Abstract
Maintenance of Kaposi's sarcoma-associated herpesvirus (KSHV) latent infection depends on the viral episomes in the nucleus being distributed to daughter cells following cell division. The latency-associated nuclear antigen (LANA) is constitutively expressed in all KSHV-infected cells. LANA binds sequences in the terminal repeat regions of the KSHV genome and tethers the viral episomes to chromosomes. To better understand the mechanism of chromosomal tethering, we performed glutathione S-transferase (GST) affinity and yeast two-hybrid assays to identify LANA-interacting proteins with known chromosomal association. Two of the interactors were the methyl CpG binding protein MeCP2 and the 43-kDa protein DEK. The interactions of MeCP2 and DEK with LANA were confirmed by coimmunoprecipitation. The MeCP2-interacting domain was mapped to the previously described chromatin binding site in the N terminus of LANA, while the DEK-interacting domain mapped to LANA amino acids 986 to 1043 in the C terminus. LANA was unable to associate with mouse chromosomes in chromosome spreads of transfected NIH 3T3 cells. However, LANA was capable of targeting to mouse chromosomes in the presence of human MeCP2 or DEK. The data indicate that LANA is tethered to chromosomes through two independent chromatin binding domains that interact with different protein partners.
The Kaposi's sarcoma-associated herpesvirus (KSHV), or human herpesvirus 8 (HHV8), is associated with all forms of Kaposi's sarcoma, primary effusion lymphoma (PEL), and some forms of multicentric Castleman's disease (6, 9, 15, 16, 48). KSHV infection is predominantly latent, and the genome is maintained as multicopied episomes in the nucleus of the infected cell. The gene coding for latency-associated nuclear antigen (LANA) is one of the few viral latency genes and is expressed from open reading frame 73 (ORF73) as a polycistronic message with the viral FLIP and cyclin homologs (44). LANA is a 222- to 234-kDa nuclear phosphoprotein (26, 41) that consists of amino-terminal and carboxy-terminal domains separated by an acidic internal repeat domain. LANA acts as a transcriptional repressor by interacting with histone deacetylase (HDAC) members (28, 45) and as a transcriptional activator of several promoters, including interleukin-6, telomerase reverse transcriptase, E2F-regulated promoters, and its own promoter (2, 27, 28, 31, 40, 43). LANA interacts with cellular proteins, including Rb, CREB-binding protein (CBP), and p53 (18, 25, 30, 40). Interaction with p53 leads to loss of p53 transcriptional activity and inhibition of apoptosis (18). LANA has oncogenic potential in that it transforms primary rat embryonic fibroblasts in cooperation with H-ras (40).
Viruses such as Epstein-Barr virus (EBV) and KSHV must distribute their episomal genomes to daughter cells during cell division to ensure the continuity of the viral life cycle. LANA's function has been compared to that of EBV nuclear antigen 1 (EBNA1), which is constitutively expressed in EBV-infected latent cells, binds the EBV genome, and is required for episomal maintenance (23, 56). EBNA1 is essential for EBV DNA association with mitotic chromosomes, and cellular EBP2 has been identified as an EBNA1-interacting protein that can mediate chromosomal tethering (46, 55). LANA can mediate the persistence of extrachromosomal KSHV DNA in uninfected lymphoblasts (4, 10) and colocalizes with viral genomes both in interphase nuclei and on mitotic chromosomes. LANA, specifically its C terminus, binds to two sites within the terminal repeat of the KSHV genome (5, 10, 21, 22). LANA has been shown to accumulate to heterochromatin-associated nuclear bodies and preferentially associates with human chromatin in human-mouse hybrids containing a single fused nucleus (45, 49). LANA associates with human mitotic chromosomes in a random, speckled fashion in infected cells (38, 49), but paints uninfected HeLa cell chromosomes (38). A chromosome binding site (CBS) has been mapped to amino acids (aa) 5 to 22 which mediate the specific interaction of LANA with mitotic chromosomes (38). Interactions with the chromatin-associated proteins Ring3, which localizes to heterochromatin, and histone H1 have also been described previously (11, 32, 39). However, the role of these proteins in LANA-mediated chromosome association is unclear.
We demonstrate here that two independent interactions with cell proteins are involved in LANA tethering to chromosomes. The first is mediated by the N terminus of LANA through the 75-kDa methyl CpG binding protein 2 (MeCP2), and the second is mediated by the C terminus of LANA through the 43-kDa DEK protein.
MATERIALS AND METHODS
Expression plasmids.
Glutathione S-transferase (GST)-LANA fusions and yeast Gal4DBD and Gal4ACT constructs were previously described (28). pDY15 expresses GFP-LANA (minus the first 2 aa) in pEGFP-C3 (Clontech). The LANA mutant mtLANA (pMW4) was made by digesting pDY15 with XhoI and AscI to delete LANA aa 1 to 15, blunt ending, and religating. Green fluorescent protein (GFP)-LANA-N (pDH389) contains LANA codons 1 to 329 cloned in pEGFP-C1 (Clontech) at BglII. GFP-LANA-C (pMW2) contains LANA codons 931 to 1164 cloned in pEGFP-C1 at BglII. LANA m1 (pMF42), LANA m2 (pMF43), and LANA m3 (pMF73) contain LANA codons 1 to 329 fused at an XbaI site to codons 928 to 1108, 928 to 1043, and 928 to 985, respectively. LANA m4 (pMF40) expresses LANA aa 1 to 329. pMF constructs have an SG5-Flag vector background. Myc-DEK (pAK7) was made by using GST-DEK (19) as a template and ligating the PCR product into pJH363 at BglII. The Myc-DEK fragment was cut from pAK7 with EcoRI and BglII and ligated into pEGFP-C2 (Clontech) at EcoRI and BamHI to obtain GFP-Myc-DEK (pAK63). DEK was also cloned into the BglII site of pSG5 (Stratagene) to make pAK6.
GST affinity assay and immunoprecipitation.
The GST assay was performed as previously described (28). Briefly, GST and GST fusion proteins were made in bacteria and bound to glutathione Sepharose 4B beads (Amersham) at 4°C overnight. The beads were washed and the amount of protein bound to beads was determined by Coomassie blue staining of proteins separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Equal amounts of each GST protein were used in the affinity assays. HeLa cells were transfected with 10 μg each of Flag-MeCP2 with calcium phosphate. Cells were harvested 48 h posttransfection, resuspended in lysis buffer, and sonicated. Supernatant from transfected cells was incubated with GST fusion proteins. The beads were washed six times and proteins were run on a SDS-PAGE gel (9% polyacrylamide) and transferred to polyvinylidene fluoride (PVDF) membrane (Bio-Rad). Interacting proteins were detected by mouse anti-Flag (Sigma) antibody (1:1,500) and visualized by using the enhanced chemiluminescence (ECL) reaction (Amersham Life Sciences). Transcription and translation of DEK were done with pAK6 in the TNT Quick Coupled System (Promega). Equal amounts of GST fusion proteins were incubated with 10 μl of the 35S-labeling reaction mixture in 600 μl of lysis buffer (0.2% NP-40, 150 mM NaCl, 1 mg of bovine serum albumin [BSA] per ml). Bound proteins were separated by SDS-PAGE and detected by autoradiography.
For immunoprecipitations, HeLa or Cos1 cells were transfected in 10-cm-diameter culture dishes with 10 μg of total DNA by using calcium phosphate (HeLa) or FuGENE6 (Roche) (Cos1) and harvested after 2 days. Cells were washed in 1x phosphate-buffered saline (PBS), lysed in 2 ml of lysis buffer (50 mM Tris [pH 7.9], 100 mM NaCl, 0.5 mM EDTA, 2% glycerol, 0.5 mM phenylmethylsulfonyl fluoride [PMSF], 0.2% NP-40, 2 μg of aprotinin per ml), and sonicated for 10 s. Extracts were precleared with Sephadex G-25 beads (Amersham) for 30 min at 4°C and then incubated with Flag antibody (Sigma) or control mouse immunoglobulin G (IgG) antibody (Santa Cruz) (5 μg per ml of extract) for 2 h. For direct precipitations, rat anti-LANA antibody (4 μg per 0.5 ml of extract; ABI) was incubated for 2 h. Protein G beads were added for 2 h. Beads were washed six times with lysis buffer, and samples (coimmunoprecipitation, 18 μl; direct precipitation, 5 μl; and input extract, 15 μl) were run on SDS-PAGE (9% polyacrylamide) gel. The amount of sample used for direct immunoprecipitations was one-fifth of the amount used for the coimmunoprecipitated sample. Western blot analysis was performed with rat anti-LANA or mouse anti-DEK (BD Transduction Laboratories) monoclonal antibody and peroxidase-conjugated antirat (Chemicon) or antimouse (Amersham) IgG secondary antibody.
Immunofluorescence assay.
As previously described (28), NIH 3T3 or HeLa cells were tranfected overnight in two-well slide chambers (LabTek) with FuGENE 6 with 2 μg of total DNA. Cells were washed in PBS, fixed in 3% paraformaldehyde-PBS for 20 min at 25°C, washed in PBS, permeabilized in 0.2% Triton X-100-PBS for 10 min at 25°C, and washed again in PBS. All staining was performed at 37°C for 1 h. Flag- and Myc-tagged proteins were detected with an anti-Flag or anti-Myc mouse antibody (Sigma) and rhodamine-conjugated donkey anti-mouse IgG secondary antibody (Jackson). LANA was detected with an anti-LANA rat antibody and either a fluorescein isothiocyanate (FITC)- or rhodamine-conjugated donkey anti-rat IgG secondary antibody (Jackson). All antibodies were used at a 1:200 dilution in 1% BSA-PBS. Cells were analyzed by confocal microscopy.
Chromosome spreads.
Ten-centimeter-diameter culture dishes of HeLa or NIH 3T3 cells were transfected with calcium phosphate (HeLa) or Mirus TransIT-LT1 (Panvera) (NIH 3T3). Twenty-four hours posttransfection, cells were treated with Colcemid (0.1 μg/ml) (Sigma) for 2 (HeLa) or 6 (NIH 3T3) h for metaphase arrest. BCBL1 cells were treated with Colcemid for 24 h. Cells were washed in 1× PBS and resuspended at 8 × 104 cells per ml in 75 mM KCl for 15 min to swell nuclei. Cells (200 μl) were cytospun for 10 min at 2,000 rpm onto Superfrost/Plus slides (Fisher) at 25°C. Slides were incubated in KCM solution (120 mM KCl, 20 mM NaCl, 10 mM Tris-HCl [pH 7.7], 0.1% Triton X-100) for 10 min. All antibodies were diluted 1:200 in KCM and incubated for 1 h at 37°C. Cells were washed three times for 3 min each in KCM solution after primary and secondary antibody incubations, fixed in 4% paraformaldehyde-PBS for 10 min at 25°C, and washed twice for 1 min each in water. Cells were mounted in Vectashield mounting solution with DAPI (4′,6′-diamidino-2-phenylindole) (Vector Labs) and analyzed by fluorescence or confocal microscopy.
Yeast assays.
The yeast two-hybrid screen was performed as previously described (28).
RESULTS
Localization of LANA on human chromosomes of KSHV-infected cells.
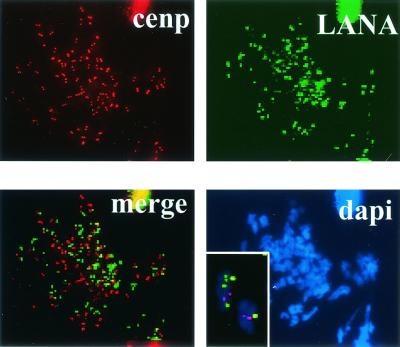
LANA colocalizes with the KSHV genome at discrete spots on chromosomes of KSHV-infected BCBL1 cells (4, 11). To investigate the relationship between the LANA spots and centromere-associated proteins, chromosome spreads were performed on BCBL1 cells (Fig. 1). Centromeres were detected with human serum containing antibodies against centromeric proteins and rhodamine-conjugated donkey anti-human immunoglobulin secondary antibody. LANA was detected with anti-LANA rat monoclonal antibody and FITC-conjugated antirat secondary antibody. Centromeres (red) appeared as discrete doublets on chromosomes. The LANA-staining spots (green) were separate from centromeres.
FIG. 1.
LANA localization on chromosomes of KSHV-infected cells. Chromosome spreads of BCBL1 cells in which the LANA punctate spots (green) localize separately from centromeres (red doublets). Inset, higher magnification of the merged image.
LANA is targeted to sites of mouse interphase and mitotic heterochromatin by human MeCP2.
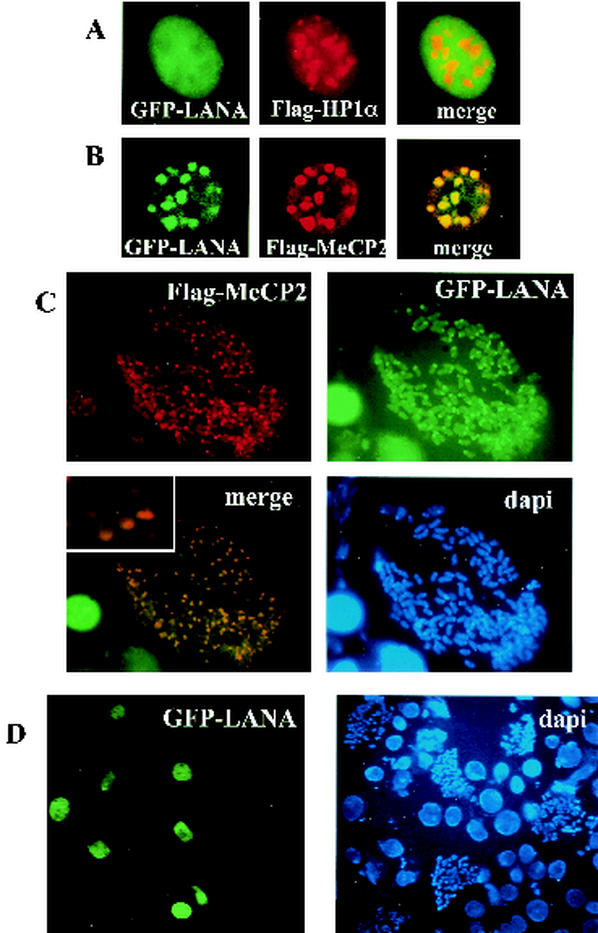
It has been observed that LANA associates with heterochromatin (11, 38, 45, 49), but the targeting mechanism has not been elucidated. Mouse genomic DNA has pericentromeric heterochromatin (PCH), which consists of transcriptionally inactive DNA regions that contain high concentrations of methylated CpGs. In studies of LANA-mediated transcriptional repression, we found that LANA interacts with the methyl CpG binding protein MeCP2 (unpublished data). We set out to determine if LANA could be targeted to heterochromatin via MeCP2. When human MeCP2 and human heterochromatin protein 1 alpha (HP1α), another marker of heterochromatin, are overexpressed in mouse cells, they each localize to PCH and appeared as nuclear spots (29, 54). Immunofluorescence assays were performed with mouse NIH 3T3 cells cotransfected with GFP-LANA and Flag-HP1α. GFP-LANA (green) and Flag-HP1α (red) did not colocalize (Fig. 2A). This indicates that LANA does not localize to mouse heterochromatin when expressed alone, and its localization is not affected by the presence of human HP1α. However, in cells cotransfected with human Flag-MeCP2, GFP-LANA (green) and Flag-MeCP2 (red) colocalized to the nuclear PCH spots (Fig. 2B). Thus, LANA is targeted to mouse PCH by human MeCP2. NIH 3T3 cells were then cotransfected with Flag-MeCP2 and GFP-LANA and then blocked with Colcemid for 6 h, and chromosome spreads were generated (Fig. 2C). Immunofluorescence assays detected Flag-MeCP2 (red) concentrated at the discrete spots of PCH on the mouse chromosome ends. GFP-LANA (green) also localized to the same regions when expressed in the presence of human MeCP2. In the absence of human MeCP2, LANA was never seen associated with NIH 3T3 chromosomes (Fig. 2D). Thus, LANA can be targeted to chromosomes by MeCP2.
FIG. 2.
MeCP2 targets LANA to sites of heterochromatin. NIH 3T3 cells were transfected with GFP-LANA and either Flag-HP1α or Flag-MeCP2. Immunofluorescence assays revealed that (A) GFP-LANA (green) did not localize to sites of mouse heterochromatin marked by Flag-HP1α staining (red). (B) GFP-LANA (green) relocalized to heterochromatin in the presence of Flag-MeCP2 (red). (C) GFP-LANA (green) localized to the Flag-MeCP2 marked pericentromeric regions of mouse NIH 3T3 chromosomes (red spots) in the presence of MeCP2. Inset, higher magnification of the merged image. (D) Chromosome spreads of NIH 3T3 cells transfected with GFP-LANA (green) reveal no association between LANA and mouse chromosomes. DNA was stained with DAPI (blue).
MeCP2 targets LANA to mouse interphase heterochromatin and mitotic chromosomes via LANA aa 1 to 15.
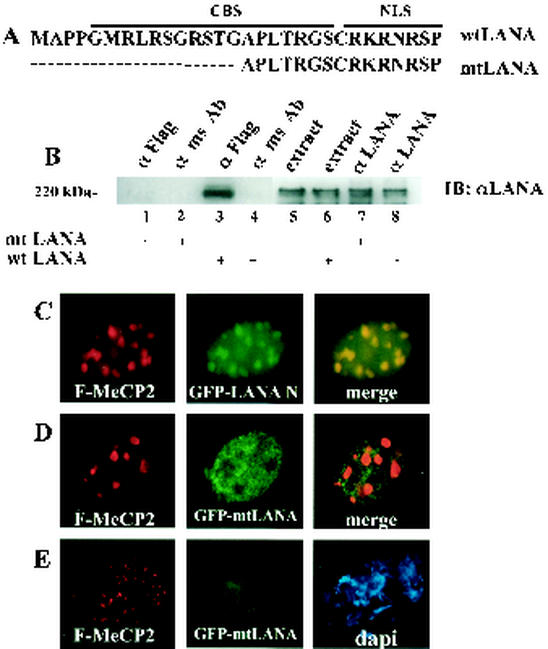
LANA contains a CBS in its N terminus, which is required for LANA's association with chromosomes (38). We made a LANA mutant (mtLANA), which has had the first 15 aa of LANA deleted, disrupting the CBS while keeping the nuclear localization signal (NLS) intact (Fig. 3A). To test the requirement for the N-terminal CBS for MeCP2 interaction, immunoprecipitation assays were performed with extracts of HeLa cells cotransfected with Flag-MeCP2 and GFP-mtLANA or GFP-wild-type LANA (Fig. 3B). Immunoprecipitated proteins were analyzed by Western blotting, and mtLANA and wild-type LANA were detected with anti-LANA monoclonal antibody. Wild-type LANA (lane 3), but not mtLANA (lane 1), coprecipitated with Flag-MeCP2 in immunoprecipitates generated by using anti-Flag antibodies. Wild-type LANA and mtLANA were present in equal amounts in the transfected-cell extracts, as indicated by direct analysis of extract (lanes 5 and 6) or immunoprecipitation with anti-LANA antibody (lanes 7 and 8).
FIG. 3.
MeCP2 targets LANA to chromosomes via the N-terminal CBS. (A) Schematic of wild-type (wt) LANA N terminus showing the CBS and NLS. The CBS is disrupted in mtLANA. (B) Immunoprecipitation assay with extracts of HeLa cells cotransfected with Flag-MeCP2 and either mtLANA or wild-type LANA. mtLANA did not coprecipitate with Flag-MeCP2 (lane 1), unlike wild-type LANA (lane 3). Neither mtLANA nor wild-type LANA was precipitated by control mouse IgG (lanes 2 and lane 4). Lanes 5 to 8 show transfected cell extracts (15 μl; lanes 5 and 6) and direct precipitation by anti-LANA antibody (lanes 7 and 8). ms Ab, mouse antibody. (C to E) Immunofluorescence assays performed with NIH 3T3 cells transfected with GFP-LANA-N or GFP-mtLANA and Flag-MeCP2. (C) LANA-N (green) localized to sites of heterochromatin in the presence of MeCP2 (red). (D) mtLANA (green) did not colocalize with heterochromatin in the presence of MeCP2 (red). (E) Chromosome spreads revealed that Flag-MeCP2 (red) localized to chromosomes, while mtLANA (green) failed to associate with chromosomes. DNA was stained with DAPI (blue).
To determine the effect of the loss of the CBS site on MeCP2-directed chomosome targeting in mouse cells, NIH 3T3 cells were cotransfected with GFP-LANA-N (LANA aa 1 to 329) or GFP-mtLANA and Flag-MeCP2, and immunofluorescence assays were performed. As demonstrated in Fig. 3C, LANA-N (green) was targeted by Flag-MeCP2 (red) to the mouse PCH nuclear spots, although diffuse nuclear staining was also apparent. Full-length LANA may make additional contacts that stabilize the heterochromatin interaction. mtLANA (green) remained nuclear diffuse in the presence of human MeCP2 and was partially excluded from the PCH regions (Fig. 3D). This result indicates that mtLANA was unable to interact with MeCP2 and is not targeted to heterochromatin. Chromosome spreads were made from the same transfection (Fig. 3E). While Flag-MeCP2 (red) again formed discrete spots on mouse chromosome ends, mtLANA was unable to associate with the mouse chromosomes. This result is consistent with mtLANA's inability to be targeted to mouse PCH by MeCP2. In summary, the previously described essential CBS of LANA is also required for MeCP2 interaction and targeting to chromosomes.
Localization of LANA C terminus.
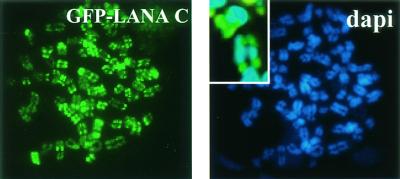
It is established that LANA has an N-terminal CBS (38). However, GFP-LANA C-terminus proteins have been expressed and show a nuclear speckled pattern in interphase nuclei (45, 49). Ballestas et al. also recognized that the LANA C terminus can independently associate with human chromosomes (M. E. Ballestas, T. Komatsu, and K. M. Kaye, 4th Int. Workshop KSHV and Related Agents, 2001). There is a cryptic NLS in the C terminus of LANA that is functional when the truncated LANA C terminus is expressed. We first demonstrated that our LANA C-terminus construction could target human chromosomes (Fig. 4). HeLa cells were transfected with GFP-LANA-C and blocked in metaphase by Colcemid for 2 h. Chromosome spreads were made, and chromosomes were viewed by immunofluorescence. LANA-C formed spots on the chromosomes, showing that LANA-C can be targeted to chromosomes in the absence of viral episomes. This confirms that a second LANA CBS and targeting mechanism exist.
FIG. 4.
LANA C-terminus associates independently with chromosomes. Immunofluorescence assay showing chromosome spreads of HeLa cells transfected with GFP-LANA-C (green spots), which associated with human chromosomes. DNA was stained with DAPI (blue). Inset, higher magnification of the merged image.
LANA interacts with DEK.
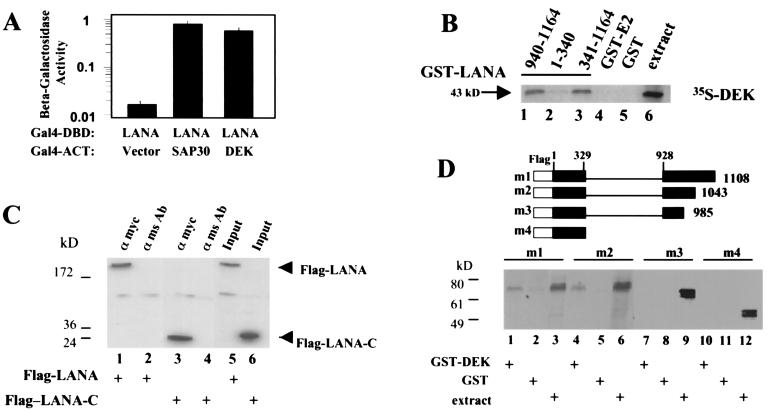
We identified DEK as a LANA-interacting protein in a yeast screen in which Gal4DBD-LANA was cotransformed into yeast with a B-cell cDNA library to seek cellular binding partners for LANA. DEK associates with and paints human chromosomes (24). Interaction between Gal4DBD-LANA and Gal4ACT-DEK is illustrated in Fig. 5A, as measured by induction of β-galactosidase activity in cotransformed yeast. A known LANA-interacting protein, SAP30, was used as a positive control in this assay.
FIG. 5.
LANA binds to DEK through a C-terminal domain. (A) Yeast two-hybrid assay in which interaction is measured by induction of β-galactosidase activity. Yeast cells were cotransformed with Gal4DBD-LANA plus Gal4ACT-DEK, Gal4ACT vector (negative control), or Gal4ACT-SAP30 (positive control). Data are averaged from two experiments, with the percent error indicated. (B) In vitro-translated [35S]methionine-labeled DEK was incubated with the indicated GST-LANA fusion proteins (lanes 1 to 3) or with control GST-EBNA2(1-58) (GST-E2) or GST proteins (lanes 4 and 5). Reaction extract (2 μl) was loaded in lane 6. Bound protein was separated by SDS-PAGE and analyzed by autoradiography for 4 h. (C) Western blot of immunoprecipitates from extracts of HeLa cells cotransfected with Flag-LANA or Flag-LANA-C and Myc-DEK and probed with anti-Flag antibody. Flag-LANA (lane 1) and Flag-LANA-C (lane 3) coprecipitated with Myc-DEK, but were not precipitated by control mouse IgG (lanes 2 and 4). Lanes 5 and 6, transfected cell extracts (15 μl). Extract in the input lanes was one-fifth of that used for coprecipitations. (D) The upper panel shows a schematic of the Flag-tagged LANA deletion mutants used in the GST affinity assay. The lower panel shows in vitro-translated, [35S]methionine-labeled LANA mutants m1 to m4 were incubated with a GST-DEK fusion protein (lanes 1, 4, 7, and 10) or with control GST protein (lanes 2, 5, 8, and 11). Extract (2 μl) was loaded in lanes 3, 6, 9, and 12. Bound protein was separated by SDS-PAGE and analyzed by autoradiography for 24 h.
We next examined LANA's ability to interact in vitro with DEK by using a GST affinity assay. Expression of GST fusion proteins was examined by SDS-PAGE and Coomassie staining, and equal amounts of protein were used in each assay (data not shown). In Fig. 5B, in vitro-transcribed and -translated DEK was labeled with [35S]methionine and incubated with the GST proteins. The 43-kDa DEK protein interacted with the GST fusions expressing LANA(940-1164) (lane 1) and LANA(341-1164) (lane 3). There was a weak and possibly indirect interaction with GST-LANA(1-340) (lane 2). No interaction was observed with control GST-EBNA2(1-58) (GST-E2) (lane 4) or with GST protein (lane 5). Extract (2 μl) was loaded in lane 6. These results mapped the DEK interaction to the C terminus of LANA. To confirm the mapping data, an immunoprecipitation assay was performed with extracts of HeLa cells cotransfected with Flag-LANA or Flag-LANA-C and Myc-DEK (Fig. 5C). Immunoprecipitated proteins were analyzed by Western blotting, and LANA proteins were detected with an anti-Flag mouse antibody. Both Flag-LANA (lane 1) and Flag-LANA-C (lane 3) coprecipitated with Myc-DEK in immunoprecipitates generated with an anti-Myc antibody, but not those generated with a control mouse Ig antibody (lanes 2 and 4).
We further defined the DEK-interacting domain of LANA by using the Flag-tagged LANA deletion mutants shown in Fig. 5D (upper). The in vitro-transcribed and -translated LANA mutants were labeled with [35S]methionine and incubated with GST-DEK or control GST protein (Fig. 5D, lower panel). The LANA mutants m1 and m2 interacted with GST-DEK (lanes 1 and 4). No interaction was observed between LANA mutants m3 and m4 and GST-DEK (lanes 7 and 10) or between the LANA mutants and the control GST proteins (lanes 2, 5, 8, and 11). Two microliters of extract was loaded for each mutant (lanes 3, 6, 9, and 12). This assay indicated that LANA aa 986 to 1043 are required for interaction with DEK.
Localization of DEK on mouse chromosomes.
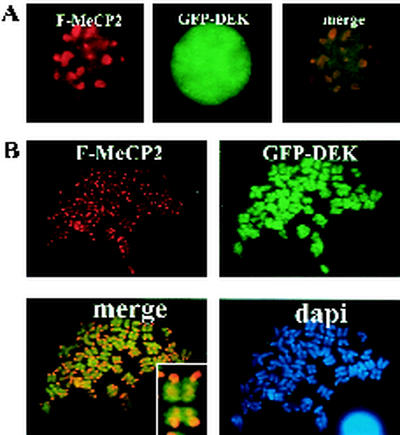
We investigated DEK's localization in mouse cells in relation to the localization of human MeCP2. NIH 3T3 cells were transfected with GFP-DEK and Flag-MeCP2, and an immunofluorescence assay was performed. GFP-DEK (green) was nuclear diffuse, in contrast to the Flag-MeCP2 nuclear spots (red) (Fig. 6A). Chromosome spreads of dually transfected cells revealed that GFP-DEK (green) painted mouse chromosomes, while Flag-MeCP2 (red) localized as before to PHC (Fig. 6B). Thus, DEK associates with mouse chromosomes, but is targeted in a different manner from MeCP2.
FIG. 6.
DEK interacts with mouse chromosomes. (A) NIH 3T3 cells were transfected with GFP-DEK and Flag-MeCP2. GFP-DEK (green) did not localize to sites of heterochromatin, unlike Flag-MeCP2 (red). (B) Chromosome spreads of NIH 3T3 cells transfected with GFP-DEK and Flag-MeCP2. GFP-DEK (green) painted mouse chromosomes and did not localize to the regions of heterochromatin marked by MeCP2 (red). Inset, higher magnification of the merged image.
LANA is targeted to mouse and human chromosomes by DEK.
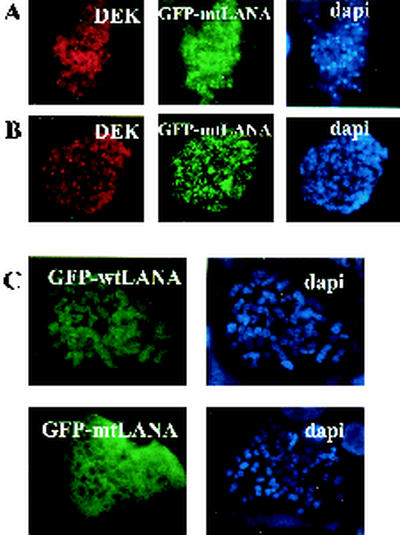
We have shown that the C terminus of LANA can interact with chromosomes and that the C terminus of LANA interacts with DEK. To investigate DEK's ability to target LANA to chromosomes, NIH 3T3 cells (Fig. 7A) or HeLa cells (Fig. 7B) were cotransfected with Myc-DEK and GFP-mtLANA lacking the N-terminal CBS and then blocked with Colcemid for 6 h, and chromosome spreads were performed. GFP-mtLANA (green) localized to mouse and human chromosomes in the presence of Myc-DEK (red). mtLANA did not associate with chromosomes when expressed alone (Fig. 7C). Taken together, the results indicate that DEK targeting through LANA aa 986 to 1043 provides a second mechanism by which LANA can bind to chromosomes. A model for LANA chromosomal tethering is presented in Fig. 8.
FIG. 7.
DEK can independently mediate LANA chromosomal tethering. Chromosome spreads of NIH 3T3 cells (A) and HeLa cells (B) transfected with GFP-mtLANA and Myc-DEK. Myc-DEK (red) targeted GFP-mtLANA (green) lacking the N-terminal CBS to mouse and human chromosomes. (C) Chromosome spreads of wild-type (wt) LANA versus mtLANA in HeLa cells. mtLANA lacking the N-terminal CBS does not associate with chromosomes. DNA was stained with DAPI (blue).
FIG. 8.
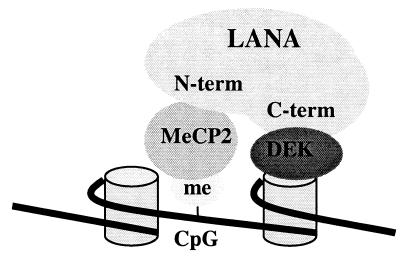
Model of LANA-mediated chromosomal tethering. LANA is targeted to chromosomes through N-terminal interactions with MeCP2, which binds methylated CpG dinucleotides in intranucleosomal linker DNA, and through C-terminal interactions with DEK, which associates with core histone proteins. Chromosomal association mediates episomal tethering through binding of the LANA C terminus to KSHV DNA (5, 10, 22).
DISCUSSION
LANA is a large multifunctional protein capable of interacting with a variety of cellular partners and playing a role in KSHV latency and KSHV-associated tumorigenesis. The colocalization of LANA with KSHV genomes on metaphase chromosomes and the requirement for LANA for episomal maintenance (4, 11) indicate that one of LANA's key functions in KSHV latency is to tether KSHV genomes to chromosomes during cell division. Szekely et al. (49) showed LANA associated with mouse chromosomes in mouse-PEL hybrids in which the human chromosomes were lost. However, we did not observe any independent LANA binding to mouse chromosomes in NIH 3T3 cells by our means of analysis and were able to use this lack of association as an assay to identify key human proteins that were necessary for chromosome tethering. We demonstrated that LANA is targeted to chromosomes via interactions with two human chromosome-associated cellular proteins, MeCP2 and DEK. Murine homologs of MeCP2 and DEK have been identified with 71 and 67% identities, respectively, to their human counterparts (42, 51). Either the murine MeCP2 and DEK homologs are poorly expressed in NIH 3T3 cells, or the association with LANA is mediated through nonconserved regions of these proteins. Previous studies identified a chromatin binding site in the LANA N terminus (38), and we now also describe a second C-terminal chromatin binding site within LANA aa 986 to 1043.
We found that LANA can be directed to mouse heterochromatin in the presence of human MeCP2. Methylation of cytosines at the carbon 5 position of CpG dinucleotides is a characteristic feature of many eukaryotic genomes. In vertebrates, somatic genomes are globally methylated, with 60 to 90% of all CpGs being methylated. This leaves a small portion of the genome, mostly consisting of CpG islands, in a methyl-free state (3). In the mouse genome, the PCH has the highest concentration of CpG methylation. The rat MeCP2 was the first protein identified to bind a single methylated CpG (29) via its N-terminus methyl binding domain (36). MeCP2 concentrates at mouse PCH while painting mouse chromosome arms at a background level. Human MeCP2 is ubiquitously expressed in adult tissues (12). Quantitative Western blots indicate ∼106 MeCP2 molecules per nucleus, while a typical diploid nucleus has ∼4 × 107 methyl CpGs, suggesting there are enough MeCP2 binding sites in vertebrate genomic DNA to saturate all MeCP2 molecules. Data suggest that MeCP2 only binds internucleosomal linker DNA by associating with methyl CpGs exposed in the major groove (8). MeCP2 appears literally as a million tiny spots along chromosome arms in mammals such as rats, hamsters, and humans, which have a broad distribution of methylated CpGs (35). LANA targeting to chromosomes via a protein with so many CBSs theoretically ensures that all episomes (estimated to be ∼25 to 80 copies per cell) (6, 7, 37) bound to LANA would be tethered to chromosomes and carried to daughter cells after cell division.
DEK was first identified in a chromosomal translocation with the CAN nucleoporin protein in a subset of acute myeloid leukemias (52). Autoantigens to DEK have been associated with several disease states, including systemic lupus erythematosus (13, 14, 53), juvenile rheumatoid arthritis (14, 34, 47, 50), and sarcoidosis (13, 14). Subsequently, DEK was described as a 43-kDa ubiquitously expressed DNA-binding phosphoprotein that recognized peri-ets sites in the human immunodeficiency virus type 2 enhancer (17, 19, 20), as a constituent of splicing complexes (33), and as a protein involved in changes of chromatin topology (1). Histones may play a supporting role in LANA tethering to chromosomes. DEK associates with histones H2A and H2B, as well as, to a lesser extent, histones H3 and H4 (1). It has been suggested that LANA tethers to chromosomes via histone H1, which is associated with heterochromatin. Interestingly, MeCP2 displaces histone H1 in order to gain access to its binding sites (35). MeCP2 and DEK both have broad distributions on human chromosomes, unlike the punctate localization of the LANA C terminus or of intact LANA on chromosomes of infected cells. The difference in LANA's chromosomal staining pattern in infected versus uninfected cells raises the possibility that episome binding may affect LANA's conformational structure and protein interactions. It is also likely that MeCP2 and DEK are part of larger functional complexes. These additional protein-protein interactions may be necessary for LANA's punctate localization on human chromosomes and the absence of such interactions in mouse cells could also account for the different distribution of LANA in those cells. Overall, the mechanism of LANA mediated chromosomal tethering appears more complex than that described for tethering by the EBV EBNA1 protein.
Acknowledgments
We thank S. Baylin for Flag-MeCP2 and Flag-HP1α plasmids, G. Anhault for sera from scleroderma patients, and D. Markovitz for GST-DEK. We thank K. Bachman, M. Rountree, and M. Strong for technical advice; Leslie Mezler at The Cell Imaging Core Facility for assistance with microscopy; M. Chiu for technical assistance; and F. Cheng for manuscript preparation.
This work was funded by National Institutes of Health award RO1 CA85151 to S.D.H. M.W. was partially supported by PHS training grant 5 T32 GM07445.
REFERENCES
- 1.Alexiadis, V., T. Waldmann, J. Andersen, M. Mann, R. Knippers, and C. Gruss. 2000. The protein encoded by the proto-oncogene DEK changes the topology of chromatin and reduces the efficiency of DNA replication in a chromatin-specific manner. Genes Dev. 14:1308-1312. [PMC free article] [PubMed] [Google Scholar]
- 2.An, J., A. K. Lichtenstein, G. Brent, and M. B. Rettig. 2002. The Kaposi sarcoma-associated herpesvirus (KSHV) induces cellular interleukin 6 expression: role of the KSHV latency-associated nuclear antigen and the AP1 response element. Blood 99:649-654. [DOI] [PubMed] [Google Scholar]
- 3.Antequera, F., and A. Bird. 1993. CpG islands, p. 169-185. In J. Jost and H. Saluz (ed.), DNA methylation: molecular biology and biological significance. Birkhauser Verlag, Basel, Switzerland.
- 4.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. [DOI] [PubMed] [Google Scholar]
- 5.Ballestas, M. E., and K. M. Kaye. 2001. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA. J. Virol. 75:3250-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Eng. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 7.Cesarman, E., P. S. Moore, P. H. Rao, G. Inghirami, D. M. Knowles, and Y. Chang. 1995. In vitro establishment and characterization of two AIDS-related lymphoma cell lines containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood 86:2708-2714. [PubMed] [Google Scholar]
- 8.Chandler, S. P., D. Guschin, N. Landsberger, and A. P. Wolffe. 1999. The methyl-CpG binding transcriptional repressor MeCP2 stably associates with nucleosomal DNA. Biochemistry 38:7008-7018. [DOI] [PubMed] [Google Scholar]
- 9.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 10.Cotter, M. A., II, C. Subramanian, and E. S. Robertson. 2001. The Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen binds to specific sequences at the left end of the viral genome through its carboxy-terminus. Virology 291:241-259. [DOI] [PubMed] [Google Scholar]
- 11.Cotter, M. A., II, and E. S. Robertson. 1999. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology 264:254-264. [DOI] [PubMed] [Google Scholar]
- 12.D'Esposito, M., N. A. Quaderi, A. Ciccodicola, P. Bruni, T. Esposito, M. D'Urso, and S. D. Brown. 1996. Isolation, physical mapping, and northern analysis of the X-linked human gene encoding methyl CpG-binding protein, MECP2. Mamm. Genome 7:533-535. [DOI] [PubMed] [Google Scholar]
- 13.Dong, X., M. A. Michelis, J. Wang, R. Bose, T. DeLange, and W. H. Reeves. 1998. Autoantibodies to DEK oncoprotein in a patient with systemic lupus erythematosus and sarcoidosis. Arthritis Rheum. 41:1505-1510. [DOI] [PubMed] [Google Scholar]
- 14.Dong, X., J. Wang, F. N. Kabir, M. Shaw, A. M. Reed, L. Stein, L. E. Andrade, V. F. Trevisani, M. L. Miller, T. Fujii, M. Akizuki, L. M. Pachman, M. Satoh, and W. H. Reeves. 2000. Autoantibodies to DEK oncoprotein in human inflammatory disease. Arthritis Rheum. 43:85-93. [DOI] [PubMed] [Google Scholar]
- 15.Dupin, N., T. L. Diss, P. Kellam, M. Tulliez, M. Q. Du, D. Sicard, R. A. Weiss, P. G. Isaacson, and C. Boshoff. 2000. HHV-8 is associated with a plasmablastic variant of Castleman disease that is linked to HHV-8-positive plasmablastic lymphoma. Blood 95:1406-1412. [PubMed] [Google Scholar]
- 16.Dupin, N., C. Fisher, P. Kellam, S. Arid, M. Tulliez, N. Franck, E. van Marck, D. Salmon, I. Gorin, J. P. Escande, R. A. Weiss, K. Alitalo, and C. Boshoff. 1999. Distribution of human herpesvirus-8 latently infected cells in Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma. Proc. Natl. Acad. Sci. USA 96:4546-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faulkner, N. E., J. M. Hilfinger, and D. M. Markovitz. 2001. Protein phosphatase 2A activates the HIV-2 promoter through enhancer elements that include the pets site. J. Biol. Chem. 276:25804-25812. [DOI] [PubMed] [Google Scholar]
- 18.Friborg, J. J., W. Kong, M. O. Hottiger, and G. J. Nabel. 1999. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature 402:889-894. [DOI] [PubMed] [Google Scholar]
- 19.Fu, G. K., G. Grosveld, and D. M. Markovitz. 1997. DEK, an autoantigen involved in a chromosomal translocation in acute myelogenous leukemia, binds to the HIV-2 enhancer. Proc. Natl. Acad. Sci. USA 94:1811-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu, G. K., and D. M. Markovitz. 1996. Purification of the pets factor. A nuclear protein that binds to the inducible TG-rich element of the human immunodeficiency virus type 2 enhancer. J. Biol. Chem. 271:19599-19605. [DOI] [PubMed] [Google Scholar]
- 21.Garber, A. C., J. Hu, and R. Renne. 2002. Latency-associated nuclear antigen (LANA) cooperatively binds to two sites within the terminal repeat, and both sites contribute to the ability of LANA to suppress transcription and to facilitate DNA replication. J. Biol. Chem. 277:27401-27411. [DOI] [PubMed] [Google Scholar]
- 22.Garber, A. C., M. A. Shu, J. Hu, and R. Renne. 2001. DNA binding and modulation of gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:7882-7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hung, S. C., M. S. Kang, and E. Kieff. 2001. Maintenance of Epstein-Barr virus (EBV) oriP-based episomes requires EBV-encoded nuclear antigen-1 chromosome-binding domains, which can be replaced by high-mobility group-I or histone H1. Proc. Natl. Acad. Sci. USA 98:1865-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kappes, F., K. Burger, M. Baack, F. O. Fackelmayer, and C. Gruss. 2001. Subcellular localization of the human proto-oncogene protein DEK. J. Biol. Chem. 276:26317-26323. [DOI] [PubMed] [Google Scholar]
- 25.Katano, H., Y. Sato, and T. Sata. 2001. Expression of p53 and human herpesvirus-8 (HHV-8)-encoded latency-associated nuclear antigen with inhibition of apoptosis in HHV-8-associated malignancies. Cancer 92:3076-3084. [DOI] [PubMed] [Google Scholar]
- 26.Kedes, D. H., M. Lagunoff, R. Renne, and D. Ganem. 1997. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi's sarcoma-associated herpesvirus. J. Clin. Investig. 100:2606-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knight, J. S., M. A. Cotter II, and E. S. Robertson. 2001. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus transactivates the telomerase reverse transcriptase promoter. J. Biol. Chem. 276:22971-22978. [DOI] [PubMed] [Google Scholar]
- 28.Krithivas, A., D. B. Young, G. Liao, D. Greene, and S. D. Hayward. 2000. Human herpesvirus 8 LANA interacts with proteins of the mSin3 corepressor complex and negatively regulates Epstein-Barr virus gene expression in dually infected PEL cells. J. Virol. 74:9637-9645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis, J. D., R. R. Meehan, W. J. Henzel, I. Maurer-Fogy, P. Jeppesen, F. Klein, and A. Bird. 1992. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell 69:905-914. [DOI] [PubMed] [Google Scholar]
- 30.Lim, C., Y. Gwack, S. Hwang, S. Kim, and J. Choe. 2001. The transcriptional activity of cAMP response element-binding protein is modulated by the latency associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Biol. Chem. 276:31016-31022. [DOI] [PubMed] [Google Scholar]
- 31.Lim, C., H. Sohn, Y. Gwack, and J. Choe. 2000. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) binds ATF4/CREB2 and inhibits its transcriptional activation activity. J. Gen. Virol. 81:2645-2652. [DOI] [PubMed] [Google Scholar]
- 32.Mattsson, K., C. Kiss, G. M. Platt, G. R. Simpson, E. Kashuba, G. Klein, T. F. Schulz, and L. Szekely. 2002. Latent nuclear antigen of Kaposi's sarcoma herpesvirus/human herpesvirus-8 induces and relocates RING3 to nuclear heterochromatin regions. J. Gen. Virol. 83:179-188. [DOI] [PubMed] [Google Scholar]
- 33.McGarvey, T., E. Rosonina, S. McCracken, Q. Li, R. Arnaout, E. Mientjes, J. A. Nickerson, D. Awrey, J. Greenblatt, G. Grosveld, and B. J. Blencowe. 2000. The acute myeloid leukemia-associated protein, DEK, forms a splicing-dependent interaction with exon-product complexes. J. Cell Biol. 150:309-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murray, K. J., W. Szer, A. A. Grom, P. Donnelly, J. E. Levinson, E. H. Giannini, D. N. Glass, and I. S. Szer. 1997. Antibodies to the 45 kDa DEK nuclear antigen in pauciarticular onset juvenile rheumatoid arthritis and iridocyclitis: selective association with MHC gene. J. Rheumatol. 24:560-567. [PubMed] [Google Scholar]
- 35.Nan, X., F. J. Campoy, and A. Bird. 1997. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell 88:471-481. [DOI] [PubMed] [Google Scholar]
- 36.Nan, X., P. Tate, E. Li, and A. Bird. 1996. DNA methylation specifies chromosomal localization of MeCP2. Mol. Cell. Biol. 16:414-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Neill, E., J. L. Douglas, M.-L. Chien, and J. V. Garcia. 1997. Open reading frame 26 of human herpesvirus 8 encodes a tetradecanoyl phorbol acetate- and butyrate-inducible 32-kilodalton protein expressed in a body cavity-based lymphoma cell line. J. Virol. 71:4791-4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piolot, T., M. Tramier, M. Coppey, J.-C. Nicolas, and V. Marechal. 2001. Close but distinct regions of human herpesvirus 8 latency-associated nuclear antigen 1 are responsible for nuclear targeting and binding to human mitotic chromosomes. J. Virol. 75:3948-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Platt, G. M., G. R. Simpson, S. Mittnacht, and T. F. Schulz. 1999. Latent nuclear antigen of Kaposi's sarcoma-associated herpesvirus interacts with RING3, a homolog of the Drosophila female sterile homeotic (fsh) gene. J. Virol. 73:9789-9795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Radkov, S. A., P. Kellam, and C. Boshoff. 2000. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells. Nat. Med. 6:1121-1127. [DOI] [PubMed] [Google Scholar]
- 41.Rainbow, L., G. M. Platt, G. R. Simpson, R. Sarid, S.-J. Gao, H. Stoiber, C. S. Herrington, P. S. Moore, and T. F. Schulz. 1997. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J. Virol. 71:5915-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reichwald, K., J. Thiesen, T. Wiehe, J. Weitzel, W. A. Poustka, A. Rosenthal, M. Platzer, W. H. Stratling, and P. Kioschis. 2000. Comparative sequence analysis of the MECP2-locus in human and mouse reveals new transcribed regions. Mamm. Genome 11:182-190. [DOI] [PubMed] [Google Scholar]
- 43.Renne, R., C. Barry, D. Dittmer, N. Compitello, P. O. Brown, and D. Ganem. 2001. Modulation of cellular and viral gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:458-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sarid, R., J. S. Wiezorek, P. S. Moore, and Y. Chang. 1999. Characterization and cell cycle regulation of the major Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter. J. Virol. 73:1438-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schwam, D. R., R. L. Luciano, S. S. Mahajan, L. Wong, and A. C. Wilson. 2000. Carboxy terminus of human herpesvirus 8 latency-associated nuclear antigen mediates dimerization, transcriptional repression, and targeting to nuclear bodies. J. Virol. 74:8532-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shire, K., D. F. J. Ceccarelli, T. M. Avolio-Hunter, and L. Frappier. 1999. EBP2, a human protein that interacts with sequences of the Epstein-Barr virus nuclear antigen 1 important for plasmid maintenance. J. Virol. 73:2587-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sierakowska, H., K. R. Williams, I. S. Szer, and W. Szer. 1993. The putative oncoprotein DEK, part of a chimera protein associated with acute myeloid leukaemia, is an autoantigen in juvenile rheumatoid arthritis. Clin. Exp. Immunol. 94:435-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos et al. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 49.Szekely, L., C. Kiss, K. Mattsson, E. Kashuba, K. Pokrovskaja, A. Juhasz, P. Holmvall, and G. Klein. 1999. Human herpesvirus-8-encoded LNA-1 accumulates in heterochromatin-associated nuclear bodies. J. Gen. Virol. 80:2889-2900. [DOI] [PubMed] [Google Scholar]
- 50.Szer, I. S., H. Sierakowska, and W. Szer. 1994. A novel autoantibody to the putative oncoprotein DEK in pauciarticular onset juvenile rheumatoid arthritis. J. Rheumatol. 21:2136-2142. [PubMed] [Google Scholar]
- 51.Takahashi, M., N. Seki, T. Ozaki, M. Kato, T. Kuno, T. Nakagawa, K. Watanabe, K. Miyazaki, M. Ohira, S. Hayashi, M. Hosoda, H. Tokita, H. Mizuguchi, T. Hayakawa, S. Todo, and A. Nakagawara. 2002. Identification of the p33(ING1)-regulated genes that include cyclin B1 and proto-oncogene DEK by using cDNA microarray in a mouse mammary epithelial cell line NMuMG. Cancer Res. 62:2203-2209. [PubMed] [Google Scholar]
- 52.von Lindern, M., M. Fornerod, S. van Baal, M. Jaegle, T. de Wit, A. Buijs, and G. Grosveld. 1992. The translocation (6;9), associated with a specific subtype of acute myeloid leukemia, results in the fusion of two genes, dek and can, and the expression of a chimeric, leukemia-specific dek-can mRNA. Mol. Cell. Biol. 12:1687-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wichmann, I., N. Respaldiza, J. R. Garcia-Lozano, M. Montes, J. Sanchez-Roman, and A. Nunez-Roldan. 2000. Autoantibodies to DEK oncoprotein in systemic lupus erythematosus (SLE). Clin. Exp. Immunol. 119:530-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wreggett, K. A., F. Hill, P. S. James, A. Hutchings, G. W. Butcher, and P. B. Singh. 1994. A mammalian homologue of Drosophila heterochromatin protein 1 (HP1) is a component of constitutive heterochromatin. Cytogenet. Cell Genet. 66:99-103. [DOI] [PubMed] [Google Scholar]
- 55.Wu, H., D. F. Ceccarelli, and L. Frappier. 2000. The DNA segregation mechanism of Epstein-Barr virus nuclear antigen 1. EMBO Rep. 1:140-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yates, J. L., N. Warren, and B. Sugden. 1985. Stable replication of plasmids derived from Epstein-Barr virus in a variety of mammalian cells. Nature (London) 313:812-815. [DOI] [PubMed] [Google Scholar]