pelleted material was analyzed by Western blotting to detect sgp130-Fc and vIL-6 (top panels). Parallel experiments were done in the presence of added recombinant soluble IL-6R (rIL-6R) (bottom panels). (Note that a nonspecific immunoreactive species migrating to a position slightly higher than vIL-6 is present on the blots [including in lanes 2, sgp130-Fc alone] and should not be confused with vIL-6.) (B) Coexpression of each of the sgp130-Fc proteins with vIL-6 was achieved by cotransfection of each of the sgp130-Fc expression vectors with pSVvIL-6. Samples of transfected cell media were used in coprecipitation assays, as before. wt, wild type.
However, although these variants can signal in the presence of IL-6R, no effect of added recombinant sIL-6R on gp130.PM5 and gp130.PM16 interactions with vIL-6 were observed in this assay. In contrast to these results, when vIL-6 was coexpressed with sgp130.PM5-Fc and sgp130.PM16-Fc, ligand-receptor interactions were detected, as they were for wild-type gp130 and the other gp130 variants (Fig. 3B). These data demonstrate that each of the gp130 variants is able to bind vIL-6 but that coexpression of vIL-6 with gp130.PM5 and gp130.PM16 is necessary for vIL-6 binding to these proteins (see below). Although the functionally defective gp130 variants bind vIL-6, their ability to dimerize in the presence of ligand may be compromised.
Effects of gp130 mutations on vIL-6 induced gp130-gp130 complexing.
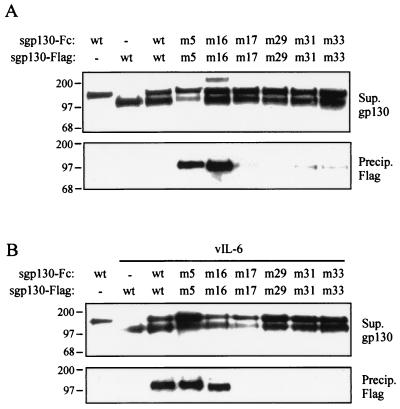
To test the possibility that the functionally altered gp130 variants were unable to dimerize, we undertook cotransfection assays in which each sgp130-Fc proteins (wild-type and variants) was cotransfected with its respective Flag-tagged counterpart and vIL-6 (or pSG5, for wild-type gp130). After protein A-agarose coprecipitation, Western analysis with Flag epitope antibody was undertaken for the detection of coprecipitated sgp130-Flag proteins. As shown in the top panel of Fig. 4A, each of the sgp130 (wild-type and variant) Fc and Flag fusion proteins was expressed and secreted in transfected HEK293-T-cell cultures. Significantly, both sgp130.PM5-Flag and sgp130.PM16-Flag were coprecipitated (by protein A-Sepharose) with their Fc-fused counterparts, demonstrating ligand-independent complexing of these variants. No evidence of gp130-gp130 dimerization (above background) was seen for wild-type sgp130 or the other gp130 variants. Coexpression of vIL-6 with the wild-type sgp130 enabled the association of sgp130-Flag with sgp130-Fc in the coprecipitation assays, but vIL-6 was unable to induce dimerization of gp130 variants PM17, PM29, PM31, and PM33 in this assay (Fig. 4B), a finding consistent with the greatly reduced abilities of these gp130 proteins to support vIL-6 signaling. Homodimerization of gp130.PM5 and gp130.PM16 was seen in the presence of vIL-6 as in its absence.
FIG. 4.
gp130 dimerization assays. (A) Fc and Flag epitope fusions of wild-type and variant sgp130 proteins were coexpressed in cotransfected cells. After protein A-Sepharose precipitation, coprecipitated sgp130-Flag was detected by Western blotting (bottom panel). Expression of each of the Fc and Flag fusion proteins in samples of transfected cell media was checked by Western blotting with gp130 antibody (top panel; the upper band corresponds to sgp130-Fc, and the lower band corresponds to sgp130-Flag). (B) Effect of vIL-6 coexpression on gp130 dimerization. Coexpression of vIL-6 with the sgp130 fusion proteins was achieved by cotransfection of pSVvIL-6 with the respective gp130 expression constructions. Ligand-induced dimerization was seen only for wild-type gp130. wt, wild type.
The combined data of Fig. 3 and 4 indicate that vIL-6, while able to bind each of the signaling-defective gp130 variants, is unable to induce their functional dimerization. This includes the PM5 and PM16 gp130 variants that are able to self-associate in the absence of ligand. Presumably, these complexes are inappropriately conformed to effect signal transduction, since we did not detect signaling by these proteins either in the presence or in the absence of vIL-6 (Fig. 1B and 2A and reference 37). Ligand-independent complexing of gp130.PM5 and gp130.PM16 explains the results shown in Fig. 3A if it is assumed that vIL-6 is unable to access the preformed gp130 dimers; coexpression of the gp130 variants with vIL-6 could allow ligand-gp130 associations prior to gp130-gp130 complex formation (a hypothesis consistent with the results in Fig. 3B).
Identification of IL-6R residues important for vIL-6 signaling.
We reported previously (37) that IL-6R could restore the ability of gp130.PM5 to respond to vIL-6, presumably by stabilizing vIL-6-gp130 interactions by formation of vIL-6-gp130-IL-6R signaling complexes analogous to those induced by hIL-6. Therefore, to investigate vIL-6-IL-6R interactions through the mutagenesis of residues of IL-6R, we used gp130.PM5 as the signaling molecule in transient-transfection assays, thereby rendering vIL-6 signaling dependent on vIL-6-IL-6R interactions. IL-6R residues previously reported to negatively affect hIL-6 interactions with IL-6R or hIL-6-IL-6R interactions with gp130, as determined by coprecipitation-based binding assays (17, 32, 38), were targeted for mutagenesis. The alterations that were made are listed in Fig. 5A. The altered amino acids correspond to gp130 residues in the E-F (PM1) and B2-C2 (PM3 and PM4) CHR loops and to IL-6R residues (PM2 and PM5) believed to lie in an IL-6R-gp130 dimerization interface (3, 32).
FIG. 5.
Functional analyses of IL-6R CHR variants. (A) Codons specifying residues within the proximal and distal domains of the CHR previously reported to affect hIL-6 binding or interactions of hIL-6-IL-6R complexes with gp130 were altered to effect amino acid substitutions, as indicated. (B) Expression constructions of each were cotransfected with pα2MCAT, pEFBOS-hgp130.PM5 (or pEFBOS, negative control), and either vIL-6 or hIL-6 expression vectors into HEK293-T cells or Hep3B cells (both used for each ligand, with indistinguishable results). Cell extracts were subsequently made and assayed for CAT activity. Each datum point is derived from either five (hIL-6) or six (vIL-6) independent transfections, and values are expressed relative to CAT activities obtained with wild-type IL-6R. WT, wild type.
The results of our cotransfection assays are shown in Fig. 5B. Each of the alterations in IL-6R had a negative effect on vIL-6 signaling through gp130.PM5, with mutations PM2 to PM5 reducing vIL-6 activation of the pα2MCAT reporter to below 30% of that obtained with wild-type IL-6R. In sharp contrast, all but one (PM5) of the IL-6R variants were able to support near-wild-type levels of signal transduction by hIL-6, despite the previously reported effects of these mutations on ligand-receptor interactions in vitro (17, 32, 38). That the effects of mutations PM1 to PM4 on hIL-6 signaling were relatively minor in our transfection assays, while clearly detectable in the previously reported binding assays, may be due to greater stability of signaling complexes in the presence of gp130 (for altered hIL-6-IL-6R interactions, PM3 and PM4) and in the context of membrane-bound receptors. Regardless, the clear effects of mutations PM2 to PM4, in addition to the PM5 alteration, on vIL-6 signaling indicate that the altered residues are important for vIL-6-induced functional complex formation, possibly reflecting their involvement in direct vIL-6-IL-6R interactions in the case of Y249 and R250/L251, and that there are differences in vIL-6 and hIL-6 interactions with IL-6R in the context of cell surface-expressed ligand-IL-6R-gp130 complexes.
Effects of IL-6R mutations on ligand-induced gp130-IL-6R complexing.
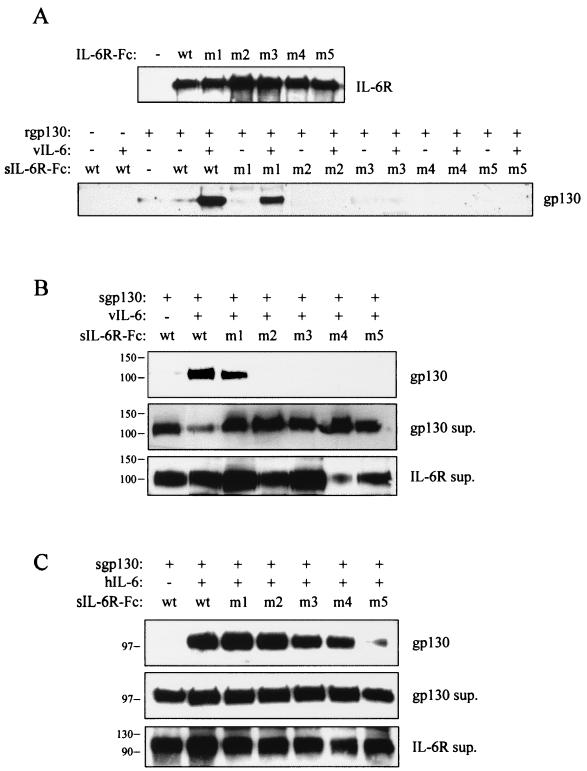
To determine the effects of introduced IL-6R mutations on the ability of IL-6R to form vIL-6-induced complexes with gp130, we used sIL-6R-Fc fusions proteins (wild type and variants PM1 to PM5) in coprecipitation assays similar to those described above. Protein components for use in the assays were derived either as recombinant protein (sgp130) or from media of cell cultures separately transfected with vIL-6 and sIL-6R-Fc expression constructions or from cells cotransfected with expression vectors for vIL-6, sgp130, and the sIL-6R-Fc fusion proteins. Appropriate negative controls were provided by using empty expression vectors in place of vIL-6 or the receptor constructions. Parallel experiments were undertaken with hIL-6 instead of vIL-6 in cotransfections. The results of the assays are shown in Fig. 6. For vIL-6, using either the in vitro mixing (Fig. 6A) or cotransfection (Fig. 6B) approaches, ligand-dependent complexing of sgp130 with sIL-6R-Fc was seen for wild-type sIL-6R and variant PM1 but not for functionally defective variants PM2, PM3, PM4, and PM5. In the case of hIL-6, gp130 complexing with sIL-6R and sIL-6R variants PM1 to PM4 could be induced efficiently by coexpression of the ligand; hIL-6-induced sgp130 complexing with sIL-6R.PM5 was barely detectable.
FIG. 6.
Effects of IL-6R CHR mutations on ligand-induced IL-6R-gp130 complexing. Expression constructions for Fc fusions of wild-type and variant sIL-6R proteins were individually transfected (A) or cotransfected (B and C) with sgp130 and ligand (vIL-6 or hIL-6) expression vectors or corresponding pEFBOS or pSG5 negative controls (−). Results of protein A-Sepharose-mediated coprecipitation assays are shown for vIL-6 (A and B) and hIL-6 (C); coprecipitation of sgp130 indicates complexing between sgp130 and sIL-6R-Fc proteins. Expression and secretion of each of the receptor components by transfected cells was checked by Western analysis of samples of culture media (“sup.”). wt, wild type.
These binding data for vIL-6 and hIL-6 are consistent with the results of the functional assays with the IL-6R variants (Fig. 5B). They indicate that the deficiencies of IL-6R variants PM2, PM3, PM4, and PM5 to support vIL-6 signaling through gp130.PM5 stem from their inabilities to form stable complexes with vIL-6 and gp130. IL-6R residues Y249 and R250/L251 (altered in variants PM3 and PM4) are likely to be involved in interactions with vIL-6, since these residues have previously been shown to be important for hIL-6 binding to IL-6R in vitro (17, 38), whereas IL-6R N230 and H280 and/or D281 (altered in variants PM2 and PM5, respectively) are probably necessary for vIL-6-induced gp130-IL-6R associations, since these residues are predicted to lie in a gp130-IL-6R dimerization interface (3, 32). IL-6R H280 and/or D281, but not N230, also appear to be centrally involved in hIL-6-induced IL-6R-gp130 complexing, since IL-6R.PM5 alone showed reduced ability to complex with gp130, and signal, in the presence of hIL-6.
Taken together, our functional and binding data implicate IL-6R residues N230, Y249, and R250/L251 as being involved in vIL-6-specific ligand-induced assembly of functional complexes of gp130 and IL-6R and indicate that IL-6R residues H280 and/or D281 are critical for both vIL-6 and hIL-6 IL-6R-dependent signaling through gp130.
DISCUSSION
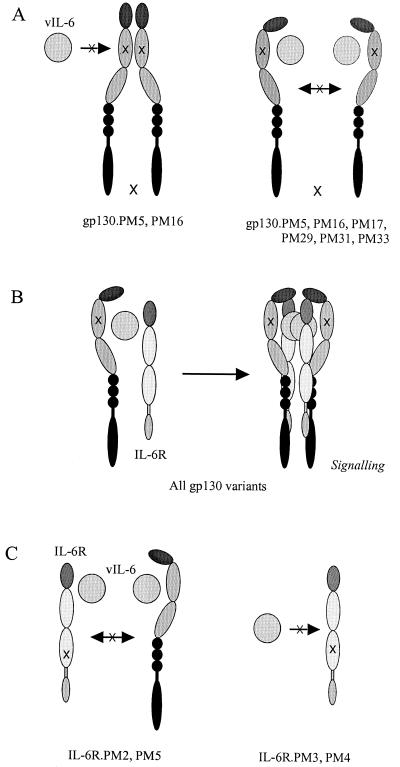
The data that we have presented here have identified specific residues within CHR of gp130 that are important for vIL-6-mediated signal transduction and gp130 binding to effect dimerization of the signal transducer. These residues are in part the same as those previously implicated in IL-6R-mediated binding to and signaling through gp130 by hIL-6. However, our data have established that residues within the CHR C-D loop, in addition to the E-F loop, are involved in functional ligand binding. Thus, alanine substitutions of residues W164, H167, and F169 within the C-D loop (variants PM29, PM31, and PM33) and residues Y190 and F191 within the E-F loop (variants PM16 and PM17) effect significantly reduced vIL-6-induced gp130 signaling, and we have determined for all except Y190 that these residues are important for vIL-6 binding to gp130, as deduced from the inability of the substitution variants to dimerize in response to vIL-6. Interestingly, the Y190A (PM16), in addition to YFV192AAA (PM5), substitution led to ligand-independent (nonfunctional) dimerization, and while vIL-6 can bind the altered gp130 protein when coexpressed with it, presumably primarily through vIL-6 site III interactions with the immunoglobulin domain of gp130, functional dimerization, requiring additionally stable site II interactions with CHR residues, apparently cannot occur. Our investigations of residues of IL-6R required for IL-6R-dependent signaling through gp130.PM5 have identified residues within equivalents of gp130 CHR E-F (S186) and B2-C2 (Y249 and R250/L251) and within the predicted gp130-IL-6R dimerization interface (N230 and H280/D281) that are involved in vIL-6-mediated signaling. The B2-C2 and receptor interface residues, at least, also appear to be involved in vIL-6-induced IL-6R-gp130 complexing. Except for H280/D281, these residues had little effect on hIL-6-mediated signaling or IL-6R-gp130 complexing in our assays, indicating differences in vIL-6 and hIL-6 binding to IL-6R. Interpretative models for our functional and binding data for vIL-6 and the receptor subunit variants are presented in Fig. 7 .
FIG. 7.
Diagrammatic summary of results and their interpretation. (A) Ligand-independent complexing occurs between gp130 proteins containing either YFV192AAA (PM5) or Y190A (PM16) substitutions (Fig. 4); Fc fusions of these proteins are unable to coprecipitate vIL-6 when mixed in vitro but can do so when coexpressed with vIL-6 in transfected cells (Fig. 3). PM5 and PM16 variants of gp130 are unable to form functional signaling complexes either in the presence (Fig. 1B) or absence (37) of vIL-6. Other functionally defective gp130 CHR variants (PM17, PM29, PM31, and PM33) are able to coprecipitate vIL-6, added either exogenously or coexpressed with the sgp130-Fc fusion proteins (Fig. 3) but are unable to undergo ligand-induced dimerization (Fig. 4). (B) Coexpression of IL-6R together with vIL-6 and functionally defective gp130 variants restores their signaling activities (Fig. 2B) (37), presumably by stabilizing ligand-induced gp130 dimerization through formation of hexameric complexes (as occurs for hIL-6). (C) IL-6R variants PM2 and PM5 containing substitutions in residues N230 and H280D281, predicted to occur in the gp130-IL-6R dimerization interface, are unable to complex with gp130 in the presence of vIL-6; Fc fusions of the sIL-6R proteins could not coprecipitate sgp130 (Fig. 6A and B). These results indicate the importance of direct gp130-IL-6R interactions for vIL-6-induced complexing of the receptor subunits. IL-6R variants PM3 and PM4 containing substitutions (Y249A and RL251SI) at positions within the distal CHR domain alsocould not coprecipitate sgp130 in the presence of vIL-6 (Fig. 6A and B); based on previous results from investigations of hIL-6-IL-6R interactions (17, 38) and the positions of these residues within the predicted structure of IL-6R, this is likely to result from effects of the substitutions on interactions of vIL-6 with IL-6R. Our data do not distinguish between the predicted effects of the IL-6R PM3 or PM4 and PM2 or PM5 alterations, and these models are therefore speculative. For hIL-6, only the HD281SV IL-6R variant (PM5) showed significantly reduced ability to support signaling (Fig. 5) and ligand-induced complexing with gp130 (Fig. 6C).
Recently, the crystallographic structure of vIL-6 complexed with gp130 domains 1 to 3 (the immunoglobulin-homology and CHR domains of gp130) has been determined (7). It is significant that these studies identified CHR residues W164, Y190, and F191 as putative vIL-6 interaction sites. Our data show that these residues of gp130 are indeed involved in functional interactions with vIL-6, confirming the importance of the C-D loop, in addition to E-F residues, of CHR in vIL-6 recognition. However, previously published data from this laboratory determined that nonconservative substitutions of putative ligand-interacting residues S251 and V252 (altered in variant gp130.PM7 and predicted to interact with vIL-6 from the crystallographic data [7]) did not affect vIL-6 signaling, ruling out the central involvement of these CHR B2-C2 loop residues in vIL-6 binding (37). Further, substitutions at positions 166 (T) and 187 (S), which potentially could contribute to vIL-6 binding to gp130 (7), did not have major effects on vIL-6 signaling (Fig. 1B). Our data suggest that if such interactions occur, they are relatively minor in comparison to the essential interactions through gp130 residues W164, Y190, and F191. Thus, our data, coupled with the crystallographic data of Chow et al. (7), have identified functionally essential residues of gp130 that are centrally involved in direct, high-affinity interactions with site II of vIL-6.
In the case of IL-6R, interpretation of our data is less clear-cut, since we do not have the benefit of corresponding structural information. However, we can speculate on the basis of the deduced structure of IL-6R (17, 32) that residues Y249 and R250/L251 are involved in functionally important interactions with vIL-6, whereas residues N230 and H280/D281 are involved in stabilization of vIL-6-induced gp130-IL-6R complexing through a receptor subunit dimerization interface. Only the H280/D281 residues were found to be important also for hIL-6-mediated signaling and gp130-IL-6R complexing in our assays, revealing differences in the relative contribution of the IL-6R residues to vIL-6- versus hIL-6-induced functional complex formation.
In summary, our data show for the first time that residues within the C-D (W164, H167, and F169) and E-F (Y190 and F191) loops of gp130 CHR are required for vIL-6 binding to the signal transducer to effect formation of functional signaling complexes and also identify residues within the IL-6R equivalent of gp130 CHR B2-C2 (Y249 and R250/L251) that are involved in IL-6R-dependent vIL-6 signaling (through gp130.PM5) and ligand-induced IL-6R-gp130 complexing. IL-6R residues (N230 and H280/D281) within the predicted IL-6R-gp130 interaction interface were also required for vIL-6-induced receptor subunit complexing and signaling. These data help define the requirements for functional vIL-6-receptor complexing and may be useful in the design of antagonists of this potential contributor to HHV-8 pathogenesis.
Acknowledgments
We thank Yvonne Jones and Oxford University Press for permission to use a modified version of a previously published figure (3) that shows the structure of sgp130.
This work was supported by grants CA-76445 and CA-83519 from the National Institutes of Health.
REFERENCES
- 1.Asou, H., J. W. Said, R. Yang, R. Munker, D. J. Park, N. Kamada, and H. P. Koeffler. 1998. Mechanisms of growth control of Kaposi's sarcoma-associated herpes virus-associated primary effusion lymphoma cells. Blood 91:2475-2481. [PubMed] [Google Scholar]
- 2.Beck, J. T., S. M. Hsu, J. Wijdenes, R. Bataille, B. Klein, D. Vesole, K. Hayden., S. Jagannath, and B. Barlogie. 1984. Alleviation of systemic manifestations of Castleman's disease by monoclonal anti-interleukin-6 antibody. N. Engl. J. Med. 330:602-605. [DOI] [PubMed] [Google Scholar]
- 3.Bravo, J., D. Staunton, J. K. Heath, and E. Y. Jones. 1998. Crystal structure of a cytokine binding region of gp130. EMBO J. 17:1665-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burger, R., F. Neipel, B. Fleckenstein, R. Savino, G. Ciliberto, J. R. Kalden, and M. Gramazki. 1998. Human herpesvirus type 8 interleukin-6 homologue is functionally active on human myeloma cells. Blood 91:1858-1863. [PubMed] [Google Scholar]
- 5.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences are present in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 6.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 265:1865-1869. [DOI] [PubMed] [Google Scholar]
- 7.Chow, D., X. He, A. L. Snow, S. Rose-John, and K. C. Garcia. 2001. Structure of an extracellular gp130 cytokine receptor signalling complex. Science 291:2150-2155. [DOI] [PubMed] [Google Scholar]
- 8.de Vos, A. M., M. Ultsch, and A. A. Kossiakoff. 1992. Human growth hormone and extracellular domain of its receptor: crystal structure of the complex. Science 255:306-312. [DOI] [PubMed] [Google Scholar]
- 9.Foussat, A., J. Wijdenes, L. Bouchet, G. Gaidano, F. Neipel, K. Balabanian, P. Galanaud, J. Couderc, and D. Emille. 1999. Human interleukin-6 is in vivo an autocrine growth factor for human herpesvirus-8-infected malignant B lymphocytes. Eur. Cytokine Netw. 10:501-508. [PubMed] [Google Scholar]
- 10.Guschin, D., N. Rogers, J. Briscoe, B. Witthuhn, D. Watling, F. Horn, S. Pellegrini, K. Yasukawa, P. Heinrich, G. R. Stark, J. N. Ihle, and I. M. Kerr. 1995. A major role for the protein tyrosine kinase JAK1 in the JAK/STAT signal transduction pathway in response to interleukin-6. EMBO J. 14:1421-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammacher, A., R. T. Richardson, J. E. Layton, D. K. Smith, L. J. Angus, D. J. Hilton, N. A. Nicola, J. Wijdenes, and R. J. Simpson. 1998. The immunoglobulin-like module of gp130 is required for signaling by interleukin-6, but not by leukemia inhibitory factor. J. Biol. Chem. 273:22701-22707. [DOI] [PubMed] [Google Scholar]
- 12.Hideshima, T., D. Chauhan, G. Teoh, N. Raje, S. P. Treon, Y.-T. Tai, Y. Shima, and K. C. Anderson. 2000. Characterization of signaling cascades triggered by human interleukin-6 versus Kaposi's sarcoma-associated herpes virus-encoded viral interleukin-6. Clin. Cancer Res. 6:1180-1189. [PubMed] [Google Scholar]
- 13.Horsten, U., G. Müller-Newen, C. Gerhatz, A. Wollmer, J. Wijdenes, P. C. Heinrich, and J. Grötzinger. 1997. Molecular modeling-guided mutagenesis of the extracellular part of gp130 leads to the identification of contact sites in the interleukin-6 (IL-6).IL-6 receptor.gp130 complex. J. Biol. Chem. 272:23748-23757. [DOI] [PubMed] [Google Scholar]
- 14.Ihle, J. N. 1996. STATs: signal transducers and activators of transcription. Cell 84:331-334. [DOI] [PubMed] [Google Scholar]
- 15.Jones, K. D., Y. Aoki, Y. Chang, P. S. Moore, R. Yarchoan, and G. Tosato. 1999. Involvement of interleukin-10 (IL-10) and viral IL-6 in the spontaneous growth of Kaposi's sarcoma herpesvirus-associated infected primary effusion lymphoma cell lines. Blood 94:2871-2879. [PubMed] [Google Scholar]
- 16.Kalai, M., F. A. Montero-Julian, J. Grötzinger, V. Fontaine, P. Vandenbussche, R. Deschuyteneer, A. Wollmer, H. Brailly, and J. Content. 1997. Analysis of the human interleukin-6/human interleukin-6 receptor binding interface at the amino acid level: proposed mechanism of interaction. Blood 89:1319-1333. [PubMed] [Google Scholar]
- 17.Kalai, M., F. A. Montero-Julian, J. Grötzinger, A. Wollmer, D. Morelle, J. Brochier, S. Rose-John, P. C. Heinrich, H. Brailly, and J. Content. 1996. Participation of two Ser-Ser-Phe-Tyr repeats on IL-6 binding sites of human interleukin-6 receptor. Eur. J. Biochem. 238:714-723. [DOI] [PubMed] [Google Scholar]
- 18.Kunkel, T. A. 1985. Rapid and efficient site-directed mutagenesis without phenotypic selection. Proc. Natl. Acad. Sci. USA 82:488-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurth, I., U. Horsten, S. Pflanz, H. Dahmen, A. Kuster, J. Grötzinger, P. C. Heinrich, and G. Müller-Newen. 1999. Activation of the signal transducer glycoprotein 130 by both IL-6 and IL-11 requires two distinct binding epitopes. J. Immunol. 162:1480-1487. [PubMed] [Google Scholar]
- 20.Miles, S. A., A. R. Rezai, J. F. Salazar-Gonzales, M. Vander Mayden, R. H. Stevens, D. M. Logan, R. T. Mitsuyasu, T. Taga, T. Hirano, T. Kishimoto, and O. Martinez-Maza. 1990. AIDS Kaposi's sarcoma-derived cells produce and respond to interleukin 6. Proc. Natl. Acad. Sci. USA 87:4068-4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mizushima, S., and S. Nagata. 1990. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 18:5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Molden, J., Y. Chang, Y. You, P. S. Moore, and M. A. Goldsmith. 1997. A Kaposi's sarcoma-associated herpesvirus-encoded cytokine homolog (vIL-6) activates signaling through the shared gp130 receptor subunit. J. Biol. Chem. 272:19625-19631. [DOI] [PubMed] [Google Scholar]
- 23.Moore, P. S., C. Boshoff, R. A. Weiss, and Y. Chang. 1996. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science 274:1739-1744. [DOI] [PubMed] [Google Scholar]
- 24.Moritz, R. L., L. D. Ward, G. F. Tu, L. J. Fabri, H. Ji, K. Yasukawa, and R. J. Simpson. 1999. The N terminus of gp130 is critical for the formation of the high affinity interleukin-6 receptor complex. Growth Factors 16:265-278. [DOI] [PubMed] [Google Scholar]
- 25.Murakami, M., M. Hibi, N. Nakagawa, T. Nakagawa, K. Yasukawa, K. Yamanishi, T. Taga, and T. Kishimoto. 1993. IL-6-induced homodimerization of gp130 and associated activation of a tyrosine kinase. Science 260:1808-1810. [DOI] [PubMed] [Google Scholar]
- 26.Nador, R. G., E. Cesarman, A. Chadburn, D. B. Dawson, M. Q. Ansari, J. Said, and D. M. Knowles. 1996. Primary effusion lymphoma: a distinct clinical entity associated with Kaposi's sarcoma-associated herpes virus. Blood 88:645-656. [PubMed] [Google Scholar]
- 27.Narazaki, M., B. A. Witthuhn, K. Yoshida, O. Silvennoinen, K. Yasukawa, J. N. Ihle, T. Kishimoto, and T. Taga. 1994. Activation of JAK2 kinase mediated by the interleukin-6 signal transducer gp130. Proc. Natl. Acad. Sci. USA 91:2285-2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicholas, J., V. Ruvolo, W. H. Burns, G. Sandford, X. Wan, D. Ciufo, S. B. Hendrickson, H.-G. Guo, G. S. Hayward, and M. S. Reitz. 1997. Kaposi's sarcoma-associated human herpesvirus-8 encodes homologues of macrophage inflammatory protein-1 and interleukin-6. Nat. Med. 3:287-292. [DOI] [PubMed] [Google Scholar]
- 29.Nishimoto, N., M. Sasai, Y. Shima, M. Nakagawa, T. Matsumoto, T. Shirai, T. Kishimoto, and K. Yoshizaki. 2000. Improvement in Castleman's disease by humanized anti-interleukin-6 receptor antibody therapy. Blood 95:56-61. [PubMed] [Google Scholar]
- 30.Oksenhendler, E., G. Carcelain, Y. Aoki, E. Boulanger, A. Maillard, J.-P. Clauvel, and F. Agbalika. 2000. High levels of human herpesvirus 8 viral load, human interleukin-6, interleukin-10, and C reactive protein correlate with exacerbation of multicentric Castleman's disease in HIV-infected patients. Blood 96:2069-2073. [PubMed] [Google Scholar]
- 31.Osborne, J., P. S. Moore, and Y. Chang. 1999. KSHV-encoded viral IL-6 activates multiple human IL-6 signaling pathways. Hum. Immunol. 60:921-927. [DOI] [PubMed] [Google Scholar]
- 32.Salvati, A. L., A. Lahm, G. Paonessa, G. Ciliberto, and C. Toniatti. 1995. Interleukin-6 (IL-6) antagonism by soluble IL-6 receptor α mutated in the predicted gp130-binding interface. J. Biol. Chem. 270:12242-12249. [DOI] [PubMed] [Google Scholar]
- 33.Schaefer, T. S., L. K. Sanders, and D. Nathans. 1995. Cooperative transcriptional activity of Jun and Stat3β, a short form of Stat3. Proc. Natl. Acad. Sci. USA 92:9097-9101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatum, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, and F. Sigaux. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 35.Stahl, N., T. G. Boulton, T. Farruggella, N. Y. Ip, S. Davis, B. A. Witthuhn, F. W. Quelle, O. Silvennoinen, G. Barbieri, S. Pellegrini, J. N. Ihle, and G. D. Yaneopoulos. 1994. Association and activation of Jak-Tyk kinases by CNTF-LIF-OSM-IL-6 β receptor components. Science 263:92-95. [DOI] [PubMed] [Google Scholar]
- 36.Taga, T., and T. Kishimoto. 1997. Gp130 and the interleukin-6 family of cytokines. Annu. Rev. Immunol. 15:797-819. [DOI] [PubMed] [Google Scholar]
- 37.Wan, X., H. Wang, and J. Nicholas. 1999. Human herpesvirus 8 interleukin-6 (vIL-6) signals through gp130 but has structural and receptor-binding properties distinct from those of human IL-6. J. Virol. 73:8268-8278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yawata, H., K. Yasukawa, S. Natsuka, M. Murakami, K. Yamanishi, M. Hibi, T. Taga, and T. Kishimoto. 1993. Structure-function analysis of human IL-6 receptor: dissociation of amino acid residues required for IL-6-binding and for IL-6 signal transduction through gp130. EMBO J. 12:1705-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoshizaki, K., T. Matsuda, N. Nishimoto, T. Kuritani, L. Taeho, K. Aozasa, T. Nakahata, H. Kawai, H. Togoh, T. Komori, S. Kishimoto, T. Hirano, and T. Kishimoto. 1989. Pathogenic significance of interleukin-6 (IL-6/BSF-2) in Castleman's disease. Blood 74:1360-1367. [PubMed] [Google Scholar]