Abstract
Phosphorylation of Ser2 of the heptapeptide repeat of the CTD of mammalian pol II by P-TEFb is associated with productive elongation of transcription of protein-coding genes. Here, we show that the CTD of pol II transcribing the human U2 snRNA genes is phosphorylated on Ser2 in vivo and that both the CDK9 kinase and cyclin T components of P-TEFb are required for cotranscriptional recognition of the 3′ box RNA 3′ end processing signal. However, inhibitors of CDK9 do not affect transcription of the U2 genes, indicating that P-TEFb functions exclusively as an RNA processing factor in expression of these relatively short, intronless genes. We also show that inhibition of CDK9 does not adversely affect either transcription of an intron-less, replication-activated histone H2b gene or recognition of the histone gene-specific U7-dependent RNA 3′ end formation signal. These results emphasize that the role of P-TEFb as an activator of transcription elongation can be separated from its role in RNA processing and that neither function is universally required for expression of mammalian pol II-dependent genes.
Keywords: H2b, processing, P-TEFb, transcription, U2
Introduction
The carboxy-terminal domain (CTD) of the large subunit of RNA polymerase II (pol II) plays a major role in coordinating transcription with cotranscriptional RNA processing in expression of protein-coding genes from yeast to man (reviewed by Zorio and Bentley, 2004). In mammalian pol II, this structure comprises 52 remarkably similar repeats of the consensus sequence tyrosine/serine/proline/threonine/serine/proline/serine (YSPTSPS), which can undergo reversible phosphorylation in vivo at the serines in positions 2 and 5 (Ser2 and Ser5). Phosphorylation of Ser5 by the cyclin dependent kinase (CDK)7 subunit of TFIIH at initiation (Trigon et al, 1998) allows the CTD to interact with and activate factors that cap the 5′ end of the nascent RNA (Ho and Shuman, 1999). Subsequent phosphorylation of Ser2 by the positive transcription elongation factor, P-TEFb, comprising CDK9 and a cyclin T subunit, is associated with productive elongation (reviewed by Price, 2000; Garriga and Grana, 2004). Accordingly, the in vivo production of full-length mammalian mRNAs can be inhibited by a range of kinase inhibitors, including 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole (DRB) and 4,5-dihydro-8-(methylthio)isoxazolo[5,4-D]benzo[c]thiophene-6-carboxamide (KM05283) (Price, 2000; Chao and Price, 2001; Medlin et al, 2003), whose targets include CDK9 (Mancebo et al, 1997; Schang, 2002). Phosphorylation also increases the ability of the CTD to activate both splicing and cleavage/polyadenylation in vitro (Hirose and Manley, 1998; Hirose et al, 1999). In turn, splicing has been shown to both recruit P-TEFb to the transcribing pol II in vitro and promote elongation (Fong and Zhou, 2001). In addition, recognition of the polyadenylation signal plays a central role in termination of transcription, which may involve loss of CTD phosphorylation (Zorio and Bentley, 2004). Thus, during elongation of transcription of at least some mammalian mRNA genes, the CTD of pol II undergoes a series of phosphorylation and dephosphorylation events that both regulate and are regulated by cotranscriptional processing.
Mammalian pol II also transcribes the short, intronless small nuclear (sn)RNA genes that encode nontranslated stable RNAs (e.g. the U1 and U2 spliceosomal RNAs). Formation of the nonpolyadenylated 3′ end of snRNAs occurs in a series of steps starting with RNA processing directed by the snRNA gene-specific 3′ box rather than a poly(A) site. Both CTD truncation and CTD kinase inhibitors, including DRB, drastically affect recognition of the 3′ box in the U2 gene, leading to ‘readthrough' of this signal (Medlin et al, 2003). Our demonstration that phospho-CTD activates 3′ box-dependent processing in vitro (Uguen and Murphy, 2003) implicates CTD phosphorylation in the cotranscriptional formation of the 3′ end of U2 gene transcripts in vivo. Here, we present evidence that the kinase activity of CDK9 is required for recognition of the U2 3′ box, that CDK9 is associated with the U2 genes in vivo and that the CTD of pol II transcribing the U2 genes is phosphorylated on Ser2. These findings support the conclusion that P-TEFb functions as an essential RNA processing factor in expression of the U2 genes and couples 3′ box-dependent processing to transcription through phosphorylation of Ser2 of the pol II CTD. However, CDK9 inhibitors, which effectively restrict pol II to the first few hundred base pairs (bp) of the β-actin gene, have no effect on transcription of the U2 genes (Medlin et al, 2003).
The protein-coding, replication-activated histone genes also lack introns, are relatively short and have a specialized U7-dependent RNA 3′ end processing signal (see Marzluff and Duronio, 2002). We find that treatment of cells with CDK9 inhibitors has no adverse effect on transcription of a human H2b gene or RNA 3′ end formation. In addition, the CTD of the pol II transcribing replication-activated genes for all four histones has a relatively low level of Ser2 phosphorylation. These results argue against a positive role of P-TEFb in expression of this type of gene.
Our findings indicate that phosphorylation of Ser2 of the pol II CTD by P-TEFb is unecessary for efficient transcription of some short intronless mammalian genes and that this modification does not automatically activate transcription elongation. The role played by P-TEFb in transcription of pol II-dependent genes is therefore likely to be dictated by the exact complement of ancillary factors.
Results
KM05283 inhibits phosphorylation of Ser2 of the pol II CTD
The CTD kinase inhibitors DRB, KM05283, 1-(5-isoquinolinylsulfonyl)-3-methylpiperazin (H-7) and N-[2-(methylamino)ethyl]-5-isoquinolinesulfonamide (H8) all inhibit 3′ end formation of transcripts from U2 snRNA genes (Medlin et al, 2003). Since these compounds each inhibit several kinases at the concentration used (Mancebo et al, 1997; Schang, 2002), we have determined the specific in vivo effect of the narrow spectrum kinase inhibitor KM05283 on CTD phosphorylation. Figure 1 shows the results of Western blot analysis of cells before and after treatment with 100 μM KM05283, using antibodies against pol II (lanes 1, 2), or antibodies specific for the CTD phosphorylated at either Ser2 (lanes 3, 4) or Ser5 (lanes 5, 6). KM05283 effectively inhibits hyperphosphorylation of pol II to the IIo form (lane 2). However, phosphorylation of Ser5 is unaffected (lane 6), indicating that CDK7 is not inhibited, consistent with the failure of this drug to effectively inhibit capping of transcripts from a U2 template (Medlin et al, 2003). In contrast, Ser2 phosphorylation is no longer detected. DRB and H8 also selectively affect Ser2 phosphorylation in vivo, although less effectively than KM05283 when used at the same concentration (data not shown).
Figure 1.
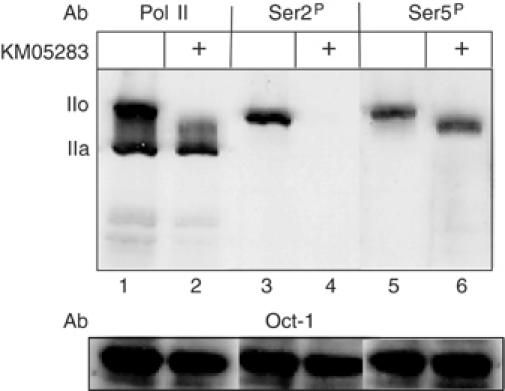
KM05283 inhibits phosphorylation of Ser2 of the pol II CTD. Western blot analysis of protein from HeLa cells before and after treatment with 100 μM KM05283 using antibodies against the N-terminus of the large subunit of Pol II (Santa Cruz, H-224) (lanes 1, 2), or the CTD repeats phosphorylated on Ser2 (Ser2P) (Covance, H5), lanes 3 and 4, or Ser5 (Ser5P) (Covance, H14), lanes 5 and 6. The positions of the hyperphosphorylated CTD form (IIo) and the hypophosphorylated CTD form (IIa) are indicated on the left. The level of the transcription factor Oct-1 (Santa Cruz, C-21 antibody) serves as a loading control.
These results implicate phosphorylation of Ser2 of the CTD, and thus CDK9 activity, in 3′ end formation of pre-U2 snRNAs.
P-TEFb is required for proper 3′ end formation of pre-U2 snRNA
Ectopic overexpression of a kinase dead, D167N, dominant-negative form of CDK9 (Garriga et al, 1996) has little effect on 3′ end formation of endogenous pre-U2 snRNA (Medlin et al, 2003). However, efficient nuclear localization of newly expressed CDK9 in 293 cells may require enhanced expression of a suitable cyclin partner (Napolitano et al, 2002). We have therefore tested the effect of coexpressing the kinase dead form of CDK9 (CDK9KD) together with each of the four cyclins known to associate with this kinase (cyclin T1, T2a, T2b, K (Peng et al, 1998; Fu et al, 1999)), on transcripts from a transfected U2G template (Figure 2A). Treatment of cells with 100 μM KM05283 causes almost total readthrough of the 3′ box in the U2G template (Medlin et al, 2003), (Figure 2B) (89% readthrough (RT), lane 2). Ectopic expression of kinase dead CDK9 (CDK9KD) increases readthrough from 16% (lane 1) to 29% (lane 4), consistent with a dominant-negative effect of the mutant, while expression of wild-type CDK9 (CDK9WT) has little effect (18% RT, lane 3). Coexpression of cyclin T1 with wild-type CDK9 causes a reduction in the level of steady-state RNA to 45% but has little effect on 3′ end formation (20% RT, lane 5). In contrast, coexpression of the kinase dead CDK9 with either of its three related cyclin T partners (see Price, 2000) causes readthrough to increase to approximately 50% (T1, lane 6, 53% RT; T2a, lane 7; 47% RT; T2b, lane 8, 55% RT). Coexpression of the much shorter cyclin K, however, has the same effect as kinase dead CDK9 alone (cyclin K, 28% RT, lane 9). In all cases, transcription is dependent on a functional PSE in the promoter (see lanes 10–13 and data not shown). Expression of kinase dead CDK9 together with cyclin T1 also affects processing of transcripts from endogenous U2 genes (Figure 2C, cf lanes 2, 3). However, the effect is low relative to the response to KM05283 (lane 4) and the effect on transcripts from transfected genes. This may indicate that CDK9 associated with these genes is not readily replaced by ectopically expressed CDK9 or that relatively few cells are transfected. Wild type and kinase dead CDK9 are expressed at approximately the same level and cyclin K is expressed at least as well as the other cyclins, as assessed by Western blot analysis (Figure 2D). Thus, the kinase dead form of CDK9 inhibits formation of the 3′ end of pre-U2 snRNA more effectively when coexpressed with an appropriate cyclin partner supporting the notion that 3′ box function requires the complete P-TEFb complex. The reduction in steady state RNA caused by excessive P-TEFb may be due to ‘squelching' of interacting factors. Since cyclin K has limited homology to the cyclin Ts (see Price, 2000) amino acids (aa) critical for selective recruitment of P-TEFb to snRNA genes, for example, may be missing.
Figure 2.
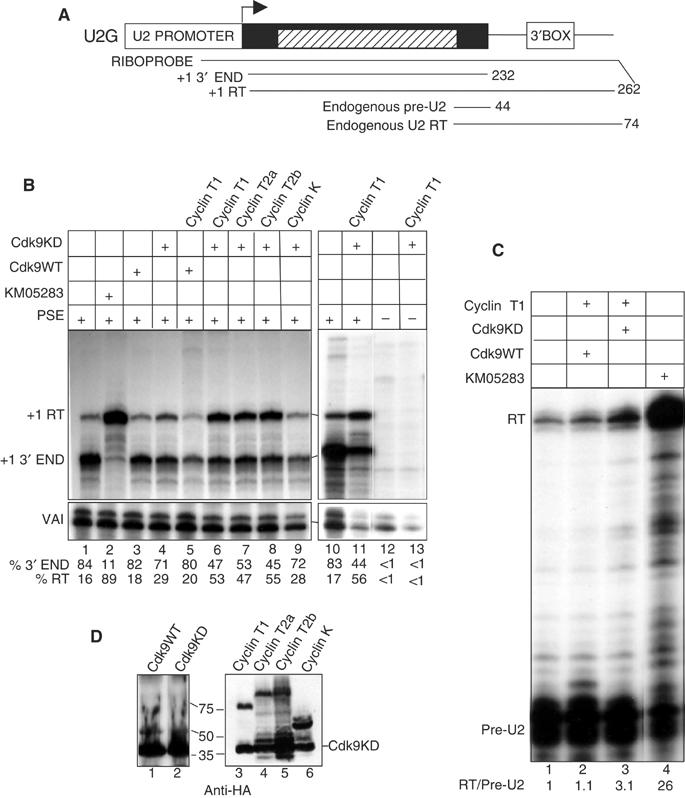
P-TEFb is required for proper 3′ end formation of U2 transcripts. (A) Structure of the U2G construct (Medlin et al, 2003) with the relative position of the riboprobe and expected RNase protection products. The hatched box corresponds to β-globin gene sequences. The size of the expected products is noted at the right. RT denotes readthrough transcripts that mismatch to the riboprobe at the 3′ end in this and subsequent figures. (B) RNAse protection analysis of RNA transcribed from U2G after ectopic expression of the wild-type CDK9 (CDK9WT) or the D167N kinase dead mutant of CDK9 (CDK9KD) together with the CDK9 cyclin partners. VAI serves as a transfection control in this and subsequent experiments. The CDK9 and cyclin construct transfected, the addition of KM05283 and the presence or absence of a functional PSE in the U2G construct is noted above each lane. The positions of the protected products are noted on the left. The relative amount of correct 3′ end and readthrough is noted below each lane. (C) RNase protection analysis of endogenous U2 gene transcripts after ectopic expression of the CDK9WT or KD together with cyclin T1. The ratio of readthrough (RT) to pre-U2 3′ end is noted below each lane. (D) Western blot analysis of cells transfected with templates encoding CDK9WT or KD and the four cyclins using an anti-HA antibody (Santa Cruz, F-7). The transfected construct is shown above each lane and the positions of size markers are noted at the left. For lanes 3–6, CDK9DN was cotransfected and the position of this protein is noted at the right.
P-TEFb interacts with the unphosphorylated CTD through cyclin T1 and this interaction is reversed by CTD phosphorylation (Taube et al, 2002; Zhang et al, 2003). Cyclin T1 also binds to CTD heptapeptide repeats with alanine substitutions at positions 2 and 5, but in this case is not readily released. This analogue effectively inhibits the function of P-TEFb when tethered to a protein-coding gene through the LexA DNA binding domain (Zhang et al, 2003) and should also inhibit 3′ processing of U2 gene transcripts if a cyclin T is involved. Accordingly, we have tested the effect of recruiting LexA/CTD analogue repeats directly to a U2 gene template containing three LexA binding sites between the start site of transcription and the 3′ box (U2LEX) (Figure 3A). Expression of the LexA binding domain alone (Lex, lane 2, 33% RT) or fused to 17 wild-type heptapeptide repeats (LexSS, lane 3, 34% RT) has little effect on the level of 3′ end formation (lane 1, 35% RT). However, recruitment of 17 heptapeptide repeats with alanine at positions 2 and 5 causes a marked inhibition of 3′ processing (LexAA, lane 4, 63% RT). Substitution of a glutamic acid phospho-mimic for serine at position 5 does not affect 3′ processing (LexSE, lane 5, 36% RT). Glutamic acid at position 2 results in a slight reduction of readthrough, (LexES, lane 6, 28% RT), suggesting that this analogue can activate 3′ end processing by mimicking the Ser2 phosphorylated CTD.
Figure 3.
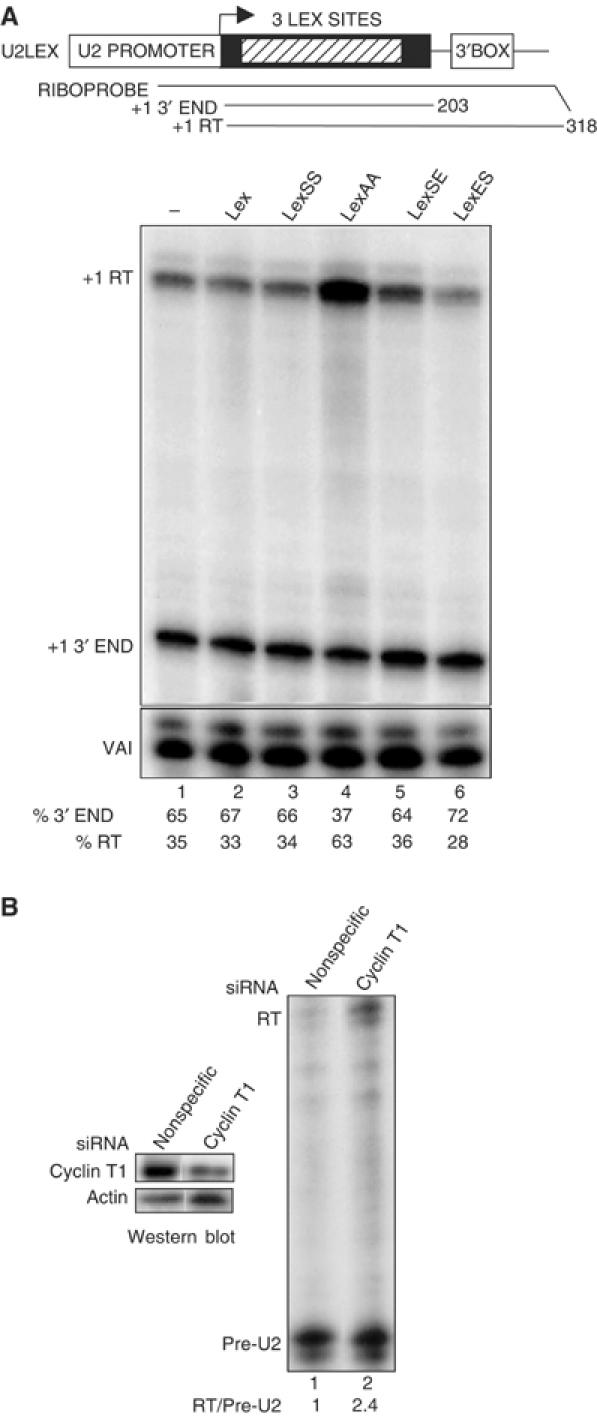
Inhibition of P-TEFb by CTD analogues or RNAi-mediated knockdown affects 3′ box-dependent processing. (A) Diagram of the U2LEX construct with the 3 lexA binding sites denoted as a hatched box (top). The relative position of the riboprobe and size of the expected RNase protection products are shown. RNase protection analysis of U2LEX transcripts after ectopic expression of the CTD analogue noted above each lane is shown below. The positions of the protected products are noted on the left. The relative amount of correct 3′ end and readthrough is shown beneath each lane. (B) RNase protection analysis of transcripts from endogenous U2 genes after transfection of a nonspecific siRNA (lane 1) or an siRNA specific for cyclin T1 (lane 2). The ratio of readthrough (RT) to pre-U2 3′ end is noted below each lane. Western blot analysis of cells treated with the same siRNAs using antibodies against cyclin T1 (Santa Cruz H245) or α-actin (Santa Cruz C-11) is shown at the left.
In addition, RNAi-mediated knockdown of cyclin T1 (Figure 3B) (left panel), and consequently also CDK9 (Chiu et al, 2004) increases the ratio of readthrough transcripts from endogenous U2 genes by 2.4-fold, as determined by RNase protection analysis (right panel, cf lanes 1, 2).
Thus, a dominant-negative form of CDK9, a CTD analogue that antagonizes P-TEFb and P-TEFb knockdown all inhibit processing directed by the 3′ box.
Mutation of Ser2 of the pol II CTD repeats causes readthrough of the U2 3′ box
Our data are consistent with a requirement for phosphorylation of Ser2 of the CTD for 3′ box-dependent processing. However, CDK9 phosphorylates several substrates in addition to the CTD during transcription of protein-coding genes. We have therefore tested the effect of selective removal of the Ser2 phospho-acceptor sites by mutation to alanine (Figure 4). Since we have shown that the first 25, largely consensus, CTD repeats support approximately 80% 3′ box-dependent processing (Medlin et al, 2003), we prepared cassette constructs encoding the α-amanitin resistant mouse pol II large subunit where the CTD has been replaced by either 25 consensus repeats (1–25 NotI) or 25 repeats with alanine at position 2 (A2) followed by an HA epitope tag and the final C-terminal 10 aa of the wild-type CTD (Figure 4A). Transcripts from the U2G template in α-amanitin-treated 293 cells cotransfected with these constructs were analysed by RNase protection (Figure 4B). In accordance with our previous results, 78% of U2G transcripts are properly 3′ processed when 25 consensus CTD repeats are present (1–25 NotI, lane 4). Mutation of all serines at position 2 to alanine reduces the proportion of proper processing to a maximum of 15% in the experiment shown (A2, lane 5), supporting the notion that Ser2 phosphorylation plays a key role in the process. Western blot analysis (Figure 4C) indicates that the A2 mutant large subunit (lane 5) is expressed at least as well as 1–25 NotI (lane 4).
Figure 4.
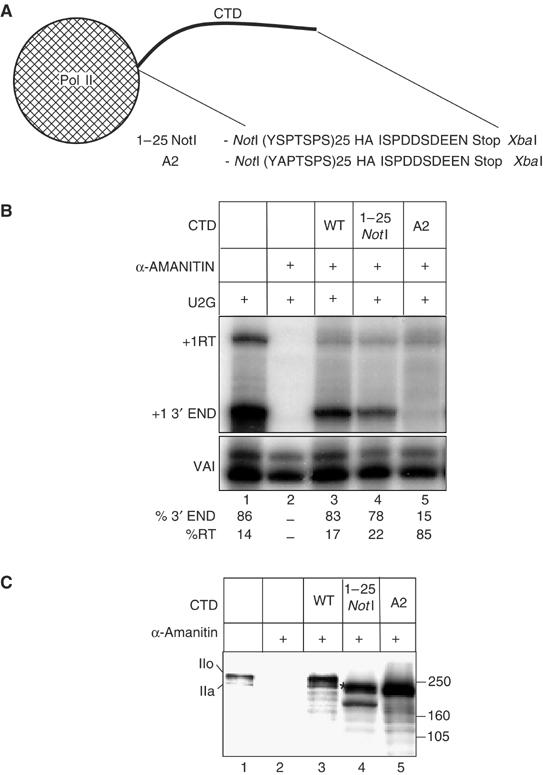
Mutation of Ser2 of the pol II CTD repeats causes readthrough of the U2 3′ box. (A) Diagram of the CTD region of the 1–25 NotI and A2 cassette constructs of the pol II large subunit showing the amino acids encoded by the region between the NotI and XbaI sites. HA and Stop denote the HA epitope tag and a stop codon respectively. (B) RNase protection analysis of RNA transcribed from U2G after ectopic expression of the α-amanitin resistant pol II large subunit constructs indicated above the lanes and treatment of cells with α-amanitin. The positions of the protected products are shown on the left. The relative amount of correct 3′ end and readthrough is noted below each lane. (C) Western blot analysis of the proteins encoded by the pol II large subunit constructs using an antibody against the large subunit of pol II (Santa Cruz, H-224). The construct transfected is shown above each lane. The position of the hyperphosphorylated form (IIo) and the hypophosphorylated form (IIa) of the endogenous and ectopically expressed pol II large subunits with a full-length CTD are noted on the left. The putative hyperphosphorylated form of 1–25 NotI is marked with an *.
P-TEFb is not required for transcription of a human H2b gene or 3′ processing of the transcripts
Transcription of the human U2 genes is unaffected by drugs such as KM05283 that inhibit pol II CTD Ser2 phosphorylation (Medlin et al, 2003). This contrasts sharply with the effect of these drugs on elongation of transcription of protein-coding genes in vitro and in vivo (Price, 2000; Chao and Price, 2001; Medlin et al, 2003) and indicates that P-TEFb does not act as a positive elongation factor in transcription of the U2 genes. This may reflect a reduced requirement for positive elongation factors in expression of short intronless genes compared to longer intron-containing genes like the β-actin gene (Medlin et al, 2003). In common with the U2 snRNA genes, the human replication-activated histone H2b genes lack introns and have a specialized RNA 3′ end formation signal within 500 bp of the transcription start site (see Figure 5A). We have therefore analysed whether phosphorylation of the pol II CTD plays a role in production of processed H2b gene transcripts. A template containing the promoter and first 231 bp of the transcribed region of an H2b gene described by Zhong et al (1983) (Accession No. X00088) followed by a 160 bp marker region from the β-globin gene and a 3′ processing signal from a second H2b gene (Collart et al, 1992; Accession No. X57985) was prepared (HISG, Figure 5A). Transcripts are readily distinguished from endogenous transcripts by RNase protection analysis using 5′ and 3′ probes that include β-globin sequences (Figure 5B and C). The majority of transcripts (97%) have the correct 5′ ends for properly initiated transcripts (Figure 5B, lane 1) and are only produced if the TATA box in the promoter is intact (Supplementary Figure 1). The major bands from analysis of the 3′ end correspond to cleavage at two potential processing sites (noted in Figure 5A) and 3′ processing is efficient (95%) (Figure 5C, lane 1). Treatment of cells with KM05283 causes an approximately two-fold increase of steady state RNA (Figure 5B, lane 2, 173%). However, 3′ end formation is 97% after drug treatment (Figure 5C, lane 2), indicating that 3′ processing of the transcripts synthesized after the addition of KM05283 occurs efficiently. Treatment of cells with DRB, H-7 and H8 also increases the level of steady-state RNA transcribed from HisG by between 1.5- and two-fold without reducing 3′ end formation (Supplementary Figure 2). Ectopic expression of kinase dead CDK9 either alone or together with cyclin T1 also causes an increase in the level of steady-state RNA (Figure 5B, lane 4, 121%; lane 6, 135%), without adversely affecting 3′ processing. Ectopic expression of wild-type CDK9 has little effect on the level of steady state RNA and causes a slight reduction in 3′ end formation to 93% (Figure 5B and C, lane 3). Coexpression of wild-type CDK9 with cyclin T1 instead reduces the level of steady state RNA to 41% (Figure 5B, lane 5) and increases the level of unprocessed transcripts to 38% (Figure 5C, lane 5). Transcripts that may correspond to transcription reading around the plasmid and back over the promoter (RA) increase from 3–28% (Figure 5B, lanes 1, 5). As expected, the readthrough products are sensitive to treatment with KM05283 (Supplementary Figure 1). However, a large proportion of the readthrough transcripts are still detected when the TATA box is deleted (Supplementary Figure 1), suggesting that they are not specific for the H2b promoter. Excess wild-type P-TEFb may therefore be activating transcription from alternative promoters in the template.
Figure 5.
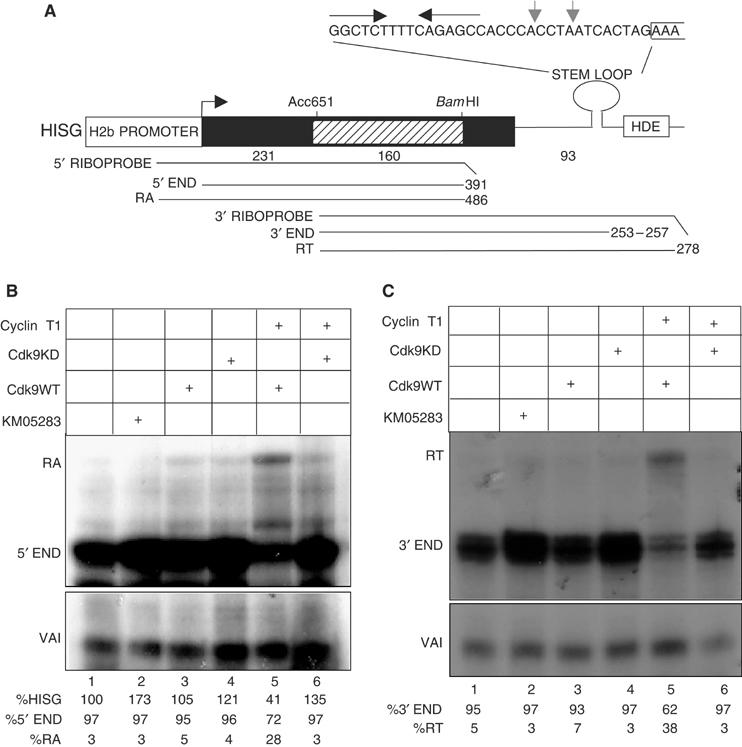
P-TEFb is not required for elongation of transcription of a transfected H2b gene or 3′ processing of the transcripts. (A) Diagram of the HISG construct with the relative position of the riboprobes and expected RNase protection products. The size of the expected products is noted at the right. The hatched box corresponds to β-globin gene sequences that allow transcripts to be distinguished from endogenous H2b mRNA and the sizes of the histone and globin sequences are noted below the diagram. The sequence of the processing signal is shown above the diagram with the stem loop denoted by convergent arrows, the start of the HDE bracketed and the positions of two potential 3′ ends marked with grey arrows. (B) 5′ RNase protection analysis of RNA transcribed from HISG after treatment of cells with KM05283 and after ectopic expression of CDK9WT or CDK9KD together with cyclin T1. The CDK9 and cyclin construct transfected and the addition of KM05283 is noted above each lane and the positions of the protected products are noted on the left in this and subsequent panels. The relative amount of expression and correct 5′ end and readaround transcription (RA) is noted below each lane. For lanes 2, 4 and 6 the average of three experiments and standard deviations were 193% and 19, 116% and 10, 129 and 16 respectively. (C) 3′ RNase protection analysis of RNA transcribed from HISG after treatment of cells with KM05283 and after ectopic expression of CDK9WT or CDK9KD together with cyclin T1. The relative amount of correct 3′ end and readthrough is noted below each lane.
Treatment of cells with CTD kinase inhibitors and ectopic expression of the dominant-negative form of P-TEFb causes an increase in the steady-state level of transcripts from the HisG template. In addition, excess P-TEFb appears to reduce specific transcription from an H2b promoter. Thus, P-TEFb may negatively regulate initiation of transcription of these genes as has been shown for the human PGC-1 gene (Sano et al, 2004).
Inhibition of phosphorylation by CDK9 either by kinase inhibitors or dominant-negative P-TEFb has no adverse effect on the function of the H2b 3′ processing signal, indicating that Ser2 phosphorylation of the CTD is not required in this case. We obtained the same results using a construct with the stem loop and HDE sequences from the same H2b gene as the promoter (data not shown), indicating that these results are not specific for one 3′ processing signal.
We have also investigated the effect of CTD kinase inhibitors on expression of an endogenous human H2b gene (Accession No. X00088) (Figure 6). Four 160 bp synthetic RNA probes were used to analyse the level of nascent transcription across this endogenous H2b gene by nuclear run-on before or after treatment of cells with KM05283 or DRB (Figure 6A). These CTD kinase inhibitors cause an increase of approximately two-fold in the level of nascent transcription across the probed regions (depicted graphically). The relative increase is very similar for each probe, suggesting that transcription initiation rather than elongation is affected.
Figure 6.
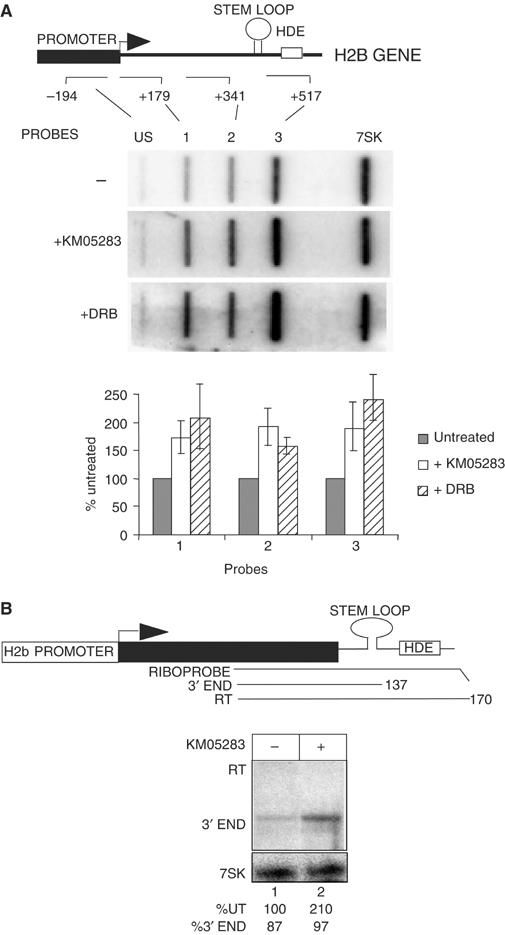
P-TEFb is not required for elongation of transcription of an endogenous H2b gene or 3′ processing of the transcripts. (A) Nuclear run-on analysis of an endogenous replication-activated H2b gene after treatment of cells with CTD kinase inhibitors. The structure of the human H2b gene analysed is shown with the relative positions of the run-on probes marked below. The numbers next to the probes indicate the 3′ boundary of the probes relative to the site of initiation. The results of run-on analysis in the absence and presence of 100 μM KM05283 or DRB are shown below each probe. The 7SK probe serves as a negative control for the effect of the kinase inhibitors. Quantitation of the results for each probe is shown below as an average of three experiments, with the signals after CTD kinase inhibitor treatment represented as a percentage of the untreated controls. The error bars indicate standard deviations. (B) RNase protection analysis of the 3′ ends of transcripts from an endogenous replication-activated H2b gene after treatment of cells with KM05283. The position of the RNase protection probe relative to the H2b gene and the sizes of the expected products are shown. The addition of inhibitor is noted above the lane and the % of the level of transcript in lane 1 and % 3′ end is noted below each lane. The 7SK RNA serves as a control for the level of RNA.
RNase protection analysis of the 3′ end of the transcripts from this endogenous gene was also carried out (Figure 6B) with 7SK RNA as a control for RNA levels. In untreated cells, 87% of the transcripts detected have the correct 3′ ends (lane 1). Treatment of cells with KM05283 increases the steady-state level of RNA with the correct 3′ ends (KM05283, 210%, lane 2) and does not inhibit 3′ end formation.
Taken together, the data in Figures 5 and 6emphasize that efficient transcription of a replication-activated H2b gene and 3′ processing of the transcripts do not require phosphorylation of either Ser2 of the pol II CTD or of any other CDK9 substrate.
The 3′ box in snRNA genes is not recognized when transcription is initiated from the promoters of protein-coding genes including the H2a promoter (Hernandez and Weiner, 1986; Neuman de Vegvar et al, 1986; Pilch and Marzluff, 1991) and the H2b promoter used here (Scurry and Murphy, unpublished observations). However, the histone processing signal is recognized in transcripts from a U1 promoter (Pilch and Marzluff, 1991) and when the H2b promoter in HisG is replaced by the U2 promoter (U2HIS, Figure 7). In contrast to the results with HISG, treatment of cells with CDK9 inhibitors (lanes 2, 3) causes readthrough of the processing signal in U2HIS transcripts. All transcripts detected are dependent on an intact PSE in the promoter (lanes 4–6). Thus, the complete lack of a requirement for CDK9 activity for efficient recognition of the histone processing signal requires the H2b promoter.
Figure 7.
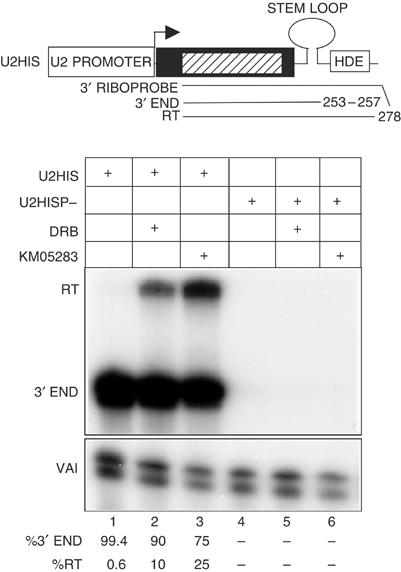
CDK9 inhibitors affect recognition of the histone processing signal in transcripts from the U2 promoter. RNase protection analysis of the 3′ ends of transcripts from constructs where the H2b promoter has been replaced by either the wild-type U2 promoter (U2HIS) or the P-mutant (U2HISP−). The constructs transfected and CDK9 inhibitor used are shown above each lane. The relative amount of correct 3′ end and readthrough is noted below each lane.
CTD Ser2 phosphorylation and CDK9 are differentially associated with U2 and H2b genes
Our results indicate a differential requirement for P-TEFb and phosphorylation of Ser2 of the pol II CTD in expression of the U2 and H2b genes. In order to determine whether this is reflected in the association of the Ser2 phosphorylated form of pol II with these genes, we have carried out ChIP on the endogenous U2 and H2b genes in HeLa cells (Figure 8). Real-time PCR analysis with primers specific for a human U2 gene (Accession No. U57614) and the H2b gene used for nuclear run-on analysis was carried out after ChIP with antibodies to the Ser2 and Ser5 phosphorylated forms of the pol II CTD (Ser2P and Ser5P) (Figure 8A). Antibodies to Ser2P immunoprecipitate greater than 10-fold more U2 gene DNA than H2b gene DNA, whereas the antibody specific for Ser5P immunoprecipitates an approximately equal amount of the two genes. Treatment of cells with KM05283 causes a marked decrease in the level of Ser2P on the U2 gene, but has no negative effect on the H2b gene ChIP by the anti-Ser2P antibody. Importantly, the level of Ser5P on either gene is not adversely affected by KM05283. In addition, the amount of pol II detected on these genes differed by only 2- to 2.5-fold in several experiments (see Figure 8C).
Figure 8.
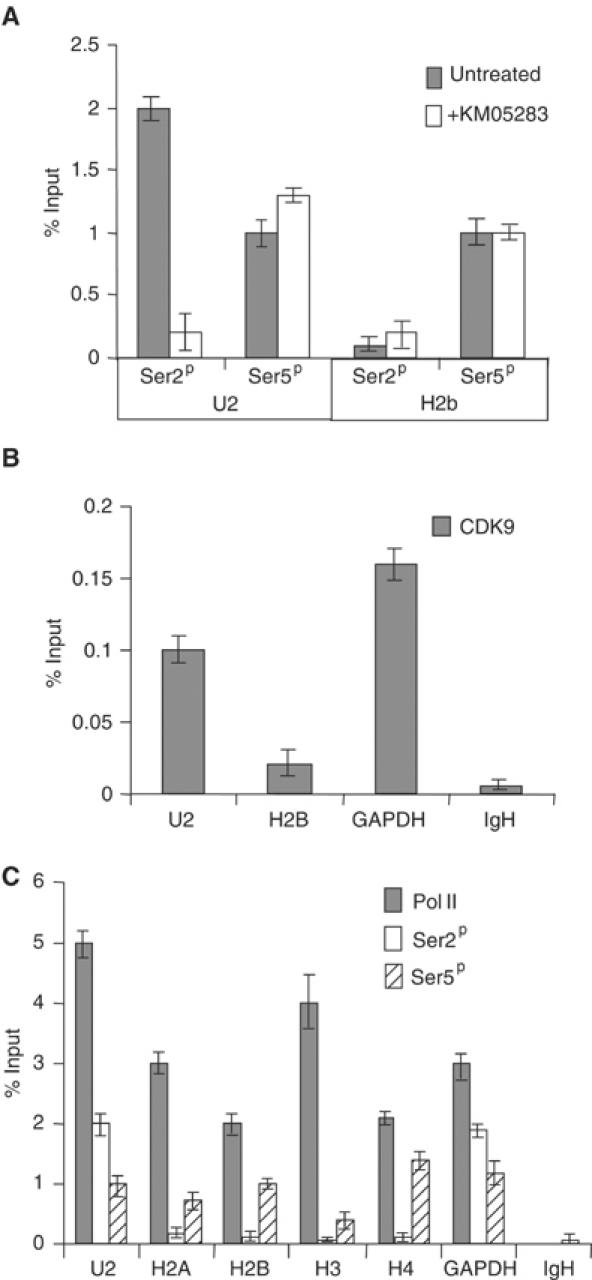
CTD Ser2 phosphorylation and CDK9 are differentially associated with U2 genes and H2b genes. (A) Real-time PCR analysis of the output from ChIP using antibodies against the pol II CTD phosphorylated on Ser2 (Ser2P) (H5, Covance) or Ser5 (Ser5P) (H14, Covance) before and after treatment of cells with KM05283. The graph shows the values obtained using primers against the U2 and H2b genes. Error bars indicate the standard deviation of real-time triplicates in this and subsequent panels. Immunoprecipitation is given as a percentage of the amount of the input DNA after subtraction of the no antibody control in this and subsequent panels. The no antibody controls were less than 0.1% of input DNA. (B) Real-time PCR analysis of the output from ChIP using antibodies against CDK9 (Santa Cruz, L-19). The graph shows the values obtained using primers against the U2, H2b, GAPDH and IgH genes. The no antibody controls were less than 0.008% of input DNA. (C) Real-time PCR analysis of the output from ChIP using antibodies against the large subunit of pol II (Santa Cruz, H-224) and the pol II CTD phosphorylated on Ser2 or Ser5. The graph shows the values obtained using primers against the U2, H2a, H2b, H3, H4, GAPDH and IgH genes. The no antibody controls were less than 0.008% of input DNA for the pol II antibody.
An antibody to CDK9 immunoprecipitates significantly less H2b gene than either the U2 or GAPDH genes (Figure 8B). The IgH gene, which is transcriptionally silent in HeLa cells, serves as a negative control.
These results suggest that the level of Ser2 phosphorylation on pol II transcribing H2b genes is significantly lower than on the U2 genes. In addition, more CDK9 is stably associated with U2 genes than H2b genes. These findings are consistent with the differential requirement for P-TEFb-mediated CTD phosphorylation in expression of these two distinct gene types.
We have also carried out ChIP analysis of replication-activated H2A, H3 and H4 histone genes to determine whether the relatively low level of Ser2P is peculiar to H2b genes. The results of this analysis (Figure 8C) indicate that Ser2P is very low relative to Ser5P on all the replication-activated genes tested. Since the primers used will amplify a region close to the transcription start site, we extended the analysis by using a second set of primers close to the 3′ processing signal for each gene to determine whether CTD Ser 2 phosphorylation can be detected further downstream. Although Ser5P is readily detected using these primers Ser2P remained at the level of the IgH gene negative control (data not shown).
These results indicate that a relatively low level of Ser2 phosphorylation of the CTD is associated with pol II transcribing genes for all four core replication-activated histones.
Discussion
CDK9 is associated with U2 snRNA genes and specific inhibition of this kinase markedly affects recognition of the U2 3′ box RNA processing signal. In addition, the CTD of pol II transcribing the U2 genes is phosphorylated on Ser2 of the heptapeptide repeat and mutation of Ser2 to alanine causes a drastic reduction in correct 3′ box-dependent processing. Taken together these results support the notion that phosphorylation of Ser2 by CDK9 plays a key role in 3′ box-dependent processing. The effect of the CTD kinase inhibitors DRB, H-7, H8 and KM05382 on formation of pre-snRNAs is therefore readily explained as the result of inhibition of P-TEFb phosphorylation of the pol II CTD.
Inhibition of P-TEFb function does not, however, adversely affect 3′ processing of the transcripts from a replication-activated H2b gene. Phosphorylation of Ser2 of the CTD is also unecessary for elongation of transcription of this gene, as we found for the U2 genes (Medlin et al, 2003). Consistent with this, relatively low levels of CDK9 and CTD Ser2 phosphorylation appear to be associated with this gene. P-TEFb is recruited to pol II-transcribed genes through interaction with a range of DNA-binding transcription factors and/or the bromodomain protein Brd4 (see Jang et al, 2005; Yang et al, 2005). It is therefore possible that factors capable of efficiently recruiting P-TEFb are not associated with the H2b gene. CDK9 inhibition increases the level of transcription suggesting that P-TEFb may negatively regulate expression of the H2b gene. However, the low level of direct association of this factor suggests this is through an indirect mechanism.
Relatively low levels of CTD Ser2 phosphorylation are also associated with replication-activated H2a, H3 and H4 genes, suggesting that this is a general feature of human replication-activated histone genes.
P-TEFb as an activator of 3′ RNA processing
Deletion of the CTD of mammalian pol II causes a major defect in capping, splicing and polyadenylation of pre-mRNAs (McCracken et al, 1997), reflecting the role of this structure in coordinating transcription with pre-mRNA processing. Likewise, the CTD is required for efficient 3′ end formation of snRNA gene transcripts (Medlin et al, 2003; Jacobs et al, 2004). Phosphorylation of Ser5 of the CTD has been unequivocaly shown to activate capping of the 5′ end of pol II transcripts by facilitating interactions between the CTD and the capping enzymes (reviewed by Zorio and Bentley, 2004). In yeast, Ser2 phosphorylation of the pol II CTD by Ctk1 instead potentiates interaction with the essential polyadenylation factor Pcf11 to activate cleavage and polyadenylation of pre-mRNAs (Barilla et al, 2001; Licatalosi et al, 2002; Ahn et al, 2004). Activation of polyadenylation in mammalian in vitro systems by phospho-CTD (Hirose and Manley, 1998) suggests that a similar mechanism operates in higher eucaryotes. The recent demonstration that P-TEFb inhibitors affect polyadenylation in Drosophila and Xenopus (Bird et al, 2004; Ni et al, 2004) supports this notion. 3′ end formation of pre-U2 snRNAs is particularly dependent on P-TEFb function, suggesting that the Ser2 phosphorylated form of the CTD is an essential component of the processing machinery. As shown for activation of capping and polyadenylation, CTD phosphorylation is likely to potentiate interactions with processing factors, resulting in increased recruitment and/or allosteric activation. 3′ processing of H2b mRNAs is at the other end of the P-TEFb-dependence spectrum, and proceeds efficiently when the CTD is not phosphorylated on Ser2. Interestingly, recognition of the histone processing signal is affected by CDK9 inhibitors if transcription is initiated from a U2 promoter. Thus, phosphorylation of a P-TEFb substrate, most likely the pol II CTD, can activate processing, possibly by helping to recruit or activate factors. The presence of the H2b promoter may therefore overcome the need for P-TEFb activity by itself helping to recruit a processing factor to the nascent transcript. The notion that processing is cotranscriptional is strengthened by our finding that antibodies to the histone mRNA processing factor SLBP immunoprecipitate an endogenous H2b gene (Medlin and Murphy, unpublished observations).
P-TEFb as an elongation factor
P-TEFb has a well-established role as an activator of elongation of transcription of mammalian protein-coding genes and the HIV genome (Price, 2000; Garriga and Grana, 2004) and CDK9 inhibitors like DRB, KM05283 and Flavopiridol can cause a drastic defect in elongation by pol II both in vitro and in vivo (Price, 2000; Chao and Price, 2001; Medlin et al, 2003; Garriga and Grana, 2004). In vitro studies have shown that pol II encounters a block to elongation caused by DSIF and NELF soon after initiation (Chodosh et al, 1989; Wada et al, 1998; Yamaguchi et al, 1999; Ivanov et al, 2000; Ping and Rana, 2001; Renner et al, 2001). Phosphorylation of the Nus-G-like N-terminus of the SPT5 subunit of DSIF by CDK9 is required both for reversal of the block and the function of SPT5 as a positive elongation factor (Ivanov et al, 2000). P-TEFb-dependent phosphorylation of the RD subunit of NELF is also required for productive elongation of HIV templates to proceed (Fuginaga et al, 2004). In addition, phosphorylation of both Ser2 and Ser5 of the pol II CTD by P-TEFb directly enhances elongation of transcription of HIV templates (Kim et al, 2002).
In transcription of U2 and H2b genes instead, inhibition of P-TEFb does not result in an elongation block within 500 bp, suggesting that the checkpoint factors DSIF and NELF do not associate with the transcription complex after initiation in all cases. The NELF-A and RD component of NELF contain RNA recognition motifs and bind a number of RNA elements including the HIV TAR (see Fujinaga et al, 2004), providing a mechanism for recruitment to specific transcripts.
Notably, Ser2 phosphorylation of the CTD of pol II transcribing the U2 genes does not activate transcription, indicating that in this case mammalian P-TEFb does not function as an elongation factor. Inhibition of P-TEFb by Flavopiridol also has a more dramatic effect on RNA 3′ end processing directed by a poly(A) site than on transcription elongation in expression of the intron-less Drosophila hsp70 gene (Ni et al, 2004). However, P-TEFb function is still required for efficient elongation to the polyadenylation signal located more than 2 kb from the transcription start site (Ni et al, 2004). The Ctk1 and Bur1 kinases in yeast are equally similar to Cdk9 and phosphorylation of Ser2 of the pol II CTD by Ctk1 activates polyadenylation (Ahn et al, 2004), while the Bur1 kinase is required for efficient transcription elongation by pol II (Keogh et al, 2003). Since Bur1 can phosphorylate Spt5, it has been suggested that these two kinases are paralogues of Cdk9, each performing one of the functions carried out in mammalian cells by P-TEFb (Keogh et al, 2003). Our results indicate that activation of transcription elongation and RNA processing by mammalian P-TEFb can also be functionally separated. The elongation function of P-TEFb in vivo may therefore depend on the association of additional positive elongation factors like Spt5 with the template or transcribed RNA and/or further modifications of the CTD. The U2 and H2b genes are short and intronless and transcription for 500 bp is sufficient to ensure a full-length pre-snRNA or H2b mRNA. Intron-containing protein-coding genes are generally much longer and are more likely to require the elongation function of P-TEFb for production of full-length transcripts. Relevant to this, splicing can activate elongation of transcription in vitro through recruitment of the transcription elongation factors Tat-SF1 and P-TEFb (Fong and Zhou, 2001), providing long, intron-containing genes with a mechanism of continued recruitment of positive elongation factors. Our preliminary results, however, suggest that an artificial intron capable of directing efficient splicing cannot efficiently activate P-TEFb-dependent elongation of transcription from an snRNA promoter (Kollakowski and Murphy, unpublished observations), implicating additional gene-specific mechanisms in the control of this process.
Materials and methods
Nuclear run-ons
Nascent transcription across the endogenous H2b gene was analysed as outlined by Furger et al (2002). CTD kinase inhibitors were added to two 150 mm plates of HeLa cells to a final concentration of 100 μM for 2 h before analysis. A 160-nt RNA was loaded into each slot; the position of the 5′ end of the US RNA and the 3′ end of the other RNAs is noted in Figure 5A. The 7SK RNA probe is complementary to sequences between +270 and +330 relative to the start of transcription of a human 7SK gene (Accession No. X05490). Quantitation was carried out using a Molecular Dynamics phosphorimager. Measurements were corrected against the level of hybridization to the 7SK RNA probe.
Constructs
The U2G construct and riboprobe have been previously described by Medlin et al (2003). The U2LEX construct was made by replacing the region of U2G between the Acc651 and BamH1 sites with a PCR fragment containing three lexA binding sites. The HISG construct has the sequence from −110 to +231 of X00088 followed by the β-globin sequence present in U2G (Medlin et al, 2003), followed by sequences +419 to +530 of X57985, taking the transcription start site as +1. The histone gene sequences are flanked at the 5′ and 3′ ends by an EcoRI site and a HindIII site, respectively. HISG T− has TATATA at −29 changed to TACGCA. In U2HIS and U2HISP− the H2b promoter of HISG is replaced by the wild-type or P-U2 promoter respectively to maintain initiation at +1.
Constructs encoding Ha-tagged cyclin T1, T2a and T2b are described by Peng et al (1998) and a construct encoding Ha-tagged cyclin K was made by cloning a PCR product from the GST-cyclin K described by Edwards et al (1998) into the HindIII and XbaI sites of Rc/CMV (Invitrogen).
To create pol II large subunit constructs 1–25 NotI and A2, a NotI site was introduced into pAT7Rpb1-25Amr (Fong and Bentley, 2001) upstream of the start of the CTD, which allows cloning of replacement heptapeptide repeats between the NotI and XbaI sites. Intermediate constructs containing 25 consensus and A2 heptapeptide repeats were produced by adding further repeats to the WT and A2 constructs with a 5′ AvaI site described by West and Corden (1995). The HA epitope and last 10 aa's of the CTD were added by PCR. In addition to changes in the CTD repeats, the arginine 5 aa's upstream from the start of the first CTD repeat is changed to alanine.
Steady-state RNA analysis
For the experiments in Figures 2, 3, 4 and 6 a 90 mm plate of 293 cells was transfected with 5 μg of U2G or HISG together with 2.5 μg. of the appropriate CDK9, cyclin or α-amanitin-resistant pol II large subunit construct and 15 ng of VA1 using Lipofectamine as recommended by Invitrogen. Total RNA was prepared 24–48 h after transfection, apart from Figure 2C where cells were collected 72 h after transfection. RNase protection was carried out as described in Medlin et al (2003). The 5′ HISG riboprobe contains the region of HisG from EcoRI to the BamHI site at the end of the β-globin sequence in PGem4. The 3′ HISG riboprobe contains the region from the Acc651 site at the 5′ end of the β-globin sequences to the HindIII in PGem4 to give a 3′ mismatch. The riboprobes were transcribed by T7 after digestion. The 7SK riboprobe was prepared from the same template used for nuclear run-on analysis. For the experiments in Figures 2 and 4, KM05283 to 100 μM was added for 4 h to a 90 mm plate of cells 24 h after transfection. RNAi-mediated knockdown was carried out as detailed in Chiu et al (2004) using anti cyclinT1 and nonspecific control siRNAs from Dharmacon.
Quantitation was carried out using a Molecular Dynamics phosphorimager. Measurements were corrected against the level of expression of the VAI cotransfection or 7SK controls. The percentage total RNA and relative correct 3′ end and readthrough were calculated by adjusting the quantitation for the number of U residues in the transcript.
Western blotting
Western blotting was carried out as described in Medlin et al (2003) with antibodies diluted 1:500. Cells were harvested after treatment with 100 μM KM05283 for 4 h. For the experiment of Figure 3C, lanes 1–3 were exposed for 1 min and lanes 4 and 5 for 1 s.
Chromatin immunoprecipitation analysis
Crosslinking and precipitations were carried out according to Boyd et al (1998). A secondary rabbit anti-mouse IgG and IgH antibody (Jackson Immunoresearch) was used with the H5 and H14 antibodies. The positions of the PCR primers for each gene relative to the +1 start site are as follows: U2: −93 to +190; H2a (Accession No. Z83741): −39 to +107, H2b: −218 to +119; H3 (Accession No. M26150): −54 to +83; H4 (Accession No. M60749): +64 to +275; IgH: (Accession No. X17116) −170 to +99; GAPDH: (Accession No. J04038) −118 to +90. Real-time PCR reactions were carried out using a Corbett Rotorgene. Reactions were run in triplicate and 45 cycles were performed using the Qiagen Quantitect Sybr Green hot start Taq mix with 0.5 μM primers.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Acknowledgments
We thank J Corden, D Price, J Garriga and S Elledge for the generous provision of constructs and Sasha Akoulitchev and Andre Furger for help and advice. JM was supported by the Wellcome Trust, AT by MRC Cooperative Group Grant No. G9826944, FZ and BMP by NIH Grant R01 AI49104.
References
- Ahn SH, Kim M, Buratowski S (2004) Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol Cell 13: 67–76 [DOI] [PubMed] [Google Scholar]
- Barilla D, Lee BA, Proudfoot NJ (2001) Cleavage/polyadenylation factor IA associates with the carboxyl-terminal domain of RNA polymerase II in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 98: 445–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird G, Zorio DA, Bentley DL (2004) RNA polymerase II carboxy-terminal domain phosphorylation is required for cotranscriptional pre-mRNA splicing and 3′-end formation. Mol Cell Biol 24: 8963–8969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd KE, Wells J, Gutman J, Bartley SM, Farnham PJ (1998) c-Myc target gene specificity is determined by a post-DNA binding mechanism. Proc Natl Acad Sci USA 95: 13887–13892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao SH, Price DH (2001) Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J Biol Chem 276: 31793–31799 [DOI] [PubMed] [Google Scholar]
- Chiu YL, Cao H, Jacque JM, Stevenson M, Rana TM (2004) Inhibition of human immunodeficiency virus type 1 replication by RNA interference directed against human transcription elongation factor P-TEFb (CDK9/CyclinT1). J Virol 78: 2517–2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodosh LA, Fire A, Samuels M, Sharp PA (1989) 5,6-Dichloro-1-beta-D-ribofuranosylbenzimidazole inhibits transcription elongation by RNA polymerase II in vitro. J Biol Chem 264: 2250–2257 [PubMed] [Google Scholar]
- Collart D, Romain PL, Huebner K, Pockwinse S, Pilapil S, Cannizzaro LA, Lian JB, Croce CM, Stein JL, Stein GS (1992) A human histone H2B.1 variant gene, located on chromosome 1, utilizes alternative 3′ end processing. J Cell Biochem 50: 374–385 [DOI] [PubMed] [Google Scholar]
- Edwards MC, Wong C, Elledge SJ (1998) Cyclin K, a novel RNA polymerase II-associated cyclin possessing both carboxy-terminal domain kinase and Cdk-activating kinase activity. Mol Cell Biol 18: 4291–4300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong N, Bentley DL (2001) Capping, splicing, and 3′ processing are independently stimulated by RNA polymerase II: different functions for different segments of the CTD. Genes Dev 15: 1783–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong YW, Zhou Q (2001) Stimulatory effect of splicing factors on transcriptional elongation. Nature 414: 929–933 [DOI] [PubMed] [Google Scholar]
- Fujinaga K, Irwin D, Huang Y, Taube R, Kurosu T, Peterlin BM (2004) Dynamics of human immunodeficiency virus transcription: P-TEFb phosphorylates RD and dissociates negative effectors from the transactivation response element. Mol Cell Biol 24: 787–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furger A, O'Sullivan JM, Binnie A, Lee BA, Proudfoot NJ (2002) Promoter proximal splice sites enhance transcription. Genes Dev 16: 2792–2799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu TJ, Peng J, Lee G, Price DH, Flores O (1999) Cyclin K functions as a CDK9 regulatory subunit and participates in RNA polymerase II transcription. J Biol Chem 274: 34527–34530 [DOI] [PubMed] [Google Scholar]
- Garriga J, Grana X (2004) Cellular control of gene expression by T-type cyclin/CDK9 complexes. Gene 337: 15–23 [DOI] [PubMed] [Google Scholar]
- Garriga J, Mayol X, Grana X (1996) The CDC2-related kinase PITALRE is the catalytic subunit of active multimeric protein complexes. Biochem J 319: 293–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez N, Weiner AM (1986) Formation of the 3′ end of U1 snRNA requires compatible snRNA promoter elements. Cell 47: 249–258 [DOI] [PubMed] [Google Scholar]
- Hirose Y, Manley JL (1998) RNA polymerase II is an essential mRNA polyadenylation factor. Nature 395: 93–96 [DOI] [PubMed] [Google Scholar]
- Hirose Y, Tacke R, Manley JL (1999) Phosphorylated RNA polymerase II stimulates pre-mRNA splicing. Genes Dev 13: 1234–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CK, Shuman S (1999) Distinct roles for CTD Ser-2 and Ser-5 phosphorylation in the recruitment and allosteric activation of mammalian mRNA capping enzyme. Mol Cell 3: 405–411 [DOI] [PubMed] [Google Scholar]
- Ivanov D, Kwak YT, Guo J, Gaynor RB (2000) Domains in the SPT5 protein that modulate its transcriptional regulatory properties. Mol Cell Biol 20: 2970–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs EY, Ogiwara I, Weiner AM (2004) Role of the C-terminal domain of RNA polymerase II in U2 snRNA transcription and 3′ processing. Mol Cell Biol 24: 846–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K (2005) The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell 19: 523–534 [DOI] [PubMed] [Google Scholar]
- Keogh MC, Podolny V, Buratowski S (2003) Bur1 kinase is required for efficient transcription elongation by RNA polymerase II. Mol Cell Biol 23: 7005–7018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y-K, Bourgeois CF, Isel C, Churcher MJ, Karn J (2002) Phosphorylation of the RNA polymerase II carboxyl-terminal domain by CDK9 is directly responsible for human immmunodeficiency virus type 1 Tat-activated transcriptional elongation. Mol Cell Biol 22: 4622–4637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licatalosi DD, Geiger G, Minet M, Schroeder S, Cilli K, McNeil JB, Bentley DL (2002) Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol Cell 9: 1101–1111 [DOI] [PubMed] [Google Scholar]
- Mancebo HS, Lee G, Flygare J, Tomassini J, Luu P, Zhu Y, Peng J, Blau C, Hazuda D, Price D, Flores O (1997) P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev 11: 2633–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF, Duronio RJ (2002) Histone mRNA expression: multiple levels of cell cycle regulation and important developmental consequences. Curr Opin Cell Biol 14: 692–699 [DOI] [PubMed] [Google Scholar]
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL (1997) The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature 385: 357–361 [DOI] [PubMed] [Google Scholar]
- Medlin J, Uguen P, Taylor A, Bentley DL, Murphy S (2003) The C-terminal domain of pol II and a DRB-sensitive kinase are required for 3′ processing of U2 snRNA. EMBO J 22: 925–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napolitano G, Licciardo P, Carbone R, Majello B, Lania L (2002) CDK9 has the intrinsic property to shuttle between nucleus and cytoplasm, and enhanced expression of cyclin T1 promotes its nuclear localization. J Cell Physiol 192: 209–215 [DOI] [PubMed] [Google Scholar]
- Neuman de Vegvar HE, Lund E, Dahlberg JE (1986) 3′ end formation of U1 snRNA precursors is coupled to transcription from snRNA promoters. Cell 47: 259–266 [DOI] [PubMed] [Google Scholar]
- Ni Z, Schwartz BE, Werner J, Suarez JR, Lis JT (2004) Coordination of transcription, RNA processing, and surveillance by P-TEFb kinase on heat shock genes. Mol Cell 13: 55–65 [DOI] [PubMed] [Google Scholar]
- Peng J, Zhu Y, Milton JT, Price DH (1998) Identification of multiple cyclin subunits of human P-TEFb. Genes Dev 12: 755–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilch DR, Marzluff WF (1991) Expression of histone-U1 snRNA chimeric genes: U1 promoters are compatible with histone 3′ end formation. Gene Exp 1: 41–53 [PMC free article] [PubMed] [Google Scholar]
- Ping YH, Rana TM (2001) DSIF and NELF interact with RNA polymerase II elongation complex and HIV-1 Tat stimulates P-TEFb-mediated phosphorylation of RNA polymerase II and DSIF during transcription elongation. J Biol Chem 276: 12951–12958 [DOI] [PubMed] [Google Scholar]
- Price DH (2000) P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol Cell Biol 20: 2629–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner DB, Yamaguchi Y, Wada T, Handa H, Price DH (2001) A highly purified RNA polymerase II elongation control system. J Biol Chem 276: 42601–42609 [DOI] [PubMed] [Google Scholar]
- Sano M, Wang SC, Shirai M, Scaglia F, Xie M, Sakai S, Tanaka T, Kulkarni PA, Barger PM, Youker KA, Taffet GE, Hamamori Y, Michael LH, Craigen WJ, Schneider MD (2004) Activation of cardiac Cdk9 represses PGC-1 and confers a predisposition to heart failure. EMBO J 23: 3559–3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schang LM (2002) Cyclin-dependent kinases as cellular targets for antiviral drugs. J Antimicrob Chemother 50: 779–792 [DOI] [PubMed] [Google Scholar]
- Taube R, Lin X, Irwin D, Fujinaga K, Peterlin BM (2002) Interaction between P-TEFb and the C-terminal domain of RNA polymerase II activates transcriptional elongation from sites upstream or downstream of target genes. Mol Cell Biol 22: 321–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trigon S, Serizawa H, Conaway JW, Conaway RC, Jackson SP, Morange M (1998) Characterization of the residues phosphorylated in vitro by different C-terminal domain kinases. J Biol Chem 273: 6769–6775 [DOI] [PubMed] [Google Scholar]
- Uguen P, Murphy S (2003) The 3′ ends of vertebrate pre-snRNAs are produced by RNA polymerase II CTD-dependent RNA processing. EMBO J 22: 4544–4554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Takagi T, Yamaguchi Y, Watanabe D, Handa H (1998) Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J 17: 7395–7403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West ML, Corden JL (1995) Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutations. Genetics 140: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y, Takagi T, Wada T, Yano K, Furuya A, Sugimoto S, Hasegawa J, Handa H (1999) NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97: 41–51 [DOI] [PubMed] [Google Scholar]
- Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q (2005) Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell 19: 535–545 [DOI] [PubMed] [Google Scholar]
- Zhang F, Barboric M, Blackwell TK, Peterlin BM (2003) A model of repression: CTD analogs and PIE-1 inhibit transcriptional elongation by P-TEFb. Genes Dev 17: 748–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Roeder RG, Heintz N (1983) The primary structure and expression of four cloned human histone genes. Nucleic Acids Res 11: 7409–7425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorio DA, Bentley DL (2004) The link between mRNA processing and transcription: communication works both ways. Exp Cell Res 296: 91–97 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2


