Abstract
The division into a latent or lytic life cycle is fundamental to all herpesviridae. In the case of Kaposi's sarcoma-associated herpesvirus (KSHV) (human herpesvirus 8), latent genes have been implicated in cell autonomous transformation, while certain lytic genes procure a tumor friendly milieu through paracrine mechanism. To query KSHV transcription, we devised and validated a high-throughput, high-specificity, high-sensitivity, real-time quantitative reverse transcription-PCR array. This novel methodology is applicable to many human pathogens. Its first use demonstrated that the mRNA levels for KSHV LANA, v-cyclin, and v-FLIP do not increase at any time after viral reactivation. The mRNA for LANA-2/vIRF-3 is similarly resistant to viral reactivation. In contrast, every other latent or lytic message was induced. Hence, LANA, v-FLIP, v-cyclin, and LANA-2 constitute a group of uniquely regulated transcripts in the KSHV genome.
By using representational difference analysis, Chang et al. (9) demonstrated the presence of a novel human virus in Kaposi's sarcoma (KS) biopsies: Kaposi’s sarcoma-associated herpesvirus (KSHV) (human herpesvirus 8). Solid epidemiological evidence has since been assembled, which links KSHV to KS (20, 21, 32, 40, 44, 73). Classic KS was first described in 1872 as a rare, disseminated sarcoma of the skin (reviewed in reference 1). KS is endemic in parts of equatorial Africa, where it is responsible for an estimated 1% of all adult tumors. In these regions, evidence for KSHV infection can be observed even before puberty (reviewed in reference 72). Concomitant, widespread human immunodeficiency virus type 1 (HIV-1) infection has since turned KS into an epidemic disease. For instance, the seroprevalence levels for KSHV reach 30% in black South African HIV patients (66), and childhood KS has become the most common neoplasm in this part of the continent. KS has also been documented in solid organ and bone marrow transplant recipients, in whom it can comprise up to 3% of all tumors depending on the regional prevalence of KSHV (38, 39, 43), and in 1981 KS was recognized as a signature pathology of AIDS. Recently, mucosal shedding of KSHV was verified for a group of men in the United States (49, 71) demonstrating that KSHV, like other members of the herpesviridae, is transmitted by saliva. KSHV can be detected in all manifestations of KS, and within the lesions all tumor cells, but not infiltrating lymphocytes, carry the viral genome (4, 68). The virus is also present in multifocal Castleman's disease (48, 67) and in primary effusion lymphoma (PEL) (8). A number of cell lines have been established from these PEL patients. Since KSHV otherwise replicates extremely poorly in culture, PEL cell lines constitute the primary system to investigate KSHV biology. Based on the complete sequence of the 137-kbp unique region, KSHV is classified as a gamma-2-herpesvirus, a member of the lymphotropic subgroup of the Herpesviridae family (25, 47, 60).
All members of the Herpesviridae display a tightly regulated program of gene expression (reviewed in reference 59). KSHV, too, conforms to this paradigm and can enter one of two modes of infection: (i) latent infection, in which the viral genome persists in its host cell but with dramatically restricted gene expression and without cell destruction (75), and (ii) lytic infection, which generates infectious progeny and destroys the host cell. Lytic infection, in turn, can be divided into alpha or immediate-early, beta or early, and gamma or late stages. By using genomic KSHV DNA fragments as probes, Sarid et al. defined three classes of differentially transcribed messages in a KSHV-infected PEL cell line (63). Class I mRNAs can be detected in untreated cultures and are not increased after 12-O-tetradecanoylphorbol-13-acetate (TPA) treatment, which reactivates the KSHV lytic replication cycle. Class II mRNAs can be detected in untreated cultures but are greatly increased by TPA treatment. Finally, class III mRNAs can only be detected in TPA-treated cultures. Translated into the customary herpesvirus classification (59), class III mRNAs correspond to lytic transcripts (alpha, beta, and gamma) and class I correspond to latent transcripts. Class II mRNAs present a conundrum, since it is not clear whether the mRNAs detected in untreated cultures stem from the few percent of cells that undergo spontaneous lytic reactivation in culture (56). These data were recently confirmed and expanded upon by using hybridization KSHV-specific cDNA arrays (28, 50). In Epstein-Barr virus, a related human gammaherpesvirus, latency-associated genes are essential for episome maintenance, as well as for host cell transformation (reviewed in reference 57). This also holds true for at least one KSHV type one latency gene: the latency-associated nuclear antigen, LANA (2, 12, 19, 53). In fact, LANA was the first latent protein of KSHV to be identified based on its immunoreactivity with patient sera (31, 33, 54). We therefore sought to determine whether there are other type one latency mRNAs in the KSHV genome and, if so, how many?
To address this question, we developed a novel methodology for scrutinizing transcription. Our genome scan is based on real-time quantitative reverse transcription-PCR (RT-PCR) (26). RT coupled to amplification by PCR is widely recognized as the most sensitive method for detecting the presence of specific RNAs. Unlike conventional DNA arrays, RT-PCR can distinguish between various spliced mRNAs through exon-specific primers. Real-time quantitative PCR measures the amount of PCR product at each cycle of the reaction either by binding of a fluorescent, double-strand-specific dye (SYBRgreen) or by hybridization to a third sequence-specific, dual-labeled fluorogenic oligonucleotide (Molecular Beacon or TaqMan). By evaluating primers specific for every predicted open reading frame (ORF) in the KSHV genome, we found that, in addition to LANA, v-cyclin, and v-FLIP—all of which are under control of a common cis-regulatory element (15, 64, 70)—LANA-2/vIRF-3 is encoded by a second latency type one mRNA in the KSHV genome. No other KSHV mRNA displays a comparable pattern of regulation. This observation establishes LANA, v-cyclin, v-FLIP, and LANA-2/vIRF-3 as markers for latent KSHV infection and implicates them in viral persistence and tumorigenesis.
MATERIALS AND METHODS
Cell lines and induction conditions.
BCBL-1 cells were cultured in RPMI 1860, 25 mM HEPES (pH 7.55), 15% fetal bovine serum, 0.05 mM 2-mercaptoethanol, 1 mM sodium pyruvate, 2 mM l-glutamine, 0.05 μg of penicillin/ml, and 50 U of streptomycin/ml (all reagents from Life Technologies, Rockville, Md.) at 37°C and in 5% CO2. Cells were split every 5 days to 2 × 105 cells/ml, and a fresh aliquot was thawed after 30 passages. Where indicated, the cells were reseeded to 2 × 105 cells/ml and induced 24 h later with 40 ng of TPA/ml (×10,000 stock in dimethyl sulfoxide; Calbiochem, San Diego, Calif.), 1 μM ionomycin (×2,000 stock in ethanol; Sigma, St. Louis, Mo.), or 0.8 mM sodium butyrate (×1,000 stock in ethanol; Sigma) for 24 h. Afterward the cells were collected by centrifugation (200 × g for 10 min at 4°C in a Hereaus Megafuge) and resuspended in an equal amount of fresh medium. Cells were collected at the indicated times after induction by centrifugation, washed once in phosphate-buffered saline, pelleted, and resuspended in 500 μl of RNAzol (Tel-Test, Inc., Friendswood, Tex.) per 107 cells.
RNA isolation and RT.
RNA was isolated by using RNAzol according to the supplier's protocol. Poly(A) mRNA was prepared by using dT-beads (Qiagen, Inc., Valencia, Calif.) according to the manufacturer's recommendations, and 500 ng of mRNA was reverse transcribed in a 20-μl reaction with 100 U of Moloney murine leukemia virus reverse transcriptase (Life Technologies), 2 mM deoxynucleoside triphosphates, 2.5 mM MgCl2, 1 U of RNasin (all from Applied Biosystems, Foster City, Calif.), and 0.5 μg of random hexanucleotide primers (Amersham Pharmacia Biotech, Piscataway, N.J.). The RT reaction was sequentially incubated at 42°C for 45 min, 52°C for 30 min, and 70°C for 10 min. The reaction was stopped by heating it to 95°C for 5 min, 0.5 μl of RNase H (Life Technologies) was added, and the reaction was incubated at 37°C for an additional 30 min. Afterward, the cDNA pool was diluted 25-fold with diethyl pyrocarbonate-treated distilled H2O and stored at −80°C.
Primer design.
The predicted KSHV ORFs were extracted from the complete KSHV sequence as described by Russo et al. (60). Quantitative real-time PCR primer pairs were designed by using Prime Express 1.5 (Applied Biosystems) to comply with TaqMan criteria (5), namely, a Tm of 59 ± 2°C for each primer, a maximal Tm difference for both primers of ≤2°C, a GC content of 20 to 80%, no GC clamp, a primer length of between 9 and 40 nucleotides, fewer than four repeated G residues per primer, no hairpins with a stem size of ≥4, and a amplicon size 50 to 150 bp. Where applicable, TaqMan probes were designed to have a Tm to be greater than 10°C compared to the flanking primer pair and no G at the 5′ end. The top 100 potential primer pairs for each KSHV ORF were visually inspected, and the highest ranking primer-probe combination was selected, unless a primer-probe combination with similar characteristics but closer to the 3′ end of the ORF could be identified. In cases where the predicted ORFs overlap (orfK3 exon 1 and orf70; orf17 and orf18; orf19, orf20, and orf21; as well as spliced forms of orfK8), primers were selected outside the region of overlap. In the absence of a complete transcript map for this virus, we cannot, however, exclude the possibility that some primer combinations were located in regions in which the 3′ untranslated region (3′UTR) or the 5′UTR segments overlap. Primer-probe combinations for known spliced transcripts were designed by using relaxed criteria in order to span splice junctions. The primer sequences are available online (http://w3.ouhsc.edu/mi/frame_faculty/dittmer.html).
Real-time quantitative PCR.
The set of forward primers was synthesized on a separate plate then the reverse primers were synthesized by using a MerMade 96-well synthesizer (BioAutomation Corp., Plano, Tex.) and stored in distilled H2O at −80°C at a concentration of 100 pmol/μl (100 μM). Primer length and purity was verified by using a P/ACE MDQ Capillary Electrophoresis System (Beckman Coulter, Inc., Fullerton, Calif.). Forward and reverse primers were combined from the master plate to yield enough primer pairs for 20 assays at a time, and their concentrations were adjusted to 1 pmol/μl (1 μM). The final PCR contained 2.5 μl of primer mix (final concentration, 166 nM), 7.5 μl of 2× SYBR PCR mix (Applied Biosystems), and 5 μl of sample. For TaqMan PCR, FAM-TAMRA-labeled probe (Applied Biosystems) was stored at 100 μM for long-term at −80°C and added to the primer mix to yield a 1 μM stock and a 166 nM final concentration. In this case the PCR contained 2.5 μl of primer-probe mix (final concentration, 166 nM), 7.5 μl of 2× TaqMan PCR mix (Applied Biosystems), and 5 μl of sample. PCR was set up in a segregated, locked, “white” room (using designated pipettes and filtered tips), in which only primers and PCR mix was handled. Here all surfaces and tools were treated with 10% bleach monthly and exposed to ceiling UV lights overnight. Designated gowns, gloves, and face masks were required for all work. Samples were prepared and added to the PCR mix in a second, “gray” room, in which no cloned KSHV plasmids were present, by using a designated set of pipettes and filtered tips. Real-time PCR was performed by using an ABI PRIZM5700 or ABI PRIZM7700 machine (Applied Biosystems) and universal cycle conditions (2 min at 50°C and 10 min at 95°C and then 40 cycles of 15 s at 95°C and 1 min at 60°C) (26). CT values were determined by automated threshold analysis by using the ABI PRIZM software. Here, the threshold was set to five times the standard deviation (SD) of the non-template control. Dissociation curves were recorded after each run, and the amplified products were routinely analyzed by agarose gel electrophoresis.
Calculations.
In order to evaluate the significance of the real-time quantitative PCR array data, some theoretical observations seem in order. (i) Prior to reaching saturation (due to exhaustion of primers and nucleotides, loss of polymerase activity, etc.), PCR amplification proceeds exponentially and can be described by Ni = N0 × (1 + k)i, where N0 represents the number of molecules in the original sample and Ni is the number of mRNA molecules at cycle i (i = 0 . . . 40). During the exponential phase the amplification efficiency k (0 ≤ k ≤ 1) of a given primer pair is constant. Real-time quantitative PCR visualizes the progressive amplification at each cycle and measures the amount of product under exponential conditions, exclusively. (ii) The amount of PCR product at each cycle is quantified by the incorporation of a fluorescent dye (SYBRgreen) or the cleavage of a TaqMan oligonucleotide (26). By either method the fluorescence intensity Rn has a logarithmic dependence on fluorophor concentration, yielding Rn = log(Ni) = log[N0 × (1 + k)i]. Real-time quantitative PCR compares two samples with the target concentrations Na and Nb by recording the cycle numbers (CT) for a and b at which the amplification product yields enough fluorescence to cross an operator determined threshold, T (set at five times the SD of the non-template control). Consequently, Rna = Rnb and log[Na × (1 + k)a] = log[Nb × (1 + k)b] or log(Na) − log(Nb) = log(1 + k)b − log(1 + k)a = log(1 + k)(b − a) (for i = 0, Ni = 0 = N0 × (1 + k)0, i.e., Ni = 0 = N0). Ideally, k = 1 and (1 + k) = 2, i.e., at each cycle two reactions products are produced per target molecule. This leads to Ni = N0 × (1 + 1)i = N0 × 2i. Assuming that log = log2, Na/Nb = 2(b − a), where Na/Nb represents the fold difference in mRNA levels of two samples, CT = a and CT = b. The algorithm used here was designed to identify primer pairs, which under universal cycling conditions (94°C for 30 s and 60°C for 60 s [40 cycles]) have an amplification frequency of 1.8 ≤(1 + k) ≤ 2. This is achieved by restricting the amplicon length to 100 ± 50 bp and the primer Tm to 60 ± 1°C for each primer. Hence, it is possible to extract the relative ratio of abundance in two samples based on this calculation. (iii) By comparison, solution hybridization-based DNA arrays have similar characteristics, since the color intensity ratio in a fluorescent Cy3/Cy5 DNA array also exhibits a logarithmic dependence on the amount of hybridized probe. This is simply the nature of fluorescent absorption (7). Analogous to the amplification efficiency k for PCR, a hybridization-efficiency K0 applies to DNA arrays, which is a function of the length and base composition of the particular cDNA fragment at a given hybridization temperature. For the purpose of this study, we therefore applied similar, rank-based statistics and cluster algorithms (17) to compare the relative ratios of the mRNA levels between different samples as determined by real-time quantitative RT-PCR.
RESULTS
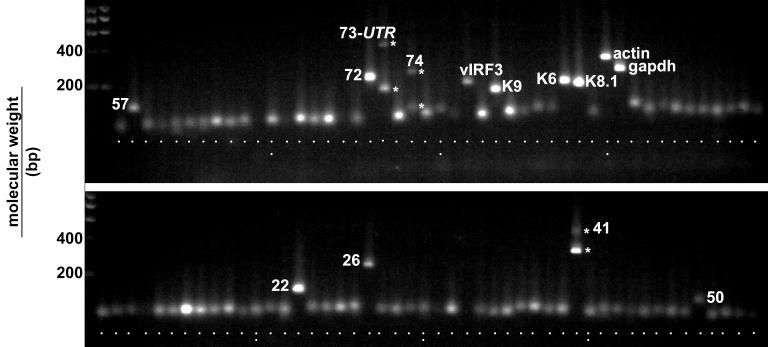
Quantitative real-time PCR primers pairs were designed for each KSHV ORF as described in Materials and Methods. Table 1 lists a subset of primer pairs for which we generated additional, sequence-specific TaqMan probes. This list compiles primers for spliced mRNAs of each KSHV kinetic class (latent, alpha, beta, and gamma). To determine the specificity of each primer pair, we analyzed the amplification products by gel electrophoresis. Figure 1 shows an ethidium bromide-stained agarose gel of the amplification products after 40 cycles with cDNA from TPA-induced BCBL-1 cells as a template. No primer dimers are visible in any of the reactions. The majority of amplicons are of a similar size (ca. 50 to 100 bp) as predicted by the design. Only a single band is amplified in each case even after 40 cycles. Exceptions with regard to size include some of the amplicons, which were designed to cross splice junctions or which required special design, namely, orf57 (115 bp), orf72/v-cyclin (192 bp), orf73/LANA 5′UTR splice (71 bp spliced and 174 bp unspliced), LANA-2/vIRF-3spliced (174 bp spliced and 268 bp unspliced), K9/vIRF-1 (141-bp latent start site and 61-bp lytic start site [10]), K6 (172 bp), K8.1 (255 bp unspliced and 161 bp spliced), actin (295 bp), GAPDH (for glyceraldehyde-3-phosphate dehydrogenase) (225 bp), K14/vGPCR (234 bp spliced), orf22 (123 bp), orf26 (223 bp), orf41 (324 bp spliced and 452 bp unspliced), and orf50 (111 bp). These results imply that unprocessed RNA or DNA species, which contain small introns (<500 bp) can be amplified under universal cycle conditions, even though this is less efficient compared to the smaller, correctly spliced variant. Significant amounts of the unspliced form only accumulated at higher cycle numbers (compare the band intensities for the spliced and unspliced forms in the case of orf73 5′UTR, vGPCR, and orf41). Since quantification by real-time PCR is obtained from the earlier, exponential phase of the reaction, these products did not affect the outcome. In the case of the LANA/orf73 we previously cloned two isoforms of its mRNA: one spliced and another unspliced in the 5′UTR (15). Therefore, both PCR bands stem from authentic mRNAs rather than from genomic DNA. With regard to orf74/vGPCR a novel mRNA was recently found which is unspliced and originates within the K14 ORF (46). This mRNA species can be amplified with our primers and might explain the upper band on the agarose gel. Large introns (>500 bp) were never amplified under universal cycling conditions, as evidenced by the absence of any high-molecular-size band in the case of orf29 (3,251-bp intron) and v-cyclin (4,037-bp intron). Primer pairs for K1 were directed against the intracellular domain of the ORF, which is conserved for all clades of KSHV (51). The primer pair for K15 did not work in any of the assays, since primer sequences were based on a BC-1-derived isolate (60) rather than BCBl-1 isolate, which differs in the K15 region (11, 24, 30, 51, 77) (a separate effort is currently under way to design primers that are specific for the different K15 spliced isoforms and different clades). Similarly, primers for K12/kaposin were omitted from the array, since kaposin mRNA varies widely between isolates (45, 61, 75). In contrast to the CT values determined during exponential amplification by real-time quantitative PCR, any differences in the amounts of product seen by gel electrophoresis were coincidental. Overall, primers that were designed based only on TaqMan criteria yielded the expected products and did not display cross-reactivity to cellular mRNAs.
TABLE 1.
Primer and probe sequences for selected KSHV latent and lytic mRNAs
| KSHV TaqMan primer-probe pair | Primer sequence
|
Probe sequence | Spliced | |
|---|---|---|---|---|
| Forward | Reverse | |||
| Human | ||||
| hu-actin | TCACCCACACTGTGCCATCTACGA | CAGCGGAACCGCTCATTGCCAATGG | ATGCCCCCCCCATGCCATC | No |
| hu-GAPDH | GAAGGTGAAGGTCGGAGTC | GAAGATGGTGATGGGATTTC | CAAGCTTCCCGTTCTCAGCC | No |
| Latent | ||||
| v-cyclin | AGCTGCGCCACGAAGCAGTCA | CAGGTTCTCCCATCGACGA | TAGCGTACTCTCGCGGCCCAGC | Yes |
| LANA-2/vIRF-3 | TCCTCAGATTCCGCGCC | TCACCTACACAGTGGGTCATCAC | CTGCGTGACCGGCACATCGC | Yes |
| Alpha/beta | ||||
| orf57 | TGGACATTATGAAGGGCATCCTA | CGGGTTCGGACAATTGCT | ACGGACGCACCGACACTGGAAGA | Yes |
| orf50 | CACAAAAATGGCGCAAGATGA | TGGTAGAGTTGGGCCTTCAGTT | AGAAGCTTCGGCGGTCCTG | Yes |
| vGPCR | TGGCCCAAACGGAGGATCCTAG | AGTTTCATTCCAGGATTCATCATC | AGAAGATGGTTAGGAAATCCTCGGC | Yes |
| k-bZIP | TGTGCCGTCGTCCGG | TGGATGGTTCCCCAGATGA | AGCAGCCAGCGTCGCCAACG | Yes |
| Gamma-2 | ||||
| orfK8.1 | AAAGCGTCCAGGCCACCACAGA | GGCAGAAAATGGCACACGGTTAC | ATGCCTTAATATCAGCCTTTTCA | Yes |
| orf29 | CCCGGAGGACGGTCCA | CCCCGAATGCTCTGTTCTTATT | CTCGCTGATGTGCGCAACATGCT | Yes |
FIG. 1.
Ethidium bromide-stained 2% agarose gel of the PCR products for each KSHV ORF after 40 cycles. The template was reverse transcribed poly(A) mRNA from BCBL-1 cells 48 h after TPA induction. Most amplicons are of the same size; exceptions are noted by name.
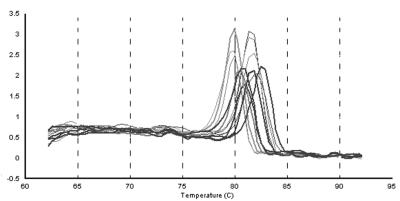
To obtain an independent measure of primer specificity, we performed melting curve analysis after the final amplification cycle. The reaction was gradually heated and actual Tm values were obtained since SYBRgreen fluorescence is lost during duplex to single-strand phase transition. Figure 2 shows the melting curves for 10 representative amplicons. In each case the single peak melting curve indicated that only a single amplification product was present in the reaction. Individual Tm values were not identical, since they depended on the length and base composition of the entire amplified sequence rather than those of the flanking primers. Every amplicon in this study yielded similar single peak melting curves (data not shown). Taken together, the results obtained by these analyses instill confidence in the specificity of the KSHV real-time quantitative PCR array.
FIG. 2.
Melting curve profile for selected KSHV ORFs after 40 cycles (amplification products for orf26, K9spl, LANA-2spl, actin, KbZIP, orf40spl, K9, LANA-2unspl, v-cyc, orf40unspl, orf57spl, orf57unspl). The template was reverse transcribed poly(A) mRNA from BCBL-1 cells 48 h after TPA induction. The temperature is shown on the horizontal axis. The vertical axis shows the value of the first derivative of the change in fluorescence intensity at each temperature. Thus, the peak identifies the phase transition.
Genome scan by real-time quantitative RT-PCR.
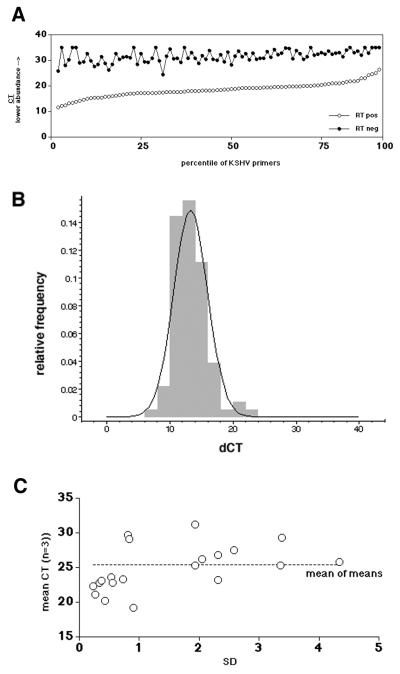
To demonstrate that the PCR signal is a measure of the amount of reverse-transcribed mRNA rather than contaminating genomic DNA, we compared RT-positive to RT-negative cDNA pools from TPA-induced BCBL-1 cells. Equal amounts of total poly(A) mRNA (as determined by absorption spectroscopy at 260 nm) served as starting material. Figure 3A plots the unmanipulated CT values for each KSHV-specific primer pair obtained under either condition (as mentioned above, CT values represent the amount of target on a log2 scale). In this case, primer pairs were sorted based on the CT of the RT-positive reaction rather than their coordinates on the KSHV genome. For each individual primer pair, RT-negative samples contained much less target (as indicated by the higher CT value) than the RT-positive samples. In all subsequent experiments we adjusted the amount of input cDNA so that no amplification could be observed with RT-negative samples. This demonstrates that our assay measures KSHV mRNA levels and not contaminating viral DNA.
FIG. 3.
(A) CT values for each primer pair in the KSHV array. Closed circles indicate the CT values if poly(A) mRNA without RT was used (RTneg); open circles indicate the CT values if the mRNA was subjected to RT (RTpos). The vertical axis indicates the CT value, and the horizontal axis indicates the percentile of primers. (B) Frequency histogram of the dCT difference between RTpos and RTneg reactions for each primer pair. Superimposed is the expected normal distribution. The relative frequency is shown on the vertical, and the dCT is shown on the horizontal axis. (C) SD (on the horizontal scale) and mean (n = 3) CT values for a group of amplicons. The dotted line indicates the mean of means.
Figure 3 also introduces the first layer of our quantitative analysis. CT values are not a linear measure of target quantity, since they depend logarithmically on the amplification efficiency k for each primer pair, as well as the initial target copy number N0. Although absolute copy numbers can be determined through dilution of an external standard, this is not practical for a 96-primer pair array. Nor is it necessary for the purpose of a genome scan experiment, which is—just like hybridization-based DNA arrays—concerned only with the differences between different treatments. Those were calculated as follows: dCT = CT (mock) − CT (treatment) for each primer pair.
At no point in this study did we attempt to compare CT values between two different primer pairs. However, we calculated the mean difference over all primer pairs between the RT-positive and RT-negative sample. Figure 3B shows the relative frequency of dCT = CT (RTneg) − CT (RTpos). The dCTs are normally distributed with mean difference of 13.25 and an SD of ±2.66. Based on an amplification efficiency of 1.8 < (1 + k) < 2 (213.25 = 9,742 and 1.813.25 = 2,412, respectively), the fraction of contaminating DNA accounted for no more than 0.05% of the signal. Figure 3B also superimposes the normal distribution based on these parameters, which fits the experimental data. The fact that CT values have a logarithmic dependence on the mRNA concentration is extremely beneficial for our statistical analysis, since the data are inherently variance stabilized (17, 23).
What is the measurement error and linear range in these experiments? To gain an understanding of the limitations of the real-time quantitative RT-PCR array, two kinds of replicate experiments were conducted. (i) We previously published the dilution profiles for several primer pairs (29). This emphasized the extraordinarily robustness of real-time quantitative PCR, since the CT values were linear dependent on input cDNA over more than 4 orders of magnitude, with as little as 103 BCBL-1 cell equivalents as input. We have obtained similar performance data for the primer pairs used here (data not shown). All data in this particular study are based upon input mRNA amounts, which were poly(dT) purified from ≥104 BCBL-1 cells per primer pair (or 106, i.e., 1 ml of culture per 96-well array). Hence, we were in the middle of the linear performance range of the assay. (ii) To determine the combined handling and instrument error, we performed triplicate measurements with a 22-primer-pair subset. Figure 3C compares the standard deviation (SD) values for each triplicate measurement (on the horizontal axis) to the mean CT value. No correlation between the dCT and the SD is evident (R < 0.4 by linear regression analysis). This establishes that the experimental error is independent of the target concentration N0 or amplification efficiency k of a given primer pair.
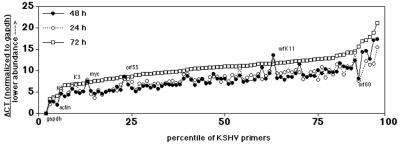
Biological variation accounts for the overwhelming majority of error in most genome array applications. To determine the contribution of changes in culture conditions to our array analysis, we prepared mRNA from mock-treated BCBL-1 cells cultured for 24, 48, or 72 h and determined the abundance of KSHV mRNAs by using the real-time RT-PCR array. Initially, we normalized each RT reaction according to the total amount of input poly(A) mRNA. Under these circumstances the level of GAPDH mRNA did not change significantly over time (CTgapdh = 12.71 at 24 h, CTgapdh = 13.58 at 48 h, and CTgapdh = 12.05 at 72 h). Since GAPDH mRNA was also the most abundant mRNA species, we normalized all individual CT values according to dCTtest = CTtest − CTgapdh. Figure 4 shows the resulting dCT values for each of the KSHV primer pairs at indicated times in culture. As a result of this normalization, higher dCT values represent lower target concentrations relative to the level of GAPDH mRNA. Primer pairs were sorted according to dCT values at t = 72 h. The dCT values at 24 and 48 h are virtually identical for every mRNA. In contrast, all values were increased after 72 h in the same culture medium, i.e., expression levels were lower due the cells entering saturation phase. Interestingly, this result was observed after we adjusted for fluctuations in GAPDH mRNA levels. This implies that the KSHV transcripts levels were depleted in the stationary phase relative to GAPDH. An exception were the mRNA levels for K3, K5, orf55, and K11 which were identical at all three time points (SD ≤ 0.5). RT-PCR was able to detect a signal for lytic KSHV mRNAs even in the absence of induction, since on average 3% of the BCBL-1 cells undergo spontaneous reactivation in culture (56). If we assume a total input of 106 cells, that amounts to ≥104 cells, which is well within the sensitivity of RT-PCR. To describe this result in quantitative terms, the normal distributions for the relative differences, ddCT = dCT(t = 72) − dCT(t = 24, t = 48), were calculated for each primer pair and used to construct frequency histograms. The mean ddCT for 48 and 24 h for all 96 primer pairs is ddCT = −0.30 ± 1.4 compared to ddCT = 2.44 ± 1.6 for 72 and 24 h and ddCT = 2.74 ± 1.3 for 72 and 48 h. The difference between the 72 h and 24 h time points was significant to P ≤ 5 × 10−7 based on a one-way analysis of variance (ANOVA), whereas the differences in KSHV mRNA levels between 48 and 24 h were insignificant. The data presented henceforth were obtained from BCBL-1 cells, which were under experimental conditions for 48 h or less. The data in Fig. 4 were used to establish the average margin of error for further studies, i.e., the minimal biologically meaningful difference, which is significant to dCT ≥ 1.50 or 21.5 ≥3-fold (5 × the mean ddCT = 0.30 for the 48- and 24-h replicate measurement of n = 96 primers).
FIG. 4.
dCT values, which were normalized to CTgapdh for each primer pair in the KSHV array. The template was poly(A) mRNA from untreated BCBL-1 cells held in culture for 24, 48, and 72 h. The vertical axis indicates the dCT value, and the horizontal axis indicates the percentile of primers.
Identification of type one latency mRNAs in the KSHV genome.
We and others previously identified a cluster of KSHV latent mRNAs, based upon in situ hybridization of KS tumor biopsies (13, 15, 54, 70). These experiments showed that the LANA/orf73, v-cyclin/orf72, and v-FLIP/orf71 mRNAs—unlike that of the latent mRNA for kaposin—were not induced upon reactivation of BCBL-1 cells. The mRNAs encoding LANA, c-cyc, and v-FLIP are coterminal, and the common promoter for these mRNAs, likewise, was resistant to TPA stimulation (29). Another latent mRNA, encoding LANA-2/vIRF-3, has recently been described, but conflicting data have been reported with regard to its expression profile (37, 58). We therefore applied the KSHV real-time quantitative PCR array to distinguish between the hypotheses (i) that LANA-1 was the only type one latent mRNA in the KSHV genome, (ii) that LANA-2 also belonged to this class, and (iii) that there might be additional type one latent messages.
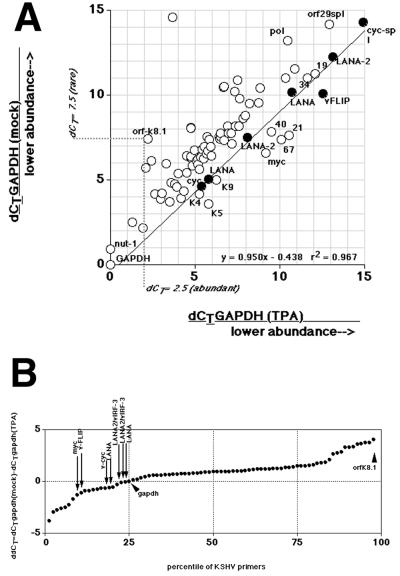
First, poly(A) mRNA was isolated either from TPA-treated or mock-treated BCBL-1 cells at 48 h after induction and subjected the real-time quantitative RT-PCR. Figure 5A shows the outcome of such an experiment. In this representation dCT values (normalized relative to GAPDH) are used as x and y coordinates and are plotted for every primer pair of the KSHV array. Hence, GAPDH marks the origin at 0,0. If two dCT values are identical under either condition, they specify a point on the 45° axis (slope of 1). If a given mRNA is induced by TPA (lower dCT), the point is above, and if it is inhibited, the point is below the 45° axis. orfK8.1 illustrates this projection: mock-treated BCBL-1 cells yielded a dCT = 7.5, which is indicated on the y axis. TPA-treated BCBL-1 cells yielded a dCT = 2.5, which is indicated on the x axis. As before, a lower dCT corresponds to higher target concentrations. Assuming an amplification efficiency of (1 + k) = 2, the induction can be approximated to 2(7.5 − 2.5 = 5) = 64-fold, which agrees with existing Northern blot and protein data (36, 52, 76). Almost every KSHV mRNA is induced at 48 h after TPA treatment, as evidenced by the location of their corresponding dCT above the 45° line. Several primer pairs in the array are specific for the two latent mRNAs encoding LANA, v-cyclin, and v-FLIP. All fall on the 45° axis, as indicated by the slope of regression m = 0.958 with R2 = 0.967. The “LANA” primer pairs are located within ORF LANA or in the LANA 5′UTR, primer pair “v-FLIP” is located within ORF v-FLIP, primer pair “cyc” is located within ORF v-cyclin, and primer pair “cyc-spl” spans the v-cyclin splice site. The different amplicons do not have identical dCTs, because the dCT values depend on (i) the level of either the tricistronic 5,300-nucleotide mRNA encoding LANA, v-cyclin, and v-FLIP or the bicistronic 1,700-nucleotide mRNA encoding v-cyclin and v-FLIP, as well as (ii) the individual amplification efficiencies k for each primer pair. The dCT values for LANA-2/vIRF-3 (two primer pairs) also fall into this group. Hence, LANA-2/vIRF-3 mRNA is another latency type one mRNA in the KSHV genome (as represented on our array). In addition to LANA and LANA-2/vIRF-3, there are other primer pairs on the 45° line, as well some primer pairs the target mRNA of which seemed inhibited by TPA (below the 45° line). However, these mRNAs increased at later time points after induction as discussed below.
FIG. 5.
(A) Plot of the dCT values (normalized to gapdh) for each primer pair in the KSHV array (circles). The vertical axis indicates the dCT value from mock-treated BCBL-1 cells, and the horizontal axis indicates the dCT value from 48-h-TPA-treated BCBL-1 cells. Also shown is the regression line, equation, and R2 value through the values for latency type one mRNAs (solid dots). The dotted line traces the values for orfK8.1 under either condition. The “LANA” primer pairs are located either within the LANA ORF or within the LANA 5′UTR Similarly, for LANA-2/vIRF-3 we designed two primer pairs, one per exon. (B) Plot of the difference for each primer pair in the KSHV array between mock- and TPA-treated BCBL-1 cells at 48 h after induction. CT values were normalized to GAPDH to yield dCT, which were then compared to yield ddCT. The vertical axis indicates the ddCT value, the horizontal axis the percentile of the total distribution. Note that the ddCT values represent a log transformation of the relative abundance of the target mRNA. A value of ddCT = 0 indicates identical abundance of the target mRNA under each condition. A positive ddCT value indicates induction, and a negative ddCT value indicates suppression. Arrows indicate the values of latency type one genes.
To gain a better perspective of the significance of these observations, we calculated the ddCT as dCTgapdh (mock) − dCTgapdh (TPA). The result is graphed in Fig. 5B. ddCT for GAPDH is located at 0; a positive ddCT indicates induction, and a negative ddCT indicates inhibition. The ddCT for GAPDH marks the 25th percentile of KSHV mRNAs on the array; in other words, 75% of the KSHV primer pairs are induced relative to GAPDH. For instance, orfK8.1 represents one of the most highly altered KSHV mRNAs. In contrast, each primer pair that measures the abundance of a latent mRNA has (i) a negative ddCT, (ii) differs by <1 ddCT unit from GAPDH, and (iii) is located in the lower 25th percentile of all data. Other mRNAs with a ddCT value in the lower 25th percentile were the ORFs 19, 21, 40, 67, 68, K5, K9, and K4. However, their ddCT values increased at later time points (see below). This demonstrates that LANA and LANA-2 are resistant to TPA induction, which is a novel, unique property among KSHV mRNAs.
To further corroborate this result, we diluted the input cDNA pool 10-fold and subjected it again to real-time quantitative PCR analysis. A similar picture ensued (data not shown) verifying that relative induction ratios did not depend upon total mRNA levels. We then calculated the difference ddCT = dCTgapdh (mock) − dCTgapdh (TPA) for each mRNA. Next, ddCT values were rank ordered and split into two groups: group 1 contained the results of primer pairs for the predicted latency type one genes (n = 8, since multiple primer pairs for the each latency type one mRNA were used), and group 2 contained all other KSHV primer pairs (n = 85). Actin, c-myc, and GAPDH were excluded. Next, the ANOVA based on ranks was calculated, which showed that both groups differed significantly (P ≤ 0.0004). This demonstrates that latency type one mRNAs are a separately regulated class of KSHV mRNAs and that both LANA and LANA-2/vIRF-3 belong to this class.
It is important to bear in mind that the magnitude of induction is dependent on the amount of mRNA in the mock control, as well as the amount in the induced sample. About 3% of BCBL-1 cells express a high enough level of lytic proteins to be detected by immunofluorescence in the absence of stimulation (56, 76). The nut-1 message is a point in case. Its levels are increased 10- to 100-fold by TPA, depending on the experimental conditions and the method of observation (28, 69, 74). Since nut-1 mRNA is also the most abundant lytic mRNA, based on absolute copy number, it is easily detected in mock-treated PEL cells (63, 75). Yet, in situ hybridization showed that it is present in only a few, lytically infected, KS and PEL tumor cells (68). As determined by real-time quantitative RT-PCR, overall nut-1 mRNA levels were as abundant as GAPDH mRNA levels and, after TPA induction, nut-1 mRNA was more abundant than GAPDH mRNA. Similarly, some early promoters might be leaky in cells even though these cells never complete the KSHV lytic cycle, resulting in a significant level of mRNA in mock-treated BSBL-1 cultures. This complicates any interpretation based solely on a single time point.
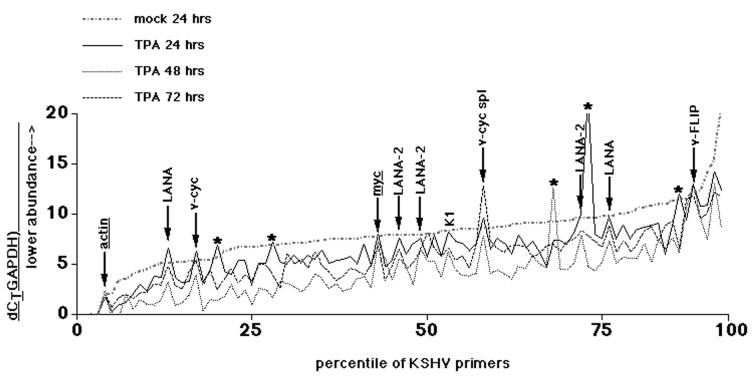
To verify that LANA-2's resistance to lytic reactivation held true over time, we performed a time course experiment. Again, BCBL-1 cells were induced by TPA, and mRNA was isolated at various times after induction and quantified by real-time RT-PCR. Figure 6 shows the outcome of the experiment. In this case, dCT values (normalized to GAPDH) were sorted based on the 24-h-mock-treated values and plotted for each primer pair. In contrast to mock treatment (see Fig. 4), dCT values (and the underlying mRNA levels) were clearly different as early as 24 h after TPA induction compared to the mock-treated control at 24 h. As expected, the real-time RT-PCR amplification for two cellular genes actin and c-myc (underlined in Fig. 6) yielded dCT values which were independent of TPA (c-myc is induced within 30 min of serum stimulation in 3T3 cells [35] but returns to steady-state levels thereafter). The primer pairs that measured mRNAs that did not respond to TPA in a consistent manner were also determined (indicated by asterisks in Fig. 6). In the extreme, these were the result of a pipetting or instrument error for that particular well and time point. In other cases, they represented mRNAs that were induced with delayed kinetics or which were unusually abundant in the mock-treated sample. In contrast, the arrows in Fig. 6 denote the results of primer pairs for latency type one messages, for which all three independent datum points—24, 48, and 72 h after TPA treatment—were nearly identical. Moreover, their dCT values were the same or higher (indicating a lower mRNA abundance) in TPA-treated BCBL-1 cells as for mock-treated BCBL-1 cells. This group of mRNAs was qualitatively and quantitatively different from the rest of the KSHV messages.
FIG. 6.
Plot of dCT (normalized to GAPDH) for each primer pair in the KSHV array for TPA-treated BCBL-1 cells at 24, 48, and 72 h after induction as well as for mock-treated cells at 24 h. The vertical axis indicates the dCT value, and the horizontal axis indicates the percentile of the total distribution. Note that the ddCT values represent a log transformation of the relative abundance of the target mRNA. The arrows indicate primer pairs for which all three time points, as well as mock-treated cells, exhibit similar dCT values, i.e., the mRNA did not change upon TPA treatment. Primer pairs which measured mRNAs that did not respond to TPA in a consistent manner are indicated with asterisks.
Figure 7 shows a conventional cluster analysis of the data. All latency type one mRNAs (LANA, LANA-2, v-cyc, and v-FLIP) group together, and the cellular genes actin, GAPDH, and myc are also part of this cluster. In contrast, immediate-early lytic mRNAs (orf50/Rta and 57/Mta), spliced forms for k-bZIP and gamma-2 mRNAs (orfK8.1 and orf29), are located elsewhere. The spliced v-cyclin mRNA signal is segregated from the other latency mRNAs, since its levels are higher in mock-treated than in TPA-treated samples. This effect is most pronounced at 72 h, suggesting that spliced v-cyclin mRNA might be rapidly degraded in lytically replicating cells. Cluster analysis revealed that orfK1 and orf75 also group with the latency type one mRNAs, whereas both mRNAs were clearly induced, as judged by a single 48-h time point comparison. It is conceivable that, by chance, a summation over the principal components (time points) for K1 and orf75 gives the same rank as a latent pattern (K1 is one of the mRNAs for which the clustering result of the two previous array analyses is discordant [28, 50]). This is one of the theoretical limitations of any cluster analysis. Since multiple mRNAs are transcribed across the K1 locus and are regulated differently (62), array analysis for this ORF is less reliable. By and large every single KSHV mRNA ordered according to real-time quantitative RT-PCR analysis follows the transcription pattern that was previously established by using the KSHV cDNA arrays (28, 50). This demonstrates that the mRNAs for LANA, v-cyclin, v-FLIP, and LANA-2 were not induced by TPA at any time after TPA induction. No other mRNA that can be measured with our array shared this property. This supports a model in which latency type one mRNA levels are regulated differently than the rest of the KSHV genome. They are resistant to the reactivation-induced genome-wide induction.
FIG. 7.

Cluster analysis with dCT (normalized to GAPDH) values of each primer pair in the KSHV array for TPA-treated BCBL-1 cells at 24, 48, and 72 h after induction, as well for as mock-treated cells at 24 h. Data were median centered by gene and time point. Hence, mRNAs that are underrepresented are in shades of green, and those that are overrepresented are in red.
DISCUSSION
Genome-wide transcriptional analyses provide valuable insights into the biology of KSHV and human pathogens in general. The first latent KSHV mRNA (kaposin) and the lytic mRNA (nut-1) were identified through reverse Northern hybridization of labeled total mRNA from mock- or TPA-treated BCBL-1 cells with a restriction digest of KSHV genomic fragments as a template (75). A whole-genome Northern blot scan for KSHV transcripts introduced the current classification into type one (latent), two (latent or lytic), and three (lytic only) mRNAs (63). Similar, more detailed data have since been obtained by using KSHV DNA gene chips, which employed full-length predicted KSHV cDNAs as a template (28, 50). The latter experiments classified KSHV transcripts into temporal classes (alpha, beta, and gamma), analogous to the other herpesviruses. Because DNA arrays are based on solution hybridization, only substantial differences in the level of each target mRNA could be discerned, and large amounts of sample [ca. 107 cells or ≥1 μg of poly(A) mRNA per routine hybridization] were needed (14, 27). A variety of template amplification methods are currently under development to address this problem (e.g., linear amplification with T7 polymerase or template switching [41, 42, 65]). However, these methods may introduce a bias because of sequence-dependent processivity of the polymerase. According to one manufacturer, the concordance of enzymatically amplified cDNA pools compared to nonamplified mRNA is estimated at no more than 80% (Clontech, Inc.). In contrast, real-time quantitative RT-PCR allows the quantification of a smaller set of mRNAs but with, in principal, single cell sensitivity (22). Since real-time quantitative PCR amplicons are on average 100 bp and all primers fit similar, highly restrictive performance criteria, enzyme processivity and secondary structures pose less of an issue. The real-time quantitative RT-PCR array introduced here can be improved to use as little as 1 ng of poly(A) mRNA or 103 cells per experiment (unpublished observations). Greater sensitivity, i.e., fewer input cells is achievable, but at the limit random target cell variation becomes a concern. While a Northern blot can distinguish between differentially spliced transcripts, DNA array experiments (with PCR-amplified DNA fragments) at present do not. Since a significant proportion of herpesvirus transcripts are spliced and many more are 3′ coterminal, a genome scan by using real-time, quantitative RT-PCR and primers designed for specific splice sites is ideally suited for investigating this genus of human pathogens.
Automated primer design based on predicted KSHV ORFs proved surprisingly successful. More than 90% of the primer pairs, which were designed for real-time, quantitative PCR under universal cycling conditions, worked without further optimization at a single primer concentration (166 nM) and Mg2+-ion concentration. Genomic DNA contamination was eliminated through poly(dT) purification of the input RNA, although limited DNase I digestion suffices for this application as well (unpublished observations). All primer pairs amplified a single KSHV-specific product, which in this study was reliably detected by using a double-strand-specific dye rather than additional, more costly fluorescently labeled probes. Whether this sensitivity and specificity is sufficient for clinical studies is currently under investigation.
Several groups previously used levels of a single KSHV mRNAs or protein as a surrogate marker for KSHV infection. Compared to the whole-genome transcript pattern, we can now evaluate the appropriateness of these choices. (i) orf29 mRNA primers were originally developed for the detection of lytically replicating KSHV by conventional RT-PCR (55). orf29 protein encodes a capsid component and, based on Northern analysis in BCBL-1 cells, the orf29 mRNA falls into the gamma-2 temporal class. Since mature orf29 mRNA is spliced by removing a 3,251-bp intron, primer pairs that are located on opposite sides of the intron never amplify contaminant DNA. Because of this property, quantification of orf29 spliced mRNA constitutes the most stringent assay for complete KSHV lytic replication. (Since a second mRNA encoding orf48 also splices into orf29 exonB, analysis of intra-exon sequences alone can result in conflicting data.) Adopting the orf29 assay for real-time quantitative RT-PCR allowed us to quantify the inhibition of KSHV de novo lytic replication in SCID-hu Thy/Liv mice by ganciclovir (16). However, CT values for spliced orf29 mRNA were consistently higher (5 to 10 cycles) than CT values of other spliced gamma-2 amplicons, such as orfK8.1. In other words, a much higher proportion of late lytically infected cells must be present in the culture to yield a significant orf29 signal. This suggests that correctly spliced orf29 mRNA is present in low quantities, late in the infectious cycle, with either a short half-life or unstable and that this assay therefore underestimates the number of lytically infected cells. (ii) Earlier on, we also developed conventional primers for spliced v-cyclin latent mRNA, as well as corresponding real-time quantitative PCR primers (16). Like the primers for orf29, the primers for v-cyclin mRNA are located on opposite sides of a large (4,037-nucleotide) intron. Hence, primers for spliced v-cyclin mRNA provide a specific probe for latently infected cells. (iii) Other real-time quantitative PCR primers for KSHV have been published, but the corresponding amplicons do not cross splice sites (3, 6, 18, 34). This has the advantage that the same primer-probe combination can be used to quantify KSHV viral load, but it relies on careful and more extensive provisions to remove contaminating viral DNA if these primers are used for transcript analysis or transmission studies. Based on these insights we compiled a set of primer pairs and corresponding TaqMan probes for high-throughput, high-sensitivity, high-specificity staging of KSHV-infected cells (see Table 1). These encompass well-characterized spliced latent, alpha, beta, and gamma mRNAs, most of which have been independently validated by protein expression studies and/or in situ analysis in KS tumors.
The principal question of this inquiry was the assignment of LANA-2/vIRF-3 as a class one latency mRNA, similar to LANA, v-cyclin and v-FLIP. LANA-2/vIRF-3 is comprised of the predicted K10.5 and K10.6 orfs and was initially identified based on its sequence similarity to viral interferon regulatory factors (vIRFs) (37, 58). Indeed, like KSHV vIRF-1/K9 and vIRF-2/K11.1, LANA-2/vIRF-3 inhibits interferon signaling in experimental systems. LANA-2/vIRF-3 stands out, however, because of its uniform, widespread expression in PEL and multifocal Castleman's disease. Overall, the transcriptional organization of the K10 region is quite complicated. LANA-2/vIRF-3 is encoded by a 1,701-bp ORF, which is readily detected in untreated PEL cell lines (37, 58). All three primer pairs, which were specific for either exon I or exon II or which spanned the splice junction in between, yielded concordant results, and these results paralleled the pattern of LANA/v-cyc/v-FLIP transcription. Hence, LANA-2/vIRF-3 is encoded by a latency type one mRNA. In contrast, vIRF-1/K9 and vIRF-2/K11.1 mRNAs are induced by TPA with early kinetics in PEL cells, as evidenced by the simultaneous comparison presented here.
Of major concern in comparing transcription patterns between various studies are the myriad technical differences such as timing, drug concentration, and single gene probe characteristics. In the case of KSHV-infected PEL cell lines, culture conditions and time in culture exert a dramatic influence on the timing and efficiency of viral reactivation. Relating the transcription profiles for all KSHV mRNAs provides a common frame of reference within in a single experiment, but this is still a reflection of the particular experimental setup.
While KSHV lytic transcription appears to be an all-or-nothing response, which proceeds to completion once orf50 is expressed, KSHV can enter very distinct latency programs in terms of temporal regulation (latency type one or two) and tissue specificity (KS tumor versus B-cell lineage). All class one latency mRNAs are resistant to the effects of other chemical inducers such as ionomycin or butyrate, as well as TPA (data not shown), whereas no other KSHV transcripts share this feature. This suggests that LANAp, the promoter for LANA, v-FLIP, and v-cyclin (15, 29), and the LANA-2/v-IRF-3 promoter share common regulatory features. The fact that latency type one mRNAs do not increase upon viral reactivation in BCBL-1 cells suggests that their expression might be latency specific and that latency type one mRNAs might be expressed in latently infected cells to the exclusion of lytic mRNAs. Alternatively, latency type one mRNAs might be constitutively transcribed, but their promoters are isolated from the orf50-mediated upregulation of neighboring lytic transcription units.
Acknowledgments
We thank Mike Lagunoff and Jeffry Martin for critical reading of the manuscript, Rebecca Hines-Boykin for invaluable technical help, and David Dyer for oligonucleotide synthesis.
This work was supported by a grant from the Oklahoma Center for the Advancement of Science and Technology (OCAST HR00-59) and NIH RR1555777.
REFERENCES
- 1.Antman, K., and Y. Chang. 2000. Kaposi's sarcoma. N. Engl. J. Med. 342:1027-1038. [DOI] [PubMed] [Google Scholar]
- 2.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. [DOI] [PubMed] [Google Scholar]
- 3.Biggar, R. J., D. Whitby, V. Marshall, A. C. Linhares, and F. Black. 2000. Human herpesvirus 8 in Brazilian Amerindians: a hyperendemic population with a new subtype. J. Infect. Dis. 181:1562-1568. [DOI] [PubMed] [Google Scholar]
- 4.Boshoff, C., T. F. Schulz, M. M. Kennedy, A. K. Graham, C. Fisher, A. Thomas, J. O. McGee, R. A. Weiss, and J. J. O'Leary. 1995. Kaposi's sarcoma-associated herpesvirus infects endothelial and spindle cells. Nat. Med. 1:1274-1278. [DOI] [PubMed] [Google Scholar]
- 5.Breslauer, K. J., R. Frank, H. Blocker, and L. A. Marky. 1986. Predicting DNA duplex stability from the base sequence. Proc. Natl. Acad. Sci. USA 83:3746-3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell, T. B., M. Borok, and L. Gwanzura. 1999. HHV-8 peripheral-blood viral load and the titer of antibodies against HHV-8. N. Engl. J. Med. 341:1241-1242. [DOI] [PubMed] [Google Scholar]
- 7.Cantor, C. R., and P. R. Schimmel. 1980. Biophysical chemistry: the behavior of biological macromolecules. W. H. Freeman & Co., New York, N.Y.
- 8.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 9.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 10.Chen, J., K. Ueda, S. Sakakibara, T. Okuno, and K. Yamanishi. 2000. Transcriptional regulation of the Kaposi's sarcoma-associated herpesvirus viral interferon regulatory factor gene. J. Virol. 74:8623-8634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi, J. K., B. S. Lee, S. N. Shim, M. Li, and J. U. Jung. 2000. Identification of the novel K15 gene at the rightmost end of the Kaposi's sarcoma-associated herpesvirus genome. J. Virol. 74:436-446. [PMC free article] [PubMed] [Google Scholar]
- 12.Cotter, M. A., II, and E. S. Robertson. 1999. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology 264:254-264. [DOI] [PubMed] [Google Scholar]
- 13.Davis, M. A., M. A. Sturzl, C. Blasig, A. Schreier, H. G. Guo, M. Reitz, S. R. Opalenik, and P. J. Browning. 1997. Expression of human herpesvirus 8-encoded cyclin D in Kaposi's sarcoma spindle cells. J. Natl. Cancer Inst. 89:1868-1874. [DOI] [PubMed] [Google Scholar]
- 14.DeRisi, J. L., V. R. Iyer, and P. O. Brown. 1997. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278:680-686. [DOI] [PubMed] [Google Scholar]
- 15.Dittmer, D., M. Lagunoff, R. Renne, K. Staskus, A. Haase, and D. Ganem. 1998. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J. Virol. 72:8309-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dittmer, D., C. Stoddart, R. Renne, V. Linquist-Stepps, M. E. Moreno, C. Bare, J. M. McCune, and D. Ganem. 1999. Experimental transmission of Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) to SCID-hu Thy/Liv mice. J. Exp. Med. 190:1857-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flore, O., S. Rafii, S. Ely, J. J. O'Leary, E. M. Hyjek, and E. Cesarman. 1998. Transformation of primary human endothelial cells by Kaposi's sarcoma-associated herpesvirus. Nature 394:588-592. [DOI] [PubMed] [Google Scholar]
- 19.Friborg, J., Jr., W. Kong, M. O. Hottiger, and G. J. Nabel. 1999. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature 402:889-894. [DOI] [PubMed] [Google Scholar]
- 20.Gao, S. J., L. Kingsley, D. R. Hoover, T. J. Spira, C. R. Rinaldo, A. Saah, J. Phair, R. Detels, P. Parry, Y. Chang, and P. S. Moore. 1996. Seroconversion to antibodies against Kaposi's sarcoma-associated herpesvirus-related latent nuclear antigens before the development of Kaposi's sarcoma. N. Engl. J. Med. 335:233-241. [DOI] [PubMed] [Google Scholar]
- 21.Gao, S. J., L. Kingsley, M. Li, W. Zheng, C. Parravicini, J. Ziegler, R. Newton, C. R. Rinaldo, A. Saah, J. Phair, R. Detels, Y. Chang, and P. S. Moore. 1996. KSHV antibodies among Americans, Italians, and Ugandans with or without Kaposi's sarcoma. Nat. Med. 2:925-928. [DOI] [PubMed] [Google Scholar]
- 22.Ghia, P., E. ten Boekel, E. Sanz, A. de la Hera, A. Rolink, and F. Melchers. 1996. Ordering of human bone marrow B lymphocyte precursors by single-cell polymerase chain reaction analyses of the rearrangement status of the immunoglobulin H and L chain gene loci. J. Exp. Med. 184:2217-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Glantz, S., and B. Slinker. 2000. Primer of applied regression and analysis of variance. McGraw-Hill Book Co., New York, N.Y.
- 24.Glenn, M., L. Rainbow, F. Aurad, A. Davison, and T. F. Schulz. 1999. Identification of a spliced gene from Kaposi's sarcoma-associated herpesvirus encoding a protein with similarities to latent membrane proteins 1 and 2A of Epstein-Barr virus. J. Virol. 73:6953-6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hayward, G. S. 1999. KSHV strains: the origins and global spread of the virus. Semin. Cancer Biol. 9:187-199. [DOI] [PubMed] [Google Scholar]
- 26.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real-time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 27.Iyer, V. R., M. B. Eisen, D. T. Ross, G. Schuler, T. Moore, J. C. F. Lee, J. M. Trent, L. M. Staudt, J. Hudson, Jr., M. S. Boguski, D. Lashkari, D. Shalon, D. Botstein, and P. O. Brown. 1999. The transcriptional program in the response of human fibroblasts to serum. Science 283:83-87. [DOI] [PubMed] [Google Scholar]
- 28.Jenner, R. G., M. M. Alba, C. Boshoff, and P. Kellam. 2001. Kaposi's sarcoma-associated herpesvirus latent and lytic gene expression as revealed by DNA arrays. J. Virol. 75:891-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeong, J., J. Papin, and D. Dittmer. 2001. Differential Regulation of the overlapping Kaposi's sarcoma-associated herpesvirus (KSHV/HHV-8) vGCR (orf74) and LANA (orf73) promoter. J. Virol. 75:1798-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kakoola, D. N., J. Sheldon, N. Byabazaire, R. J. Bowden, E. Katongole-Mbidde, T. F. Schulz, and A. J. Davison. 2001. Recombination in human herpesvirus-8 strains from Uganda and evolution of the K15 gene. J. Gen. Virol. 82:2393-2404. [DOI] [PubMed] [Google Scholar]
- 31.Kedes, D. H., M. Lagunoff, R. Renne, and D. Ganem. 1997. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi's sarcoma-associated herpesvirus. J. Clin. Investig. 100:2606-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kedes, D. H., E. Operskalski, M. Busch, R. Kohn, J. Flood, and D. Ganem. 1996. The seroepidemiology of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus): distribution of infection in KS risk groups and evidence for sexual transmission. Nat. Med. 2:918-924. (Erratum, 2:1041.) [DOI] [PubMed] [Google Scholar]
- 33.Kellam, P., C. Boshoff, D. Whitby, S. Matthews, R. A. Weiss, and S. J. Talbot. 1997. Identification of a major latent nuclear antigen, LNA-1, in the human herpesvirus 8 genome. J. Hum. Virol. 1:19-29. [PubMed] [Google Scholar]
- 34.Lallemand, F., N. Desire, W. Rozenbaum, J. C. Nicolas, and V. Marechal. 2000. Quantitative analysis of human herpesvirus 8 viral load using a real-time PCR assay. J. Clin. Microbiol. 38:1404-1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lau, L. F., and D. Nathans. 1987. Expression of a set of growth-related immediate early genes in BALB/c 3T3 cells: coordinate regulation with c-fos or c-myc. Proc. Natl. Acad. Sci. USA 84:1182-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li, M., J. MacKey, S. C. Czajak, R. C. Desrosiers, A. A. Lackner, and J. U. Jung. 1999. Identification and characterization of Kaposi's sarcoma-associated herpesvirus K8.1 virion glycoprotein. J. Virol. 73:1341-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lubyova, B., and P. M. Pitha. 2000. Characterization of a novel human herpesvirus 8-encoded protein, vIRF-3, that shows homology to viral and cellular interferon regulatory factors. J. Virol. 74:8194-8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luppi, M., P. Barozzi, G. Santagostino, R. Trovato, T. F. Schulz, R. Marasca, D. Bottalico, L. Bignardi, and G. Torelli. 2000. Molecular evidence of organ-related transmission of Kaposi sarcoma-associated herpesvirus or human herpesvirus-8 in transplant patients. Blood 96:3279-3281. [PubMed] [Google Scholar]
- 39.Luppi, M., P. Barozzi, T. F. Schulz, G. Setti, K. Staskus, R. Trovato, F. Narni, A. Donelli, A. Maiorana, R. Marasca, S. Sandrini, G. Torelli, and J. Sheldon. 2000. Bone marrow failure associated with human herpesvirus 8 infection after transplantation. N. Engl. J. Med. 343:1378-1385. [DOI] [PubMed] [Google Scholar]
- 40.Martin, J. N., D. E. Ganem, D. H. Osmond, K. A. Page-Shafer, D. Macrae, and D. H. Kedes. 1998. Sexual transmission and the natural history of human herpesvirus 8 infection. N. Engl. J. Med. 338:948-954. [DOI] [PubMed] [Google Scholar]
- 41.Matz, M., D. Shagin, E. Bogdanova, O. Britanova, S. Lukyanov, L. Diatchenko, and A. Chenchik. 1999. Amplification of cDNA ends based on template-switching effect and step-out PCR. Nucleic Acids Res. 27:1558-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matz, M. V., and S. A. Lukyanov. 1998. Different strategies of differential display: areas of application. Nucleic Acids Res. 26:5537-5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mendez, J. C., G. W. Procop, M. J. Espy, T. F. Smith, C. G. McGregor, and C. V. Paya. 1999. Relationship of HHV8 replication and Kaposi's sarcoma after solid organ transplantation. Transplantation 67:1200-1201. [DOI] [PubMed] [Google Scholar]
- 44.Moore, P. S., L. A. Kingsley, S. D. Holmberg, T. Spira, P. Gupta, D. R. Hoover, J. P. Parry, L. J. Conley, H. W. Jaffe, and Y. Chang. 1996. Kaposi's sarcoma-associated herpesvirus infection prior to onset of Kaposi's sarcoma. Aids 10:175-180. [DOI] [PubMed] [Google Scholar]
- 45.Muralidhar, S., A. M. Pumfery, M. Hassani, M. R. Sadaie, M. Kishishita, J. N. Brady, J. Doniger, P. Medveczky, and L. J. Rosenthal. 1998. Identification of kaposin (open reading frame K12) as a human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus) transforming gene. J. Virol. 72:4980-4988. (Erratum, 73:2568, 1999.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nador, R. G., L. L. Milligan, O. Flore, X. Wang, L. Arvanitakis, D. M. Knowles, and E. Cesarman. 2001. Expression of Kaposi's sarcoma-associated herpesvirus G protein-coupled receptor monocistronic and bicistronic transcripts in primary effusion lymphomas. Virology 287:62-70. [DOI] [PubMed] [Google Scholar]
- 47.Neipel, F., J. C. Albrecht, and B. Fleckenstein. 1997. Cell-homologous genes in the Kaposi's sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J. Virol. 71:4187-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Leary, J., M. Kennedy, D. Howells, I. Silva, V. Uhlmann, K. Luttich, S. Biddolph, S. Lucas, J. Russell, N. Bermingham, M. O'Donovan, M. Ring, C. Kenny, M. Sweeney, O. Sheils, C. Martin, S. Picton, and K. Gatter. 2000. Cellular localisation of HHV-8 in Castleman's disease: is there a link with lymph node vascularity? Mol. Pathol. 53:69-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pauk, J., M. L. Huang, S. J. Brodie, A. Wald, D. M. Koelle, T. Schacker, C. Celum, S. Selke, and L. Corey. 2000. Mucosal shedding of human herpesvirus 8 in men. N. Engl. J. Med. 343:1369-1377. [DOI] [PubMed] [Google Scholar]
- 50.Paulose-Murphy, M., N. K. Ha, C. Xiang, Y. Chen, L. Gillim, R. Yarchoan, P. Meltzer, M. Bittner, J. Trent, and S. Zeichner. 2001. Transcription program of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus). J. Virol. 75:4843-4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Poole, L. J., J. C. Zong, D. M. Ciufo, D. J. Alcendor, J. S. Cannon, R. Ambinder, J. M. Orenstein, M. S. Reitz, and G. S. Hayward. 1999. Comparison of genetic variability at multiple loci across the genomes of the major subtypes of Kaposi's sarcoma-associated herpesvirus reveals evidence for recombination and for two distinct types of open reading frame K15 alleles at the right-hand end. J. Virol. 73:6646-6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raab, M. S., J. C. Albrecht, A. Birkmann, S. Yaguboglu, D. Lang, B. Fleckenstein, and F. Neipel. 1998. The immunogenic glycoprotein gp35-37 of human herpesvirus 8 is encoded by open reading frame K8.1. J. Virol. 72:6725-6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Radkov, S. A., P. Kellam, and C. Boshoff. 2000. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene hras transforms primary rat cells. Nat. Med. 6:1121-1127. [DOI] [PubMed] [Google Scholar]
- 54.Rainbow, L., G. M. Platt, G. R. Simpson, R. Sarid, S. J. Gao, H. Stoiber, C. S. Herrington, P. S. Moore, and T. F. Schulz. 1997. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J. Virol. 71:5915-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Renne, R., D. Blackbourn, D. Whitby, J. Levy, and D. Ganem. 1998. Limited transmission of Kaposi's sarcoma-associated herpesvirus in cultured cells. J. Virol. 72:5182-5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Renne, R., W. Zhong, B. Herndier, M. McGrath, N. Abbey, D. Kedes, and D. Ganem. 1996. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 2:342-346. [DOI] [PubMed] [Google Scholar]
- 57.Rickenson, A. B., and E. Kief. 2001. Epstein-Barr virus, p. 2575-2628. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa. [Google Scholar]
- 58.Rivas, C., A. E. Thlick, C. Parravicini, P. S. Moore, and Y. Chang. 2001. Kaposi's sarcoma-associated herpesvirus LANA2 is a B-cell-specific latent viral protein that inhibits p53. J. Virol. 75:429-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roizman, B, and D. M. Knipe. 2001. Herpesviridae, p. 2399-2460. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed., vol. 2. Lippincott-Raven Publishers, Philadelphia, Pa. [Google Scholar]
- 60.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sadler, R., L. Wu, B. Forghani, R. Renne, W. Zhong, B. Herndier, and D. Ganem. 1999. A complex translational program generates multiple novel proteins from the latently expressed kaposin (K12) locus of Kaposi's sarcoma-associated herpesvirus. J. Virol. 73:5722-5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Samaniego, F., S. Pati, J. Karp, O. Prakash, and D. Bose. 2001. Human herpesvirus 8 k1-associated nuclear factor-κB-dependent promoter activity: role in Kaposi's sarcoma inflammation? J. Natl. Cancer Inst. Monogr. 2001:15-23. [DOI] [PubMed] [Google Scholar]
- 63.Sarid, R., O. Flore, R. A. Bohenzky, Y. Chang, and P. S. Moore. 1998. Transcription mapping of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1). J. Virol. 72:1005-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sarid, R., J. S. Wiezorek, P. S. Moore, and Y. Chang. 1999. Characterization and cell cycle regulation of the major Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter. J. Virol. 73:1438-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shagin, D. A., K. A. Lukyanov, L. L. Vagner, and M. V. Matz. 1999. Regulation of average length of complex PCR product. Nucleic Acids Res. 27:23.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sitas, F., H. Carrara, V. Beral, R. Newton, G. Reeves, D. Bull, U. Jentsch, R. Pacella-Norman, D. Bourboulia, D. Whitby, C. Boshoff, and R. Weiss. 1999. Antibodies against human herpesvirus 8 in black South African patients with cancer. N. Engl. J. Med. 340:1863-1871. [DOI] [PubMed] [Google Scholar]
- 67.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, et al. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood 86:1276-1280. [PubMed] [Google Scholar]
- 68.Staskus, K. A., W. Zhong, K. Gebhard, B. Herndier, H. Wang, R. Renne, J. Beneke, J. Pudney, D. J. Anderson, D. Ganem, and A. T. Haase. 1997. Kaposi's sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J. Virol. 71:715-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun, R., S. F. Lin, K. Staskus, L. Gradoville, E. Grogan, A. Haase, and G. Miller. 1999. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J. Virol. 73:2232-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Talbot, S. J., R. A. Weiss, P. Kellam, and C. Boshoff. 1999. Transcriptional analysis of human herpesvirus-8 open reading frames 71, 72, 73, K14, and 74 in a primary effusion lymphoma cell line. Virology 257:84-94. [DOI] [PubMed] [Google Scholar]
- 71.Vieira, J., M. L. Huang, D. M. Koelle, and L. Corey. 1997. Transmissible Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in saliva of men with a history of Kaposi's sarcoma. J. Virol. 71:7083-7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weiss, R. A., D. Whitby, S. Talbot, P. Kellam, and C. Boshoff. 1998. Human herpesvirus type 8 and Kaposi's sarcoma. J. Natl. Cancer Inst. Monogr. 1998:51-54. [DOI] [PubMed] [Google Scholar]
- 73.Whitby, D., M. R. Howard, M. Tenant-Flowers, N. S. Brink, A. Copas, C. Boshoff, T. Hatzioannou, F. E. Suggett, D. M. Aldam, A. S. Denton, et al. 1995. Detection of Kaposi sarcoma-associated herpesvirus in peripheral blood of HIV-infected individuals and progression to Kaposi's sarcoma. Lancet 346:799-802. [DOI] [PubMed] [Google Scholar]
- 74.Zhong, W., and D. Ganem. 1997. Characterization of ribonucleoprotein complexes containing an abundant polyadenylated nuclear RNA encoded by Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8). J. Virol. 71:1207-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhong, W., H. Wang, B. Herndier, and D. Ganem. 1996. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc. Natl. Acad. Sci. USA 93:6641-6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zoeteweij, J. P., S. T. Eyes, J. M. Orenstein, T. Kawamura, L. Wu, B. Chandran, B. Forghani, and A. Blauvelt. 1999. Identification and rapid quantification of early- and late-lytic human herpesvirus 8 infection in single cells by flow cytometric analysis: characterization of antiherpesvirus agents. J. Virol. 73:5894-5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zong, J. C., C. Metroka, M. S. Reitz, J. Nicholas, and G. S. Hayward. 1997. Strain variability among Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genomes: evidence that a large cohort of United States AIDS patients may have been infected by a single common isolate. J. Virol. 71:2505-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]