Abstract
The positive transcription elongation factor b (P-TEFb), a complex of Cdk9 and cyclin T1/T2, stimulates transcription by phosphorylating RNA polymerase II. The 7SK small nuclear RNA, in cooperation with HEXIM1 protein, functions as a general polymerase II transcription regulator by sequestering P-TEFb into a large kinase-inactive 7SK/HEXIM1/P-TEFb complex. Here, determination and characterization of the functionally essential elements of human 7SK snRNA directing HEXIM1 and P-TEFb binding led to a new model for the assembly of the 7SK/HEXIM1/P-TEFb regulatory complex. We demonstrate that two structurally and functionally distinct protein binding elements located in the 5′- and 3′-terminal hairpins of 7SK support the in vivo recruitment of HEXIM1 and P-TEFb. Consistently, a minimal regulatory RNA composed of the 5′ and 3′ hairpins of 7SK can modulate polymerase II transcription in HeLa cells. HEXIM1 binds independently and specifically to the G24-C48/G60-C87 distal segment of the 5′ hairpin of 7SK. Binding of HEXIM1 is a prerequisite for association of P-TEFb with the G302-C324 apical region of the 3′ hairpin of 7SK that is highly reminiscent of the human immunodeficiency virus transactivation-responsive RNA.
Cyclic phosphorylation of the tandemly repeated YSPTSPS heptapeptide motif in the carboxy-terminal domain (CTD) of the largest subunit of RNA polymerase II (Pol II) is crucial for stimulating mRNA production (9, 14). For example, during the early stage of transcription, phosporylation of the CTD at serine 2 by the positive transcription elongation factor b (P-TEFb) is essential for the transition from abortive to productive transcription elongation (5, 7, 30, 35). P-TEFb is a general transcription factor that facilitates the production of full-length mRNAs of most, if not all, protein-coding genes and also stimulates the Pol II-mediated synthesis of human immunodeficiency virus (HIV) transcripts from the 5′ long terminal repeat of the integrated proviral genome (5, 29).
P-TEFb is composed of a cyclin-dependent kinase, Cdk9, and the regulatory subunit cyclin (Cyc) T1, T2, or K (22, 33, 41; reviewed in reference 23). In human HeLa cells, about half of P-TEFb is associated with large ribonucleoprotein (RNP) complexes which also contain the 7SK small nuclear RNA (snRNA) and the HEXIM1 or HEXIM2 protein (3, 17, 20, 36, 37, 39). In contrast to its free form, the 7SK/HEXIM1-associated fraction of P-TEFb shows little CTD kinase activity, indicating that the 7SK snRNA, in collaboration with HEXIM1, functions as an inhibitory factor of P-TEFb. Association of P-TEFb with 7SK/HEXIM1 is specific and reversible. Inhibition of transcription by chemical or UV treatment induces dissociation of P-TEFb from the kinase-inactive 7SK/HEXIM1/P-TEFb complex (17, 20, 36, 37). Consequently, increased accumulation of free P-TEFb facilitates CTD phosphorylation and mRNA production. Likewise, depletion of 7SK snRNA increases the CTD kinase activity of P-TEFb and stimulates transcription from Pol II-specific promoters, including the HIV long terminal repeat (20, 36). In cardiac myocytes, hypertrophic signals can activate P-TEFb by facilitating its dissociation from 7SK snRNA and HEXIM1, leading to increased mRNA and protein synthesis, which finally can promote cardiac hypertrophy (28).
7SK is an abundant, 331-nucleotide-long, Pol III-synthesized snRNA that shows high sequence conservation in vertebrates (11, 15, 19). In human HeLa cells, about 30% of 7SK snRNA is sequestered into 7SK/HEXIM/P-TEFb RNP. The molecular interactions governing the dynamic and regulated assembly of 7SK/HEXIM1/P-TEFb that eventually controls the availability of active P-TEFb for Pol II transcription are poorly understood. HEXIM1 has been demonstrated to specifically bind to 7SK snRNA via a stretch of 18 basic amino acids reminiscent of the arginine-rich RNA recognition motif present in many RNA binding proteins (16, 37). In vitro reconstitution experiments showed that P-TEFb can also interact with 7SK snRNA (37, 38). Reversible phosphorylation of CDK9, most probably at the conserved Thr-186 in the T loop, is required for in vitro association of P-TEFb and 7SK snRNA (6). This might explain why in vitro reconstitution performed with recombinant P-TEFb failed to confirm a direct interaction between P-TEFb and 7SK (16). Finally, yeast two-hybrid assays and in vitro pull-down experiments showed that the C-terminal domain of HEXIM1 can specifically interact with CycT1, and most probably this interaction is responsible for inactivation of P-TEFb (16, 17). However, HEXIM1 can interact with P-TEFb and can inhibit its CTD kinase activity only in the presence of 7SK snRNA, suggesting that 7SK provides the structural scaffold for the assembly of the kinase-inactive 7SK/HEXIM1/P-TEFb complex (16, 38).
In contrast to its central role in the regulation of cellular RNA production, nothing is known about the structural determinants of 7SK snRNA supporting its regulatory function. In this study, we have identified and characterized the elements of human 7SK snRNA that are required for binding of HEXIM1 and P-TEFb in living cells. Based on our results, we propose a model for the assembly of the CTD kinase-inactive 7SK/HEXIM1/P-TEFb complex.
MATERIALS AND METHODS
General procedures.
Unless otherwise stated, all manipulations were performed according to standard laboratory protocols (27). Human HeLa and G3H (21) (provided by Q. Zhou, University of California, Berkeley) cells were grown in Dulbecco's modified Eagle medium supplemented with 10% fetal calf serum (Invitrogen). A standard calcium phosphate transfection protocol was used to introduce expression vectors into HeLa and G3H cells.
Expression constructs.
To generate the p7SK expression construct, the human 7SK snRNA gene carrying 244-bp upstream and 5-bp downstream flanking sequences was PCR amplified by using oligonucleotides A (TGGAGACTGCAGTATTTAGC) and B (ATAGAATTCGGGTCAAAAGAAAGGCAGAC) as 5′- and 3′-specific primers, respectively, and human genomic DNA as a template. The nucleotide sequence of the human 7SK gene was obtained from GenBank (accession number X05490). The amplified DNA was digested with PstI and EcoRI and inserted into the same sites of pBluescript II KS(+) (Stratagene). The same approach was used for construction of p7SKd6, p7SKd17 to p7SKd19, p7SKm10, p7SKm13, p7SKm14, p7SK5′hp, p7SKm11/m11c, and p7SKm12/m12c, except that appropriate mutagenic oligonucleotides were used as 3′-specific primers and p7SK was used as a template. For construction of all other 7SK expression vectors, a two-step PCR amplification approach was used. In the first reaction, either the 5′ or the 3′ half of the human 7SK gene was amplified by using an appropriate mutagenic internal primer and oligonucleotide A or B. The resulting DNA products were used as megaprimers in the second amplification step in combination with the appropriate terminal primer (oligonucleotide A or B). The amplified products were digested with PstI and EcoRI and cloned into the same sites of pBluescribe. The sequences of the utilized mutagenic oligonucleotide primers are available upon request. To generate pHA-HEXIM1, hemagglutinin (HA) epitope was added to the N-terminal region of HEXIM1 by PCR amplification using oligonucleotides H1 (ACCCAAGCTTACTCTACTAGCCATGTACCCATACGACGTCCCAGACTACGCTGCCGAGCCATTCTTGTCAGA) and H2 (TCAGCGGCCGCAAGTTTCAGTCTAGTCTCCA) as primers and a full-length HEXIM1 cDNA clone as a template (Invitrogen). The PCR product was digested with HindIII and NotI and inserted into the same sites of the pcDNA3 expression vector (Invitrogen). The identity of each construct was verified by sequence analysis.
IP and in vitro reconstitution.
About 5 × 106 cells were washed in ice-cold TBS buffer (150 mM NaCl, 40 mM Tris-HCl, pH 7.4). Cells were resuspended in 500 μl of cold NET-2 buffer (150 mM NaCl, 0.05% NP-40, 50 mM Tris-HCl, pH 7.4) and sonicated for 3 × 30 s with 60-s intervals at setting 3 with a Branson sonifier. Cell debris was removed by centrifugation, and the supernatant was incubated with 20 μl of protein A-agarose beads (Sigma) swollen in NET-2 buffer and saturated with anti-HA antibody (12CA5; Roche). After immunoprecipitation (IP), the beads were washed five times with 1 ml of NET-2 buffer and RNAs were extracted by phenol-chloroform extraction.
For in vitro reconstitution experiments, about 30 μl of protein A-agarose beads coated with an anti-CycT1 antibody (ab2098; Abcam) was incubated with a G3H cell extract under the conditions described above, except that the extract had been pretreated with 50 units of micrococcal nuclease (Amersham Biosciences) for 20 min at 30°C. After immunoprecipitation of P-TEFb, the beads were saturated with 5 μg of Escherichia coli tRNA, washed five times with NET-2 buffer, and incubated for 1 h at 4°C with a mixture of about 200 pg of in vitro-transcribed, internally labeled mutant or wild-type 7SK RNAs and about 1.5 pmol of purified His-tagged recombinant HEXIM1 protein (17). The beads were washed five times with NET-2 buffer, binding of 7SK RNAs was tested by polyacrylamide gel electrophoresis, and retention of HA-tagged CycT1 and His-tagged HEXIM1 was verified by Western blot analysis performed with anti-HA (12CA5; Roche) and anti-His antibodies (ab1187; Abcam). To generate template DNA for in vitro transcription of 7SK, 7SK5′+3′hp, 7SKd6, and 7SKd12 RNAs, the 7SK-coding regions of p7SK, p7SK5′+3′hp, p7SKd6, and p7SKd12 were PCR amplified by using appropriate oligonucleotide primers carrying the T7 RNA polymerase promoter.
RNA analysis.
For Northern blots, RNAs were separated on a 6% sequencing gel and electrotransferred onto a Hybond-N membrane (Amersham). For detection of wild-type and mutant 7SK snRNAs, oligonucleotides P1 (AGGCAGACTGCCACATGCAG), P2 (GTGTCTGGAGTCTTGGAAGC), and P3 (CATGGAGCGGTGAGGGAGGA), complementary to different regions of 7SK, were used as probes after being terminally labeled with T4 polynucleotide kinase and [γ-32P]ATP (3,000 Ci/mmol; Amersham). The RNase A/T1 mapping procedure was described earlier (10). To synthesize sequence-specific antisense RNA probes, the appropriate 7SK expression constructs were linearized by PstI and used as templates for transcription by T7 RNA polymerase. To generate an RNA probe for mapping of luciferase mRNA, a 150-bp fragment of the luciferase gene was PCR amplified by using oligonucleotides L1 (TGGAGACTGCAGGCGCCATTCTATCCTCTAGA) and L2 (ATAGAATTCAACGGACATTTCGAAGTATTCC). The amplified fragment was inserted into the PstI and EcoRI sites of pBST, and the resulting pLUC plasmid, after linearization with PstI, was transcribed by T7 polymerase. Production of an antisense RNA probe for U6-5.8 RNA has been reported (8).
Luciferase reporter gene assay.
Human HeLa cells were cotransfected with 1 μg of luciferase reporter plasmid, pSV4LUC (4), carrying the luciferase coding region under the control of the simian virus 40 (SV40) promoter with 5 μg of pU6-5.8 and 10 μg of pBST, p7SK, p7SKd12, or p7SK5′+3′hp; 48 h later, transfected cells were washed with phosphate-buffered saline and lysed in reporter lysis buffer, and luciferase activity was determined by standard chemiluminescence detection procedures using a Sirius luminometer (2). Values were normalized to the total cell protein content.
Fluorescence in situ hybridization.
Synthesis of amino-modified oligonucleotides, chemical conjugation with FluoroLink Cy3 monofunctional reactive dye (Amersham Biosciences), fluorescence in situ hybridization of human HeLa cells, and image acquisition and processing were performed according to the protocols of the laboratory of R. Singer (http://singerlab.aecom.yu.edu). The following oligonucleotides were used to detect transiently expressed 7SK5′+3′hp (TT*ATGCAGCGCCTCATTT*CGCACATGGAGCGGTT*T) and wild-type 7SK (TT*GGATGTGTCTGGAGTCTT*GGAAGCTTGACTACT*T) snRNAs. Amino-allyl-modified T residues are indicated by asterisks. Nuclear DNA was stained with 0.1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI).
Nucleotide sequence accession numbers.
The following GenBank accession numbers were obtained for the sequences of chicken (AJ890101), zebrafish (AJ890102), Tetraodon nigrovidis (AJ890103), and Fugu rubripes (AJ890104) 7SK RNAs identified in our laboratory (unpublished results).
RESULTS
Two distinct regions of 7SK snRNA are required for in vivo binding of P-TEFb.
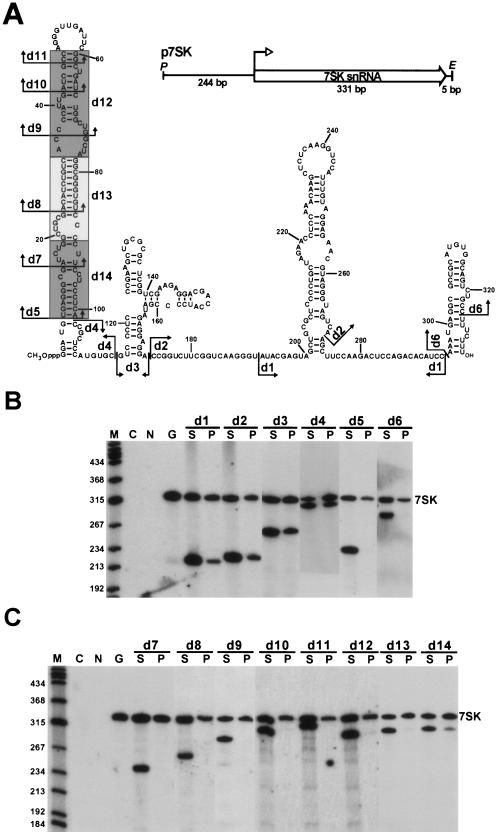
To define the structural elements of human 7SK snRNA responsible for either direct (6, 37) or indirect (16) binding of P-TEFb in living cells, a series of mutant 7SK RNAs carrying nested internal deletions were transiently expressed in human G3H cells expressing an HA epitope-tagged version of CycT1 (21). The P-TEFb binding capacity of each ectopically expressed RNA was determined by co-IP with the HA-CycT1/CDK9 complex, hereafter called HA-P-TEFb. To generate the p7SK expression plasmid, the human 7SK gene was PCR amplified and inserted into a plasmid vector (Fig. 1A). The internal deletions d1 to d14 introduced into the coding region of p7SK are shown on a previously proposed secondary structure of human 7SK snRNA (31). Since the Pol III-mediated synthesis of 7SK snRNA is controlled by upstream promoter elements, deletions in the coding region were not expected to alter the transcriptional activity of the truncated 7SK genes (15, 19).
FIG. 1.
Two distinct regions of 7SK snRNA are required for in vivo binding of P-TEFb. (A) Structures of transiently expressed internally truncated 7SK snRNAs. The schematic structure of the p7SK expression construct and the proposed secondary structure of human 7SK snRNA (31) are shown. The 7SK coding region (open arrow), the transcription initiation site (open arrowhead), and the relevant restriction sites (P, PstI; E, EcoRI) are indicated. Internal deletions are indicated by arrows (d1 to d11) or shaded boxes (d12 to d14). (B) Localization of P-TEFb binding elements in 7SK snRNA. Mutant 7SK snRNAs, indicated above the lanes, were transiently expressed in G3H cells expressing an HA-tagged version of CycT1 (21). Cellular extracts were incubated with protein A-agarose saturated with anti-HA antibody. After centrifugation, RNAs extracted from the agarose beads (P) and supernatants (S) were fractionated in 6% denaturing polyacrylamide gels and probed with terminally labeled oligonucleotides complementary to the human 7SK snRNA. The endogenous wild-type 7SK snRNA is shown (7SK). As controls, RNAs immunoselected from nontransfected G3H (G) and HeLa (N) cells were also probed. Lane C represents a control blot with E. coli tRNA that was used to facilitate RNA precipitation. Lane M, size markers in nucleotides (terminally labeled HaeIII- and TaqI-digested pBR322). (C) Identification of P-TEFb binding elements in the 5′-terminal hairpin of 7SK snRNA. For details, see the legend to panel B.
The d1-to-d6 deletions tested in the first experiment covered the entire coding region of the 7SK gene except for the 5′-terminal nucleotides G1 to G5, which may influence transcription initiation, and the 3′-terminal T-rich track that serves as a terminator for Pol III. Expression of the truncated 7SK RNAs and their association with HA-P-TEFb were monitored by IP with an anti-HA monoclonal antibody, followed by Northern blotting (Fig. 1B). Probing of RNAs recovered from the supernatant (Fig. 1B, lanes S) of each IP reaction revealed that all mutant 7SK RNAs accumulated efficiently in G3H cells. Blotting of the immunoprecipitated RNAs (lanes P) showed that the 7SKd1, 7SKd2, 7SKd3, and 7SKd4 RNAs, like the endogenous wild-type 7SK snRNA, coprecipitated with HA-P-TEFb, indicating that the middle part of 7SK from C102 to C295 carries no crucial elements for P-TEFb binding. It is important to note, however, that each mutant RNA showed a slightly reduced P-TEFb binding affinity compared to the endogenous 7SK snRNA. Most likely, the central region of 7SK contributes to P-TEFb binding by facilitating the correct folding of the full-length RNA. As expected, the mutant 7SKd1, 7SKd2, 7SKd3, and 7SKd4 RNAs were also recovered in IP experiments performed with an antibody directed against Cdk9 (data not shown). More importantly, the 7SKd5 and 7SKd6 RNAs lacking the 5′- or 3′-teminal hairpin of the wild-type RNA, respectively, were fully inactive in P-TEFb binding, as demonstrated by both anti-HA (Fig. 1B) and anti-CycT1 (data not shown) IPs. These results demonstrate that the elements crucial for P-TEFb binding are contained in the two terminal hairpins of human 7SK.
To delimit more precisely the RNA sequences that constitute the P-TEFb binding motif in the long 5′ hairpin of 7SK, additional internally truncated RNAs, 7SKd7 to 7SKd14 (see Fig. 1A), were expressed in G3H cells and their HA-P-TEFb binding capacities were tested (Fig. 1C). We found that removal of the terminal loop and two adjacent basepairs from the top of the 5′ hairpin abolished the HA-P-TEFb binding capacity of the resulting 7SKd11 RNA. As expected, 7SKd7, 7SKd8, 7SKd9, and 7SKd10 RNAs featuring more extensive 5′-hairpin truncations also failed to associate with HA-P-TEFb. The same results were obtained upon removal of the d12 and d13 internal segments of the 5′ hairpin; the resulting 7SKd12 and 7SKd13 RNAs did not interact with HA-P-TEFb. Removal of the d14 basal segment of the 5′ hairpin of 7SK reduced but did not fully abolish the HA-P-TEFb binding capacity of 7SKd14, indicating that the proximal part of the 5′ hairpin contains no crucial elements for P-TEFb binding. In summary, we conclude that the 3′-terminal hairpin and the distal part of the 5′ hairpin of 7SK contain all the elements that are fundamental for in vivo recruitment of P-TEFb.
The distal part of the 5′-terminal hairpin of 7SK directs HEXIM1 binding.
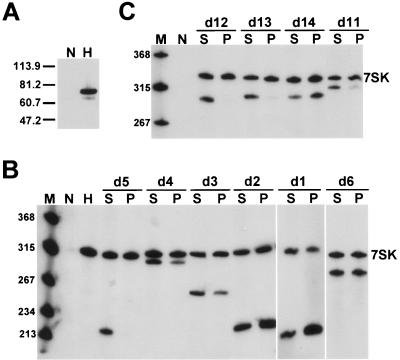
The 7SK-dependent inhibition of the CTD kinase activity of P-TEFb also requires HEXIM1, which, besides interacting with CycT1, directly binds to 7SK snRNA (16, 38). To define the HEXIM1 binding elements of 7SK snRNA, an HA-tagged version of human HEXIM1 (HA-HEXIM1) was transiently expressed in HeLa cells (Fig. 2A). Interaction of HA-HEXIM1 with coexpressed mutant 7SK RNAs carrying the d1, d2, d3, d4, d5, or d6 internal deletion was investigated by IP with anti-HA antibody (Fig. 2B). Northern blot analysis showed that all mutant 7SK RNAs, except 7SKd5, coprecipitated with HA-HEXIM1, indicating that the 5′-terminal hairpin carries the HEXIM1 binding elements of 7SK.
FIG. 2.
Elements directing HEXIM1 binding are confined to the distal portion of the 5′-terminal hairpin of 7SK. (A) Expression of HA-tagged HEXIM1. Accumulation of HA-HEXIM1 in HeLa cells either transfected (H) or untransfected (N) with the pHA-HEXIM1 expression construct was monitored by Western blotting with an anti-HA antibody. Molecular mass markers (kDa) are shown. (B) Determination of 7SK snRNA regions supporting in vivo binding of HEXIM1. (C) Mapping of HEXIM1 binding elements in the 5′ hairpin of 7SK. Details are given in the legend to Fig. 1.
Next, we investigated the HA-HEXIM1 binding abilities of 7SK RNAs lacking the d11, d12, d13, or d14 5′-hairpin fragment (Fig. 2C). While 7SKd14 RNA formed a strong interaction with HA-HEXIM1, neither 7SKd12 nor 7SKd13 RNA was detectable in the pellets from the IP reactions, demonstrating that the d12 and d13 internal regions are crucial for HEXIM1 binding. Likewise, a very weak HA-HEXIM1 interaction was detected for 7SKd11, indicating that the d11 apical stem-loop largely contributes to the HEXIM1 binding capacity of 7SK snRNA. These results demonstrate that the HEXIM1 binding elements of 7SK snRNA are located in the distal portion of the 5′-terminal hairpin. Since this region is also crucial for P-TEFb binding (Fig. 1C), we conclude that elements directing P-TEFb and HEXIM1 binding largely, if not fully, overlap in the 5′ hairpin of 7SK snRNA.
Inhibition of SV40-driven luciferase expression by a minimal 7SK RNA.
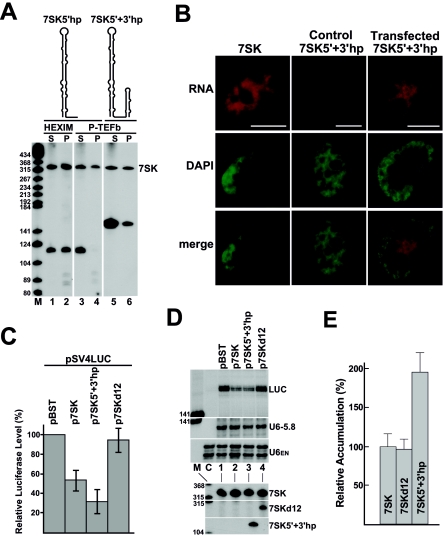
The experiments presented thus far indicate that the RNA elements crucial for in vivo recruitment of HEXIM1 and P-TEFb are located exclusively in the 5′ and 3′ hairpins of 7SK snRNA. To confirm this notion, a minimal 7SK RNA, 7SK5′+3′hp, carrying the 5′- and 3′-terminal hairpins but lacking the U116-C295 internal fragment of 7SK, was transiently expressed in HeLa and G3H cells (Fig. 3A). Another truncated RNA, 7SK5′hp, composed of the 5′-terminal hairpin (G1 to G111) and the U-rich transcription terminator (U325 to U331) of 7SK, was also expressed as a control. Northern analysis showed that both 7SK5′+3′hp and 7SK5′hp accumulated efficiently in transfected cells (Fig. 3A, lanes 1, 3, and 5). IP performed with anti-HA antibody revealed that the 7SK5′hp control RNA associated with HA-HEXIM1 (lane 2) but, as expected, failed to bind HA-P-TEFb (lane 4). This demonstrates that in living cells, binding of HEXIM1 to 7SK snRNA is independent of P-TEFb recruitment. In contrast to 7SK5′hp, 7SK5′+3′hp was able to bind both HA-P-TEFb (lane 6) and HA-HEXIM1 (data not shown), further corroborating the idea that the internal part of 7SK possesses no vital elements for HEXIM1 and P-TEFb binding.
FIG. 3.
Expression of 7SK5′+3′hp represses luciferase expression controlled by the SV40 promoter. (A) Expression of 7SK5′hp and 7SK5′+3′hp RNAs. The predicted schematic structures of 7SK5′hp and 7SK5′+3′hp RNAs coexpressed with HA-HEXIM1 in HeLa cells or with HA-P-TEFb in G3H cells are indicated. Interaction of 7SK5′hp and 7SK5′+3′hp RNAs with HA-HEXIM1 and HA-P-TEFb was tested by co-IP followed by Northern analysis. For other details, see the legend to Fig. 1. (B) In situ localization of 7SK5′+3′hp. HeLa cells transfected or untransfected (control) with p7SK5′+3′hp were hybridized with a fluorescent oligonucleotide probe complementary to the junction region of the 5′ and 3′ hairpins of 7SK5′+3′hp. Wild-type 7SK snRNA was localized in untransfected HeLa cells by using a sequence-specific fluorescent probe. Nuclei were stained with DAPI. The bars represent 10 μm. (C) Luciferase gene expression driven by the SV40 promoter. HeLa cells were cotransfected with the pSV4LUC luciferase reporter plasmid and the indicated expression and control plasmids. The cells were lysed in reporter lysis buffer, and the protein content and luciferase activity were determined. The activities of lysates obtained from cells transfected with pSV4LUC and pBST were set as 100%. The average values (with standard deviations) of six independent experiments, each performed in quintuplicate, are shown. (D) RNA analysis. RNAs were isolated from HeLa cells cotransfected with pSV4LUC and the indicated plasmids. Accumulation of luciferase mRNA (LUC), the RNA Pol III-transcribed U6-5.8 transfection control RNA, and the endogenous U6 snRNA was measured by RNase A/T1 mapping. Lane C represents control mapping with untransfected HeLa RNAs. Accumulation of 7SK, 7SKd12, and 7SK5′-3′hp was monitored by Northern blot analysis. (E) Relative accumulation of ectopically expressed 7SK, 7SKd12, and 7SK5′+3′hp. The intensities of 7SK RNAs were quantified by phosphorimager. Overaccumulation of the wild-type 7SK snRNA was determined by subtraction of the level of endogenous 7SK measured in control cells transfected with pBST. The relative levels of 7SK, 7SKd12, and 7SK5′+3′hp were normalized to the transiently expressed U6-5.8 RNA. The relative accumulation of ectopically expressed 7SK was considered 100%. Standard deviations are indicated.
In vitro inhibition of the kinase activity of P-TEFb necessitates only two trans-acting factors, the 7SK snRNA and HEXIM1 (16, 17, 37, 38). Therefore, we tested whether 7SK5′+3′hp, which binds both P-TEFb and HEXIM1, is capable of modulating transcription elongation in living cells. Fluorescence in situ hybridization microscopy revealed identical subcellular localizations for the ectopically expressed 7SK5′+3′hp RNA and the wild-type 7SK snRNA (Fig. 3B). Both RNAs showed a slightly speckled nucleoplasmic localization pattern with predominant exclusion from the nucleolar territories, suggesting that 7SK5′+3′hp is faithfully localized in transfected HeLa cells. Next, the p7SK5′+3′hp construct and the pSV4LUC transcription reporter plasmid expressing luciferase under the control of the P-TEFb-dependent SV40 promoter were cotransfected into HeLa cells (4, 36). As controls, an empty plasmid (pBST) or an expression plasmid encoding either the wild-type RNA (p7SK) or a mutant 7SK RNA lacking both HEXIM1 and P-TEFb binding capacities (p7SKd12) was transfected along with pSV4LUC. The transfection efficiency was controlled by inclusion of the pU6-5.8S expression plasmid carrying a tagged version of the RNA Pol III-transcribed human U6 snRNA gene (8). The transfected cells were lysed, and the luciferase activities of the lysates were determined (Fig. 3C). Cells transfected with p7SK or p7SK5′+3′hp showed strongly reduced luciferase activities compared to control cells carrying either pBST or p7SKd12. Moreover, RNase protection demonstrated that the compromised luciferase activities of the p7SK5′+3′hp- and p7SK-containing cells were accompanied by a reduced accumulation of luciferase mRNA (Fig. 3D, lanes 2 and 3), since the control cells accumulated luciferase mRNA about two times more efficiently (lanes 1 and 4). The relative accumulation levels of the ectopically expressed 7SK5′+3′hp, 7SKd12, and 7SK RNAs were determined by phosphorimager quantification and normalized to the U6-5.8 transfection control RNA (Fig. 3E). The 7SK5′+3′hp RNA accumulated about twofold more efficiently than the 7SK and 7SKd12 RNAs. Consistent with this, luciferase expression was inhibited almost two times more efficiently in p7SK5′+3′hp-transfected cells. We concluded that 7SK5′-3′hp, like wild-type 7SK, is capable of inhibiting transcription from the luciferase-linked SV40 promoter and that the 5′- and 3′-terminal hairpins of 7SK snRNA contain all the elements required for modulation of Pol II-specific transcription in living cells. Finally, the observation that overexpression of 7SK snRNA can inhibit Pol II transcription demonstrates that 7SK is limiting for the assembly of the 7SK/HEXIM1/P-TEFb regulatory complex (see Discussion).
The HEXIM1 and P-TEFb binding elements of 7SK show strong evolutionary conservation.
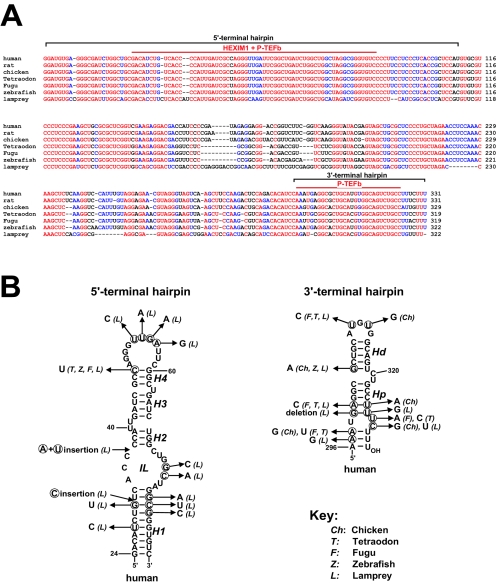
Consistent with its important function in transcription regulation, the 7SK snRNA shows striking sequence conservation in vertebrates. A sequence alignment of the previously characterized human, rat, and lamprey 7SK snRNAs (11, 15, 18, 24), along with four novel 7SK RNAs identified in our laboratory from chicken, Tetraodon nigrovidis, fugu, and zebra fish (unpublished data), revealed 59% sequence identity for the seven vertebrate RNAs (Fig. 4A). In general, the 5′- and 3′-terminal regions show the highest conservation, while sequences in the internal part of 7SK are more variable. Importantly, the HEXIM1 and P-TEFb binding elements defined by our analysis are located within the most conserved regions of 7SK snRNA.
FIG. 4.
Evolutionary conservation of 7SK elements required for HEXIM1 and P-TEFb binding. (A) Alignment of vertebrate 7SK snRNA sequences. The sequences of human (GenBank accession number NR_001445), rat (K02909), and lamprey (Lampetra fluviatilis) (11) 7SK snRNAs have been reported. The sequences of chicken (AJ890101), zebra fish (AJ890102), Tetraodon nigrovidis (AJ890103), and Fugu rubripes (AJ890104) 7SK RNAs were identified in our laboratory (unpublished results). Invariant nucleotides are indicated in red, while nucleotides conserved in all but one sequence are in blue. The 5′- and 3′-hairpin areas and regions essential for HEXIM1 and P-TEFb binding are indicated. (B) Proposed secondary structures of human 7SK elements directing HEXIM1 and P-TEFb binding. The proposed stem (H1, H2, H3, H4, Hd, and Hp) and internal-loop (IL) structures are indicated. The nucleotide changes in vertebrate 7SK RNAs are shown.
The proposed secondary structure of the 5′- and 3′-end regions of human 7SK snRNA implicated in HEXIM1 and P-TEFb binding, together with the nucleotide alterations found in vertebrate 7SK RNAs, are shown in Fig. 4B. The correctness of these structures was supported by in vitro and in vivo structure probing (31). The distal region of the 5′ hairpin required for both HEXIM1 and P-TEFb binding is predicted to contain four helices (H1 to H4), a terminal and an internal loop (IL), and two bulges (U63 and 40-UU-41). This structure is preserved in all 7SK snRNAs, except that a shorter H1 helix can be folded for lamprey 7SK. The 3′-terminal hairpin is composed of a small terminal loop and two conserved helices (Hd and Hp) separated by the bulged 320-CU-321 residues. Formation of a previously proposed short helix at the base of the 3′ hairpin is not supported by phylogenetic conservation. Otherwise, the structure of the upper part of the 3′ hairpin is invariant, supporting the notion that 7SK elements directing recruitment of P-TEFb and HEXIM1 show strong structural conservation during evolution.
The RNA elements directing HEXIM1 or P-TEFb binding to the 5′ hairpin are indistinguishable.
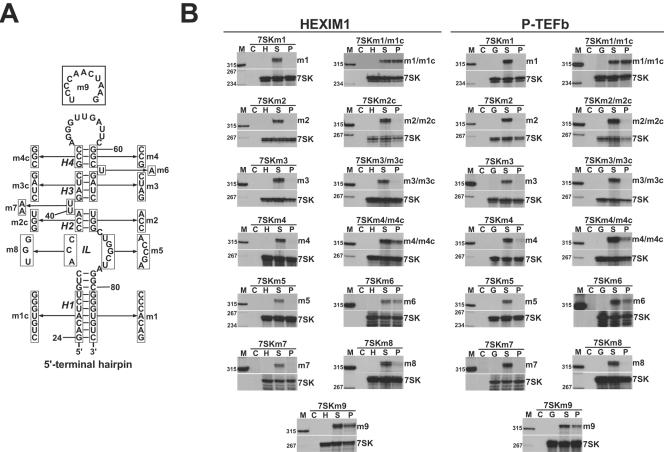
To further characterize the HEXIM1 and P-TEFb binding elements in the 5′ hairpin of 7SK, the predicted H1, H2, H3, and H4 stems were destroyed by substitution of their descending (3′-side) strands (Fig. 5A). The resulting 7SKm1, 7SKm2, 7SKm3, and 7SKm4 RNAs were coexpressed with HA-P-TEFb and HA-HEXIM1, and their P-TEFb and HEXIM1 binding capacities were assayed (Fig. 5B). We found that the m1, m2, m3, and m4 mutations completely abolished both the HEXIM1 and P-TEFb binding abilities of 7SK snRNA. However, when the H1 and H4 stems were restored by replacement of the 5′ sides of these stems with complementary sequences (m1c and m4c), the HEXIM1 and P-TEFb binding capacities of the 7SKm1/m1c and 7SKm4/m4c double-mutant RNAs were largely restored. Two major conclusions can be drawn from these results. First, the H1 and H4 helices indeed exist in 7SK snRNA, and more importantly, these helices support HEXIM1 and P-TEFb binding in a sequence-independent manner. Our attempts to restore the H2 and H3 stems in 7SKm2/m2c and 7SKm3/m3c RNAs, however, failed to rescue the HEXIM1 and P-TEFb binding abilities of these RNAs. This may indicate that the sequence composition of the H2 and H3 stems, if they are really formed, is crucial for 7SK function. Likewise, alteration of the internal-loop (IL) sequences or changing the bulged 40-UU-41 residues abolished association of 7SKm5, 7SKm8, and 7SKm7 RNAs with both HEXIM1 and P-TEFb. In contrast, replacement of the bulged U63 with an A residue in 7SKm6 or alteration of the terminal-loop sequence of 7SKm9 reduced but did not fully eliminate the HEXIM1 and P-TEFb binding capacities of these RNAs, indicating that these sequences are not fundamental to 7SK function. Thus, we propose that a segment of the 5′ hairpin delimited by the H1 and H4 stems contains all the elements essential for HEXIM1 and P-TEFb binding. While the nucleotide composition of the H1 and H4 helices is not critical, the internal sequences are fundamental for binding of both HEXIM1 and P-TEFb. It is important to note that mutational analysis of the 5′ hairpin of 7SK failed to identify distinct binding elements for HEXIM1 and P-TEFb. With no exception, all structural alterations of 7SK which abolished or compromised HEXIM1 binding also prevented or reduced P-TEFb association. These results, taken together with the facts that CycT1 can interact with HEXIM1 (16, 17) and that HEXIM1 alone can bind to 7SK snRNA (Fig. 3A), indicate that P-TEFb is recruited to the 5′ hairpin of 7SK through an interaction formed with HEXIM1.
FIG. 5.
Structural requirements of HEXIM1 and P-TEFb binding to the 5′-terminal hairpin of human 7SK snRNA. (A) Sequence alterations introduced into the 5′-hairpin region of the p7SK expression construct. The proposed helices (H1 to H4) and the internal loop (IL) are indicated. (B) Interactions of transiently expressed mutant 7SK RNAs with HA-tagged HEXIM1 and P-TEFb. The transiently expressed and endogenous 7SK RNAs were detected by RNase A/T1 mapping with antisense RNA probes specific for the ectopically expressed mutant 7SK RNAs. For other details, see the legends to Fig. 1 and 2.
Integrity of the 3′ hairpin is essential for P-TEFb binding.
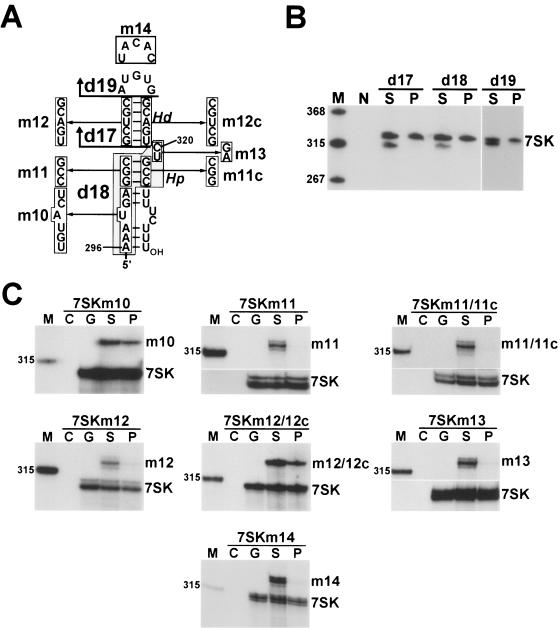
To define the structural determinants of P-TEFb binding to the 3′ hairpin, we performed a detailed mutational analysis of this region of 7SK (Fig. 6A). Upon removal of the distal (d17) or proximal (d18) region of the 3′ hairpin or elimination of its terminal loop (d19), the mutant 7SKd17, 7SKd18, and 7SKd19 RNAs failed to interact with P-TEFb (Fig. 6B). Likewise, destruction of the distal (Hd) or proximal (Hp) stems by replacement of their 5′-side sequences with noncomplementary sequences (m12 or m11) fully obliterated the P-TEFb binding capacities of 7SKm12 and 7SKm11 RNAs. Next, we attempted to reestablish the P-TEFb binding abilities of 7SKm12 and 7SKm11 by restoring the Hd and Hp helices through replacement of their descending strands with the m12c or m11c complementary sequences. Indeed, the 7SKm12/m12c RNA regained its P-TEFb binding ability, leading to the conclusion that the Hd stem, although its nucleotide composition is not critical, plays an essential role in P-TEFb binding. In contrast, the 7SKm11/m11c RNA remained inactive in P-TEFb binding, suggesting that the nucleotide composition of the Hp stem is crucial for P-TEFb recruitment. The same results were obtained when the terminal-loop nucleotides or the C320 and U321 bulged residues were replaced (m14 and m13); neither 7SKm13 nor 7SKm14 could interact with P-TEFb. Finally, as suggested by our phylogenetic comparison (Fig. 4), alteration of the evolutionarily variable 296-AAAUGA-301 sequences at the base of the 3′-hairpin did not inhibit the P-TEFb binding capacity of 7SKm10. These results demonstrate that in the 3′-terminal hairpin of 7SK, the G302-to-C324 region, including the Hd and Hp helices, the terminal loop, and the bulged 320-CU-321 residues, constitutes the minimal structure required for establishment of an in vivo interaction with P-TEFb.
FIG. 6.
Elements directing P-TEFb binding to the 3′ hairpin of 7SK snRNA. (A) Structures of modified 7SK RNAs assayed for P-TEFb binding ability. Deleted (d17, d18, and d19) and altered sequences are shown. (B) Association of internally truncated 7SK RNAs with HA-P-TEFb. The endogenous (7SK) and transiently expressed 7SK RNAs were detected by Northern analysis. (C) Association of 7SK RNAs carrying altered 3′-hairpin sequences with HA-P-TEFb. 7SK RNAs were detected by RNase protection analysis performed with probes complementary to the mutant RNAs.
In vitro assembly of 7SK/HEXIM1/P-TEFb.
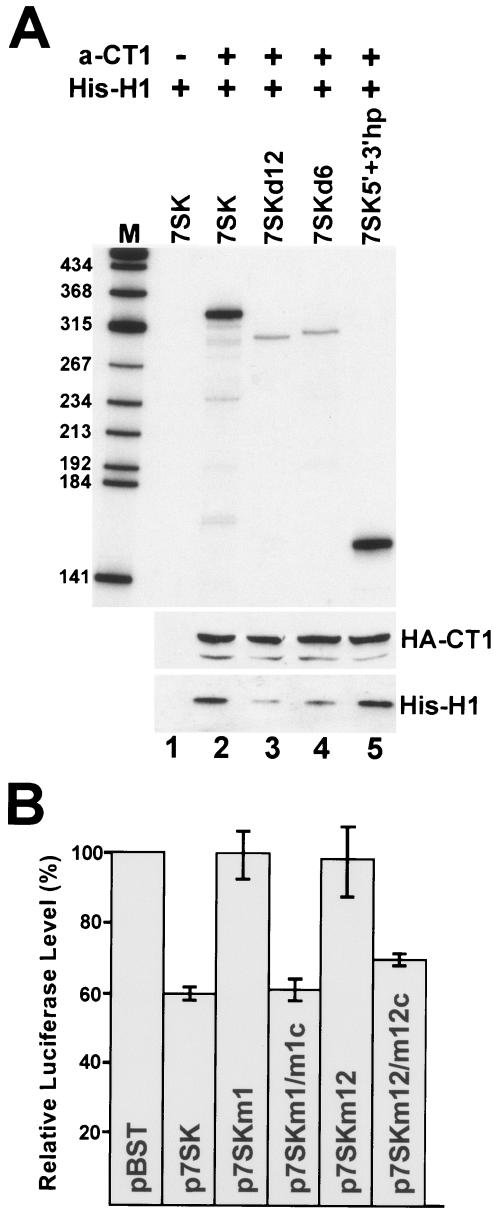
We next tested whether 7SK elements directing in vivo recruitment of HEXIM1 and P-TEFb are also important for assembly of the 7SK/HEXIM1/P-TEFb complex in the test tube. Previous reconstitution experiments showed that phosphorylation of Cdk9 is crucial for 7SK/P-TEFb interaction (6). Therefore, P-TEFb was immunoprecipitated by an anti-CycT1 antibody from G3H cell extract that had been treated with micrococcal nuclease to eliminate endogenous 7SK and to destroy 7SK/HEXIM1/P-TEFb complexes. Beads retaining P-TEFb were incubated with purified His-tagged recombinant HEXIM1 protein (17) in the presence of in vitro-synthesized internally labeled wild-type or mutant 7SK RNAs (Fig. 7A). After extensive washing, binding of 7SK RNAs was tested by polyacrylamide gel electrophoresis, and retention of HA-CycT1 and His-HEXIM1 was controlled by Western blot analysis. While the wild-type 7SK and the 7SK5′+3′hp RNAs were able to bind to P-TEFb in the presence of His-HEXIM1 (lanes 2 and 5), the 7SKd12 and 7SKd6 RNAs lacking putative HEXIM1 and P-TEFb binding elements, respectively, failed to stably associate with immobilized P-TEFb. This demonstrates that the 5′- and 3′-terminal hairpin elements of 7SK snRNA are also crucial for in vitro assembly of 7SK/HEXIM1/P-TEFb.
FIG. 7.
Elements directing in vivo binding of HEXIM1 and P-TEFb are crucial for in vitro assembly of 7SK/HEXIM1/P-TEFb and in vivo inhibition of Pol II transcription. (A) In vitro reconstitution of 7SK/HEXIM1/P-TEFb particle. From a G3H cell extract, P-TEFb and HA-P-TEFb were immobilized on protein A-agarose beads either coated (+) with an anti-CycT1 (a-CT1) antibody or uncoated (−). The beads were incubated with purified His-tagged HEXIM1 protein (His-H1) and in vitro-transcribed labeled 7SK RNAs as indicated. Retention of HA-CycT1 (HA-CT1), His-HEXIM1 (His-H1), and 7SK RNAs was tested by Western blot analysis and polyacrylamide gel electrophoresis, respectively. (D) In vivo inhibition of luciferase gene expression with coexpression of mutant 7SK RNAs. For details, see the legend to Fig. 3.
Finally, to confirm the functional importance of the newly determined HEXIM1 and P-TEFb binding sites of 7SK snRNA, we assayed the transcriptional-regulatory capacities of some representative mutant 7SK RNAs (Fig. 7B). As was predictable, 7SKm1 and 7SKm12 RNAs lacking HEXIM1 and/or P-TEFb binding abilities were unable to inhibit SV40 promoter-driven luciferase expression in transiently transfected HeLa cells. However, restoration of the HEXIM1 and P-TEFb binding capacities of these RNAs reestablished the transcriptional-inhibitory functions of 7SKm1/m1c and 7SKm12/m12c RNAs.
DISCUSSION
Controlling the nuclear level of free P-TEFb through sequestering into kinase-inactive 7SK/HEXIM1/P-TEFb complexes is an important mechanism to regulate Pol II transcription, cell growth, and proliferation (5, 20, 28, 34, 36). In this process, the 7SK snRNA has been proposed to play a central role by providing the structural platform for the coordinated assembly of HEXIM1 and P-TEFb. To understand the molecular mechanism underlying the transcriptional-regulatory function of 7SK, we determined and characterized the elements of human 7SK snRNA directing recruitment of P-TEFb and HEXIM1 in living cells. We showed that two distinct regions of 7SK, namely, the G24-C48/G60-C87 distal segment of the 5′-terminal hairpin and the G306-C324 apical domain of the 3′-terminal hairpin, contain all the elements that are fundamental for in vivo assembly of 7SK/HEXIM1/P-TEFb and inhibition of Pol II transcription. While the binding element in the 5′ hairpin of 7SK is essential for recruitment of both P-TEFb and HEXIM1, the 3′ hairpin functions only in P-TEFb binding. Sequences outside these two regions, although they can slightly facilitate P-TEFb and HEXIM1 binding, carry no fundamental elements for HEXIM1 and P-TEFb binding. In agreement with this conclusion, a transiently expressed minimal 7SK RNA composed of the 5′ and 3′ hairpins of human 7SK can both bind and inactivate P-TEFb in HeLa cells. The functions of the internal sequences located between the terminal hairpins of 7SK remain unknown, but it is important to note that the strong sequence conservation of some internal regions (e.g., G115-C150 and G190-A219) points to the possible functional importance of this part of 7SK.
Previous work established HEXIM1 as a crucial adapter protein that either bridges P-TEFb to 7SK (16, 17) or facilitates formation of a direct interaction between P-TEFb and 7SK (37, 38). In this respect, HEXIM1 is highly reminiscent of the HIV transactivator protein Tat, which recruits P-TEFb to the apical hairpin of the transactivation-responsive (TAR) RNA formed by the 5′-terminal region of the newly synthesized viral transcript (1, 32). Both Tat and HEXIM1 can specifically interact with CycT1, and they possess similar arginine-rich RNA binding motifs, which are responsible for binding of the viral TAR RNA or the 7SK snRNA, respectively (13, 16, 17, 38). These facts led to the intriguing concept that the TAR/Tat/P-TEFb and 7SK/HEXIM1/P-TEFb ternary complexes share similar structural organizations (16, 38). However, given that the apical-hairpin element of the TAR RNA alone can bind both Tat and P-TEFb (13), our finding that recruitment of HEXIM1 and P-TEFb to 7SK requires two structurally and functionally distinct snRNA elements indicates that assembly of the TAR/Tat/P-TEFb and 7SK/HEXIM1/P-TEFb complexes follows different architectural plans.
HEXIM1 is a genuine RNA binding protein that has been shown to specifically associate with 7SK snRNA through an arginine-rich RNA binding motif located close to the center of the protein (16, 38). Here, we demonstrated that HEXIM1 binds to the G24-C48/G60-C87 segment of the 5′ hairpin of 7SK (Fig. 4B). Introduction of compensatory sequence confirmed that the H1 and H4 helices that represent the distal and proximal ends of the HEXIM1 binding domain are formed in 7SK snRNA and also showed that they function in a sequence-independent manner. The central part of the HEXIM1 binding motif of 7SK shows great sensitivity to nucleotide alterations. Unfortunately, due to the high degree of sequence conservation, evolutionary comparison of vertebrate 7SK snRNAs failed to detect sequence covariations, and thus, to provide support for the structural organization of the inner part of the HEXIM1 binding motif of 7SK snRNA. In the future, further studies will be required for an understanding of the detailed structural determinants of the specific interaction of 7SK and HEXIM1.
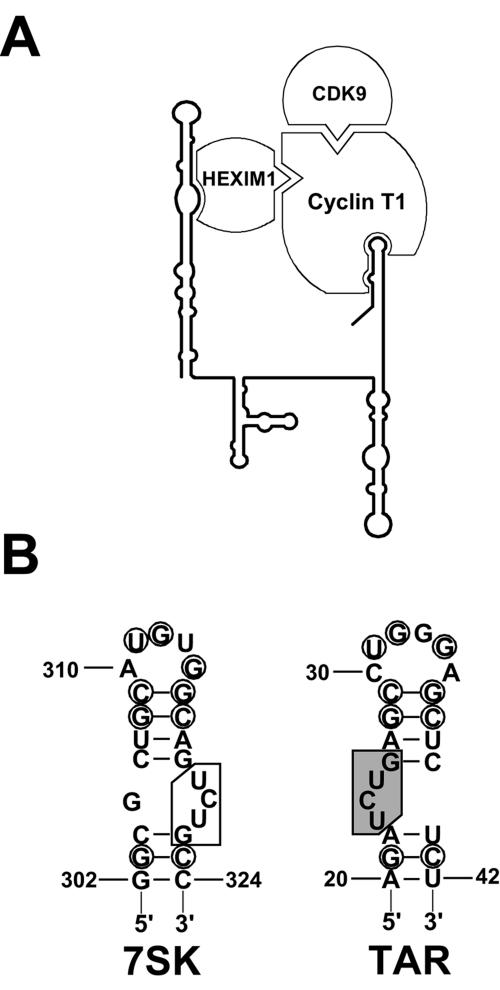
The finding that 3′-end-truncated 7SK RNAs lacking P-TEFb binding capacity can still bind HEXIM1 clearly demonstrated that in living cells, association of HEXIM1 with 7SK does not depend on P-TEFb binding and that HEXIM1 alone possesses the capacity to specifically and efficiently interact with 7SK snRNA. A systematic mutational analysis of the HEXIM1 binding domain of 7SK showed that RNA alterations abolishing or compromising the HEXIM1 binding affinity of 7SK also obliterate or inhibit the recruitment of P-TEFb. In contrast, our analyses failed to identify RNA elements in the 5′ hairpin of 7SK which would specifically interfere with P-TEFb binding without affecting recruitment of HEXIM1. The most obvious interpretation of these results is that binding of P-TEFb to the 5′ hairpin of 7SK is mediated by HEXIM1. This conclusion is in good agreement with the results of recent in vitro reconstitution studies suggesting that binding of HEXIM1 to 7SK snRNA is a prerequisite for P-TEFb recruitment (16). Along these lines, two hybrid assays and in vitro pull-down experiments showed that the C-terminal domain of HEXIM1 and the N-terminal cyclin homology domain of CycT1 can form a specific interaction that most probably renders P-TEFb kinase inactive (16, 17, 38). Interestingly, under in vitro conditions, full-length HEXIM1 seems to possess a structural hindrance to association with CycT1. Removal of the putative “regulatory domain” (amino acids 150 to 181) encompassing the 7SK binding motif of HEXIM1 or inclusion of in vitro-synthesized 7SK snRNA can restore the interaction of HEXIM1 and CycT1. This may indicate that binding of 7SK snRNA provokes remodeling of the C-terminal domain of HEXIM1, thereby making it competent for CycT1 binding (16). Taken together, the available data are most consistent with a model in which tethering of P-TEFb to the 5′-terminal hairpin of 7SK is mediated by a protein-protein interaction formed between the 7SK-associated HEXIM1 and CycT1 (Fig. 8A), although at the moment, we cannot rule out the formal possibility that P-TEFb might also directly interact with the 5′ hairpin of 7SK.
FIG. 8.
7SK-dependent regulation of the CTD kinase activity of P-TEFb. (A) Structural organization of the kinase-inactive 7SK/HEXIM1/P-TEFb complex. The schematic structure of human 7SK snRNA, together with associated proteins, is shown. Our experiments do not exclude the possibility that HEXIM1 binds to 7SK snRNA as a homodimer (39). (B) Structural comparison of the HIV TAR RNA and the P-TEFb binding motif of the 3′ hairpin of human 7SK snRNA. The conserved nucleotides are circled. Nucleotides essential for Tat binding are shaded (13). The “inverted Tat binding-like” motif of 7SK snRNA is boxed.
We also demonstrated that in vivo recruitment and inactivation of P-TEFb necessitates another intermolecular interaction formed between P-TEFb and the G306-C324 3′-terminal hairpin region of 7SK snRNA. Former in vitro reconstitution experiments showed that under certain conditions, immobilized FLAG-tagged P-TEFb obtained from HeLa cells can directly and specifically interact with 7SK snRNA even in the absence of HEXIM1 (6, 37). In this study, the finding that mutant 7SK RNAs defective in HEXIM1 binding are unable to recruit P-TEFb demonstrated that binding of P-TEFb to the 3′-terminal hairpin of 7SK is a HEXIM1-dependent process in living cells. An interaction between HEXIM1 and CycT1 may position the RNA binding motif of CycT1 close to the 3′-terminal hairpin of 7SK. In fact, the internal 7SK truncations d1, d2, and d14, although they did not affect HEXIM1 binding, largely compromised P-TEFb recruitment, suggesting that the 3′-terminal hairpins of these RNAs occupy unfavorable positions for P-TEFb interaction (Fig. 1 and 2). Finally, it is also conceivable that an interaction with HEXIM1 can enhance the binding affinity of P-TEFb to the 3′-terminal hairpin of 7SK.
The P-TEFb binding motif in the 3′ hairpin of 7SK is composed of two short helices separated by two bulged residues and topped by a 5-nucleotide terminal loop. While the nucleotide sequences of the terminal loop, the proximal helix, and the internal bulge are critical, the base composition of the distal helix, given that base pairings are maintained, is less important for P-TEFb binding. We noticed that the P-TEFb binding motif of 7SK shares intriguing structural similarities with the extensively characterized Tat/P-TEFb binding motif of the HIV TAR RNA (Fig. 8B). The TAR RNA is composed of a 6-nucleotide loop and a 3-nucleotide bulge that separates two short stems. Binding of P-TEFb to TAR RNA requires the virally encoded Tat protein, and it is a highly cooperative process (13, 40). Interaction of CycT1 and Tat enhances the TAR binding affinity of Tat, and in return, TAR RNA further enhances the association of Tat and CycT1. While the 23-UCU-25 bulge directs the specific binding of Tat, the terminal-loop region of the TAR RNA has been implicated in CycT1 binding (13, 25, 26). Consistent with this, RNA cross-linking and footprinting experiments indicated that residues between 252 and 261 of CycT1 directly contact nucleotides in the terminal loop of TAR (26). Interestingly, an overlapping region of CycT1 located between residues 255 and 333 was found to contribute to the in vitro binding of P-TEFb to 7SK snRNA (6). Moreover, some TAR RNA loop nucleotides, such as G32 and G34, essential for in vitro formation of a stable TAR/Tat/CycT1 complex, are also conserved in the central loop of the 3′-terminal hairpins of 7SK snRNAs (25). Thus, these results together suggest that binding of P-TEFb to the TAR RNA and the 3′ hairpin of 7SK may share some common structural determinants. Although the available data favor the idea that P-TEFb, with the assistance of HEXIM1, can directly bind to 7SK, at present, we cannot unambiguously rule out the formal possibility that another cellular protein, functionally similar to Tat, might facilitate the binding of P-TEFb to the 3′ hairpin of 7SK in living cells. In this context, we have to note that the 3′ side of the P-TEFb binding motif of 7SK snRNA may carry a “Tat binding-like” element (319-UCU-321) (Fig. 8B) that, like the authentic Tat binding motif of TAR, is located 4 basepairs below the central loop of the 3′ hairpin.
Finally, the observation that overexpression of 7SK snRNA can inhibit Pol II transcription from the P-TEFb-dependent SV40 promoter indicates that 7SK, contrary to previous belief, is limiting for the formation of the 7SK/HEXIM1/P-TEFb complex. This is an unexpected finding, since 7SK is present in the nucleus in great excess compared to HEXIM1 and P-TEFb and only about 30% of 7SK is associated with HEXIM1 and P-TEFb (12). An interpretation of these results is that the major portion of 7SK is sequestered into another RNP complex, which prevents its interaction with HEXIM1 and/or P-TEFb. Regulation of the nuclear concentration of 7SK snRNA competent in HEXIM1 and P-TEFb binding might provide a mechanism to control cellular P-TEFb activity. Consistent with this hypothesis, in HeLa nuclei, about 50% of P-TEFb remains in free form, contrary to the great excess of 7SK snRNA (20, 36). Therefore, 7SK snRNA, besides providing a scaffold for the dynamic assembly of kinase-inactive 7SK/HEXIM1/P-TEFb complex, may play a more complex role in the regulation of Pol II transcription.
Acknowledgments
We are grateful to Q. Zhou (University of California, Berkeley) and O. Bensaude (Ecole Normale Supérieure, Paris, France) for providing us with G3H cells and the pPET21-MAQ1 (HEXIM1) expression construct, respectively.
S.E. and E.V.H. were funded by the Ministère de l'Education Nationale, de la Recherche et de la Technologie; la Ligue Nationale Contre le Cancer; and L'Association pour la Recherche sur le Cancer. Our work was supported by the Centre Nationale de la Recherche Scientifique and la Ligue Nationale Contre le Cancer.
REFERENCES
- 1.Berkhout, B., R. H. Silverman, and K. T. Jeang. 1989. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell 59:273-282. [DOI] [PubMed] [Google Scholar]
- 2.Brasier, A. R., and J. J. Fortin. 1994. Nonisotopic assays for reporter gene activity, p. 9.7.12-9.7.21. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 1. John Wiley & Sons, Inc., New York, N.Y. [Google Scholar]
- 3.Byers, S. A., J. P. Price, J. J. Cooper, Q. Li, and D. H. Price. 2005. HEXIM2, a HEXIM1 related protein, regulates P-TEFb through association with 7SK. J. Biol. Chem. 280:16360-16367. [DOI] [PubMed] [Google Scholar]
- 4.Callige, M., I. Kieffer, and H. Richard-Foy. 2005. CSN5/Jab1 is involved in ligand-dependent degradation of ERa by the proteasome. Mol. Cell. Biol. 11:4349-4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chao, S.-H., and D. H. Price. 2001. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J. Biol. Chem. 276:31793-31799. [DOI] [PubMed] [Google Scholar]
- 6.Chen, R., Z. Yang, and Q. Zhou. 2004. Phosphorylated positive transcription elongation factor b (P-TEFb) is tagged for inhibition through association with 7SK snRNA. J. Biol. Chem. 279:4153-4160. [DOI] [PubMed] [Google Scholar]
- 7.Cho, E. J., M. S. Kobor, M. Kim, J. Greenblatt, and S. Buratowski. 2001. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev. 15:3319-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ganot, P., B. E. Jady, M. L. Bortolin, X. Darzacq, and T. Kiss. 1999. Nucleolar factors direct the 2′-O-ribose methylation and pseudouridylation of U6 spliceosomal RNA. Mol. Cell. Biol. 19:6906-6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garriga, J., and X. Grana. 2004. Cellular control of gene expression by T-type cyclin/CDK9 complexes. Gene 337:15-23. [DOI] [PubMed] [Google Scholar]
- 10.Goodall, G. J., K. Wiebauer, and W. Filipowicz. 1990. Analysis of pre-mRNA processing in transfected plant protoplasts. Methods Enzymol. 181:148-161. [DOI] [PubMed] [Google Scholar]
- 11.Gursoy, H. C., D. Koper, and B. J. Benecke. 2000. The vertebrate 7S K RNA separates hagfish (Myxine glutinosa) and lamprey (Lampetra fluviatilis). J. Mol. Evol 50:456-464. [DOI] [PubMed] [Google Scholar]
- 12.Haaland, R. E., C. H. Herrmann, and A. P. Rice. 2003. Increased association of 7SK snRNA with Tat cofactor P-TEFb following activation of peripheral blood lymphocytes. AIDS 17:2429-2436. [DOI] [PubMed] [Google Scholar]
- 13.Karn, J. 1999. Tackling Tat. J. Mol. Biol. 293:235-254. [DOI] [PubMed] [Google Scholar]
- 14.Kobor, M. S., and J. Greenblatt. 2002. Regulation of transcription elongation by phosphorylation. Biochim. Biophys. Acta 1577:261-275. [DOI] [PubMed] [Google Scholar]
- 15.Kruger, W., and B. J. Benecke. 1987. Structural and functional analysis of a human 7 S K RNA gene. J. Mol. Biol. 195:31-41. [DOI] [PubMed] [Google Scholar]
- 16.Michels, A. A., A. Fraldi, Q. Li, T. E. Adamson, F. Bonnet, V. T. Nguyen, S. C. Sedore, J. P. Price, D. H. Price, L. Lania, and O. Bensaude. 2004. Binding of the 7SK snRNA turns the HEXIM1 protein into a P-TEFb (CDK9/cyclin T) inhibitor. EMBO J. 23:2608-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michels, A. A., V. T. Nguyen, A. Fraldi, V. Labas, M. Edwards, F. Bonnet, L. Lania, and O. Bensaude. 2003. MAQ1 and 7SK RNA interact with CDK9/cyclin T complexes in a transcription-dependent manner. Mol. Cell. Biol. 23:4859-4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murphy, S., F. Altruda, E. Ullu, M. Tripodi, L. Silengo, and M. Melli. 1984. DNA sequences complementary to human 7 SK RNA show structural similarities to the short mobile elements of the mammalian genome. J. Mol. Biol. 177:575-590. [DOI] [PubMed] [Google Scholar]
- 19.Murphy, S., C. Di Liegro, and M. Melli. 1987. The in vitro transcription of the 7SK RNA gene by RNA polymerase III is dependent only on the presence of an upstream promoter. Cell 51:81-87. [DOI] [PubMed] [Google Scholar]
- 20.Nguyen, V. T., T. Kiss, A. A. Michels, and O. Bensaude. 2001. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature 414:322-325. [DOI] [PubMed] [Google Scholar]
- 21.O'Keeffe, B., Y. Fong, D. Chen, S. Zhou, and Q. Zhou. 2000. Requirement for a kinase-specific chaperone pathway in the production of a Cdk9/cyclin T1 heterodimer responsible for P-TEFb-mediated tat stimulation of HIV-1 transcription. J. Biol. Chem. 275:279-287. [DOI] [PubMed] [Google Scholar]
- 22.Peng, J., N. F. Marshall, and D. H. Price. 1998. Identification of a cyclin subunit required for the function of Drosophila P-TEFb. J. Biol. Chem. 273:13855-13860. [DOI] [PubMed] [Google Scholar]
- 23.Price, D. H. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 20:2629-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reddy, R., D. Henning, C. S. Subrahmanyam, and H. Busch. 1984. Primary and secondary structure of 7-3 (K) RNA of Novikoff hepatoma. J. Biol. Chem. 259:12265-12270. [PubMed] [Google Scholar]
- 25.Richter, S., H. Cao, and T. M. Rana. 2002. Specific HIV-1 TAR RNA loop sequence and functional groups are required for human cyclin T1-Tat-TAR ternary complex formation. Biochemistry 41:6391-6397. [DOI] [PubMed] [Google Scholar]
- 26.Richter, S., Y. H. Ping, and T. M. Rana. 2002. TAR RNA loop: a scaffold for the assembly of a regulatory switch in HIV replication. Proc. Natl. Acad. Sci. USA 99:7928-7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Sano, M., M. Abdellatif, H. Oh, M. Xie, L. Bagella, A. Giordano, L. H. Michael, F. J. DeMayo, and M. D. Schneider. 2002. Activation and function of cyclin T-Cdk9 (positive transcription elongation factor-b) in cardiac muscle-cell hypertrophy. Nat. Med. 8:1310-1317. [DOI] [PubMed] [Google Scholar]
- 29.Shim, E. Y., A. K. Walker, Y. Shi, and T. K. Blackwell. 2002. CDK-9/cyclin T (P-TEFb) is required in two postinitiation pathways for transcription in the C. elegans embryo. Genes Dev. 16:2135-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wada, T., T. Takagi, Y. Yamaguchi, D. Watanabe, and H. Handa. 1998. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 17:7395-7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wassarman, D. A., and J. A. Steitz. 1991. Structural analyses of the 7SK ribonucleoprotein (RNP), the most abundant human small RNP of unknown function. Mol. Cell. Biol. 11:3432-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei, P., M. E. Garber, S. M. Fang, W. H. Fischer, and K. A. Jones. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92:451-562. [DOI] [PubMed] [Google Scholar]
- 33.Wei, W. Z., R. F. Gill, R. F. Jones, D. Lichlyter, and J. P. Abastado. 1996. Induction of cytotoxic T lymphocytes to murine mammary tumor cells with a Kd-restricted immunogenic peptide. Int. J. Cancer 66:659-663. [DOI] [PubMed] [Google Scholar]
- 34.Wittmann, B. M., N. Wang, and M. M. Montano. 2003. Identification of a novel inhibitor of breast cell growth that is down-regulated by estrogens and decreased in breast tumors. Cancer Res. 63:5151-5158. [PubMed] [Google Scholar]
- 35.Yamaguchi, Y., T. Takagi, T. Wada, K. Yano, A. Furuya, S. Sugimoto, J. Hasegawa, and H. Handa. 1999. NELF, a multisubunit complex containing RD, cooperates with DSIF to repress RNA polymerase II elongation. Cell 97:41-51. [DOI] [PubMed] [Google Scholar]
- 36.Yang, Z., Q. Zhu, K. Luo, and Q. Zhou. 2001. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 414:317-322. [DOI] [PubMed] [Google Scholar]
- 37.Yik, J. H., R. Chen, R. Nishimura, J. L. Jennings, A. J. Link, and Q. Zhou. 2003. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol. Cell 12:971-982. [DOI] [PubMed] [Google Scholar]
- 38.Yik, J. H., R. Chen, A. C. Pezda, C. S. Samford, and Q. Zhou. 2004. A human immunodeficiency virus type 1 Tat-like arginine-rich RNA-binding domain is essential for HEXIM1 to inhibit RNA polymerase II transcription through 7SK snRNA-mediated inactivation of P-TEFb. Mol. Cell. Biol. 24:5094-5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yik, J. H., R. Chen, A. C. Pezda, and Q. Zhou. 2005. Compensatory contributions of HEXIM1 and HEXIM2 in maintaining the balance of active and inactive P-TEFb complexes for control of transcription. J. Biol. Chem. 280:16368-16376. [DOI] [PubMed] [Google Scholar]
- 40.Zhang, J., N. Tamilarasu, S. Hwang, M. E. Garber, I. Huq, K. A. Jones, and T. M. Rana. 2000. HIV-1 TAR RNA enhances the interaction between Tat and cyclin T1. J. Biol. Chem. 275:34314-34319. [DOI] [PubMed] [Google Scholar]
- 41.Zhu, Y., T. Peery, J. Peng, Y. Ramanathan, N. F. Marshall, T. Marshall, B. Amendt, B. Mathews, and D. H. Price. 1997. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 11:2622-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]