Abstract
Import of core histones into the nucleus is a prerequisite for their deposition onto DNA and the assembly of chromatin. Here we demonstrate that nucleosome assembly protein 1 (Nap1p), a protein previously implicated in the deposition of histones H2A and H2B, is also involved in the transport of these two histones. We demonstrate that Nap1p can bind directly to Kap114p, the primary karyopherin/importin responsible for the nuclear import of H2A and H2B. Nap1p also serves as a bridge between Kap114p and the histone nuclear localization sequence (NLS). Nap1p acts cooperatively to increase the affinity of Kap114p for these NLSs. Nuclear accumulation of histone NLS–green fluorescent protein (GFP) reporters was decreased in Δnap1 cells. Furthermore, we demonstrate that Nap1p promotes the association of the H2A and H2B NLSs specifically with the karyopherin Kap114p. Localization studies demonstrate that Nap1p is a nucleocytoplasmic shuttling protein, and genetic experiments suggest that its shuttling is important for maintaining chromatin structure in vivo. We propose a model in which Nap1p links the nuclear transport of H2A and H2B to chromatin assembly.
Keywords: histone/importin/karyopherin/nuclear transport/nucleosome assembly
Introduction
Chromatin assembly is a complex process occurring in all eukaryotes, which integrates DNA replication, transcription, DNA repair and the regulation of cell cycle progression (reviewed in Verreault, 2000). The process of chromatin assembly involves the initial deposition of a tetramer of histones H3 and H4 onto DNA, followed by the addition of H2A–H2B heterodimers to complete the histone octamer (Adams and Kamakaka, 1999; Verreault, 2000). A host of chromatin assembly factors and histone chaperones have been identified that facilitate nucleosome assembly, but the mechanisms governing how they function and their coordinate regulation remain poorly understood (Ito et al., 1997; Adams and Kamakaka, 1999; Verreault, 2000).
Several chromatin assembly factors (CAFs), such as CAF-1, nuclear assembly protein 1 (Nap1p) and replication coupled assembly factor (RCAF), are conserved from yeast to man (Ito et al., 1996; Verreault et al., 1996; Tyler et al., 1999). Both CAF-1 and RCAF associate preferentially with newly synthesized H3–H4, whereas Nap1p appears to associate preferentially with H2A and H2B (Ishimi et al., 1987; Verreault et al., 1996; Chang et al., 1997; Tyler et al., 1999; Mosammaparast et al., 2001). The precise function of Nap1p is not known, but one possibility is that it serves as a histone chaperone involved in the nuclear import of histones H2A and H2B (Ito et al., 1996; Chang et al., 1997). Previous work has demonstrated that in Drosophila, Nap1p is imported into the nucleus specifically during S phase, coincident with chromatin assembly (Ito et al., 1996). Nap2, a human Nap1p homolog, is predominantly nuclear in cells during S phase but is excluded from the nucleus during the rest of the cell cycle (Rodriguez et al., 1997). Though Nap1p has histone chaperone activity in vitro, it is generally not sufficient for the formation of periodic nucleosome arrays seen in vivo (Ito et al., 1996). Evidence suggests that chromatin assembly is a multistep process, where Nap1p is involved in an intermediate step during the process (Ito et al., 1996). Recent work also suggests that Nap1p may facilitate transcriptional activation by playing a role in chromatin remodeling (Walter et al., 1995; Ito et al., 2000; Shikama et al., 2000).
While biochemical approaches clearly suggest that Nap1p has a role in chromatin assembly, genetic studies reveal other functions for this protein. A knockout of Nap1l2, a Nap1p homolog in mouse, causes embryonic lethality due to defects in neurulation (Rogner et al., 2000). Surprisingly, genetic experiments in yeast have shown that Nap1p has a role in cell cycle progression during G1 and mitosis (Kellogg and Murray, 1995; Zimmerman and Kellogg, 2001). Δnap1 strains have a delay in mitosis and develop a long bud phenotype (Kellogg and Murray, 1995). Nap1p is associated with a number of cellular factors such as the B-type cyclin, Clb2p, the Gin4p kinase and Sda1p, consistent with its role in cell cycle progression (Kellogg et al., 1995; Altman and Kellogg, 1997; Zimmerman and Kellogg, 2001).
Histones, like other nuclear cargo, are imported into the nucleus by association with specific nuclear import receptors, named karyopherins (Kaps) or importins (Pemberton et al., 1998; Wozniak et al., 1998; Macara, 2001). These receptors recognize nuclear localization signals (NLSs) in their cargos and traverse the nuclear envelope by associating with the components of the nuclear pore complex (Bagley et al., 2000; Rout et al., 2000; Macara, 2001). Upon entering the nucleus, the Kap associates with Ran, a GTPase which regulates Kap–cargo association (Macara, 2001). The association of the Kap with RanGTP results in the release of cargo, and the Kap cycles back to the cytoplasm (Macara, 2001). While it is clear that a number of Kaps are involved in histone import, specific Kaps serve as the primary or predominant receptors in yeast (Mosammaparast et al., 2001, 2002). Histones H2A and H2B utilize Kap114p as their primary import receptor (Mosammaparast et al., 2001), while Kap123p serves this role for histones H3 and H4 (Mosammaparast et al., 2002). Kap121p appears to function as a secondary receptor for H2A–H2B as well as H3–H4 (Mosammaparast et al., 2001, 2002). Recent evidence also suggests that numerous Kaps may also be involved in the import of histones in mammalian cells (Muhlhausser et al., 2001). The mechanism via which specific Kaps appear to be ‘preferred’ transport receptors for different histones, or other import cargos, remains unclear. Furthermore, any functional link between the nuclear import of the core histones and their downstream deposition to form chromatin remains unexplored.
Here we demonstrate that Nap1p serves as a cofactor in the nuclear import of histones H2A and H2B. We demonstrate that yeast Nap1p is a nucleocytoplasmic shuttling protein that associates directly with Kap114p, and that a Kap114p–Nap1–histone complex can be formed. The affinity of Kap114p for the NLSs of H2A and H2B is increased in the presence of Nap1p. Interestingly, Nap1p also serves as a Kap specificity determinant by inhibiting the association of the H2A and H2B NLSs with Kaps other than Kap114p. We propose a model in which Nap1p links the nuclear import of H2A and H2B to their deposition on DNA to form chromatin.
Results
Nap1p interacts with Kap114p both in vitro and in vivo
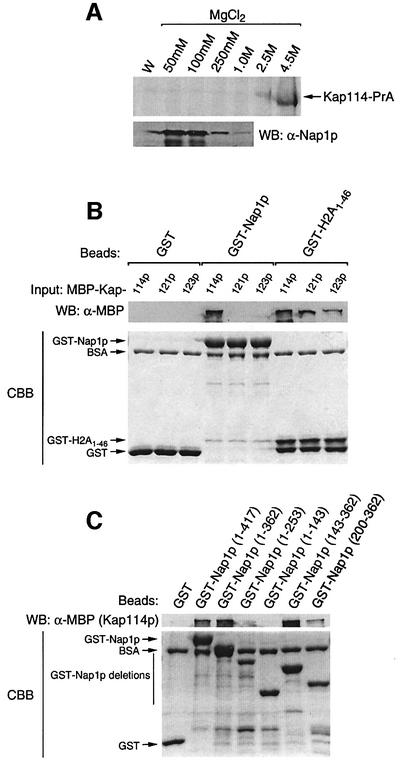
We previously isolated Nap1p with histones H2A and H2B from yeast cytosol with Kap114p (Mosammaparast et al., 2001). To test the association between Kap114p and Nap1p, cytosol from a Kap114-PrA strain was incubated with IgG–Sepharose; associated proteins were eluted with a MgCl2 step gradient and analyzed by SDS–PAGE. As we previously reported, Kap114-PrA elutes predominantly at the 4.5 M MgCl2 fraction and associated proteins elute at lower MgCl2 concentrations (Figure 1A; Mosammaparast et al., 2001). The same fractions were blotted and probed with a Nap1p-specific antibody. A strong signal corresponding to Nap1p indicated that this protein is associated with Kap114p in yeast cytosol (Figure 1A).
Fig. 1. Kap114p binds to Nap1p both in vivo and in vitro. (A) Kap114-PrA and associated proteins were isolated from cytosol. Kap114-PrA was visualized by Coomassie Blue staining, while Nap1p was visualized by western blotting (WB) and probing with an anti-Nap1p antibody. W, final wash fraction. (B) GST (1.9 µM), GST–Nap1p (675 nM) or GST–H2A1–46 (1.6 µM) were immobilized on glutathione–Sepharose, and binding of MBP–Kap114p, MBP–Kap121p or MBP–Kap123p (60 nM of each as input) was tested. Bound fractions were separated by SDS–PAGE, and the bottom portion of the gel was stained with Coomassie Blue (CBB). Bound MBP–Kaps were visualized by western blotting and probing with an anti-MBP antibody. (C) Different portions of Nap1p fused to GST (1 µM) were tested for binding to MBP–Kap114p as in (B).
To test for a direct interaction between Nap1p and Kap114p, an in vitro binding assay using recombinant proteins was performed. GST, GST–Nap1p or GST– H2A1–46 (the H2A NLS) were incubated with maltose-binding protein (MBP)-tagged Kap114p, Kap121p and Kap123p in the presence of glutathione–Sepharose. We previously reported that each of these Kaps interacts with GST–H2A1–46 (Mosammaparast et al., 2001) and, as expected, binding was observed for all three MBP–Kaps with GST–H2A1–46 but not with GST, indicating that the recombinant Kaps are functional (Figure 1B). However, only MBP–Kap114p was able to bind to immobilized GST–Nap1p (Figure 1B). We also determined that Kap95p did not bind to immobilized GST–Nap1p (data not shown). This suggested a direct and specific interaction between Nap1p and Kap114p.
Previous studies have shown that there are several domains within Nap1p which are required for histone binding (Fujii-Nakata et al., 1992). We investigated which domains of Nap1p were required for binding to Kap114p. GST–Nap1p deletions were purified and tested for binding to Kap114p. This analysis revealed that the minimal domain of Nap1p tested that was sufficient for binding to Kap114p was located within residues 200–362 (Figure 1C). This same domain of Nap1p is not sufficient for histone binding and chromatin assembly (Fujii-Nakata et al., 1992) and, although the binding domains may overlap, it is possible that Nap1p could bind to Kap114p and histones simultaneously.
Kap114p contains distinct sites for Nap1p and histone binding
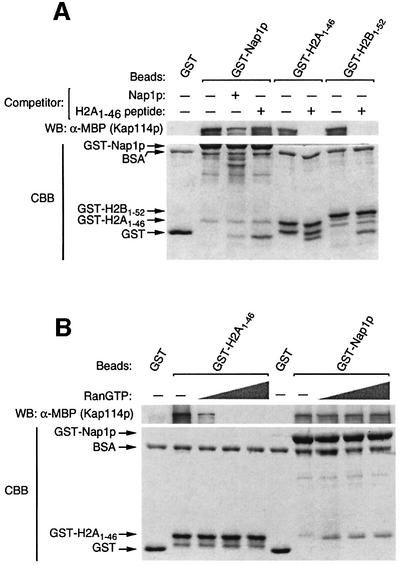
As Kap114p bound H2A, H2B and Nap1p, we wanted to determine whether each interacted via a common cargo-binding site within Kap114p. If Kap114p has a common binding site for Nap1p and histones, the binding of Nap1p and histones to Kap114p should be competitive. When a recombinant peptide consisting of the H2A NLS (H2A1–46) was pre-incubated with MBP–Kap114p, the binding of this Kap to immobilized GST–H2A1–46 was inhibited significantly, as was the binding to GST–H2B1–52 (the H2B NLS; Figure 2A). This suggested that the H2A- and H2B-binding sites on Kap114p are overlapping. However, the binding of MBP–Kap114p to GST–Nap1p was not inhibited by this peptide (Figure 2A). As a control, we showed that tag-free Nap1p could reduce the binding of MBP–Kap114p to GST–Nap1p (Figure 2A). These results suggested that Kap114p contains at least two distinct sites for binding to Nap1p and histones.
Fig. 2. Kap114p contains a distinct, RanGTP-insensitive binding site for Nap1p. (A) GST, GST–Nap1p, GST–H2A1–46 or GST–H2B1–52 (0.5 µM of each) were immobilized on glutathione–Sepharose. Binding of Kap114p (120 nM) was then tested in the presence or absence of free Nap1p (4 µM) or an H2A1–46 NLS peptide (15 µM) as competitor. Bound fractions were analyzed as described. (B) MBP–Kap114p (60 nM) was pre-incubated with or without human Ran Q69L (at 10, 20 or 50 µM), and binding was tested with immobilized GST (0.75 µM), GST–H2A1–46 (0.75 µM) or GST–Nap1p (0.65 µM). Bound fractions were analyzed as described.
Nap1p association with Kap114p is insensitive to RanGTP
The above experiments suggested that Nap1p may be a cargo for Kap114p, and as such this interaction should be sensitive to RanGTP addition, as with the Kap114p–histone NLS interactions (Mosammaparast et al., 2001). MBP–Kap114p was pre-incubated with increasing concentrations of RanQ69L, a GTPase- deficient Ran mutant, and binding to immobilized GST–H2A1–46 or GST–Nap1p was tested. The interaction between MBP–Kap114p and GST–H2A1–46 was highly sensitive to RanGTP (Figure 2B). However, the association of MBP–Kap114p with GST–Nap1p was insensitive to RanGTP, even at 50 µM RanGTP (Figure 2B). It was possible that Nap1p competed with RanGTP for binding to Kap114p, leading to the apparent insensitivity to RanGTP of the Kap114p–Nap1p interaction. To eliminate this possibility, we tested the binding of MBP–Kap114p to RanQ69L in the presence of increasing concentrations of Nap1p, and found no discernible change in RanGTP binding even at very high concentrations of Nap1p (data not shown). Thus, the association of Kap114p with Nap1p occurs at a distinct, RanGTP-insensitive site.
Nap1p can bridge the interaction between Kap114p and histone NLSs
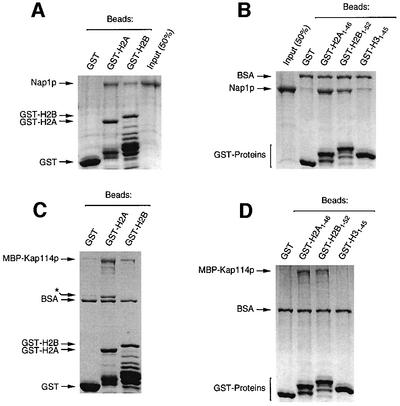
We hypothesized that Nap1p may not merely be a cargo for Kap114p, but may play a direct role in histone import. However, we previously showed that Kap114p can bind directly to the H2A and H2B NLSs, and that Nap1p is not required for this interaction (Mosammaparast et al., 2001). To understand its role further, we investigated how Nap1p interacted with H2A and H2B. We demonstrated that purified Nap1p was capable of binding to GST–H2A and –H2B (Figure 3A). Published studies using trypsinized histones indicated that the histone N-termini are required for the Nap1p–histone interaction (McQuibban et al., 1998). We tested whether Nap1p associated directly with the N-terminal domains on H2A and H2B. We found that Nap1p associated with the N-termini of H2A and H2B but not with H3 (Figure 3B). Kap114p and Nap1p were found to have similar binding sites within the histones, as the N-termini of H2A and H2B were also sufficient for Kap114p binding (Figure 3C and D).
Fig. 3. Nap1p and Kap114p bind directly to the N-termini of H2A and H2B. (A) Immobilized GST (4 µM), GST–H2A (1 µM) or GST–H2B (4 µM) were incubated with recombinant Nap1p (500 nM). The purified GST–histones, especially GST–H2B, contained degradation products that retained the GST tag. Bound fractions were analyzed by SDS–PAGE and Coomassie staining. (B) Analysis of Nap1p binding to GST–histone N-terminal tails (1 µM of each) as in (A). (C) MBP–Kap114p (200 nM) binding to GST–histones as in (A). The asterisk indicates a bacterial protein, probably the product of the dnaK gene, which co-purified with GST–H2A. (D) MBP–Kap114p (200 nM) binding to GST–histone N-terminal tails as in (B).
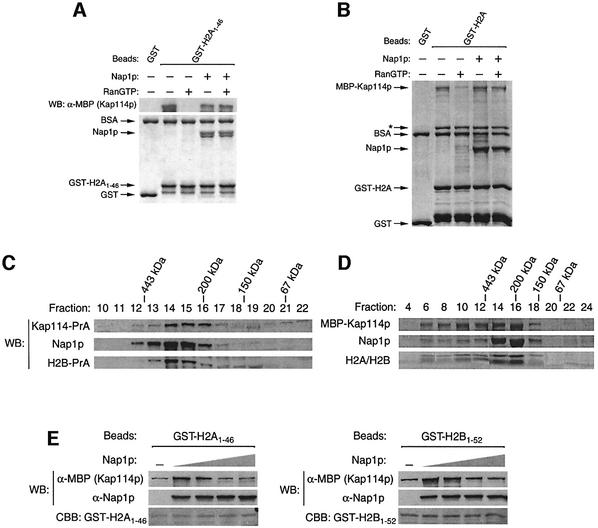
If Nap1p and Kap114p recognize the same domain within H2A and H2B, they could compete for binding to these histones. However, as Nap1p can bind directly to Kap114p, it may bridge the interaction between this Kap and the histone NLSs. If this was true, in light of our previous results, the association of Kap114p with an immobilized histone NLS would become insensitive to RanGTP in the presence of Nap1p. We tested MBP–Kap114p binding to immobilized GST–H2A1–46 in the presence or absence of RanGTP and Nap1p. As expected, in the absence of Nap1p, the binding of this Kap was sensitive to RanGTP (Figure 4A). However, in the presence of Nap1p, Kap114p binding to H2A was insensitive to RanGTP (Figure 4A). This same observation was made using full-length GST–H2A (Figure 4B). This suggested that Kap114p can bind to the histone NLS via Nap1p, and thus Nap1p serves to bridge the Kap–histone interaction. This model does not preclude the possibility that Kap114p also contacts the histones directly in this co-complex.
Fig. 4. A co-complex consisting of Kap114p, Nap1p and histones. (A) MBP–Kap114p was pre-incubated with or without human Ran Q69L (20 µM), and binding was then tested with immobilized GST or GST–H2A1–46 (0.75 µM) in the presence or absence of Nap1p (1.5 µM). Unless indicated by WB (western blot), samples were analyzed by Coomassie staining. (B) Analysis of MBP–Kap114p binding as in (A) except that GST–H2A (full-length; 1 µM) was substituted for GST–H2A1–46, Ran Q69L was used at 10 µM and Nap1p was used at 1 µM. (C) Yeast cytosol was prepared from a Kap114-PrA/H2B-PrA strain and analyzed by FPLC using a Superdex 200 column. The positions of protein standards are indicated above. (D) Recombinant MBP–Kap114p (0.75 µM), Nap1p (0.75 µM) and H2A–H2B dimer (3.6 µM) were pre-incubated at 4°C for complex formation and analyzed by FPLC as in (C). (E) GST–H2A1–46 (50 nM) or GST–H2B1–52 (100 nM) were immobilized on glutathione–Sepharose, and binding to MBP–Kap114p (100 nM) was tested in the presence or absence of recombinant Nap1p (20, 100 and 500 nM, or 1.25 µM).
As a Kap114p–Nap1p–histone co-complex was suggested from the above experiments, we tested whether these proteins co-fractionated in vivo. Yeast cytosol extract was analyzed by size exclusion chromatography using a Superdex 200 column. Our analysis revealed that a portion of Kap114p, Nap1p and H2B co-elute in fractions 14–16, which would correspond to a complex of ∼250 kDa (Figure 4C). Indeed, using only recombinant proteins, we observed a similar pattern of fractionation consistent with a ∼250 kDa complex consisting of Kap114p, Nap1p and H2A/H2B (Figure 4D).
Nap1p increases the affinity of Kap114p for H2A and H2B
If Nap1p was bridging this interaction, it may serve to increase the affinity of Kap114p for the histone NLSs. We initially performed binding assays with Kap114p and limiting amounts of immobilized GST–H2A1–46 and GST–H2B1–52, in the absence of Nap1p, to determine the minimal concentration of immobilized NLS needed to detect binding to Kap114p (Figure 4E; data not shown). This ensured that we were in the linear concentration range of substrate/NLS binding for Kap114p. When recombinant Nap1p was present in these binding reactions, the amount of Kap114p bound to the NLSs was significantly increased (Figure 4E). This suggested that Nap1p could facilitate the binding of Kap114p to these NLSs. At higher concentrations of Nap1p, this stimulation was not seen, most probably because distinct Kap114p–Nap1p and Nap1p–histone NLS complexes were formed which no longer bound cooperatively.
Decreased nuclear accumulation of H2A and H2B NLSs in a Δnap1 strain
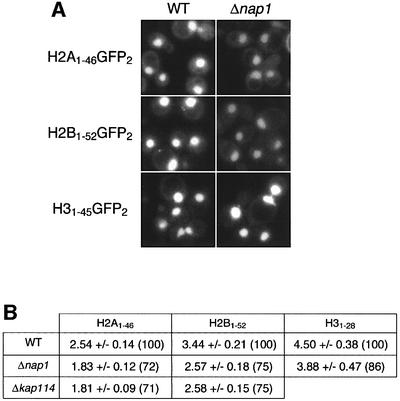
To determine whether Nap1p plays a role in H2A and H2B nuclear transport, we compared localization of the H2A and H2B NLS–green fluorescent protein (GFP) reporters in wild-type and an isogenic Δnap1 strain. NLS reporters were used instead of full-length histone constructs to avoid heterodimerization, which could mask a subtle phenotype. A small decrease in the nuclear accumulation of these two reporters was seen in the Δnap1 strain (Figure 5). Quantitation of the mean nuclear to cytoplasmic ratios (N:C) of GFP fluorescence intensities from digital images confirmed that this decrease in nuclear accumulation is similar to that seen in a Δkap114 strain (Mosammaparast et al., 2001; Figure 5A). The N:C ratios observed for H2A and H2B in the Δnap1 strain relative to wild-type were 72 and 75%, respectively (Figure 5). Analysis of the H3 NLS–GFP reporter revealed a smaller defect in the Δnap1 strain (N:C ratio of 86% compared with wild-type; Figure 5), suggesting that Nap1p probably does not play as important a role in the nuclear transport of this histone.
Fig. 5. In vivo analysis of histone NLS–GFP reporters. (A) Wild-type or Δnap1 cells containing the indicated reporter were analyzed by fluorescence microscopy. (B) Mean nuclear:cytoplasmic fluorescence intensity ratios ± SD were determined from digital images using the indicated combinations of strains and NLS–GFP reporters. Numbers in parentheses indicate the mean nuclear:cytoplasmic ratio relative to wild-type cells expressed as a percentage.
Nap1p determines karyopherin specificity for histones H2A and H2B in vitro
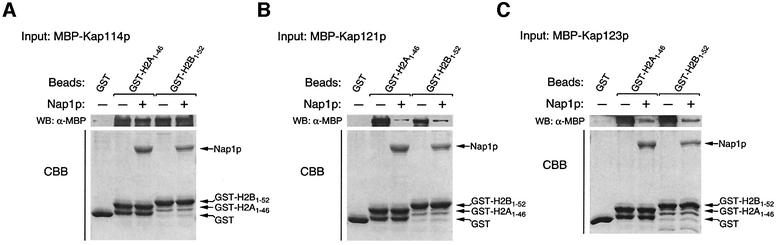
We had demonstrated previously that in vitro a number of other, more abundant Kaps, such as Kap121p or Kap123p, can also associate with the histone NLSs (Mosammaparast et al., 2001). It was possible that Nap1p, by its ability to interact only with Kap114p, could direct histones H2A and H2B specifically to this Kap. To test this, we bound recombinant Kap114p, Kap121p or Kap123p to immobilized H2A and H2B NLSs in the presence or absence of Nap1p (Figure 6). In agreement with our previous reports (Mosammaparast et al., 2001), all three Kaps associated with the histone NLSs in the absence of Nap1p. However, unlike Kap114p (Figure 6A), the binding of Kap121p and Kap123p was significantly inhibited in the presence of Nap1p (Figure 6B and C). These results suggested that Nap1p can serve as a chaperone which only promotes Kap114p association with H2A/H2B.
Fig. 6. Nap1p is a karyopherin specificity determinant in vitro. Binding of recombinant MBP–Kap114p (A), MBP–Kap121p (B) or MBP–Kap123p (C) (120 nM each) was tested on immobilized GST, GST–H2A1–46 and GST–H2B1–52 (1.6 µM) in the presence or absence of recombinant Nap1p (0.8 µM). The bound fractions were analyzed as described.
Nap1p is a karyopherin specificity determinant in vivo
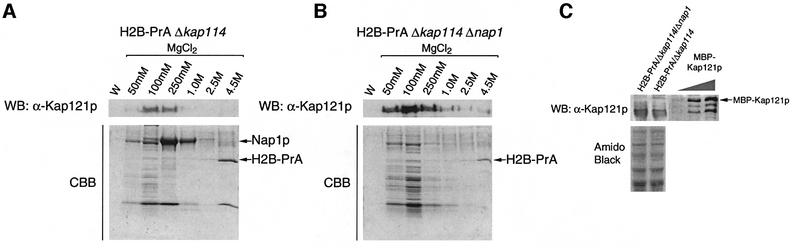
If Nap1p is responsible for Kap114p specificity in vivo, the amount of a Kap such as Kap121p associated with either of these histones in cytosol might be increased in the absence of Nap1p. We therefore constructed two yeast strains, an H2B-PrA/Δkap114 strain and an H2B-PrA/Δkap114/Δnap1 strain. We had shown previously that in the former strain some Kap121p is associated with H2B-PrA in cytosol (Mosammaparast et al., 2001). Using equal amounts of cytosol extract, we found that the amount of Kap121p associated with H2B-PrA is increased significantly in the absence of Nap1p, although the total amount of Kap121p present in these two strains is similar (Figure 7). This suggested that in the absence of Nap1p, Kap121p could bind more effectively to the histone NLSs.
Fig. 7. Nap1p is a karyopherin specificity determinant in vivo. H2B-PrA/Δkap114 and H2B-PrA/Δkap114/Δnap1 strains were created. (A and B) H2B-PrA and associated proteins were isolated from each strain using equivalent amounts of cytosol (standardized for total protein), separated by SDS–PAGE and stained with Coomassie Blue or western blotted with an anti-Kap121p antibody. W, final wash fraction. Fractions in (A) and (B) were run on the same gel and blot and thus represent the same staining and western blot. (C) Western blot analysis of whole-cell extracts from these strains probed for Kap121p. Recombinant MBP–Kap121p (1.5, 3.0 or 6.0 nmol) was used as a reference.
Nap1p contains sequences that mediate both nuclear and cytoplasmic localization
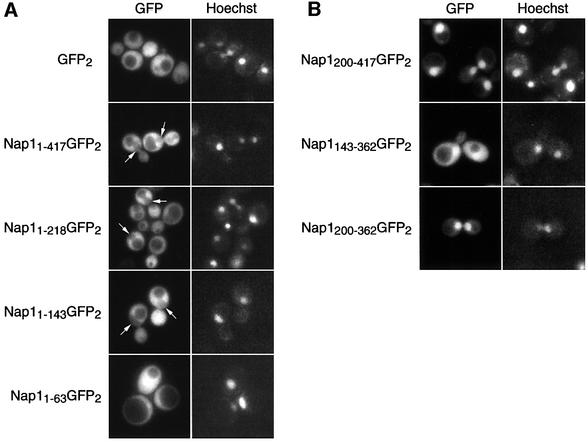
The above studies suggested that Nap1p is a cofactor in the Kap114p-mediated nuclear import of H2A and H2B. Various reports have suggested that there are both nuclear and cytoplasmic pools of Nap1p, although in yeast Nap1p has been shown to be predominantly cytoplasmic (Kellogg et al., 1995). We determined the localization of Nap1p in vivo, by expressing Nap1p as a fusion to two tandem copies of GFP (GFP2) in wild-type yeast. We found that Nap1p–GFP2 (1–417) was predominantly cytoplasmic and appeared to be partially excluded from the nucleus at steady state (Figure 8A; see arrowed ‘holes’). This contrasted with GFP2 alone, which was distributed equally between the nuclear and cytoplasmic compartments (Figure 8A).
Fig. 8. Distinct domains of Nap1p mediate cytoplasmic or nuclear localization of Nap1p. (A and B) Nap1p (Nap11–417) or various deletions of Nap1p (as indicated by amino acid numbers) were expressed as fusions to GFP2 in wild-type yeast. The coincident Hoechst staining is shown, and the positions of nuclei are arrowed.
It was possible that yeast Nap1p is a nucleocytoplasmic shuttling protein. We set out to determine whether different domains of Nap1p specify import and export by deletion analysis. Expression of amino acids 1–218 or 1–143 of Nap1p resulted in predominantly cytoplasmic localization, as was seen with full-length Nap1p (Figure 8A). However, the localization of residues 1–63 of Nap1p resembled that of GFP2 alone. The N-terminal domain was removed and fragments corresponding to amino acids 200–417, 143–362 and 200–362 were tested. These fragments specified dramatic nuclear accumulation of the reporter (Figure 8B), and demonstrated that yeast Nap1p could enter the nucleus. These results suggested that the N-terminus specified export, whilst a central/C-terminal domain contained sequences specific for Nap1p import. The import domain overlapped with the Kap114p-binding domain, implicating Kap114p in this process (amino acids 200–362; see Figure 1C).
Nap1p contains a leucine-rich NES
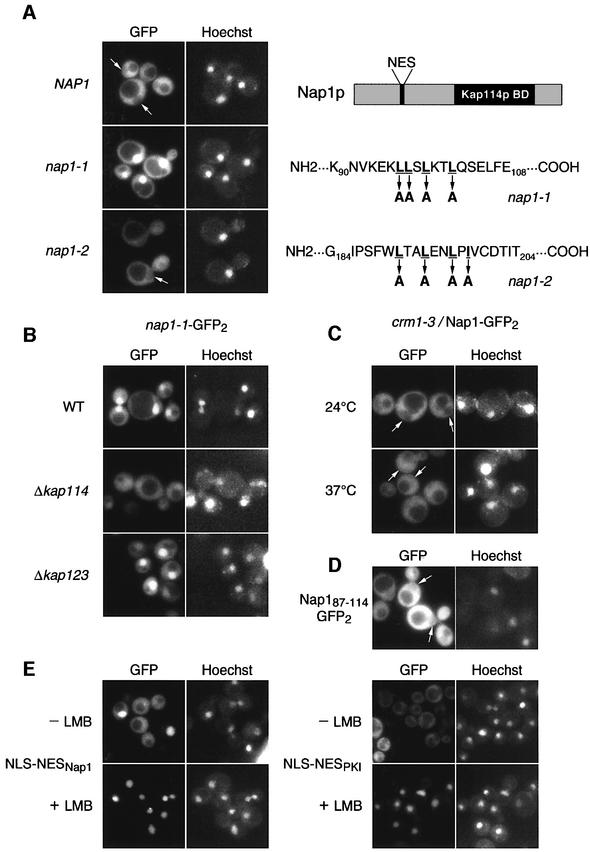
Analysis of the Nap1p N-terminal sequences revealed two conserved leucine-rich domains which may serve as leucine-rich nuclear export sequences (NESs; see schematic in Figure 9A). We mutated each of the leucine-rich sequences in Nap1p in the context of the GFP2 reporter; in both cases, four of the leucines or isoleucines were changed to alanines (these mutants hereafter will be referred to as nap1-1 and nap1-2; see Figure 9A). When expressed in wild-type yeast, the localization of nap1-1–GFP2 was clearly nuclear, while the localization of nap1-2–GFP2 resembled that of wild-type Nap1p–GFP2 (Figure 9A). Thus residues 96–102 of Nap1p represented an NES.
Fig. 9. Nucleocytoplasmic shuttling of Nap1p. (A) Two putative NES mutants (nap1-1 and nap1-2) were created in Nap1p and expressed in wild-type yeast as GFP2 fusions. The underlined residues were changed to alanine in these two mutants. A schematic of yeast Nap1p is shown on the right. Kap114p BD = Kap114p-binding domain. (B) nap1-1–GFP2 was expressed in the indicated strains. (C) Nap1–GFP2 was expressed in crm1-3 grown at 24°C or shifted to 37°C for 1 h. (D) The NES of Nap1p (residues 87–114) fused to GFP2 was analyzed in wild-type yeast. (E) The Nap1p NES or PKI NES was fused to an exogenous NLS and GFP2, and analyzed in an LMBs strain with or without LMB treatment (200 nM) for 30 min. The coincident Hoechst staining is shown, and positions of nuclei are arrowed.
Mechanism of Nap1p nucleocytoplasmic shuttling
The above results provided evidence for Nap1p nucleocytoplasmic shuttling. To determine whether Nap1p import depended on Kap114p, we expressed the nuclear mutant, nap1-1–GFP2, in a Δkap114 strain. As shown in Figure 9B, the nuclear localization of this export-deficient mutant was somewhat decreased in the Δkap114 strain but not in the Δkap123 strain. This suggested that Kap114p plays a role in the import phase of Nap1p shuttling, although it is likely that Nap1p can also enter the nucleus independently of Kap114p.
Our analysis of Nap1p revealed the presence of a leucine-rich NES located near the N-terminus of Nap1p. It has been shown that leucine-rich NESs are recognized by the export karyopherin, Crm1p (Xpo1p). We therefore tested whether Nap1p was exported from the nucleus by Crm1p in vivo. We expressed Nap1p–GFP2 in a crm1 temperature-sensitive strain, crm1-3 (Neville et al., 1997). Nuclear exclusion of Nap1p–GFP2 was slightly decreased in this strain at the non-permissive temperature (Figure 9C). Because the nuclear accumulation of this reporter in the crm1-3 strain was much less dramatic than that observed with the nap1-1–GFP2 NES mutant (Figure 9A), we hypothesized that a second pathway of export exists for Nap1p. To test directly whether the leucine-rich NES mediated export, residues 87–114 were fused to GFP2 and the localization examined. The fusion protein was excluded from the nucleus, suggesting that it was actively exported (Figure 9D). Since it was possible that this fusion protein was not actively gaining access to the nucleus, we constructed an NLS– NESNAP1–GFP2 fusion protein, which localized to both the cytoplasm and nucleus (Figure 9E). This reporter and another utilizing the Crm1-dependent NES from PKI (NLS–NESPKI–GFP2) were expressed in a leptomycin B (LMB)-sensitive crm1 mutant strain. After LMB addition, both of these reporters rapidly accumulated in the nucleus (Figure 9E). This suggested that the identified Nap1p NES mediated export via Crm1p in vivo.
Nucleocytoplasmic shuttling of Nap1p is important for Ty1 element transcription
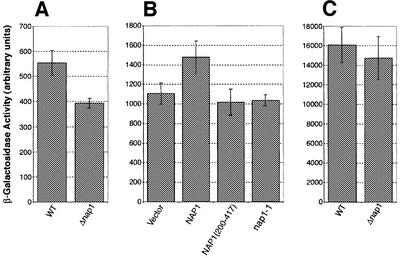
It was likely that the nucleocytoplasmic shuttling of Nap1p would be crucial for its proposed function in histone deposition. An in vivo assay that is sensitive to the levels of histones in the cell is the measurement of transcription levels from the yeast Ty1 element (reviewed in Yamaguchi et al., 2001). It has been shown that deletion of the HTA1 and HTB1 alleles results in an SPT phenotype (suppression of the Ty1 element), which presumably is caused by decreasing the total cellular H2A–H2B levels (Clark-Adams et al., 1988). We used a Ty1–lacZ fusion integrated at the HIS4 locus, and transcription was measured by assaying β-galactosidase activity (Dudley et al., 1999). Comparison of an otherwise wild-type strain with one lacking NAP1 showed that deletion of NAP1 resulted in an ∼29% decrease in β-galactosidase activity relative to wild-type (Figure 10A). To test whether the shuttling of Nap1p was important for Ty1 transcription, we used a plasmid-based β-galactosidase activity assay. A Δnap1 deletion strain expressing the Ty1–lacZ fusion was transformed with empty vector or plasmids expressing either wild-type Nap1p or the non-shuttling nuclear mutants (Nap1p200–417 or the NES mutant, nap1-1) under the control of the GAL1 promoter. After galactose induction, β-galactosidase activity was assayed. A decrease of ∼20–25% was observed in the absence of Nap1p compared with wild-type, similar to that described above (Figure 10B). Both mutants also showed a similar decrease compared with wild-type. Analysis of transcription from a CYC1 promoter in the Δnap1 strain showed little decrease in β-galactosidase activity compared with wild-type (Figure 10C). This suggested that decreased transcription from the Ty1 promoter observed in the Δnap1 strain did not reflect a general defect in transcription. These data indicated that while Nap1p itself was important for Ty1 transcription, so was the shuttling function of Nap1p. The observed phenotype may reflect a defect in histone import or deposition in the absence of Nap1p shuttling.
Fig. 10. Nucleocytoplasmic shuttling of Nap1p is necessary for the proper transcription of the Ty1 element. (A) β-galactosidase activity, as a measurement of Ty1–LacZ transcription, was measured in a wild-type (WT) and an isogenic Δnap1 strain. (B) β-galactosidase assays were performed in a Δnap1 strain after expression of wild-type NAP1, NAP1 residues 200–417 and the NAP1 NES mutant, nap1-1 or vector alone. (C) Transcription assays were performed using a non-Ty1 (CYC1-lacZ) reporter system in a wild-type (WT) and an isogenic Δnap1 strain.
Discussion
In this report, we demonstrate that yeast Nap1p provides a link between the nuclear transport of histones H2A and H2B and their downstream deposition to form chromatin. We have identified a mechanism through which this may occur by showing that Nap1p specifically promotes the interaction of histones H2A and H2B with their preferred transport factor, Kap114p. This would ensure the efficient import of these histones into the nucleus.
Our results demonstrate for the first time that yeast Nap1p is a nucleocytoplasmic shuttling protein. We do not know if all of Nap1p continuously shuttles or whether this represents a small pool. However, as the Nap1p NES mutant led to high levels of nuclear accumulation, it is likely that the entire pool of yeast Nap1p can shuttle. In other eukaryotes, both cytoplasmic and nuclear localization of Nap1p have been observed (Ito et al., 1996; Chang et al., 1997; Rodriguez et al., 1997), and in certain species the cell cycle-regulated nuclear import of Nap1p has been shown (Ito et al., 1996; Rodriguez et al., 1997; Rogner et al., 2000). Collectively, these data suggest that all Nap1p orthologs and isoforms may be able to shuttle, but their residence time in each compartment may be regulated differentially. We currently do not have any evidence that yeast Nap1p localization is cell cycle regulated.
Putative NES and NLS consensus sequences have been observed in Nap1p in several species (Ito et al., 1996). Our results suggest that at least the conserved NES is functional and, as expected for a leucine-rich NES, when added to a reporter it can confer Crm1p-dependent export. We have analyzed the export of full-length Nap1p in several crm1 mutant strains, including the LMB-sensitive strain, and seen negligible or very small defects in nuclear export. This suggests that, in addition to Crm1p, Nap1p can exit the nucleus via another factor. We have tested mutants of CSE1, LOS1, MSN5 and MEX67 as candidate export factors, but detected no effect on Nap1p export. It is possible that multiple or undiscovered proteins can contribute to Nap1p export, and it also remains to be determined whether they also recognize leucine-rich sequences. Interestingly, the protein Upf3p recently was shown to have a Crm1p-independent leucine-rich NES (Shirley et al., 2002).
We have also demonstrated that Nap1p contains a central domain that mediates direct interaction with Kap114p and hence this domain could constitute an NLS. However, as the Nap1p–Kap114p interaction is not sensitive to RanGTP, it suggests that the domain is not a typical NLS. We did not observe complete inhibition of import of the Nap1p NES mutant from the nucleus in the absence of Kap114p, demonstrating that Nap1p has more than one route to the nucleus. As Nap1p does not appear to interact with other Kaps that we have tested, it is unclear which other proteins would mediate Nap1p import. It is possible that in the absence of Kap114p, Nap1p may piggy-back into the nucleus with another binding partner.
Our gel filtration experiments are consistent with Kap114p, Nap1p, H2A and H2B forming a complex. The observation that the H2A and H2B N-terminal tails can bind directly to Kap114p and Nap1p, and that Nap1p binds directly to Kap114p, suggests that a number of different complexes may also be possible in vivo. It is likely that a small fraction of histones bind directly to Kap114p and to other Kaps that can mediate their nuclear import. Clearly, secondary nuclear transport pathways exist for H2A and H2B (Mosammaparast et al., 2001), and redundant factors probably exist for chromatin assembly (Adams and Kamakaka, 1999), which explains why both Kap114p and Nap1p are not essential in yeast (Kellogg et al., 1995; Ito et al., 1996). Our data suggest that a co-complex of H2A, H2B, Nap1p and Kap114p can be formed. The fact that Nap1p addition renders the Kap114p–histone association RanGTP insensitive suggests that Nap1p bridges this interaction. However, in this configuration, direct contact between Kap114p and the histone NLS is likely, resulting in an increased binding affinity of Kap114p for the histones. As a result, Kap114p may be the favored pathway for transport due to its increased affinity for histones. In addition, if Nap1p binds the histones as soon as they are synthesized, Kap114p will be able to join the complex whereas other Kaps would have to compete with Nap1p for binding to the histone NLSs.
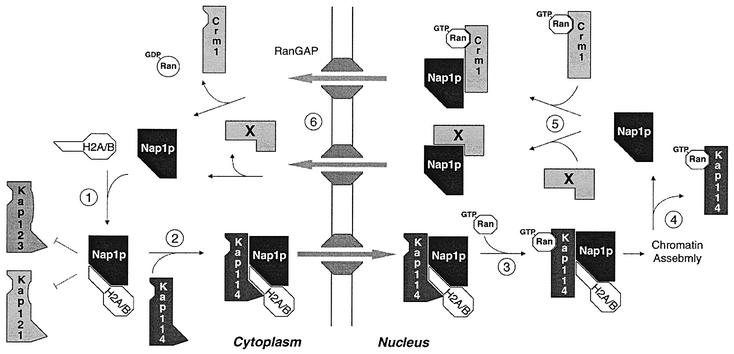
From the above results, we propose a model for the function of Nap1p in the nuclear transport of histones H2A and H2B (Figure 11). Newly synthesized H2A and H2B associate with free Nap1p in the cytoplasm via their N-terminal tails. The association of Nap1p with H2A and H2B prevents the association of Kaps, such as Kap121p and Kap123p, with H2A/H2B. Kap114p joins the Nap1p–histone complex, which translocates into the nucleus. In the nucleus, Kap114p associates with RanGTP, which dissociates Kap114p from the H2A and H2B N-terminal tails. However, Kap114p remains bound to the Nap1p–histone complex via its RanGTP-insensitive Nap1p-binding domain. Nap1p is then poised for chromatin assembly, as it is the only component in the complex that remains bound to the histones.
Fig. 11. Model for the function of Nap1p in the nuclear transport of histones H2A and H2B.
The mechanism by which the Kap114p–Nap1p complex dissociates in the nucleus in not clear. However, if the complex did not dissociate, the localization of Kap114p and Nap1p would be interdependent. Preliminary experiments suggest that the localization of Kap114p–GFP does not change in a Δnap1 strain, or when overexpressing nap1-1 (N.Mosammaparast and L.F.Pemberton, unpublished data). The fact that the localization of Nap1p and Kap114p is independent suggests that the two proteins dissociate in the nucleus. In order to dissociate this Ran-GTP-insensitive complex, interaction of Nap1p with other chromatin assembly factors or the deposition of bound histones onto DNA could trigger a conformational change in Kap114p and/or Nap1p that allows dissociation to occur. This may be analogous to the DNA-stimulated RanGTP-mediated dissociation of TATA box-binding protein (TBP) from Kap114p (Pemberton et al., 1999). Alternatively, a factor that acts further downstream of chromatin assembly may be involved. If the latter is true, the binding of Nap1p to its export factor (e.g. Crm1p or another factor) could be responsible for the dissociation (Figure 11). Once in the cytoplasm, Nap1p is freed for another round of nuclear import and histone deposition.
Our model predicts that the presence of Nap1p, as well as its ability to shuttle, are important for its function. Quantitation of the N:C ratios of H2A and H2B NLS–GFP reporters in strains lacking Nap1p indicated that there is a small decrease in nuclear accumulation of these histone reporters in the absence of Nap1p. This defect was very similar to that reported previously in strains lacking Kap114p (Mosammaparast et al., 2001). If Kap114p and Nap1p were working by the same pathway, they would be predicted to have similar effects on import of these histones. Our analysis of Ty1 element transcription suggests that NAP1 may be an SPT gene. Although the molecular mechanism of Ty1 element transcription suppression is not understood precisely, all the SPT genes discovered so far have been involved directly in chromatin metabolism or transcriptional initiation (Yamaguchi et al., 2001). Interestingly, deletion of one copy of H2A and H2B, and the consequent decrease in H2A and H2B protein levels in the cell, also causes an SPT phenotype (Clark-Adams et al., 1988). Deletion of NAP1 may result in a decrease in the relative amounts of properly deposited H2A–H2B, resulting in an SPT phenotype. Expression of constitutively nuclear Nap1p mutants also result in decreased Ty1 transcription. This result allows us to hypothesize that Nap1p shuttling is necessary for maintaining proper chromatin structure at this locus, possibly by ensuring that H2A and H2B are imported into the nucleus efficiently.
The ability of Nap1p to inhibit the binding of certain Kaps to the NLS sequences of H2A and H2B defines it as a Kap specificity determinant. This may give histones a dedicated import Kap, allowing their rapid import during synthesis in S phase. Like Kaps, Nap1p may also function as a chaperone to shield positively charged domains of the histones while they are in transit (Jäkel et al., 2002). By coupling H2A–H2B transport to a single specific Kap, transport of cargos by other Kaps remains undisturbed. Another possible advantage may be the simultaneous nuclear import of H2A–H2B and Nap1p, by Kap114p. Thus, Kap114p serves as the import factor for H2A–H2B, while simultaneously serving the same function for the assembly cofactor involved in H2A–H2B deposition.
The involvement of Nap1p in the nuclear transport of H2A–H2B by Kap114p and the RanGTP-insensitive association between Kap114p and Nap1p in the nucleus allow the intriguing possibility that Kap114p is involved with Nap1p in the deposition of H2A–H2B onto chromatin. Yeast Nap1p also plays an important role in a signaling network during mitosis and in G1 (Kellogg et al., 1995; Kellogg and Murray, 1995; Zimmerman and Kellogg, 2001). How can this function be reconciled with reports that Nap1p can assemble histones onto DNA, and the activity as a Kap specificity factor described here? It is possible that the cell cycle- and chromatin-associated activities link a common chaperone or nuclear import function of Nap1p. Factors involved in chromatin assembly obviously need to be imported into the nucleus, but there are also reports that cyclins and their associated proteins shuttle between the nucleus and cytoplasm (Pines and Hunter, 1991; Yang and Kornbluth, 1999), and hence Nap1p may serve to regulate their function in this respect.
It is clear from this and other recent studies that the mechanisms of nuclear transport are far more complex than initially thought. A host of cofactors involved in both the import and export of various cargos are beginning to be discovered, and their mechanisms of action are being unveiled. Our work suggests that characterization of cofactors involved in transport may provide links to the downstream function of transport cargos.
Materials and methods
Yeast strains
The yeast strains in this study were derived from DF5 (Finley et al., 1987) and manipulated as previously described (Sherman et al., 1986). The Δkap114, Δkap123 and crm1-3 strains have been described previously (Neville et al., 1997; Rout et al., 1997; Pemberton et al., 1999). The H2B-PrA Δkap114 Δnap1 and the Kap114-PrA H2B-PrA strains were made by mating, sporulating and dissecting the relevant haploids. For β-galactosidase assays, strain FY1472 (provided by Fred Winston, Harvard Medical School) was used as the parent strain (Dudley et al., 1999); nap1 was deleted in this strain by integration of URA3. The LMB-sensitive crm1 mutant (KWy175) was a kind gift of Karsten Weis (UC Berkeley).
Plasmids
For overexpression of MBP-tagged Kaps, the open reading frames (ORFs) corresponding to Kap114p, Kap121p and Kap123p were cloned into pMAL-c2x (NEB). GST tagging of all proteins in this study was performed by cloning the relevant sequences into pGEX-4T1 or pGEX-2T1 (Stratagene). For plasmid-based β-galactosidase assays, full-length NAP1, NAP1 (200–417) and nap1-1 were cloned into pRS424-GAL1 (ATCC). All reporter constructs were made by cloning the relevant sequences into pGFP2-C-FUS (Mosammaparast et al., 2001). The nap1-1 and nap1-2 mutants were constructed by oligonucleotide site-directed mutagenesis. The NLS–NES reporters were constructed by cloning the H4 NLS (residues 1–29) and the relevant NES into pGFP2-C-FUS. The CYC1-lacZ reporter has been described previously (Keleher et al., 1992).
Cytosol preparation
Post-ribosomal cytosol was prepared from 1 l of each indicated PrA-tagged strain as previously described (Aitchison et al., 1996). Kap114-PrA or H2B-PrA and their associated proteins were isolated using IgG–Sepharose as described (Aitchison et al., 1996; Pemberton et al., 1997). The Nap1p (Santa Cruz Biotechnology, Inc.) and Kap121p antibodies have been described previously (Marelli et al., 1998). All antibody interactions were visualized with horseradish peroxidase (HRP)-conjugated secondary antibodies and enhanced chemiluminescence (ECL; Amersham Pharmacia Biotech).
Purification of recombinant proteins and in vitro binding assays
MBP-tagged Kaps were purified by overexpression in Escherichia coli strain BL21 and purified on amylose–agarose according to the manufacturer’s instructions (NEB). All GST-tagged proteins were purified as previously described (Mosammaparast et al., 2001). Tag-free Nap1p and the H2A1–46 NLS peptide were purified initially as GST-tagged fusions and the GST tag was removed by thrombin (Sigma). Purified His6-tagged human RanQ69L (from Ian Macara, University of Virginia) was added at the concentrations indicated in the figure legends. The purified recombinant untagged histone H2A–H2B dimer was a kind gift of Robert Dutnall (UCSD). All binding assays were performed in transport buffer (TB) (Mosammaparast et al., 2001) using 10 µl of glutathione–Sepharose beads. The beads were pre-blocked with 10% bovine serum albumin (BSA) and each binding assay contained a final concentration of 3% BSA. All binding assays were performed in a total volume of 100 µl using the indicated amounts of each protein. Binding reactions were incubated at 4°C for 1 h. Beads were then washed extensively in TB, and a final wash was performed with 1× phosphate-buffered saline (PBS). Bound material was eluted with SDS loading buffer, analyzed by SDS–PAGE and stained with Coomassie Brilliant Blue or western blotted, as indicated. For detection of MBP-tagged Kaps, an anti-MBP antibody was used (NEB).
Gel filtration
Yeast cytosol from a Kap114-PrA/H2B-PrA strain was separated by gel filtration on a Superdex 200 column using TB/5% glycerol buffer. After precipitation in methanol, fractions were analyzed by SDS–PAGE and western blotting with a Nap1p-specific antibody which also recognized the PrA tags. Recombinant MBP–Kap114p, Nap1p and H2A–H2B dimer were incubated together for 20 min for complex formation, and separated similarly.
Cell culture and microscopy
All cell culture methods and microscopy have been described previously (Mosammaparast et al., 2001). The GFP images in each figure panel were acquired using identical exposure settings, and manipulated identically for each reporter shown, using Adobe Photoshop. Quantitation of N:C ratios from captured images was described previously (Mosammaparast et al., 2001). Temperature-sensitive and LMB-sensitive strains were grown as described in the figure legends.
β-galactosidase assays
Each strain to be assayed was grown under selection in glucose, then grown in raffinose for 8–10 h and induced overnight in galactose. Whole-cell lysates were assayed for β-galactosidase activity as previously described (Moretti et al., 1994). Each strain was assayed using six individual replicas, and the mean ± SD of the replicas was calculated. Each experiment was repeated at least four times, and a single representative experiment is shown.
Acknowledgments
Acknowledgements
We thank those individuals cited in the text for strains and antibodies. We particularly wish to thank Dr Robert Dutnall (UCSD) for purified histones, Karsten Weis (UC Berkeley) for the LMB-sensitive strain and Sarah Wilkinson and Jennifer Hodges for technical help. N.M. was supported by the NIH Cell and Molecular Biology Training Grant and the Medical Scientist Training Program. This work was supported in part by Research Grant 5-FY00-563 from the March of Dimes Birth Defects Foundation and Research Grant R01 GM65385 from the NIH.
References
- Adams C.R. and Kamakaka,R.T. (1999) Chromatin assembly: biochemical identities and genetic redundancy. Curr. Opin. Genet. Dev., 9, 185–190. [DOI] [PubMed] [Google Scholar]
- Aitchison J.D., Blobel,G. and Rout,M.P. (1996) Kap104p: a karyopherin involved in the nuclear transport of messenger RNA binding proteins. Science, 274, 624–627. [DOI] [PubMed] [Google Scholar]
- Altman R. and Kellogg,D. (1997) Control of mitotic events by Nap1 and the Gin4 kinase. J. Cell Biol., 138, 119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagley S., Goldberg,M.W., Cronshaw,J.M., Rutherford,S. and Allen,T.D. (2000) The nuclear pore complex. J. Cell Sci., 113, 3885–3886. [DOI] [PubMed] [Google Scholar]
- Chang L., Loranger,S.S., Mizzen,C., Ernst,S.G., Allis,C.D. and Annunziato,A.T. (1997) Histones in transit: cytosolic histone complexes and diacetylation of H4 during nucleosome assembly in human cells. Biochemistry, 36, 469–480. [DOI] [PubMed] [Google Scholar]
- Clark-Adams C.D., Norris,D., Osley,M.A., Fassler,J.S. and Winston,F. (1988) Changes in histone gene dosage alter transcription in yeast. Genes Dev., 2, 150–159. [DOI] [PubMed] [Google Scholar]
- Dudley A.M., Gansheroff,L.J. and Winston,F. (1999) Specific components of the SAGA complex are required for Gcn4- and Gcr1-mediated activation of the his4-912δ promoter in Saccharomyces cerevisiae. Genetics, 151, 1365–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D., Ozkaynak,E. and Varshavsky,A. (1987) The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation and other stresses. Cell, 48, 1035–1046. [DOI] [PubMed] [Google Scholar]
- Fujii-Nakata T., Ishimi,Y., Okuda,A. and Kikuchi,A. (1992) Functional analysis of nucleosome assembly protein, NAP-1. The negatively charged COOH-terminal region is not necessary for the intrinsic assembly activity. J. Biol. Chem., 267, 20980–20986. [PubMed] [Google Scholar]
- Jäkel S., Mingot,J.M., Schwarzmaier,P., Hartmann,E. and Gorlich,D. (2002) Importins fulfil a dual function as nuclear import receptors and cytoplasmic chaperones for exposed basic domains. EMBO J., 21, 377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimi Y., Kojima,M., Yamada,M. and Hanaoka,F. (1987) Binding mode of nucleosome-assembly protein (AP-I) and histones. Eur. J. Biochem., 162, 19–24. [DOI] [PubMed] [Google Scholar]
- Ito T., Bulger,M., Kobayashi,R. and Kadonaga,J.T. (1996) Drosophila NAP-1 is a core histone chaperone that functions in ATP-facilitated assembly of regularly spaced nucleosomal arrays. Mol. Cell. Biol., 16, 3112–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Tyler,J.K. and Kadonaga,J.T. (1997) Chromatin assembly factors: a dual function in nucleosome formation and mobilization? Genes Cells, 2, 593–600. [DOI] [PubMed] [Google Scholar]
- Ito T., Ikehara,T., Nakagawa,T., Kraus,W.L. and Muramatsu,M. (2000) p300-mediated acetylation facilitates the transfer of histone H2A–H2B dimers from nucleosomes to a histone chaperone. Genes Dev., 14, 1899–1907. [PMC free article] [PubMed] [Google Scholar]
- Keleher C.A., Redd,M.J., Schultz,J., Carlson,M. and Johnson,A.D. (1992) Ssn6-Tup1 is a general repressor of transcription in yeast. Cell, 68, 709–719. [DOI] [PubMed] [Google Scholar]
- Kellogg D.R. and Murray,A.W. (1995) NAP1 acts with Clb1 to perform mitotic functions and to suppress polar bud growth in budding yeast. J. Cell Biol., 130, 675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg D.R., Kikuchi,A., Fujii-Nakata,T., Turck,C.W. and Murray,A.W. (1995) Members of the NAP/SET family of proteins interact specifically with B-type cyclins. J. Cell Biol., 130, 661–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macara I.G. (2001) Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev., 65, 570–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marelli M., Aitchison,J.D. and Wozniak,R.W. (1998) Specific binding of the karyopherin Kap121p to a subunit of the nuclear pore complex containing Nup53p, Nup59p and Nup170p. J. Cell Biol., 143, 1813–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuibban G.A., Commisso-Cappelli,C.N. and Lewis,P.N. (1998) Assembly, remodeling and histone binding capabilities of yeast nucleosome assembly protein 1. J. Biol. Chem., 273, 6582–6590. [DOI] [PubMed] [Google Scholar]
- Moretti P., Freeman,K., Coodly,L. and Shore,D. (1994) Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev., 8, 2257–2269. [DOI] [PubMed] [Google Scholar]
- Mosammaparast N., Jackson,K.R., Guo,Y., Brame,C.J., Shabanowitz,J., Hunt,D.F. and Pemberton,L.F. (2001) Nuclear import of histone H2A and H2B is mediated by a network of karyopherins. J. Cell Biol., 153, 251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosammaparast N., Guo,Y., Shabanowitz,J., Hunt,D.F. and Pemberton,L.F. (2002) Pathways mediating the nuclear import of histones H3 and H4 in yeast. J. Biol. Chem., 277, 862–868. [DOI] [PubMed] [Google Scholar]
- Muhlhausser P., Muller,E.C., Otto,A. and Kutay,U. (2001) Multiple pathways contribute to nuclear import of core histones. EMBO Rep., 2, 690–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neville M., Stutz,F., Lee,L., Davis,L.I. and Rosbash,M. (1997) The importin-β family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr. Biol., 7, 767–775. [DOI] [PubMed] [Google Scholar]
- Pemberton L.F., Rosenblum,J.S. and Blobel,G. (1997) A distinct and parallel pathway for the nuclear import of an mRNA-binding protein. J. Cell Biol., 139, 1645–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton L.F., Blobel,G. and Rosenblum,J.S. (1998) Transport routes through the nuclear pore complex. Curr. Opin. Cell Biol., 10, 392–399. [DOI] [PubMed] [Google Scholar]
- Pemberton L.F., Rosenblum,J.S. and Blobel,G. (1999) Nuclear import of the TATA-binding protein: mediation by the karyopherin Kap114p and a possible mechanism for intranuclear targeting. J. Cell Biol., 145, 1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines J. and Hunter,T. (1991) Human cyclins A and B1 are differentially located in the cell and undergo cell cycle-dependent nuclear transport. J. Cell Biol., 115, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez P. et al. (1997) Functional characterization of human nucleosome assembly protein-2 (NAP1L4) suggests a role as a histone chaperone. Genomics, 44, 253–265. [DOI] [PubMed] [Google Scholar]
- Rogner U.C., Spyropoulos,D.D., Le Novere,N., Changeux,J.P. and Avner,P. (2000) Control of neurulation by the nucleosome assembly protein-1-like 2. Nat. Genet., 25, 431–435. [DOI] [PubMed] [Google Scholar]
- Rout M.P., Blobel,G. and Aitchison,J.D. (1997) A distinct nuclear import pathway used by ribosomal proteins. Cell, 89, 715–725. [DOI] [PubMed] [Google Scholar]
- Rout M.P., Aitchison,J.D., Suprapto,A., Hjertaas,K., Zhao,Y. and Chait,B.T. (2000) The yeast nuclear pore complex: composition, architecture and transport mechanism. J. Cell Biol., 148, 635–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Fink,G.R. and Hicks,J.B. (eds) (1986) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Shirley R.L., Ford,A.S., Richards,M.R., Albertini,M. and Culbertson,M.R. (2002) Nuclear import of Upf3p is mediated by importin-α/-β and export to the cytoplasm is required for a functional nonsense-mediated mRNA decay pathway in yeast. Genetics, 161, 1465–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikama N., Chan,H.M., Krstic-Demonacos,M., Smith,L., Lee,C.W., Cairns,W. and La Thangue,N.B. (2000) Functional interaction between nucleosome assembly proteins and p300/CREB-binding protein family coactivators. Mol. Cell. Biol., 20, 8933–8943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler J.K., Adams,C.R., Chen,S.R., Kobayashi,R., Kamakaka,R.T. and Kadonaga,J.T. (1999) The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature, 402, 555–560. [DOI] [PubMed] [Google Scholar]
- Verreault A. (2000) De novo nucleosome assembly: new pieces in an old puzzle. Genes Dev., 14, 1430–1438. [PubMed] [Google Scholar]
- Verreault A., Kaufman,P.D., Kobayashi,R. and Stillman,B. (1996) Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell, 87, 95–104. [DOI] [PubMed] [Google Scholar]
- Walter P.P., Owen-Hughes,T.A., Cote,J. and Workman,J.L. (1995) Stimulation of transcription factor binding and histone displacement by nucleosome assembly protein 1 and nucleoplasmin requires disruption of the histone octamer. Mol. Cell. Biol., 15, 6178–6187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wozniak R.W., Rout,M.P. and Aitchison,J.D. (1998) Karyopherins and kissing cousins. Trends Cell Biol., 8, 184–188. [DOI] [PubMed] [Google Scholar]
- Yamaguchi Y., Narita,T., Inukai,N., Wada,T. and Handa,H. (2001) SPT genes: key players in the regulation of transcription, chromatin structure and other cellular processes. J. Biochem., 129, 185–191. [DOI] [PubMed] [Google Scholar]
- Yang J. and Kornbluth,S. (1999) All aboard the cyclin train: subcellular trafficking of cyclins and their CDK partners. Trends Cell Biol., 9, 207–210. [DOI] [PubMed] [Google Scholar]
- Zimmerman Z.A. and Kellogg,D.R. (2001) The Sda1 protein is required for passage through start. Mol. Biol. Cell, 12, 201–219. [DOI] [PMC free article] [PubMed] [Google Scholar]