Abstract
Studies in mutant organisms deficient in RNA interference (RNAi) and related post-transcriptional gene silencing implicated a role for a single class of RNA-dependent RNA polymerases (RdRp). Nevertheless, sequence homologs to these RdRps have not been found in coelomate organisms such as Drosophila or mammals. This lack of homologous sequences does not exclude that an RdRp functions in RNAi in these organisms because an RdRp could be acquired by horizontal transfer from an RNA virus. In fact, such a sequence is found in mice (Aquarius) and we observe that it is expressed in mouse oocytes and early embryos, which exhibit RNAi. We report here that cordycepin, an inhibitor of RNA synthesis, does not prevent Mos double-strand RNA (dsRNA) to target endogenous Mos mRNA in mouse oocytes and that targeting a chimeric Mos–EGFP mRNA with dsRNA to EGFP does not reduce the endogenous Mos mRNA, but does target the chimeric mRNA. These results indicate that an RdRp is not involved in dsRNA-mediated mRNA degradation in mammalian oocytes, and possibly in mammals in general, and therefore that only homologous sequences to the dsRNA are targeted for degradation.
Keywords: RNA interference, RNA-dependent RNA polymerase, mouse oocyte
INTRODUCTION
In most eukaryotic organisms, double-strand RNA (dsRNA) mediates sequence-specific RNA degradation termed RNA interference (RNAi). The mechanism of this pathway has been deciphered mainly in lower organisms (for review, see Hannon 2002; Hutvagner and Zamore 2002). Much less is known, however, about the RNAi pathway in mammals, except that it operates and that dsRNA is processed into 21–23 nucleotide short interfering RNAs (siRNAs) similar to other RNAi-exhibiting organisms (Billy et al. 2001; Svoboda et al., unpubl.). Besides human and mouse Dicer orthologs, none of the genes involved in RNAi in mammals has been identified and cloned (Bernstein et al. 2001; Nicholson and Nicholson 2002; P. Svoboda, P. Stein, and R.M. Schultz, unpubl.). Three components of RNAi, namely, Dicer, argonaute/piwi/paz family, and RNA-dependent RNA polymerases (RdRps), are very well conserved between distant taxa exhibiting RNAi and related phenomena.
RdRp proteins have been identified in a variety of organisms that exhibit RNAi-like phenomena and include EGO-1, RRF-1, and RRF-3 in Caenorhabditis elegans (Smardon et al. 2000; Sijen et al. 2001; Simmer et al. 2002); SGS2/SDE1 in Arabidopsis (Dalmay et al. 2000; Mourrain et al. 2000); QDE-1 in Neurospora (Cogoni and Macino 1999); and RrPA in Dictyostelium (Martens et al. 2002). Two recent reports specified a function for an RdRp activity in RNAi in C. elegans (Sijen et al. 2001) and Drosophila (Lipardi et al. 2001) such that the siRNAs prime an RdRp for RNA synthesis to produce more dsRNA, thereby amplifying the response. There are, however, no known homologs of RdRps involved in RNAi in coelomates whose genomes have been sequenced completely (Drosophila, human, and mouse). Furthermore, an RdRp-based model for “random degradative PCR” in Drosophila (Lipardi et al. 2001) is not consistent with a previous report that the targeted mRNA is cleaved only within the region of homology with the dsRNA in vitro (Zamore et al. 2000) and that RNAi is exon-specific in vivo (Celotto and Graveley 2002). Therefore, the role of RdRp in RNAi in Drosophila, the closest organism to mammals in which RdRp was studied, remains unclear.
Current data imply that RdRps involved in dsRNA-mediated post-transcriptional silencing evolved once. Many of the aforementioned RdRp proteins were independently found mutated in organisms with impaired RNAi or some related silencing mechanism, and they exhibit sequence similarity, indicating that they evolved from a common ancestral gene. Moreover, there is no evidence for another class of RdRps involved in dsRNA-mediated post-transcriptional silencing despite the plausible hypothesis that such an RdRp could be repeatedly acquired horizontally from an RNA virus. Viral RdRps probably participate in RNAi-like antiviral response in plants (Dalmay et al. 2000) and therefore complement endogenous RdRp. Horizontal transfer would explain the lack of RdRp orthologs in Drosophila and mammals if RdRp activity is essential for RNAi. It is also possible that the RdRp component of RNAi became nonessential and was lost during the evolution of true coelomates.
Aquarius, a novel mammalian gene, may be distantly related to RdRps, because it has a conserved RdRp active site motif and a weak homology to the RdRp of murine hepatitis viruses (Sam et al. 1998), suggesting its origin by horizontal transfer. The observation that Aquarius transcripts are detected by RT-PCR in mouse oocytes and preimplantation embryos (P. Svoboda, P. Stein, and R.M. Schultz, unpubl.) led us to test whether or not an RdRp activity is involved in RNAi in mammals and we have obtained two lines of evidence that it is not. After this work was submitted, Schwarz et al. (2002) reached a similar conclusion using Drosophila embryo and HeLa cell extracts.
RESULTS
The “random degradative PCR” hypothesis (Lipardi et al. 2001) predicts that an RdRp transcribes sequences upstream of the targeted region to give rise to dsRNA that is fed back into the RNAi pathway such that sequences in the targeted mRNA, but not present in the original dsRNA, are now targeted. To test this hypothesis in vivo, we designed two experiments. In the first experiment, we tested the efficiency of RNAi in the presence of cordycepin (3′-deoxyadenosine), a chain terminator of RNA elongation. Therefore, if an RdRp is an integral component of the RNAi pathway, cordycepin should inhibit the degradation of the targeted mRNA.
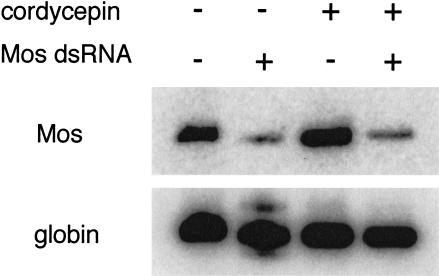
Fully-grown, germinal vesicle (GV) intact mouse oocytes were microinjected with ∼1 × 106 molecules of Mos dsRNA, incubated for 24 h in the presence or absence of 2 mM cordycepin, and then Mos mRNA levels were determined (Fig. 1 ▶). Results of these experiments indicated that Mos mRNA was decreased by ∼80% in both instances, and this decrease was similar to that described previously (Svoboda et al. 2000). Under these conditions, cordycepin, which inhibits polyadenylation in one-cell embryos (Fuchimoto et al. 2001), inhibited [3H]uridine incorporation into acid-insoluble material by 85%–95%.
FIGURE 1.
RT-PCR analysis of Mos mRNA in oocytes injected with Mos dsRNA and treated with cordycepin. The RT-PCR assays were conducted as described previously under conditions that reflect changes in the relative amounts of transcript (Svoboda et al. 2000). Oocytes were microinjected with ∼106 molecules of Mos dsRNA and the relative Mos mRNA levels were assayed 20 h later in either untreated control oocytes or cordycepin-treated (2 mM) oocytes; cordycepin was present from the initial time of oocyte collection. Rabbit globin mRNA was used as an external standard to allow comparison of relative differences in Mos mRNA levels (Svoboda et al. 2000).
In the second experimental approach, we addressed the amplification of sequences upstream of the targeted region by generating a chimeric mRNA composed of Mos sequence at the 5′ end and enhanced green fluorescence protein (EGFP) sequence at the 3′ end (Fig. 2 ▶). We microinjected mouse oocytes with this chimeric transcript and either EGFP siRNA or EGFP dsRNA. This co-injection should generate dsRNA from sequences upstream of EGFP and the subsequent targeting of the endogenous Mos mRNA if an RdRp-dependent amplification mechanism was operative.
FIGURE 2.
Schematic diagram of the chimeric Mos–EGFP mRNA and EGFP dsRNA and siRNA used to target the EGFP sequence. The dsRNA covered the entire coding region of EGFP exactly from the fusion point and the siRNA corresponded to bases 81–103 of the EGFP sequence from the fusion point.
MAP kinase activity, which is an indirect measure of Mos mRNA, was assayed to test the efficiency of Mos mRNA targeting in oocytes microinjected with either chimeric transcript or EGFP dsRNA, or both (Fig. 2 ▶). The translationally dormant Mos mRNA is recruited during oocyte maturation and results in the activation of MAP kinase that can be assayed in individual oocytes. Fully-grown GV-intact oocytes were microinjected with ∼7.5 × 106 molecules of the chimeric mRNA ± EGFP dsRNA and then cultured for 20 h in the presence of 0.2 mM 3-isobutyl-1-methyl-xanthine (IBMX). IBMX inhibits resumption of meiosis, which allows time to target the endogenous Mos mRNA (Svoboda et al. 2000). After 20 h, the oocytes were transferred to IBMX-free medium and matured in vitro for an additional 18 h, after which time the oocytes were assayed for MAP kinase activity.
We observed the normal maturation-associated increase in MAP kinase activity in all microinjected oocytes (Fig. 3 ▶), which indicates that the endogenous Mos mRNA was not efficiently targeted. Experiments using EGFP siRNAi yielded identical negative results (data not shown). In these experiments, we microinjected up to two orders of magnitude more dsRNA and chimeric message than Mos dsRNA needed to observe a consistent decrease of MAP kinase activity in microinjected oocytes. In fact, three orders of magnitude less dsRNA can efficiently induce Mos mRNA degradation (Svoboda et al. 2000). Therefore, an RdRp-mediated RNA synthesis upstream of the targeted sequence was either not efficient or not involved in RNAi in mouse oocytes.
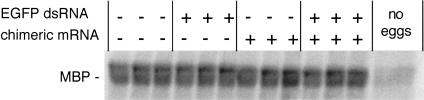
FIGURE 3.
MAP kinase activity in oocytes microinjected with chimeric mRNA and EGFP dsRNA and matured in vitro to metaphase II. MAP kinase activity was assayed in single eggs using myelin basic protein (MBP) as a substrate. Similar results were obtained when EGFP siRNA was injected. The experiment was performed three times and at least 15 individual oocytes were analyzed in each group. Shown are the results of a representative experiment.
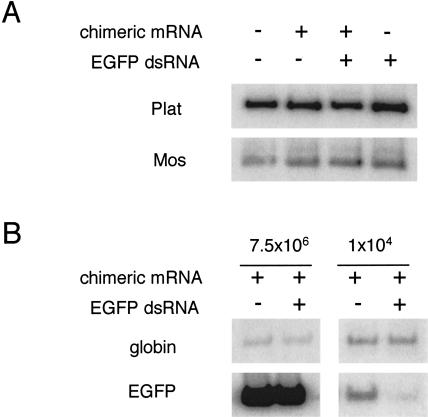
MAP kinase activity, however, is only an indirect measure of Mos mRNA level and therefore it was possible that the Mos mRNA was inefficiently targeted so that no detectable change in MAP kinase activity was observed. Therefore, we measured the relative abundance of Mos transcript in microinjected oocytes to test if there was any detectable change in Mos mRNA level (Fig. 4A ▶). As described above, ∼7.5 × 106 molecules of the chimeric mRNA, dsRNA or both of them were microinjected into fully-grown, GV-intact oocytes that were incubated for 20 h in the presence of IBMX, after which time the relative amount of Mos mRNA was determined as described previously (Svoboda et al. 2000); the presence of fluorescence in the injected oocytes indicated that the injected chimeric mRNA was translated (data not shown). Results of these experiments did not reveal any decrease in the relative abundance of either Mos mRNA or the nontargeted Plat mRNA in any of the microinjected samples.
FIGURE 4.
RT-PCR analysis of Mos mRNA in oocytes injected with EGFP dsRNA and chimeric Mos–EGFP mRNA. (A) Absence of Mos mRNA degradation in oocytes microinjected with chimeric transcript and/or EGFP dsRNA. GV-intact oocytes were microinjected with ∼7.5 × 106 molecules of chimeric transcript and/or EGFP dsRNA and then cultured in the presence of IBMX for 20 h. RNA was then isolated and the relative amount of Mos and Plat transcripts were determined by semi-quantitative RT-PCR as described previously (Svoboda et al. 2000). The amount of the nontargeted Plat PCR product permits relative comparison of microinjected samples (Svoboda et al. 2000). (B) EGFP dsRNA induces degradation of the chimeric transcript. Relative amounts of the chimeric transcript in microinjected oocytes were estimated based on co-injection of rabbit globin mRNA (10 ng/μL). High amounts (∼7.5 × 106 molecules) of microinjected chimeric message do not allow detection of degradation of the chimeric mRNA (left panels), whereas dramatic reduction (right panels) of chimeric mRNA is observed in oocytes microinjected with lower amounts (∼104 molecules).
It was formally possible, however, that the microinjected chimeric mRNA was not targeted by the EGFP siRNA or the EGFP dsRNA and therefore the siRNAs required to prime an RdRp were not produced. The large number of injected chimeric mRNA molecules prohibited detecting any decrease in the abundance of the injected chimeric mRNA by RT-PCR (Fig. 4B ▶). The activity of the RNAi pathway degrades endogenous transcripts at a rate of ∼0.5–1 × 103 transcripts/h (Svoboda et al. 2000). Therefore, in the 20-h incubation, only ∼1% of the injected chimeric mRNA would be degraded. To verify that the chimeric mRNA was targeted by the EGFP dsRNA, we decreased the amount of microinjected chimeric message to ∼104 molecules. In this case, a detectable and specific targeting of the chimeric mRNA was observed (Fig. 4B ▶). The absence of targeting of the endogenous Mos mRNA, therefore, was most likely attributable to the absence or the inefficiency of RdRp amplification.
DISCUSSION
One of the proposed roles for RdRp in RNAi is to amplify the RNAi response (Zamore et al. 2000; Hutvagner and Zamore 2002). Consistent with a loss of RdRp, RNAi in mouse oocytes exhibits slower kinetics and lower efficiency compared with that in C. elegans (Svoboda et al. 2000; Ui-Tei et al. 2000). The absence of an RdRp is probably only one of the reasons that the RNAi efficiency is lower in mouse oocytes, because RNAi in Drosophila is much more efficient and yet Drosophila likely lacks RdRp. We have observed that mouse preimplantation embryo lysates exhibit much lower Dicer activity when compared with equivalent protein amounts of Drosophila embryo extract (data not shown), which may account for the apparent lower activity of the RNAi pathway in mouse oocytes and preimplantation embryos.
The observation that RNAi functions in mouse oocytes in the presence of cordycepin indicated that an RdRp amplification mechanism doesn’t operate. Moreover, the results of the chimeric mRNA injection experiments indicate that even if it does function, it is not of sufficient activity to promote the degradation of the endogenous Mos mRNA. The conclusion that an RdRp amplification mechanism does not function in mouse oocytes is consistent with our previous results. We have shown previously that injection of stoichiometric amounts of Mos dsRNA (∼104 copies) results in targeting the endogenous Mos mRNA for which there are ∼104 copies of the mRNA. Injection of 102 molecules of Mos dsRNA, however, does not result in any detectable decrease in the amount of endogenous Mos mRNA. For Mos to be targeted efficiently by dsRNA produced by an RdRp, about 103−104 dsRNA molecules would be needed. Nevertheless, when stochiometric amounts of chimeric mRNA plus an excess of dsRNA or an excess of chimeric mRNA and dsRNA (∼7.5 × 106 molecules) were provided, we did not detect any decrease in MAP kinase activity or the amount of Mos mRNA, that is, any potential RdRp activity present in the oocyte is not efficient enough to generate enough dsRNA to trigger RNAi. The fact that we do not see targeting under these conditions implies an absence of efficient amplification of sequences upstream to the dsRNA because an endogenous mechanism would likely operate with dsRNA molecules <104.
Our findings have several important implications. First, the mammalian RNAi pathway likely differs from that of C. elegans, plants, and fungi that all employ RdRps to amplify the RNAi response. Our data indicate that in response to dsRNA, an RdRp activity is not detectably involved in mammalian oocytes, and perhaps in mammals in general. An RdRp, however, could have a role in mammalian RNAi to generate trigger dsRNA from “aberrant transcripts” that are targeted by RNAi. Our experiments provide no information regarding such a function, and because the class of “aberrant transcripts” that would trigger RNAi is not characterized and the mechanism is unknown, we cannot test this possibility at this time. Second, it would be interesting to try to enhance the RNAi pathway in mammals by expressing RdRp. In fact, transgenic mice expressing catalytically active tomato RdRp and dsRNA in erythroid tissue were generated but no RNAi effect was observed in this model system (de Wit et al. 2002). It will also be interesting to see if tomato RdRp increases the sensitivity to dsRNA in mouse oocytes, which respond to dsRNA. Last, and most important, the lack of RdRp in mammals permits the specific targeting of other RNAs that share identical or highly similar sequences, as well as the specific targeting of alternatively spliced forms.
MATERIALS AND METHODS
Preparation of EGFP dsRNA and chimeric mRNA
EGFP dsRNA was prepared by in vitro transcription of an EGFP coding sequence in both directions and subsequent annealing. The plasmid used for in vitro transcription was generated by inserting a BamHI/NotI fragment from pEGFP-N2 plasmid (Clontech, CA) into BamHI and NotI sites of pCRII plasmid (Invitrogen, CA). pCRII, which contains SP6 and T7 promoters for in vitro transcription, was linearized using BamHI or NotI. About 5 μg of the template was in vitro-transcribed, annealed, and purified as described previously (Svoboda et al. 2000). This purification included treatment with RNase T1, which removes short single-strand overhangs produced during the generation of dsRNA in plasmids like pCRII. EGFP dsRNA starts immediately at the fusion point with the Mos fragment to assure synthesis of the Mos sequence even in the presence of an RdRp activity with very low processivity.
The plasmid construct to generate the chimeric mRNA was prepared as follows: a 535-bp fragment of Mos was amplified from oocyte cDNA by Pfu polymerase in a PCR reaction using the following primers: forward 5′-GCTCGAGTAATACGACTCAC TATAGGGAGACCATCAAGCAAGTAAACAAG -3′, reverse 5′-G GGATCCAGGGTGATTCCAAAACAGTA-3′. The forward primer contains T-7 promoter sequence and an XhoI site to facilitate cloning, and the reverse primer carries a BamHI site. The XhoI/BamHI-digested PCR product was cloned into XhoI/BamHI sites of pEHGP-N2 plasmid (Clontech) generating pMos/EGFP. The globin 3′ UTR and poly A tail in pMos/EGFP were generated by replacing the NotI/AflII blunted fragment with a NotI/BamHI blunted fragment from pXT7. For in vitro transcription, 2 μg of the plasmid was linearized using SalI. T7 mMessage mMachine kit (Ambion, TX) was used for preparation of the chimeric mRNA according to the manufacturer’s protocol. RNA was purified by phenol/chloroform extraction and dissolved in sterile RNase-free water. The quality and the amount were estimated by spectrophotometry and gel electrophoresis, respectively.
Oocyte collection, microinjection, and culture
Fully grown, GV-intact oocytes were obtained from pregnant mare’s serum gonadotropin (PMSG)-primed 6-week-old female CF-1 mice (Harlan) and freed of attached cumulus cells, as described previously (Schultz et al. 1983). The collection medium was bicarbonate-free minimal essential medium (Earle’s salt) supplemented with polyvinylpyrrolidone (3 mg/mL) and 25 mM HEPES, pH 7.3. Germinal vesicle breakdown was inhibited by including 0.2 mM 3-isobutyl-1-methyl-xanthine (IBMX). The oocytes were transferred to CZB medium (Chatot et al. 1989) containing 0.2 mM IBMX (CZB+IBMX) and cultured in an atmosphere of 5% CO2 in air at 37°C. Oocytes were microinjected in Whitten’s medium (Whitten 1971) containing 5 mM NaHCO3, 15 mM HEPES, 0.01% polyvinyl alcohol (PVA), and 0.2 mM IBMX; the injections were performed as described previously (Kurasawa et al. 1989). In the experiments where the effect of cordycepin was tested, both culture and microinjection of oocytes were done in the presence of 2 mM cordycepin (Sigma). In experiments in which mRNA levels were measured, the oocytes were kept in CZB+ IBMX for 20 h until they were collected and processed for RNA isolation. In experiments in which MAP kinase activity was assayed, microinjected oocytes were cultured in CZB+IBMX for 20 h. They were then washed through 10 drops of IBMX-free CZB and matured in vitro in CZB for 18 h.
RNA isolation and semi-quantitative RT-PCR
RNA isolation and semiquantitative RT-PCR were performed as described previously (Svoboda et al. 2000). Mos primers used in the chimeric mRNA experiments were from outside the Mos sequence in the chimeric message (Svoboda et al. 2001). Mos primers and PCR conditions in the cordycepin experiments were corresponding to the targeted region as described previously (Svoboda et al. 2000).
MAP kinase assay
MAP kinase activity, assayed by ability to phosphorylate myelin basic protein (MBP), was determined in individual oocytes as described previously (Svoboda et al. 2000). This enzyme activity, which increases during oocyte maturation, serves as an indirect measure of the levels of MOS protein (Svoboda et al. 2000).
[3H]Uridine incorporation
Oocytes were cultured in CZB containing 1 mCi/mL [5,6-3H]uridine (32 Ci/mmole, Amersham) for 2 h in the presence of 2 mM cordycepin or its absence, after which the oocytes were processed for acid-insoluble and acid-soluble radioactivity as described previously (Brower et al. 1981). Percent incorporation was calculated as the ratio of acid-insoluble cpm/(acid-insoluble cpm + acid-soluble cpm).
Acknowledgments
The authors thank Greg Hannon for providing the EGFP siRNA (GCCCUCGCCGGACACGCUGAAC and UCAG CGUGUCCGG CGAGGGCGA). This work was supported by a grant from the NIH (HD 22681 to R.M.S.). Portions of this work are being submitted by P. Svoboda in partial fulfillment for the Ph.D. requirements at the University of Pennsylvania.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2860603.
REFERENCES
- Bernstein, E., Caudy, A.A., Hammond, S.M., and Hannon, G.J. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366. [DOI] [PubMed] [Google Scholar]
- Billy, E., Brondani, V., Zhang, H., Muller, U., and Filipowicz, W. 2001. Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc. Natl. Acad. Sci. 98: 14428–14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brower, P.T., Gizang, E., Boreen, S.M., and Schultz, R.M. 1981. Biochemical studies of mammalian oogenesis: Synthesis and stability of various classes of RNA during growth of the mouse oocyte in vitro. Dev. Biol. 86: 373–383. [DOI] [PubMed] [Google Scholar]
- Celotto, A.M. and Graveley, B.R. 2002. Exon-specific RNAi: A tool for dissecting the functional relevance of alternative splicing. RNA 8: 718–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatot, C.L., Ziomek, C.A., Bavister, B.D., Lewis, J.L., and Torres, I. 1989. An improved culture medium supports development of random-bred 1-cell mouse embryos in vitro. J. Reprod. Fertil. 86: 679–688. [DOI] [PubMed] [Google Scholar]
- Cogoni, C. and Macino, G. 1999. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature 399: 166–169. [DOI] [PubMed] [Google Scholar]
- Dalmay, T., Hamilton, A., Rudd, S., Angell, S., and Baulcombe, D.C. 2000. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101: 543–553. [DOI] [PubMed] [Google Scholar]
- de Wit, T., Grosveld, F., and Drabek, D. 2002. The tomato RNA-directed RNA polymerase has no effect on gene silencing by RNA interference in transgenic mice. Transgenic Res. 11: 305–310. [DOI] [PubMed] [Google Scholar]
- Fuchimoto, D., Mizukoshi, A., Schultz, R.M., Sakai, S., and Aoki, F. 2001. Posttranscriptional regulation of cyclin A1 and cyclin A2 during mouse oocyte meiotic maturation and preimplantation development. Biol. Reprod. 65: 986–993. [DOI] [PubMed] [Google Scholar]
- Hannon, G.J. 2002. RNA interference. Nature 418: 244–251. [DOI] [PubMed] [Google Scholar]
- Hutvagner, G. and Zamore, P.D. 2002. RNAi: Nature abhors a double-strand. Curr. Opin. Genet. Dev. 12: 225–232. [DOI] [PubMed] [Google Scholar]
- Kurasawa, S., Schultz, R.M., and Kopf, G.S. 1989. Egg-induced modifications of the zona pellucida of mouse eggs: Effects of microinjected inositol 1,4,5-trisphosphate. Dev. Biol. 133: 295–304. [DOI] [PubMed] [Google Scholar]
- Lipardi, C., Wei, Q., and Paterson, B.M. 2001. RNAi as random degradative PCR: siRNA primers convert mRNA into dsRNAs that are degraded to generate new siRNAs. Cell 107: 297–307. [DOI] [PubMed] [Google Scholar]
- Martens, H., Novotny, J., Oberstrass, J., Steck, T.L., Postlethwait, P., and Nellen, W. 2002. RNAi in Dictyostelium: The role of RNA-directed RNA polymerases and double-stranded RNase. Mol. Biol. Cell 13: 445–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain, P., Beclin, C., Elmayan, T., Feuerbach, F., Godon, C., Morel, J.B., Jouette, D., Lacombe, A.M., Nikic, S., Picault, N., et al. 2000. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101: 533–542. [DOI] [PubMed] [Google Scholar]
- Nicholson, R.H. and Nicholson, A.W. 2002. Molecular characterization of a mouse cDNA encoding Dicer, a ribonuclease III ortholog involved in RNA interference. Mamm. Genome 13: 67–73. [DOI] [PubMed] [Google Scholar]
- Sam, M., Wurst, W., Kluppel, M., Jin, O., Heng, H., and Bernstein, A. 1998. Aquarius, a novel gene isolated by gene trapping with an RNA-dependent RNA polymerase motif. Dev. Dyn. 212: 304–317. [DOI] [PubMed] [Google Scholar]
- Schultz, R.M., Montgomery, R.R., and Belanoff, J.R. 1983. Regulation of mouse oocyte meiotic maturation: implication of a decrease in oocyte cAMP and protein dephosphorylation in commitment to resume meiosis. Dev. Biol. 97: 264–273. [DOI] [PubMed] [Google Scholar]
- Schwarz, D.S., Hutvágner, G., Haley, B., and Zamore, P.D. 2002. Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Mol. Cell 10: 537–548. [DOI] [PubMed] [Google Scholar]
- Sijen, T., Fleenor, J., Simmer, F., Thijssen, K.L., Parrish, S., Timmons, L., Plasterk, R.H., and Fire, A. 2001. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107: 465–476. [DOI] [PubMed] [Google Scholar]
- Simmer, F., Tijsterman, M., Parrish, S., Koushika, S., Nonet, M., Fire, A., Ahringer, J., and Plasterk, R. 2002. Loss of the putative RNA-directed RNA Polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr. Biol. 12: 1317–1319. [DOI] [PubMed] [Google Scholar]
- Smardon, A., Spoerke, J.M., Stacey, S.C., Klein, M.E., Mackin, N., and Maine, E.M. 2000. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr. Biol. 10: 169–178. [DOI] [PubMed] [Google Scholar]
- Svoboda, P., Stein, P., Hayashi, H., and Schultz, R.M. 2000. Selective reduction of dormant maternal mRNAs in mouse oocytes by RNA interference. Development 127: 4147–4156. [DOI] [PubMed] [Google Scholar]
- Svoboda, P., Stein, P., and Schultz, R.M. 2001. RNAi in mouse oocytes and preimplantation embryos: Effectiveness of hairpin dsRNA. Biochem. Biophys. Res. Comm. 287: 1099–1104. [DOI] [PubMed] [Google Scholar]
- Ui-Tei, K., Zenno, S., Miyata, Y., and Saigo, K. 2000. Sensitive assay of RNA interference in Drosophila and Chinese hamster cultured cells using firefly luciferase gene as target. FEBS Lett. 479: 79–82. [DOI] [PubMed] [Google Scholar]
- Whitten, W.K. 1971. Nutrient requirements for the culture of preimplantation mouse embryo in vitro. Adv. Biosci. 6: 129–139. [Google Scholar]
- Zamore, P.D., Tuschl, T., Sharp, P.A., and Bartel, D.P. 2000. RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101: 25–33. [DOI] [PubMed] [Google Scholar]