Abstract
MicroRNAs (miRNAs) represent a new class of noncoding RNAs encoded in the genomes of plants, invertebrates, and vertebrates. MicroRNAs regulate translation and stability of target mRNAs based on (partial) sequence complementarity. Although the number of newly identified miRNAs is still increasing, target mRNAs of animal miRNAs remain to be identified. Here we describe 31 novel miRNAs that were identified by cloning from mouse tissues and the human Saos-2 cell line. Fifty-three percent of all known mouse and human miRNAs have homologs in Fugu rubripes (pufferfish) or Danio rerio (zebrafish), of which almost half also have a homolog in Caenorhabditis elegans or Drosophila melanogaster. Because of the recurring identification of already known miRNAs and the unavoidable background of ribosomal RNA breakdown products, it is believed that not many more miRNAs may be identified by cloning. A comprehensive collection of miRNAs is important for assisting bioinformatics target mRNA identification and comprehensive genome annotation.
Keywords: microRNA, hairpin RNA
INTRODUCTION
MicroRNAs (miRNAs) represent a class of noncoding RNAs encoded in the genomes of plants and invertebrate and vertebrate animals. Mature miRNAs are ∼21 nt long and excised from 60- to 80-nt double-stranded RNA foldbacks (dsRNA hairpins) by Dicer RNase III (Grishok et al. 2001; Hutvágner et al. 2001; Ketting et al. 2001; Park et al. 2002; Reinhart et al. 2002). The processing reaction is generally asymmetric, and only one of the strands from the hairpin is accumulating and is referred to as the mature miRNA (Lee et al. 1993; Reinhart et al. 2000; Lagos-Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001). The hairpins themselves are excised from longer primary transcripts, some of which have intron/exon pre-mRNA structure, whereby the miRNA precursors are located in either exon or intron sequence (Lagos-Quintana et al. 2001, 2002; Lau et al. 2001; Lee et al. 2002). Clusters of miRNAs are also expressed as long primary transcripts (Lee et al. 2002). Trimming of the primary transcripts to the miRNA precursor hairpin takes place in the nucleus (Lee et al. 2002). Subsequently, the hairpin is exported to the cytoplasm, and Dicer excises the mature miRNA (Lee et al. 2002).
Our understanding of miRNA function originates from studies of the developmentally regulated miRNAs lin-4 and let-7 in Caenorhabditis elegans (for reviews, see Slack and Ruvkun 1997; Ambros 2000; Rougvie 2001; Pasquinelli and Ruvkun 2002). Biochemical characterization of the lin-4 miRNA of C. elegans revealed that miRNAs are able to block protein synthesis after transcription initiation or possibly regulate protein stability (Wightman-etal-1993; Olsen and Ambros 1999; Seggerson et al. 2002). The lin-4 miRNA recognizes complementary sequences within the 3′-UTR of its targets by forming an imperfect, bulged RNA duplex structure. The evolutionarily conserved let-7 miRNA of C. elegans also regulates the expression of downstream genes based on partial sequence complementarity between the miRNA and the target 3′-UTR (Reinhart et al. 2000). The targets for lin-4 and let-7 miRNAs were first identified genetically, and only after visual inspection of the sequences was the partial sequence complementarity detected. Intriguingly, the 5′-ends of some Drosophila melanogaster miRNAs were found to be complementary to 3′-UTR sequence motifs already known to mediate negative posttranscriptional regulation (Lai 2002). Furthermore, single nucleotide polymorphisms in the 3′-UTR of certain mRNAs have been associated with increased risk for certain types of cancer, indicating possible interference with miRNA regulation (Conne et al. 2000; Hayward 2000; Mendell and Dietz 2001; Kiyohara et al. 2002).
In contrast to animal miRNAs, almost all identified plant miRNAs show near-perfect complementarity to subsets of mRNAs that are the likely targets for these miRNAs (Rhoades et al. 2002). Arabidopsis thaliana miR-171 , which is also conserved in Nicotiana benthamiana and rice, was shown experimentally to specifically target SCL6-III and SCL-IV mRNAs for degradation (Llave et al. 2002b), indicating that plant miRNAs can act as small interfering RNAs (siRNAs) that guide target RNA cleavage (Elbashir et al. 2001) rather than mediating translational control. At the same time, it was observed that endogenous human let-7 miRNA is able to guide target RNA cleavage as long as a target RNA was introduced that carried a fully complementary segment of let-7 miRNA (Hutvágner and Zamore 2002). Argonaute proteins that are associated with single-stranded siRNAs in the target RNA-cleaving RNA-induced silencing complex (RISC; Hammond et al. 2001; Martinez et al. 2002) were found to be associated with mature miRNAs (Mourelatos et al. 2002; Schwarz et al. 2002). Together, these data indicate that miRNAs and siRNAs are present in similar RNP complexes, but that their function depends on the specific base-pairing structure formed between the small RNA and the target mRNA. Prediction of the targets of animal miRNAs remains difficult because these miRNAs show no more matches to mRNAs than random sequences, so that it must be assumed that few, if any, of the miRNAs will recognize their respective targets with near-perfect complementarity (Rhoades et al. 2002).
To date, nearly 200 miRNAs have been described from C. elegans, D. melanogaster, human, mouse, and A. thaliana (Lagos-Quintana et al. 2001, 2002; Lau et al. 2001; Lee and Ambros 2001; Llave et al. 2002a; Mourelatos et al. 2002; Park et al. 2002; Reinhart et al. 2002). However, not all tiny RNAs qualify as miRNAs, and care has to be taken not to annotate breakdown products of abundant RNAs like rRNAs or tRNAs as miRNAs. Important criteria for a small RNA to qualify as an miRNA are 20–30-bp hairpin precursor structures, phylogenetic conservation in multiple species, and experimental evidence for their existence by cDNA cloning or Northern blot analysis (see Annotation guide for miRNAs, V. Ambros, B. Bartel, D.P. Bartel, C.B. Burge, J.C. Carrington, X. Chen, G. Dreyfuss, S.R. Eddy, S. Griffiths-Jones, M. Matzke, et al., in prep.). The 21-nt sequences that have been cloned but do not match to the genome of the respective organism or a closely related species do not qualify as miRNAs.
Small cloned RNAs may originate from transcribed repetitive heterochromatic sequences, recently identified in the fission yeast (Reinhart and Bartel 2002). These small RNAs, also referred to as heterochromatic siRNAs (Reinhart and Bartel 2002), provide a link to chromatin silencing and DNA methylation, likely by nucleating histone H3 lysine 9 methylation (Hall et al. 2002; Volpe et al. 2002). Other small RNAs that were derived from retro-elements have been detected in D. melanogaster (Elbashir et al. 2001) and A. thaliana (Hamilton et al. 2002; Llave et al. 2002a).
MicroRNAs are expressed constitutively or in a developmentally regulated manner, and may accumulate in certain tissues while being excluded from others (Lee et al. 1993; Reinhart et al. 2000; Lagos-Quintana et al. 2001, 2002; Lau et al. 2001; Lee and Ambros 2001; Llave et al. 2002a; Park et al. 2002; Reinhart et al. 2002). To function, the miRNAs need to be coexpressed with their respective target mRNAs. We have generated a comprehensive collection of mammalian miRNAs by cloning them from various mouse tissues and human cell lines to provide a starting point for their functional characterization. This report describes another 31 novel miRNAs, more than half of which are conserved between other vertebrate animals, and discusses some features of the more interesting miRNAs. We have now reached what we believe are the limits of miRNA isolation from somatic tissues or cells by conventional RNA isolation and cloning methods.
RESULTS AND DISCUSSION
We have continued the characterization of mouse miRNAs by cloning the ∼21-nt RNAs from lung, kidney, skin, testis, ovary, thymus, spinal cord, and eye of 18.5-week-old adult mice using the method described previously (Lagos-Quintana et al. 2001, 2002). Additionally, the miRNAs expressed in the human osteoblast sarcoma cell line Saos-2 were cloned. In total, ∼600 clones of miRNAs were obtained, 91% of which represent previously identified miRNAs and 9% as-yet unidentified miRNAs. The newly identified miRNAs are listed in Table 1 ▶. The novel miRNAs were identified in the various genome databases by BLAST sequence analysis. It was then confirmed that the flanking sequences can base-pair to the cloned sequence, forming the typical hairpin precursor structures (data not shown). When human and mouse orthologous miRNAs differed slightly in sequence, the species name is indicated before the miRNA name. In all, 31 novel miRNAs were identified. From all the newly identified mouse miRNAs, only one could not be identified within the human genome. Vice versa, three miRNAs cloned from human could not be identified in the mouse genome sequence, although one of them was identified in zebrafish. Because the majority (91%) of sequenced miRNAs matched to previously identified miRNAs, we believe that we have reached near-complete identification of miRNA genes expressed in somatic mouse or human cells. Although we cannot exclude the possibility that certain rare cell types or tissues, such as stem cells, may express distinct sets of miRNAs, it remains impractical to identify these miRNAs by the conventional cloning protocols, because the amount or the purity of RNA that can be obtained is insufficient.
TABLE 1.
Novel mouse and human miRNA sequences
| Number of clones | ||||||||||||
| miRNA | Sequence (5′ to 3′) | ht | ln | lv | sp | kd | sk | ts | ov | thy | eye | S |
| miR-10b | CCCUGUAGAACCGAAUUUGUGU | 1 | 3 | |||||||||
| miR-129b | CUUUUUGCGGUCUGGGCUUGUU | 1 | 1 | 1 | ||||||||
| miR-181 | AACAUUCAACGCUGUCGGUGAGU | 1 | 1 | 2 | ||||||||
| miR-182 | UUUGGCAAUGGUAGAACUCACA | 1 | ||||||||||
| miR-183 | UAUGGCACUGGUAGAAUUCACUG | 1 | ||||||||||
| miR-184 | UGGACGGAGAACUGAUAAGGGU | 2 | ||||||||||
| miR-185 | UGGAGAGAAAGGCAGUUC | 1 | ||||||||||
| miR-186 | CAAAGAAUUCUCCUUUUGGGCUU | 1 | 1 | |||||||||
| miR-187 | UCGUGUCUUGUGUUGCAGCCGG | 1 | ||||||||||
| miR-188 | CAUCCCUUGCAUGGUGGAGGGU | 1 | ||||||||||
| miR-189 | GUGCCUACUGAGCUGACAUCAGU | 1 | ||||||||||
| miR-190 | UGAUAUGUUUGAUAUAUUAGGU | 2 | ||||||||||
| miR-191 | CAACGGAAUCCCAAAAGCAGCU | 2 | 1 | |||||||||
| miR-192 | CUGACCUAUGAAUUGACA | 2 | 1 | |||||||||
| miR-193 | AACUGGCCUACAAAGUCCCAG | 1 | ||||||||||
| miR-194 | UGUAACAGCAACUCCAUGUGGA | 1 | ||||||||||
| miR-195 | UAGCAGCACAGAAAUAUUGGC | 2 | 1 | 1 | ||||||||
| miR-196 | UAGGUAGUUUCAUGUUGUUGG | 1 | 1 | |||||||||
| miR-197 | UUCACCACCUUCUCCACCCAGC | 3 | ||||||||||
| miR-198 | GGUCCAGAGGGGAGAUAGG | 2 | ||||||||||
| miR-199-s | CCCAGUGUUCAGACUACCUGUU | 2 | ||||||||||
| miR-199-as | UACAGUAGUCUGCACAUUGGUU | 1 | ||||||||||
| miR-200a | UAACACUGUCUGGUAACGAUG | 1 | ||||||||||
| miR-200b | UAAUACUGCCUGGUAAUGAUGAC | 2 | 1 | |||||||||
| miR-201 | UACUCAGUAAGGCAUUGUUCU | 1 | ||||||||||
| miR-202 | AGAGGUAUAGCGCAUGGGAAGA | 1 | ||||||||||
| miR-203 | UGAAAUGUUUAGGACCACUAG | 2 | 1 | |||||||||
| miR-204 | UUCCCUUUGUCAUCCUAUGCCUG | 1 | ||||||||||
| miR-205 | UCCUUCAUUCCACCGGAGUCUG | 1 | ||||||||||
| miR-206 | UGGAAUGUAAGGAAGUGUGUGG | 2 | ||||||||||
| miR-207 | GCUUCUCCUGGCUCUCCUCCCUC | 1 | ||||||||||
| miR-208 | AUAAGACGAGCAAAAAGCUUGU | 1 | ||||||||||
The number of clones identified from the indicated mouse tissues or the human osteoblast sarcoma cell line Saos-2 is presented. Abbreviations: ht, heart; ln, lung; lv, liver; sp, spleen; kd, kidney; sk, skin; ts, testis; ov, ovary; thy, thymus; S, Saos-2. The suffixes -s and -as indicate that the RNA is derived from either the 5′ half or the 3′ half of an miRNA precursor. Comprehensive tables for mouse and human miRNAs, miRNA precursors, and their respective tissue distribution are provided online at http://www.mpibpc.gwdg.de/abteilungen/100/105/mirna.html.
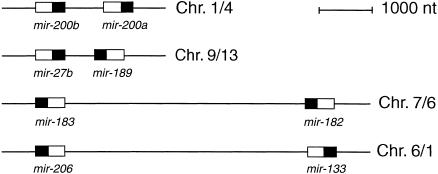
One of the features of miRNAs is that they can occur in clusters that are coexpressed (Lagos-Quintana et al. 2001, 2002; Lau et al. 2001; Lee et al. 2002). Seven of the newly identified miRNAs are found in clusters (Fig. 1 ▶), and the distance between the clustered miRNAs varies between 500 and 5000 bp.
FIGURE 1.
MicroRNA gene clusters. The precursor structure is indicated as a box, and the location of the miRNA within the precursor is shown in black. The clusters are transcribed from left to right. To the right, the chromosome location is indicated for human/mouse. The cluster of mir-183 and mir-182 is also conserved in zebrafish.
It was previously noted that some miRNAs were strongly enriched in specific tissues, such as miR-1 variants in heart, miR-122 variants in liver, or miR-124 variants in brain (Lee and Ambros 2001; Lagos-Quintana et al. 2002). Here, we find that the miRNA profiles of eye and spinal cord were very similar to those observed from brain tissues, in which miR-124 variants are strongly enriched, presumably reflecting the high neuronal cell content. None of the other examined tissues or cell lines showed a strong dominance of a specific miRNA. Variants of let-7 miRNA were fairly abundant in all examined somatic tissues or cells. Similar to the ubiquitous let-7 distribution, we find miR-16, miR-26a, miR-27a, and miR-143a in almost all somatic cells or tissues. miR-21 is also ubiquitously expressed but excluded from neuronal tissue. The exclusion of miR-21 and the presence of neuron-specific miR-124 variants from neuronal tissue may contribute to neuronal cell specification.
To establish tissue culture systems for analyzing miRNA function, we have previously cloned the miRNAs from the cervical cancer cell line HeLa SS3 (Lagos-Quintana et al. 2001). To examine the miRNA profiles between different cell lines in culture, we cloned the miRNAs from the osteoblast sarcoma cell line Saos-2, another commonly used laboratory cell line. Six new miRNAs (miR-129b, miR-186, miR-196 to miR-199 ) were cloned from this cell line, albeit at such low frequency that it cannot be stated with certainty that these miRNAs are expressed cell-line-specifically. The expression pattern of the most abundant miRNAs (let-7a , miR-16, and miR-21) was very similar between Saos-2 and HeLa cells.
We have analyzed the genomic position of 54 mouse (60 human) miRNAs and find that 31 (33) are localized in intergenic regions. Of the remaining miRNAs, 11 (13) are found in sense orientation within introns of coding transcripts, 7 (7) in sense orientation within introns of noncoding genes, and 5 (7) in the reverse orientation within an intronic region. Therefore, miRNAs are either transcribed from their own promoters or derived from a pre-mRNA that frequently codes for an additional gene product. An example of an intronic miRNA is miR-186 : Human and mouse miR-186 are both located in intron 8 of the pre-mRNA of the zinc finger protein 265; both introns are ∼2 kb, but only the miRNA precursors and ∼50 nt of flanking sequences are conserved between the two species. An example of intronic miRNAs located within a non-protein-coding transcript are miR-15a and miR-16 : These miRNAs are clustered and located within the intron of a transcribed region referred to as LEU2. The LEU2 gene lies within the deleted minimal region (DMR) of the B-cell chronic lymphocytic leukemia (B-CLL) tumor suppressor locus (Bullrich et al. 2001; Migliazza et al. 2001), for which the tumor suppressor gene yet has to be identified; miR-15a and miR-16 are strong candidates.
Some miRNAs are found in interesting genomic locations. For example, miR-10 , which was originally identified by cloning from D. melanogaster embryos (Lagos-Quintana et al. 2001), is located in the Hox gene cluster between Dfd (Hox4) and Scr (Hox5). miR-10 also appears to be conserved in the Hox cluster of Anopheles gambiae (mosquito) and Tribolium castaneum (red flour beetle). miR-10 is also preserved in the Hox clusters of zebrafish, pufferfish, mouse, and human. In mouse and human the mir-10 gene has been duplicated and is now present in the form of two variants. miR-10a is located between Hox4b and Hox5b in mouse and human, whereas miR-10b is found within intron 4 of Hox4d in mouse, and between Hoxd4 and Hoxd8 in human. Considering the spatial and temporal colinearity of Hox gene expression and the positional conservation of miR-10 within the Hox gene clusters, it is conceivable that miR-10 is important for regulating developmental events. The evolutionary conserved miRNAs, such as miR-10 , are probably the most interesting starting points for functional analysis of this vast gene family (Table 2 ▶).
TABLE 2.
miRNA and miRNA families conserved between vertebrates and invertebrates
| miRNA family | H. sap. | M. mus. | F. rub. | D. rerio | D. mel. | C. elegans |
| let-7/miR-98 | p c n | p c n | p | p n | p c n | p c n |
| lin-4/miR-125 | p | p c n | p | p n | p c n | |
| miR-1 | p c n | p c n | p | p c n | p c n | |
| miR-4/75/79/131 | p c | p c | p | p | p c n | p |
| miR-7 | p | p | p | p c n | ||
| miR-8/141 | p | p c | p c n | |||
| miR-9 | p | p c | p | p c n | ||
| miR-10/99/100 | p c | p c | p | p | p c n | |
| miR-29/83 | p c | p c | p | p | p c | |
| miR-31/73 | p c | p | p | p | ||
| miR-34 | p | p | p c | |||
| miR-25/92 | p c | p c | p | p | p c | |
| miR-124 | p | p c n | p | p | p | p |
| miR-133 | p | p c | p | p | ||
| miR-184 | p | p c | p | p | p |
p) A predicted miRNA identified by BLAST searching of a cloned miRNA in another species. The flanking sequences of the predicted miRNA have the capacity to fold with the miRNA into a hairpin precursor. (c) The miRNA was cloned from the respective organism. (n) The miRNA expression was confirmed by Northern blot analysis (Lagos-Quintana et al. 2001, 2002; Lee et al. 2002). Abbreviations: H. sap., Homo sapiens; M. mus., Mus musculus; F. rub. Fugu rubripes; D. rerio, Danio rerio; D. mel., Drosophila melanogaster; C. elegans, Caenorhabditis elegans.
In summary, 31 novel mammalian miRNAs were identified in an effort to provide a comprehensive list of these regulatory molecules. The identification of functional RNA genes is important for obtaining fully annotated genome sequences in order to assist researchers determined to identify genes linked to a particular disease and to understand posttranscriptional regulation of gene expression.
MATERIALS AND METHODS
Total RNA isolation, cloning, and bioinformatics analysis
RNA preparation and cloning of miRNAs was performed as last described (Lagos-Quintana et al. 2002). Total RNA from Saos-2 cells was isolated from a cell line that was stably transfected with an inducible p53 gene. No significant differences in miRNA composition were observed between RNA preparations from p53-induced or p53-non-induced cells. For sequence searches and analysis, we used the Ensembl database (http://www.ensembl.org), NCBI online resources (http://www.ncbi.nlm.nih.gov), the DOE Joint Genome Institute Web site (http://www.jgi.doe.gov), the UCSC Genome Bioinformatics Web site (http://genome.ucsc.edu), and the mfold Web server (http://www.bioinfo.rpi.edu/applications/mfold/old/rna).
Acknowledgments
We thank S.M. Elbashir and W. Lendeckel for the preparation of mouse tissues, G. Dowe for some of the sequencing, A. Aravin and T. Fischer for bioinformatics assistance and program development, Y. Dorsett and J. Martinez for critical comments on the manuscript, and R. Lührmann for support. This work was funded by a Bundesministerium für Bildung und Forschung (BMBF) Biofuture grant, number 0311856.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘‘advertisement’’ in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.2146903.
REFERENCES
- Ambros, V. 2000. Control of developmental timing in Caenorhabditis elegans. Curr. Opin. Genet. Dev. 10: 428–433. [DOI] [PubMed] [Google Scholar]
- Bullrich, F., Fujii, H., Calin, G., Mabuchi, H., Negrini, M., Pekarsky, Y., Rassenti, L., Alder, H., Reed, J.C., Keating, M.J., et al. 2001. Characterization of the 13q14 tumor suppressor locus in CLL: Identification of ALT1, an alternative splice variant of the LEU2 gene. Cancer Res. 61: 6640–6648. [PubMed] [Google Scholar]
- Conne, B., Stutz, A., and Vassalli, J.D. 2000. The 3′ untranslated region of messenger RNA: A molecular ‘hotspot’ for pathology? Nat. Med. 6: 637–641. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Lendeckel, W., and Tuschl, T. 2001. RNA interference is mediated by 21 and 22 nt RNAs. Genes & Dev. 15: 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok, A., Pasquinelli, A.E., Conte, D., Li, N., Parrish, S., Ha, I., Baillie, D.L., Fire, A., Ruvkun, G., and Mello, C.C. 2001. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106: 23–34. [DOI] [PubMed] [Google Scholar]
- Hall, I.M., Shankaranarayana, G.D., Noma, K.I., Ayoub, N., Cohen, A., and Grewal, S.I. 2002. Establishment and maintenance of a heterochromatin domain. Science 297: 2232–2237. [DOI] [PubMed] [Google Scholar]
- Hamilton, A., Voinnet, O., Chappell, L., and Baulcombe, D. 2002. Two classes of short interfering RNA in RNA silencing. EMBO J. 21: 4671–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond, S.M., Boettcher, S., Caudy, A.A., Kobayashi, R., and Hannon, G.J. 2001. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293: 1146–1150. [DOI] [PubMed] [Google Scholar]
- Hayward, N. 2000. New developments in melanoma genetics. Curr. Oncol. Rep. 2: 300–306. [DOI] [PubMed] [Google Scholar]
- Hutvágner, G. and Zamore, P.D. 2002. A microRNA in a multiple-turnover RNAi enzyme complex. Science 297: 2056–2060. [DOI] [PubMed] [Google Scholar]
- Hutvágner, G., McLachlan, J., Bálint, É., Tuschl, T., and Zamore, P.D. 2001. A cellular function for the RNA interference enzyme Dicer in small temporal RNA maturation. Science 93: 834–838. [DOI] [PubMed] [Google Scholar]
- Ketting, R.F., Fischer, S.E., Bernstein, E., Sijen, T., Hannon, G.J., and Plasterk, R.H. 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes & Dev. 15: 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyohara, C., Otsu, A., Shirakawa, T., Fukuda, S., and Hopkin, J. 2002. Genetic polymorphisms and lung cancer susceptibility: A review. Lung Cancer 37: 241. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Lendeckel, W., and Tuschl, T. 2001. Identification of novel genes coding for small expressed RNAs. Science 294: 853–858. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana, M., Rauhut, R., Yalcin, A., Meyer, J., Lendeckel, W., and Tuschl, T. 2002. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12: 735–739. [DOI] [PubMed] [Google Scholar]
- Lai, E.C. 2002. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet. 30: 363–364. [DOI] [PubMed] [Google Scholar]
- Lau, N.C., Lim, L.P., Weinstein, E.G., and Bartel, D.P. 2001. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294: 858–862. [DOI] [PubMed] [Google Scholar]
- Lee, R.C. and Ambros, V. 2001. An extensive class of small RNAs in Caenorhabditis elegans. Science 294: 862–864. [DOI] [PubMed] [Google Scholar]
- Lee, R.C., Feinbaum, R.L., and Ambros, V. 1993. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854. [DOI] [PubMed] [Google Scholar]
- Lee, Y., Jeon, K., Lee, J.T., Kim, S., and Kim, V.N. 2002. microRNA maturation: Stepwise processing and subcellular localization. EMBO J. 21: 4663–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave, C., Kasschau, K.D., Rector, M.A., and Carrington, J.C. 2002a. Endogenous and silencing-associated small RNAs in plants. Plant Cell 14: 1605–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llave, C., Xie, Z., Kasschau, K.D., and Carrington, J.C. 2002b. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297: 2053–2056. [DOI] [PubMed] [Google Scholar]
- Martinez, J., Patkaniowska, A., Urlaub, H., Lührmann, R., and Tuschl, T. 2002. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110: 563–574. [DOI] [PubMed] [Google Scholar]
- Mendell, J.T. and Dietz, H.C. 2001. When the message goes awry: Disease-producing mutations that influence mRNA content and performance. Cell 107: 411–414. [DOI] [PubMed] [Google Scholar]
- Migliazza, A., Bosch, F., Komatsu, H., Cayanis, E., Martinotti, S., Toniato, E., Guccione, E., Qu, X., Chien, M., Murty, V.V., et al. 2001. Nucleotide sequence, transcription map, and mutation analysis of the 13q14 chromosomal region deleted in B-cell chronic lymphocytic leukemia. Blood 97: 2098–2104. [DOI] [PubMed] [Google Scholar]
- Mourelatos, Z., Dostie, J., Paushkin, S., Sharma, A., Charroux, B., Abel, L., Rappsilber, J., Mann, M., and Dreyfuss, G. 2002. miRNPs: A novel class of ribonucleoproteins containing numerous microRNAs. Genes & Dev. 16: 720–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, P.H. and Ambros, V. 1999. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 216: 671–680. [DOI] [PubMed] [Google Scholar]
- Park, W., Li, J., Song, R., Messing, J., and Chen, X. 2002. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12: 1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli, A.E. and Ruvkun, G. 2002. Control of developmental timing by microRNAs and their targets. Annu. Rev. Cell Dev. Biol. 18: 495–513. [DOI] [PubMed] [Google Scholar]
- Reinhart, B.J. and Bartel, D.P. 2002. Small RNAs correspond to centromere heterochromatic repeats. Science 297: 1831. [DOI] [PubMed] [Google Scholar]
- Reinhart, B.J., Slack, F.J., Basson, M., Pasquinelli, A.E., Bettinger, J.C., Rougvie, A.E., Horvitz, H.R., and Ruvkun, G. 2000. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403: 901–906. [DOI] [PubMed] [Google Scholar]
- Reinhart, B.J., Weinstein, E.G., Rhoades, M.W., Bartel, B., and Bartel, D.P. 2002. MicroRNAs in plants. Genes & Dev. 16: 1616–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades, M.W., Reinhart, B.J., Lim, L.P., Burge, C.B., Bartel, B., and Bartel, D.P. 2002. Prediction of plant microRNA targets. Cell 110: 513–520. [DOI] [PubMed] [Google Scholar]
- Rougvie, A.E. 2001. Control of developmental timing in animals. Nat. Rev. Genet. 2: 690–701. [DOI] [PubMed] [Google Scholar]
- Schwarz, D.S., Hutvágner, G., Haley, G., and Zamore, P.D. 2002. siRNAs function as guides, not primers, in the RNAi pathway in Drosophila and human cells. Mol. Cell (in press). [DOI] [PubMed]
- Seggerson, K., Tang, L., and Moss, E.G. 2002. Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev. Biol. 243: 215–225. [DOI] [PubMed] [Google Scholar]
- Slack, F. and Ruvkun, G. 1997. Temporal pattern formation by heterochronic genes. Annu. Rev. Genet. 31: 611–634. [DOI] [PubMed] [Google Scholar]
- Volpe, T.A., Kidner, C., Hall, I.M., Teng, G., Grewal, S.I., and Martienssen, R.A. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837. [DOI] [PubMed] [Google Scholar]
- Wightman, B., Ha, I., and Ruvkun, G. 1993. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75: 855–862. [DOI] [PubMed] [Google Scholar]