Abstract
We find that immediately following transcript initiation, RNA polymerase II pauses at several locations even in the presence of relatively high (200 μM) levels of nucleoside triphosphates. Strong pauses with half-lives of >30 s were observed at +7, +18/19, and about +25 on the template used in these experiments. We show that the strong pause at +7, after the synthesis of 5′-ACUCUCU, leads to repeated cycles of upstream slippage of the RNA-DNA hybrid followed by re-pairing with the DNA and continued RNA synthesis. The resulting transcripts are 2, 4, and 6 bases longer than predicted by the template sequence. Slippage is efficient when transcription is primed with the +1/+2 (ApC) dinucleotide, and it occurs at even higher levels with the +2/+3 primer (CpU). Slippage can occur at high levels with ATP initiation, but priming with CpA (−1/+1) supports very little slippage. This latter result is not simply an effect of transcript length at the point of pausing. Slippage can also occur with a second template on which the polymerase can be paused after synthesizing ACUCU. Slippage is not reduced by an ATP analog that blocks promoter escape, but it is inhibited by substitution of 5Br-U for U in the RNA. Our results reveal an unexpected flexibility of RNA polymerase II ternary complexes during the very early stage of transcription, and they suggest that initiation at different locations within the same promoter gives rise to transcription complexes with different properties.
An RNA polymerase II transcription cycle consists of several distinct steps: recruitment to assemble the preinitiation complex, open-complex formation, initiation, promoter escape and clearance, transcript elongation, and termination. It is now clear that regulation may be imposed at steps after initiation (reviewed in references 20, 26, 31 and 36). In particular, many transcription units contain newly initiated RNA polymerases that are stalled from 20 to 40 nucleotides downstream of transcription start (reviewed in reference 24). The molecular basis for the regulated pausing and release of RNA polymerase II from promoter-proximal locations has not been established, in part because the transition from the well-characterized preinitiation complex to the stable transcript elongation complex is poorly understood.
Rate-limiting steps early in transcription should be particularly interesting, since they correspond to likely regulatory targets and could identify important structural transitions in the transcription complex. This idea is strongly supported by the recent results of Kugel and Goodrich (18), who placed the rate-limiting step for in vitro transcription from a nonregulated promoter somewhere between the formation of the 4th and 14th bonds. In light of this report, we began the current work by preparing gently washed RNA polymerase II preinitiation complexes, which were then incubated with nonlimiting (i.e., 200 μM) levels of labeled nucleoside triphosphates (NTPs). This experiment revealed that transcription immediately downstream of initiation does not proceed uniformly. Instead, we observed a series of very discrete pauses with half-lives at least 20 times longer than the average bond formation rate under the conditions we employed. The first of these pauses occurs at +7, after the synthesis of the transcript 5′-ACUCUCU, on the template we are studying. We were surprised to discover that the +7 pause allowed the RNA-DNA hybrid to shift upstream by 2 bp and then re-pair with the template, leading to the synthesis of a transcript 2 bases longer than that predicted by the template sequence. Multiple rounds of this upstream slippage and reextension event can take place.
The transcript slippage reaction we describe here is interesting for a number of reasons. It can occur 7 bases downstream of initiation, at which point the transcription complex has almost completed assembly of the 8 to 9-bp RNA-DNA hybrid (16) that characterizes the final transcript elongation complex. Particularly in light of the tight pairing of the 3′-most bases of the transcript with the template (8), the ability of a 7-mer complex to disrupt the RNA-DNA hybrid and displace it upstream is surprising. Although transcript slippage at our promoter is affected by NTP concentration, it is most sensitive to the exact location at which transcription begins. Slippage is easily observed with the normal initiation substrate (ATP) or with a +1/+2 primer. However, priming with the −1/+1 dinucleotide does not support significant transcript slippage, while priming with the +2/+3 dinucleotide results in over half of the initiations going through at least one round of slippage even at near-physiological (800 μM) NTP levels. Our results with transcript slippage reveal an unexpected flexibility in the early RNA polymerase II transcription complex, and they also suggest that not all potentially available initiation sites give rise to transcription complexes with identical properties.
MATERIALS AND METHODS
Reagents.
Ultrapure NTPs were obtained from Pharmacia Biotechnology, ApC and CpU dinucleotides were obtained from Sigma, 32P-labeled NTPs (800 Ci/mmole) were obtained from NEN, Deep Vent DNA polymerase and restriction enzymes were obtained from New England Biolabs, RNase T1 was obtained from Boehringer Mannheim, and streptavidin-coated magnetic beads were obtained from Promega Biotech.
Plasmids.
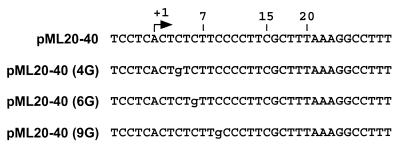
All plasmids used in this study are based on the adenovirus major late (AdML) promoter. Plasmid pML20-40 is identical to the previously- described pML20-42 construct (34) with the exception of a single nucleotide change of T to G at position +16 on the nontemplate strand (Fig. 1). Plasmids pML20-40(4G), pML20-40(6G), and pML20-40(9G) were constructed by replacing the BssHII-StuI fragment of pML20-40 with a synthetic DNA fragment carrying a G instead of a C at position +4, +6, or +9 on the nontemplate strand (Fig. 1). All constructs were verified by DNA sequencing.
FIG. 1.
Sequences of the nontemplate strands of the initially transcribed regions of the plasmids used in these studies. The single G residue which differs from the pML20-40 sequence in the 4G, 6G, and 9G templates is shown in each instance in lowercase type.
Template preparation for in vitro transcription reactions.
Template DNAs were made by PCR using the forward primer 5′-GGCATCAAGGAAGGTGATTG-3′ (biotinylated at the 5′ end) and the reverse primer 5′-CAGTGCCAAGCTTGCATG-3 (a HindIII site is underlined). The 190-bp product thus obtained has the transcription start site 96 bp downstream of the biotinylated end. After purification using the Concert cleanup kit (Bethesda Research Laboratories) and digestion with HindIII to reduce transcription arrest before the end of the template (12), the DNA was extracted with phenol-chloroform and precipitated with ethanol.
In vitro transcription reaction on attached templates.
Preinitiation complexes were assembled using HeLa cell nuclear extract and biotinylated template DNA immobilized on streptavidin-coated magnetic beads as described previously (29, 34). Initial complex assembly was carried out at either 25 or 30°C for 25 min. Bead-attached complexes were washed twice in 150 μl of BC100 (20 mM Tris [pH 7.9], 8 mM MgCl2, 100 mM KCl, 1 mM dithiothreitol, 20% glycerol, 0.2 mM EDTA), resuspended in BC100, and stored on ice until used. Transcription reaction mixtures contained 1 mM dinucleotide primer (ApC, CpA, or CpU), dATP (usually 20 μM) to satisfy the energy requirement for transcript initiation, and other nucleotides at the concentrations indicated in the figure legends. The labeling nucleotides had specific activities of 800 Ci/mmol unless otherwise indicated. Reaction times are given in the legends. When ATP was used to initiate transcription instead of a dinucleotide, it was present at 100 μM and dATP was absent. Only 5 μM dATP was used for the reactions in Fig. 8B and whenever transcription was primed with CpU. Reactions were stopped by adding an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1). The aqueous phase from the initial extraction was reextracted with chloroform-isoamyl alcohol (24:1). Reaction mixtures involving synthesis of 15-nucleotide or longer RNAs were usually ethanol precipitated before analysis. For shorter RNAs, the aqueous phase was lyophilized, resuspended in water, and treated with calf intestinal phosphatase (0.1 U/μl for 10 min at 30°C). RNase T1 digestions were performed in 10 μl at 37°C for 10 min with 1 or 2 U of T1 and 0.25 μg of yeast tRNA as carrier. RNAs were resuspended either in 50% formamide or 8 M urea before resolution on 25% acrylamide-3% bisacrylamide gels containing 7 M urea. Gels were imaged, and individual bands were quantified using a PhosphorImager and ImageQuant software (Molecular Dynamics). RNA length markers were made by partial resection of the transcript of a particular halted complex in the presence of 2 mM sodium pyrophosphate at 37°C for 2, 4, or 8 min.
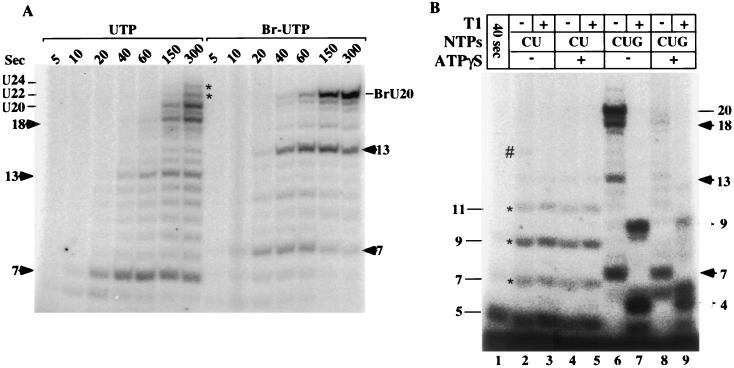
FIG. 8.
Effects of 5-Br UTP and ATPγS on transcript slippage. (A) RNA synthesis was initiated on the pML20-40 template at 25°C using 1 mM ApC, 20 μM dATP, 2 μM [α-32P]CTP, 20 μM GTP, and either 20 μM UTP or 20 μM 5Br-UTP (as marked) and continued for the indicated times. The lengths of the major paused RNAs are indicated by solid arrowheads, as well as the lengths of the A-stop RNA and slippage transcripts. Slippage products are indicated by asterisks. (B) Transcription was carried out on the 6G template with 1 mM ApC, 5 μM dATP, 2 μM [α-32P]UTP, and 20 μM CTP for 40 s at 30°C (lane 1). Aliquots of this reaction mixture were then made 20 μM in nonlabeled UTP, with (lanes 4 and 5) or without (lanes 2 and 3) 100 μM ATPγS; incubation was continued at 30°C for an additional 2 min. One-half of each of these reaction mixtures was treated with RNase T1 as indicated. To other aliquots of the 40-s reaction mixture (lane 1), 20 μM GTP was added without (lanes 6 and 7) or with (lanes 8 and 9) 100 μM ATPγS. One-half of each of these reaction mixtures was also treated with T1. Slippage products are indicated by asterisks, and RNAs produced by readthrough of the G-stop are marked by #.
RESULTS
RNA polymerase II exhibits distinct kinetic pauses in the early stage of RNA synthesis.
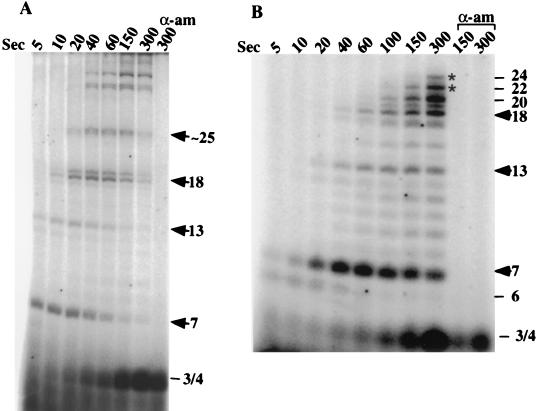
The template we chose for our study, called pML20-40 (Fig. 1), is based on the AdML promoter. In our earlier work we had used similar templates to purify and characterize transcription complexes stalled at various locations from 13 nt downstream of the transcription start (23, 33, 34). Our most recent studies revealed unexpectedly strong pausing when Sarkosyl-rinsed transcription complexes with RNAs as short as 15 nt were allowed to resume transcript elongation (29; see also reference 18). We were therefore interested in determining what other pauses might occur during the very first steps in transcription on the AdML promoter. We assembled preinitiation complexes with HeLa nuclear extracts on bead-attached pML20-40 templates, rinsed them gently with transcription buffer (without Sarkosyl) and then initiated RNA synthesis with 1 mM ApC primer and 200 μM each NTP. This concentration of NTPs supports transcript elongation at 1 to 2 nt/s at 30°C with promoter-distal RNA polymerase II transcription complexes (29). The transcripts were uniformly labeled by including both [α-32P]CTP and [α-32P]UTP in the reaction mixtures. Total RNA was isolated from 5 to 300-s time points and resolved on a high-percentage polyacrylamide gel. Since all of the NTPs were present at the same relatively high concentration, one might have expected to observe a zone of transcripts moving uniformly up the gel. However, as shown in Fig. 2A. most of the RNA appeared at a series of very discrete pause sites, most prominently at +7, +18/19, and roughly +25. A less pronounced pause occurred at +13. We repeated the experiment in Fig. 2A with limiting NTP concentrations (1 mM ApC, 2 μM [α-32P]UTP and 20 μM each CTP and GTP); in this case ATP was left out to stall transcription at +20 (Fig. 2B). With the NTP levels reduced by either 10- or 100-fold relative to the reaction in Fig. 2A, the residence time at the pause sites in Fig. 2B was considerably longer but the locations of the major pauses were unchanged. These results strongly suggest that RNA polymerase II is undergoing a series of relatively slow conformational transitions during the synthesis of the first 25 bases of the transcript. We will return to this point in Discussion.
FIG. 2.
RNA polymerase II pauses at distinct locations in the early phase of RNA synthesis on the pML20-40 template. (A) RNA synthesis performed at 30°C with 1 mM ApC and 200 μM each ATP, GTP, CTP, and UTP. The CTP and UTP were each 32P labeled (final specific activity, 40 Ci/mmol). The reactions were stopped at the indicated times by pipetting the mixtures directly into phenol-chloroform-isoamyl alcohol. (B) RNA synthesis performed at 25°C with 1 mM ApC, 20 μM each CTP, GTP, and dATP, and 2 μM [α-32P]UTP. Transcripts longer than predicted by the template sequence are marked by asterisks. For both panels, lanes marked α-am contained 2 μg of α-amanitin per ml. Solid arrows indicate sites of pausing.
Transcript slippage by RNA polymerase II.
A surprising outcome of these experiments can be seen in Fig. 2B, particularly in the 150- and 300-s lanes. In these reactions, no RNAs longer than 20 nt should have been made. However, a pair of bands reproducibly appeared above the 20-mer, whose length we verified with size markers (see Fig. 3A). The RNAs longer than 20 nt should not have resulted simply from readthrough of the A-stop at +20, since we saw no such readthrough on other, comparable templates at similar nucleotide concentrations (data not shown here, but see for example Fig. 4). When we ran RNAs from reactions like those in Fig. 2B along with length markers, it was apparent that the unexpected RNAs were 2, 4, and 6 nt longer than the 20-mer (see asterisk bands in Fig. 3A) whereas readthrough due to very low levels of contaminating ATP should have resulted in production of low levels of 21-nt and even lower levels of 22-nt RNAs (Fig. 1).
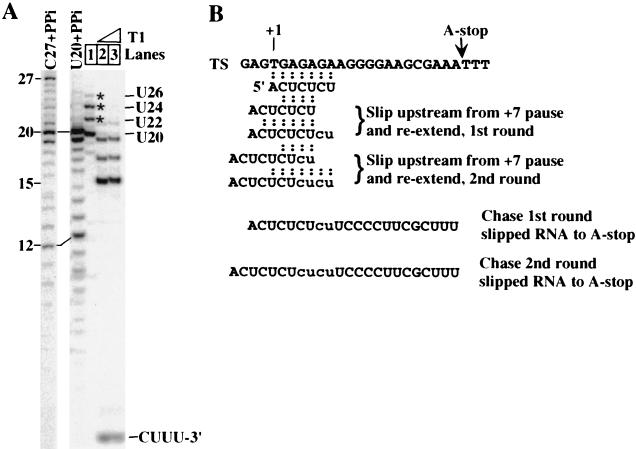
FIG. 3.
Transcript slippage on the pML20-40 template. (A) In lanes 1 to 3, RNA was synthesized with 1 mM ApC, 2 μM [α-32P]UTP, and 20 μM each CTP, GTP, and dATP at 30°C for 4 min; 20 μM nonlabeled UTP was then added, and the reaction was continued for 4 min. Transcripts were purified (lane 1) and then digested with 1 U (lane 2) or 2 U (lane 3) of RNase T1 per 10 μl of reaction mixture. Length standards were generated by pyrophosphorolysis (PPi) of transcripts halted at C27 or U20 on template pML20-40 (4G). The single 4-nt T1 cleavage product (5′-CUUU-3′) from the 3′ termini of the RNAs is indicated (lanes 2 and 3). The slippage-reextension products are marked by asterisks. (B) Schematic of the first and second round of transcript slippage and reextension on the pML20-40 template. TS, template strand; base pairing of the transcripts before and after slipping is indicated.
FIG. 4.
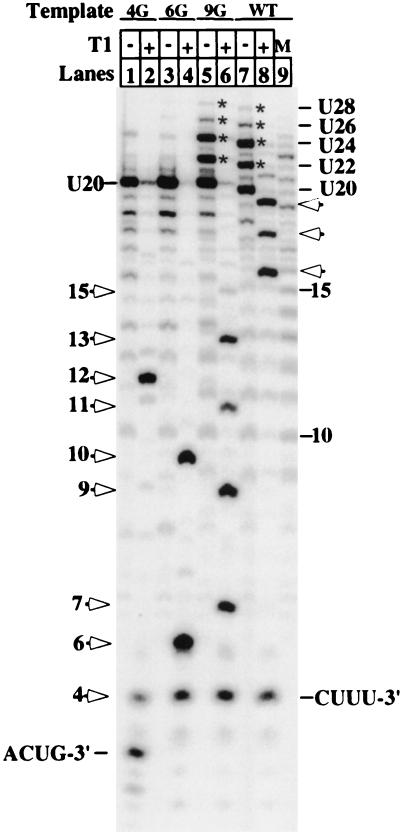
Under free-running transcription conditions on the pML20-40 template, transcript slippage occurs mostly from complexes paused at +7. RNA was synthesized on the indicated templates (WT is pML20-40) with ApC primers using the same protocol given in the legend of Fig. 3. One-half of each sample was digested with RNase T1. Slippage products are indicated by asterisks. Open arrowheads indicate T1 cleavage products; lengths are given to the right of the gel for the pML20-40 transcripts and to the left of the gel for the 4G, 6G, and 9G transcripts. Lane 9 (M) shows length standards generated by pyrophosphorolysis of A-stop RNA (U20 and longer slippage products) formed on the pML20-40 template. Note that transcripts of identical length from the pML20-40 and 4G, 6G, and 9G templates are expected to migrate slightly differently due to different sequence.
Note that transcripts from the pML20-40 template begin 5′-ACUCUCU (Fig. 1). This suggested to us that the unanticipated 22-nt RNA in Fig. 2B could result from upstream slippage of the initial RNA-DNA hybrid by 2 nt followed by re-pairing of the RNA with the template and resumption of RNA synthesis. The 24-nt RNA could result from two cycles of this process, and so forth. This hypothesis is diagrammed in Fig. 3B. We propose position +7 as the most likely location for slippage to begin, for two reasons. First, there is a strong natural pause at this location during transcription (Fig. 2), and second, this position marks the end of the three CU repeat elements and thus offers the maximum number of bases hybridized at the 3′ end (four) if slippage takes place.
If hybrid slipping and reextension occurred as diagrammed in Fig. 3B, cleavage of the transcripts in Fig. 3A with RNase T1 should lead to the production of CUUU from the 3′ ends of all of the RNAs, with bands of 16, 18, 20, and 22 nt generated from the 5′ ends. The results of T1 digestion (Fig. 3A) were consistent with this expectation. Also, if our slippage model is correct, changes in the template which interrupt the CU repeats should prevent stable extended hybrid formation after slippage and thus block the slippage-reextension reaction. We generated three variants of our initial template, which we designated pML20-40(4G), pML20-40(6G), and pML20-40(9G). As shown in Fig. 1, each template differed from the original by a single C-to-G change on the nontemplate strand. None of these changes alter the strength of the RNA-DNA hybrid compared to the original template. We confirmed (data not shown) that all of these templates support the identical pausing pattern during early RNA synthesis, including the prominent pause at +7, which we observed with the pML20-40 template. Note that when the polymerase pauses at +7 on either the 4G or 6G template, the RNA-DNA hybrid cannot slip 2 nt upstream while retaining 4 bp at the 3′ end. However, slippage from the 7-mer should be unaffected on the 9G template.
When we tested these templates under reaction conditions like those used in the experiment in Fig. 3, we obtained the expected results. RNAs longer than 20 nt were produced as efficiently on the 9G template as on pML20-40, but these RNAs were obtained at a much lower level on the 4G and 6G templates (Fig. 4). RNase T1 digestion of the RNA produced on the 9G template gave, in addition to the expected 4-, 7-, and 9-nt products, RNAs of 11 and 13 nt, consistent with two rounds of slippage and re-pairing as diagramed in Fig. 3B.
We observed a strong pause at +7 even under free-running transcription conditions with 200 μM NTPs (Fig. 2A). However, the detailed characterization of the slipping reaction was done with more limiting NTP concentrations, particularly for the 32P-labeled NTP. To explore the dependence of transcript slipping on transcription conditions, we transcribed the pML20-40 and 9G templates with ApC primers and different concentrations of CTP, UTP, and GTP, ranging from the labeling NTP at 1 μM to all NTPs at 200 μM. We resolved the RNAs on high-percentage acrylamide gels as in the experiment in Fig. 4 and determined the relative yields of 20-, 22-, 24-, and 26-nt RNA. We also performed ApC-primed transcriptions on pML20-40 with CTP and UTP at 815 μM. In this last case, the expected product was 15 nt long and the slippage products were 17- and 19-mers. The results are shown in Table 1. Not surprisingly, the amounts of slippage-reextension products were larger when transcription was performed at low NTP levels. However, slipped RNAs represented 16% of the total transcript with all NTPs at 200 μM, and this percentage was essentially the same (18%) when 815 μM NTPs were used. Thus, transcript slippage is not an artifact of low NTP concentrations. As expected, slippage-reextension transcripts were made with equal efficiency on the pML20-40 and 9G templates when reaction conditions were identical.
TABLE 1.
Relative yields of normal transcripts and slippage-reextension products as a function of NTP concentrationa
| Template | [NTP] (μM)b | Intensity of transcript (%)c
|
||||||
|---|---|---|---|---|---|---|---|---|
| 15 nt | 17 nt | 19 nt | 20 nt | 22 nt | 24 nt | 26 nt | ||
| pML20-40 | 20 C, 20 G, (1 + 20) U | 57.9 ± 5.3 | 20.6 ± 0.9 | 19.3 ± 3.1 | 3.0 ± 1.3 | |||
| 20 C, 20 G, U | 69.1 ± 6.6 | 18.3 ± 2.4 | 11.6 ± 4.6 | 1.4 ± 0.4 | ||||
| 200 C, 200 G, 200 U | 83.9 ± 1.7 | 10.4 ± 1.6 | 5.3 ± 1.1 | |||||
| 9G | 20 C, 20 G, (1 + 20) U | 63.6 ± 2.1 | 20.4 ± 1.2 | 15.6 ± 0.1 | 1.6 ± 0.8 | |||
| pML20-40 | 815 C, 815 U | 82 | 8 | 10 | ||||
Transcription was performed on the indicated templates with 1 mM ApC and the indicated concentrations of CTP, UTP, and GTP at 30°C for 20 min (top four rows) or with 815 μM CTP and UTP for 10 min (last row).
(1 + 20) U indicates that the reaction mixture contained 1 μM 32P-labeled UTP for 5 min followed by addition of nonlabeled 20 μM UTP and incubation for an additional 15 min.
The intensities of nonslipped (20 or 15 nt) and slipped (22, 24, and 26 nt, or 17 and 19 nt) transcripts were quantified with a PhosphorImager (Molecular Dynamics) and expressed as a percentage of the total transcript of 20 nt and longer. Values were corrected for different numbers of U residues in the various transcripts. Standard deviations for the top four rows were calculated from at least three independent experiments. The experiment in the last row was done only once under these conditions; it was repeated at 25°C with very similar results.
Effect of the initiating nucleotide and site of initiation on transcript slippage.
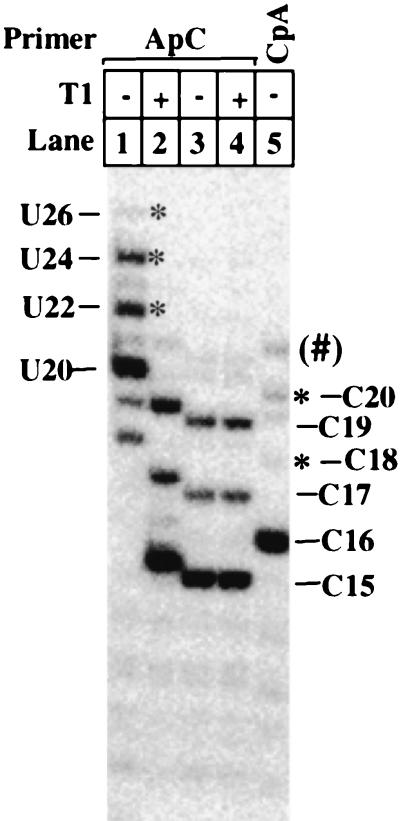
A number of earlier studies have reported slippage and resumption of RNA synthesis during transcript initiation by Escherichia coli RNA polymerase (2, 35). It was suggested that the lack of slippage for certain transcripts reflected a stabilization of the ternary complex by interaction of the 5′ end of the transcript with an upstream RNA binding site. We therefore compared the extent of slippage on the pML20-40 template in the ApC-initiated reaction (priming at +1/+2 [Fig. 1]) with that seen with CpA priming (−1/+1). The results are shown in Fig. 5. In lanes 1 and 2, RNA was generated with ApC priming, CTP, UTP, and GTP, as in lane 7 of Fig. 4. Slippage products of 22, 24, and (very faint) 26 nt were obtained (Fig. 5, lane 1). These RNAs were shortened by 4 nt, as expected, after treatment with RNase T1 (lane 2). When a similar reaction was done without GTP, the G-stop band at 15 nt and the slippage bands at 17, 19, and (faintly) 21 nt were also observed (lane 3). These latter bands all corresponded to stops before the incorporation of the G at +16, since they were unaffected by RNase T1 (lanes 3 and 4). Interestingly, when G-less reactions were carried out using a CpA primer, much less slippage was observed (lane 5). Since we have hypothesized that slippage with the ApC primer depends on the strong pause at +7, at the end of the three CU repeats, it was important to establish that this pause also occurred with CpA-mediated initiation. Kinetic assays (as in Fig. 2B) demonstrated that for CpA initiations, a pause of similar duration occurred at the same template location as seen with ApC priming (data not shown). That is, an 8-nt RNA transiently accumulated in the CpA-primed reactions.
FIG. 5.
The initiating dinucleotide can strongly influence the efficiency of transcript slippage. Reactions used the pML20-40 template and ApC or CpA primers at 1 mM in the indicated lanes. Other transcription conditions were those given in the legend of Fig. 3, except that no GTP was added in lanes 3 to 5. Transcripts were digested with RNase T1 as indicated. Transcripts generated by slippage in lanes 1 and 5 are marked by asterisks. Lengths of nondigested ApC-primed RNAs made in the presence of GTP are given to the left of the gel, and lengths of the ApC-primed (C15, C17, and C19) and CpA-primed (C16, C18, and C20) RNAs made in the absence of GTP are given to the right of the gel. An apparently 21-nt RNA, presumed to be generated by readthrough of the G-stop in the CpA primed reaction, is indicated by (#). This was not confirmed by T1 digestion for in this particular experiment. See Fig. 6 and 7B for other examples of the low level of CpA-primed slippage and Fig. 7 for other examples of readthrough products.
We also assayed transcription of the pML20-40 template without dinucleotide primers, using ATP, CTP, and UTP to support RNA synthesis. This resulted in detectable slippage products, but the level of slippage was lower than we observed with ApC priming. However, when we performed kinetic experiments like those in Fig. 2 with ATP-initiated reactions, we found that the pause at +7 was significantly shorter (data not shown). We therefore attribute the lower level of slippage in ATP-initiated reactions, relative to similar reactions primed with ApC, to the shorter residence time at the critical +7 position. On other templates which permitted pause times to be manipulated, high levels of slippage after initiation with ATP could be observed (see Fig. 7), but we were unable under any experimental conditions to obtain high levels of slippage in CpA-primed reactions (see Fig. 7).
FIG. 7.
Transcript initiation with the +2/+3 primer CpU promotes efficient slippage. (A) The 6G template was transcribed at 25°C with 1 mM CpU, 2 μM [α-32P]UTP, 5 μM dATP, and 20 μM CTP; reactions were stopped at the indicated times (lanes 3 to 8). Reactions in lanes 1 and 2 were done without the CpU primer. An aliquot of the 240-s reaction mixture was digested with RNase T1 (lane 8). Another aliquot of the 240-s reaction mixture was chased with 200 μM each NTP (lane 9); RNA from an equivalent reaction mixture was digested with T1 (lane 10). Slippage products are marked with asterisks, and the lengths of these RNAs are indicated to the left of the gel. (B) Transcription on the 6G template was initiated either with 1 mM dinucleotide or with 0.1 mM ATP, as indicated on the figure, along with 2 μM [α-32P]UTP, 20 μM CTP, and, in dinucleotide-containing reactions, 5 μM dATP, at 30°C for 2 min. One-half of each reaction mixture was digested with RNase T1 as indicated. For lanes 9 and 10, an ApC-primed reaction mixture otherwise identical to that in lane 3 was supplemented with 20 μM GTP; one-half of this reaction mixture was also digested with T1 as indicated (lane 10). Slippage products are marked by asterisks; transcripts resulting from readthrough of the G stop are marked by #. The lengths of the nonslipped G-stop RNAs for the CpA-, ApC-, and ATP-initiated reactions are marked on the gel. (C) The pML20-40 template was transcribed with 1 mM dinucleotide or 1 mM ATP plus 800 μM each CTP and UTP and 15 μM each [α-32P]CTP and [α-32P]UTP at 30°C for 10 min. Reaction products were ethanol precipitated, and the indicated samples were digested with RNase T1. Slippage products and RNAs resulting from readthrough of the G stop are marked by asterisks and #, respectively.
Transcript slippage may occur upstream of +7 if pausing is induced at the upstream location.
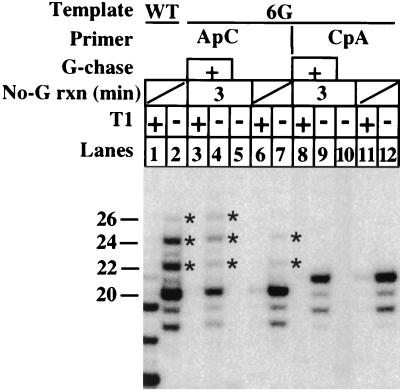
As noted above, our results are consistent with a model in which the transcription complex paused at +7 is the primary precursor of transcript slippage. Another potential location for transcript slippage would be +5, after the synthesis of ACUCU. Upstream slippage in this case would leave 2 bases hybridized at the 3′ end of the RNA (Fig. 1). Since the 6G template supported very little transcript slippage when GTP was present in the reaction mixture (Fig. 4), it is possible that only 2 bp is insufficient to stabilize the RNA-DNA hybrid during slippage and re-pairing. Alternatively, since transcription does not pause significantly after synthesis of the 5-mer on the 6G template (data not shown), there may be insufficient time for slippage to take place. The sequence of the 6G template allows us to discriminate between these two explanations. We compared the level of slippage-reextension products in ApC-primed transcriptions on 6G, either in the continuous presence of CTP, UTP, and GTP or with only CTP and UTP (which should stall transcription at +5), followed by the addition of GTP 3 min later. The results are shown in Fig. 6. As expected, the plus-G reaction led to only low levels of RNA longer than 20 nt (lane 7). The level of slippage products was considerably increased if the addition of GTP was delayed by 3 min (lane 4). However, this reaction still produced much lower levels of slippage-reextension RNAs than did a comparable reaction on the original pML20-40 template (lane 2). Thus, 2 bp of hybrid at the 3′ end are sufficient to allow re-pairing of the transcript and continued transcription after upstream slippage. However, reextension is more efficient if 4 bp of hybrid can be formed. It is important to note that CpA-initiated transcripts showed no evidence of upstream slippage in comparable transcription reactions on the 6G template, including those in which elongation was paused for 3 min at +5 (lanes 8 to 10). Therefore, pausing and the ability to pair at least 2 bases at the 3′ end after slippage are necessary but not sufficient conditions for efficient upstream slippage, re-pairing, and continued transcription to take place.
FIG. 6.
Transcript slippage by RNA polymerase II paused at +5. Transcription was performed on the 6G or pML20-40 (WT) templates as indicated. Reactions contained 1 mM either ApC or CpA (as indicated) plus 2 μM [α-32P]UTP and 20 μM CTP and dATP. For the reactions in lanes 1 and 2, 6 and 7, and 11 and 12, 20 μM GTP was also added and transcription proceeded for 4 min at 30°C. Reactions in lanes 3 to 5 and 8 to 10 proceeded for 3 min without GTP: 20 μM GTP was then added, and the mixtures were incubated for a further 4 min. RNAs in the indicated lanes were digested with RNase T1. The much shorter T1 digestion products from the 6G template are not shown here (see Fig. 4). The lengths of the nondigested RNAs made on pML20-40 in the presence of GTP are given to the left of the gel. Slippage products are marked with asterisks.
The fact that only 2 bp at the 3′ end of the RNA is needed after upstream slippage for transcription to resume may explain the low level of slippage products we see with the 6G template (Fig. 4). Even though there is no discernable pause at +5, the residence time at this position may be long enough for some slippage to take place. Upstream slippage by 2 bases from the normal pause site at +7 seems less likely for 6G transcripts, since this would result in an attempt to pair two G residues. On the other hand, slipping upstream from the pause at +7 may account for the low level of slippage we observed with the 4G template. In this case, translocation upstream by 2 bases would result in the pairing of UCU in the RNA with ACA in the DNA. Only 2 bp are formed, but the middle position in this case would not result in a potential purine-purine clash.
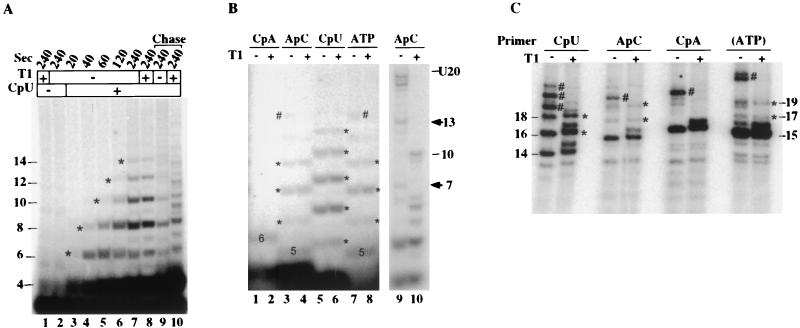
The results in Fig. 6 indicated that only two of the CU repeats need be present at the 3′ end of the transcript at the point of pausing for slippage to take place. This finding allowed us to test another dinucleotide primer, CpU, for its ability to support slippage. CpU can prime transcription on our templates from two locations, −3/−2 and +2/+3 (Fig. 1). Preinitiation complexes on the 6G template incubated with CpU, CTP, and radiolabeled UTP should give rise to only one labeled product, the CUCU tetramer initiated from the +2/+3 position (Fig. 1). Note that this RNA could slip upstream by 2 nt and re-pair in a manner analogous to that just described for ApC initiation in the absence of GTP on the 6G template. As shown in Fig. 7A, CpU initiation from +2/+3 yielded not only the expected 4-nt RNA but also RNAs that are 2, 4, 6, 8, and, at a low level, even 10 nt longer. None of these RNAs represented readthrough of the G stop, since none of them were sensitive to RNase T1 (lanes 7 and 8). High levels of the slippage-reextension products did not accumulate until the reaction had proceeded for at least 40 s. Most of the slippage products remained in ternary complex after 4 min because they could be chased into longer RNAs (lane 9). Thus, the slippage reaction not only occurs with a +2/+3 primer but is even more efficient than with the +1/+2 primer, ApC.
The ability to stall the RNA polymerase indefinitely at +5 on the 6G template allowed us to assay for slippage with all of the initiation substrates under comparable reaction conditions (Figs. 7B and C). In Fig. 7B, each 2-min reaction mixture contained CTP and radiolabeled UTP in addition to the initiating nucleotide or dinucleotide. Transcripts attributable to multiple rounds of slippage were observed with both ApC and CpU priming, but not with CpA priming, as shown above. (Note that the CpA-primed 6-mer is the normal G-stop transcript and not a slippage product.) Interestingly, when the RNA polymerase was forced to pause at +5 for an extended period, products consistent with at least three rounds of slippage could also be observed in ATP-initiated reactions (lanes 7 and 8). All of the RNAs marked with asterisks in lanes 1 to 8 of Fig. 7B should represent slippage products and not readthrough of the G stop, since these RNAs were not sensitive to RNase T1. If readthrough had taken place, the resulting transcripts should have accumulated at +15, before the next G stop. Low levels of such RNAs were observed in the ATP and ApC-initiated reactions (lanes 3 and 7); these transcripts were sensitive to RNase T1 (lanes 4 and 8). As a second positive control for activity of the T1, a parallel ApC-primed reaction was run which did include GTP. These transcripts gave the expected fragments on treatment with T1 (lanes 9 and 10).
In light of the efficient slippage supported by the CpU primer, we decided to test for slippage in CpU-initiated reactions with high levels of NTPs. For these experiments, we transcribed the pML20-40 template with 1 mM CpU and 815 μM CTP and UTP. With these substrates, transcription should proceed freely through the region in which slippage could occur and stall at +15 before the G stop (Fig. 1). As controls, we also initiated transcription on pML20-40 with ApC or CpA primers, or with ATP, in the presence of 815 μM CTP and UTP. The results are shown in Fig. 7C. The CpU primer supported synthesis of the expected 14-mer in addition to equivalent or greater amounts of 16- and 18-nt RNAs which are presumably slippage products, since they were not sensitive to RNase T1. Since high levels of NTPs were used in this assay, it is not surprising that substantial readthrough of the G stop was also observed; these readthrough RNAs were cleaved by T1. In the CpU-primed reaction in Fig. 7C, the 14-mer (nonslipped) RNA was 34% of the nonreadthrough transcript, while the RNA produced by one round of slippage (16-mer) represented 44% of the total and the 18-mer transcript from two rounds of slippage represented 22%. Thus, remarkably, when NTP levels are nearly physiological, the majority of CpU-primed initiations at the pML20-40 promoter go through at least one cycle of slippage. RNAs produced by slippage represented 18% of the total nonreadthrough transcript for the ApC-primed reaction (8% 17-mers, from one round of slippage, and 10% 19-mers, from two rounds [Table 1]) and 10% of the total for the ATP-initiated reactions (about 5% each for one-round and two-round products). However, we failed to detect slippage products for the CpA-primed reaction, consistent with our other studies at lower NTP levels.
Roles of RNA-DNA hybrid strength and helicase activity in transcript slippage.
Slippage requires the RNA-DNA hybrid to be transiently disrupted in order to allow re-pairing at a more upstream location. This predicts that strengthening the RNA-DNA hybrid would inhibit the slippage reaction. To test this, we substituted 5Br-UTP for UTP in ApC-primed transcription reactions with the pML20-40 template. As shown in Fig. 8A, the slippage products longer than 20 nt observed with UTP were completely absent (even at long exposure) when 5Br-UTP was substituted. The presence of 5Br-U also reduced the pause time at +7 compared to that in the UTP reaction, but the 40 to 60-s residence time at this location was sufficient to support easily detectable slippage in other experiments (data not shown) using UTP.
The location from which slippage initiates, as far downstream as +7, raises the question whether the TFIIH initiation factor might be involved in this process, since it has been established that inhibiting the XPB helicase activity of TFIIH with ATPγS blocks promoter escape (5–7) and inhibits the accumulation of transcripts longer than 9 or 10 nt. Earlier work (38) indicated that TFIIH should still be present in our early elongation complexes, which are substrates for slippage. We performed a series of transcription reactions on the 6G template with an ApC primer, CTP, and radiolabeled UTP, plus 5 μM dATP to support promoter unwinding. After 40 s, the expected 5-mer had accumulated, plus very low levels of slippage products (Fig. 8B, lane 1). When transcription was continued for 2 min without any additions (lanes 2 and 3), 7-, 9-, and 11-nt RNAs accumulated, as expected from the results in Fig. 7B. Also as expected from the results in Fig. 4, continuation of the 40-s reaction in the presence of GTP (lanes 6 and 7) led primarily to the accumulation of 20-nt RNA with only low levels of slippage. When ATPγS was added at 30 s along with GTP, the accumulation of the 7-nt RNA was not strongly affected but very little RNA longer than 7 nt was made (lanes 8 and 9), consistent with earlier reports (6, 7). Significantly, slippage products accumulated in the presence of ATPγS to essentially the same levels as they did in the absence of the inhibitor (compare lanes 2 and 4), while synthesis of a low level of 15-mer readthrough RNA was blocked. Thus, the XPB helicase is not required for transcript slippage.
DISCUSSION
We have studied the kinetics of the early postinitiation phase of transcription by native RNA polymerase II complexes. For the promoter used in this study, strong pauses were seen at +7, +18/19, and about +25, with a weaker stop at +13 (Fig. 2A). The average rate of bond formation by RNA polymerase II was 1 to 2 nt/s under the conditions used for the experiment in Fig. 2A, but the polymerase paused for 30 s or more at the three major promoter-proximal pause sites. Note that in the Fig. 2A experiment all of the NTPs were present at 200 μM, which is at or close to the Ks values determined for the four NTPs when calf thymus RNA polymerase II transcribes dC-tailed templates (180 μM for CTP, 200 μM for GTP, 400 μM for UTP, and 480 μM for ATP [15]). This suggests that these pauses are not simply the result of NTP limitation but instead mark important structural transitions within the early postinitiation transcription complex. It is useful to consider what these transitions might be, as an introduction to the discussion of the remarkable capacity for newly initiated RNA polymerase II complexes to allow upstream slipping and re-pairing of the transcript with the DNA template.
Cross-linking and biochemical studies have identified three essential features of the multisubunit RNA polymerases responsible for the continued elongation competence of ternary complexes: an RNA-DNA hybrid of about 8 bp (16, 28), the interaction of RNA immediately upstream of the hybrid with an RNA binding site on the RNA polymerase (the RNA exit channel [1, 26, 37]), and the interaction of the polymerase with about 9 bp of double-stranded DNA downstream of the transcription bubble (19, 26, 27). RNase protection studies with RNA polymerase II showed that 17 to 19 nt of transcript resists digestion in ternary elongation complexes (9), consistent with the presence of 8 to 9 nt of RNA in the RNA exit channel immediately upstream of the RNA-DNA hybrid (see also reference 30). The model constructed from biochemical evidence is consistent with the results of recent crystallographic studies with both E. coli RNA polymerase (17, 39) and yeast RNA polymerase II (8).
In light of these findings, it is tempting to suggest that the +7 pause is related to the breaking of the RNA-DNA hybrid and the initial threading of the transcript into the exit channel. The RNA-DNA hybrid reported for the yeast RNA polymerase II ternary complex is 9 bp long (8), but it has been suggested that this complex is in a paused configuration (21). Perhaps the approach of the upstream end of the growing RNA-DNA hybrid to the “rudder,” which is thought to be at least partially responsible for separating the transcript from the template (8), provides some resistance to translocation along the template. Such resistance could be particularly deleterious to elongation when consecutive U residues are being added to the transcript, as is the case at +7 on the pML20-40 template. This explanation is consistent with the reduction in the +7 pause which occurred when 5Br-U was substituted for U (Fig. 8). The +18/19 pause also corresponds to a location in which consecutive U residues are being added to the growing RNA chain, and this pause is also reduced in the presence of 5Br-U (Fig. 8). As with the +7 pause, there may be some resistance to continuing transcription past +18/+19 which is exaggerated by the formation of a weak hybrid at the 3′ end of the transcript. A transcript length of 18 to 19 nt marks the point at which the RNA exit channel should be filled, as noted above (30).
It is not clear why there should be resistance to continuing transcription at this point. It is possible that there is a transient unfavorable interaction of the transcript and the polymerase at the point where the 5′ end emerges from the exit channel (29). It is worth noting that interpretation of the effects of NTP analogs must be undertaken with caution, given the fact that pausing and termination efficiencies at various sites have been reported to increase or decrease, depending on the site, in response to the substitution of 5-BrU for U (4) or I for G (22).
We were quite surprised to discover that, as a result of the strong pause at +7, the RNA-DNA hybrid in the RNA polymerase II ternary complex can translocate, or slip, upstream by 2 bases, reanneal to the DNA template, and support continued transcript elongation. Although there have been many reports of transcript slippage during the early stages of transcription, we would emphasize that the phenomenon we report here is mechanistically unique. For E. coli RNA polymerase, slippage has been observed beginning at +4 to +6, at the end of a stretch of either U or A residues in the transcript, leading to the introduction of long runs of U or A in the RNA (10, 13, 14, 25). This prevented the synthesis of longer, correctly templated RNAs at the pyrB1 promoter (25). A second class of slippage events includes cases in which 3-nt (2) or 4-nt (35) initial transcripts slipped upstream by 3 bases, re-paired with the template, and supported continued transcription. None of the examples just cited correspond to the process we have documented with RNA polymerase II. The generation of long A or U polymers (10, 25) is presumed to occur in a stuttering mode that requires only a single-base upstream slippage at the 3′ end of the transcript and thus does not entail large-scale disruption of the RNA-DNA hybrid as an initiating event. Unlike some of the stuttering reactions, slippage on pML20-40 does not remove RNA polymerase II from the normal mode of productive transcription. The 22-, 24-, and 26-nt RNAs produced by slippage on pML20-40 in the absence of ATP were as likely to be elongated in subsequent chase reactions as were the RNAs which had not slipped (data not shown in this case [see also Fig. 7]). In one case, the stuttering reaction was found to be stimulated by high levels of the NTP incorporated in the polymer (25), whereas in our case, slippage is most pronounced at low NTP concentrations (Table 1). The examples with upstream slippage of short oligonucleotides (2, 35) more closely resemble the reaction we report here. However, the longest RNA which was translocated by slippage was 4 nt, and neither of these events could be observed with all NTPs present. Slippage in these cases required that the polymerase be halted at the appropriate point early in transcription by leaving some NTPs out of the reaction mixture. In contrast, slippage in our case is easily observed even with NTPs at 815 μM (Fig. 7C). With CpU priming, the majority of initiations undergo at least one round of slippage at these high NTP concentrations (Fig. 7C).
Slippage by RNA polymerase II during promoter-directed initiation has been reported in only one other instance of which we are aware. Cowie et al. (3) showed that the sequences of a significant proportion of the 5′ ends of polyomavirus early transcripts generated either in vivo or in vitro could not be explained without invoking upstream slippage and re-pairing during the synthesis of the initial three bonds. As in our case, this reaction apparently proceeds at physiological NTP concentrations. However, as with the E. coli RNA polymerase examples cited above, the transcript which undergoes slippage is much shorter than the 7-mer which can be the precursor to slippage in our case. In our view, the ability to reposition a 7-bp RNA-DNA hybrid by 2 bases is quite unanticipated in light of the detailed structure of the yeast RNA polymerase II ternary complex bearing a 15-nt transcript (8). In that structure, the 3-bp sequence at the 3′ end of the RNA-DNA hybrid is in a tight heteroduplex and is in close contact with the RNA polymerase. It is difficult to imagine how this intimate association could be disrupted to allow displacement upstream. However, it is important to note that the yeast ternary complex contains a 15-nt RNA and thus has passed through the promoter escape transition, while complexes paused in our experiments at +7 would not have escaped the promoter (reference 5 and references therein). Our laboratory had attempted to induce slippage at promoter-distal locations by introducing the sequence (AC)10 into the template strand downstream of one of our AdML promoter-based constructs. We transcribed this sequence with very limiting ATP and CTP levels, and we also caused RNA polymerase to resume transcription within this sequence after upstream translocation from an arrest site. In no case did we obtain any evidence of transcript slippage, in experiments where 1% slippage should have been detected (32). All of these results, taken together, suggest that prior to escape, the clamp domain may not be tightly closed over the RNA-DNA hybrid. This more open configuration would allow the flexibility needed for slippage (8). This idea can be tested by moving the repetitive element in our template further downstream from transcription start, with the expectation that at some point before +15 this element will no longer cause upstream slipping.
A particularly interesting and unanticipated aspect of the slippage reaction is its dependence on the site of transcript initiation. Initiation with the CpA primer (−1/+1) leads to almost no slippage, while initiation at +1 (ATP) or with ApC (+1/+2) supports slippage. Slippage occurs even more efficiently when transcription is primed with CpU (+2/+3). One might argue that CpA-initiated transcripts, which are 8 bases long at the +7 stop, are long enough to be stabilized (for example, because the RNA-DNA hybrid is complete) and thus resist slippage, in contrast to the 7-nt ApC-initiated transcripts. However, recall that CpA-initiated transcripts made in the absence of GTP on the 6G template, which are only 6 nt long, support almost no slippage (Fig. 7B). These observations suggest that the newly initiated transcription complex is qualitatively different when initiation is primed with CpA from when it is primed with ApC or CpU. A mechanistic basis for this difference may lie in the relative positions of the 5′ ends of the transcripts and the transcription bubble. Holstege et al. have shown that the initial melted region for an AdML-based promoter very similar in sequence to the one used here is −9 to +2 (11). The leading edge of the transcription bubble advances in synchrony with transcription, but the upstream edge remains fixed, until the 10th bond is made. At this point, the upstream edge abruptly shifts to +3. Since the upstream edge of the bubble does not move during the synthesis of the transcripts which are precursors to slippage, it seems possible that different locations of the 5′ end of the RNA relative to the fixed upstream bubble edge could result in somewhat different structures for the various transcription complexes and thus lead to different stabilities of the RNA-DNA hybrids within these complexes. This idea is reinforced by the observation that the closure of the clamp structure of RNA polymerase, which must move inward to retain the nucleic acids during transcript elongation, depends on the interaction of so-called switch regions at the base of the clamp with the RNA-DNA hybrid (8, 21).
An idea put forward in earlier work on transcript slippage by E. coli RNA polymerase is that slipping should be inhibited once the transcript is long enough to reach an upstream RNA binding site (2, 35). Current models for RNA polymerase II would place the entry of the transcript into the exit channel at some point immediately upstream of the end of the RNA-DNA hybrid, i.e., at 10 to 11 nt from of the point of bond formation (8, 21). In the case of ApC-primed transcription of the pML20-40 plasmid, two rounds of slippage occur with relatively high efficiency while much lower levels of third- and fourth-round products can be observed (Fig. 4). The precursors for the first and second rounds of slipping are 7- and 9-mers, respectively (schematic in Fig. 3), while the much less prevalent subsequent rounds would begin with 11- and 13-mers. This would be consistent with the idea that transcripts longer than 9 nt could begin to fill the exit channel and thus be stabilized against slippage. However, it could also simply be a kinetic effect, since the polymerases in the reaction in Fig. 4 were free to continue transcription past the pause site at template position +7. In the reaction on the 6G template in Fig. 7A, where RNA polymerase was forced to stall after synthesizing the transcript CUCU, five rounds of slipping and reextension can be detected. The precursors to the last two rounds were 10 and 12 nt long, respectively. It should be possible to extend the reaction shown in Fig. 7A to much longer times, to determine if there is a maximum transcript length which can lead to slippage.
We cannot say that the path that RNA takes upstream of the RNA-DNA hybrid in complexes which have undergone multiple rounds of slippage is identical to that which would be taken in complexes where no slippage had occurred. However, we emphasize that the large majority of the slipped and reextended RNAs in the CpU-primed transcription experiment, even those which have gone through four or five rounds of slippage, appear to remain in active transcription complexes, since they chase with high efficiency (Fig. 7A).
In summary, we have shown that the process of early transcript elongation by RNA polymerase II is nonuniform. There are a number of extended pauses during the synthesis of the first 25 bases of the transcript, which probably reflects the structural transitions that the polymerase must make as it assumes the transcript elongation configuration. A particularly surprising consequence of the earliest of these pauses on the template we used is the ability of a 7-nt transcript to slip upstream, reanneal with the template, and be efficiently reextended by the RNA polymerase. Our studies cannot address the issue of whether slippage occurs in vivo from the AdML promoter, although easily detectable levels of slippage did take place in ATP-initiated reactions at near-physiological NTP levels (Fig. 7C). It is interesting to recall that slippage at the polyomavirus early promoter was discovered by examination of viral RNA (3). Regardless of whether slippage plays an important role during initiation in vivo, the existence of this reaction provides useful insight into the mechanism by which RNA polymerase II passes from initiation into the stable elongation state.
Acknowledgments
This work was supported by grant GM 29487 from the National Institutes of Health.
REFERENCES
- 1.Artsimovitch, I., and R. Landick. 1998. Interaction of a nascent RNA structure with RNA polymerase is required for hairpin-dependent transcriptional pausing but not for transcript release. Genes Dev. 12:3110–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borukhov, S., V. Sagitov, C. A. Josaitis, R. L. Gourse, and A. Goldfarb. 1993. Two modes of transcription initiation in vitro at the rmB P1 promoter of Escherichia coli. J. Biol. Chem. 268:23477–23482. [PubMed] [Google Scholar]
- 3.Cowie, A., P. Jat, and R. Kamen. 1982. Determination of sequences at the capped 5′ ends of polyoma virus early region transcripts synthesized in vivo and in vitro demonstrates an unusual microheterogeneity. J. Mol. Biol. 159:225–255. [DOI] [PubMed] [Google Scholar]
- 4.Dedrick, R. L., C. M. Kane, and M. J. Chamberlin. 1987. Purified RNA polymerase II recognizes specific termination sites during transcription in vitro. J. Biol. Chem. 262:9098–9108. [PubMed] [Google Scholar]
- 5.Dvir, A., J. W. Conaway, and R. C. Conaway. 2001. Mechanism of transcription initiation and promoter escape by RNA polymerase II. Curr. Opin. Genet. Dev. 11:209–214. [DOI] [PubMed] [Google Scholar]
- 6.Dvir, A., R. C. Conaway, and J. W. Conaway. 1996. Promoter escape by RNA polymerase II—a role for an ATP cofactor in suppression of arrest by polymerase at promoter-proximal sites. J. Biol. Chem. 271:23352–23356. [DOI] [PubMed] [Google Scholar]
- 7.Dvir, A., R. C. Conaway, and J. W. Conaway. 1997. A role for TFIIH in controlling the activity of early RNA polymerase II elongation complexes. Proc. Natl. Acad. Sci. USA 94:9006–9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gnatt, A. L., P. Cramer, J. Fu, D. A. Bushnell, and R. D. Kornberg. 2001. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 Å resolution. Science 292:1876–1882. [DOI] [PubMed] [Google Scholar]
- 9.Gu, W. G., M. Wind, and D. Reines. 1996. Increased accommodation of nascent RNA in a product site on RNA polymerase II during arrest. Proc. Natl. Acad. Sci. USA 93:6935–6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo, H.-C., and J. W. Roberts. 1990. Heterogeneous initiation due to slippage at the bacteriophage 82 late gene promoter in vitro. Biochemistry 29:10702–10709. [DOI] [PubMed] [Google Scholar]
- 11.Holstege, F. C. P., U. Fiedler, and H. T. M. Timmers. 1997. Three transitions in the RNA polymerase II transcription complex during initiation. EMBO J. 16:7468–7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Izban, M. G., I. Samkurashvili, and D. S. Luse. 1995. RNA polymerase II ternary complexes may become arrested after transcribing to within 10 bases of the end of linear templates. J. Biol. Chem. 270:2290–2297. [DOI] [PubMed] [Google Scholar]
- 13.Jacques, J.-P., and M. M. Susskind. 1990. Pseudo-templated transcription by Escherichia coli RNA polymerase at a mutant promoter. Genes Dev. 4:1801–1810. [DOI] [PubMed] [Google Scholar]
- 14.Jin, D. J. 1994. Slippage synthesis at the galP2 promoter of Escherichia coli and its regulation by UTP concentration and cAMP · cAMP receptor protein. J. Biol. Chem. 269:17221–17227. [PubMed] [Google Scholar]
- 15.Kadesch, T. R., and M. J. Chamberlin. 1982. Studies of in vitro transcription by calf thymus RNA polymerase II using a novel duplex DNA template. J. Biol. Chem. 257:5286–5295. [PubMed] [Google Scholar]
- 16.Kireeva, M. L., N. Komissarova, D. S. Waugh, and M. Kashlev. 2000. The 8-nucleotide-long RNA-DNA hybrid is a primary stability determinant of the RNA polymerase II elongation complex. J. Biol. Chem. 275:6530–6536. [DOI] [PubMed] [Google Scholar]
- 17.Korzheva, N., A. Mustaev, M. Kozlov, A. Malhotra, V. Nikiforov, A. Goldfarb, and S. A. Darst. 2000. A structural model of transcription elongation. Science 289:619–625. [DOI] [PubMed] [Google Scholar]
- 18.Kugel, J. F., and J. A. Goodrich. 2000. A kinetic model for the early steps of RNA synthesis by human RNA polymerase II. J. Biol. Chem. 275:40483–40491. [DOI] [PubMed] [Google Scholar]
- 19.Landick, R. 1997. RNA polymerase slides home: Pause and termination site recognition. Cell 88:741–744. [DOI] [PubMed] [Google Scholar]
- 20.Landick, R. 1999. Transcription—shifting RNA polymerase into overdrive. Science 284:598–599. [DOI] [PubMed] [Google Scholar]
- 21.Landick, R. 2001. RNA polymerase clamps down. Cell 105:567–570. [DOI] [PubMed] [Google Scholar]
- 22.Levin, J. R., and M. J. Chamberlin. 1987. Mapping and characterization of transcriptional pause sites in the early genetic region of bacteriophage T7. J. Mol. Biol. 196:61–84. [DOI] [PubMed] [Google Scholar]
- 23.Linn, S. C., and D. S. Luse. 1991. RNA polymerase II elongation complexes paused after the synthesis of 15- or 35-base transcripts have different structures. Mol. Cell. Biol. 11:1508–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lis, J. 1998. Promoter-associated pausing in promoter architecture and postinitiation transcriptional regulation. Cold Spring Harbor Symp. Quant. Biol. 63:347–356. [DOI] [PubMed] [Google Scholar]
- 25.Liu, C., L. S. Heath, and C. L. Turnbough, Jr. 1994. Regulation of pyrBI operon expression in Escherichia coli by UTP-sensitive reiterative RNA synthesis during transcriptional initiation. Genes Dev. 8:2904–2912. [DOI] [PubMed] [Google Scholar]
- 26.Nudler, E. 1999. Transcription elongation: structural basis and mechanisms. J. Mol. Biol. 288:1–12. [DOI] [PubMed] [Google Scholar]
- 27.Nudler, E., E. Avetissova, V. Markovtsov, and A. Goldfarb. 1996. Transcription processivity: protein-DNA interactions holding together the elongation complex. Science 273:211–217. [DOI] [PubMed] [Google Scholar]
- 28.Nudler, E., A. Mustaev, E. Lukhtanov, and A. Goldfarb. 1997. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell 89:33–41. [DOI] [PubMed] [Google Scholar]
- 29.Pal, M., D. McKean, and D. S. Luse. 2001. Promoter clearance by RNA polymerase II is an extended, multistep process strongly affected by sequence. Mol. Cell. Biol. 21:5815–5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reeder, T. C., and D. K. Hawley. 1996. Promoter proximal sequences modulate RNA polymerase II elongation by a novel mechanism. Cell 87:767–777. [DOI] [PubMed] [Google Scholar]
- 31.Reines, D., R. C. Conaway, and J. W. Conaway. 1999. Mechanism and regulation of transcriptional elongation by RNA polymerase II. Curr. Opin. Cell. Biol. 11:342–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rudd, M. D. 1997. Ph.D. thesis. University of Cincinnati College of Medicine, Cincinnati, Ohio.
- 33.Samkurashvili, I., and D. S. Luse. 1996. Translocation and transcriptional arrest during transcript elongation by RNA polymerase II. J. Biol. Chem. 271:23495–23505. [DOI] [PubMed] [Google Scholar]
- 34.Samkurashvili, I., and D. S. Luse. 1998. Structural changes in the RNA polymerase II transcription complex during transition from initiation to elongation. Mol. Cell. Biol. 18:5343–5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Severinov, K., and A. Goldfarb. 1994. Topology of the product binding site in RNA polymerase revealed by transcript slippage at the phage lambda PL promoter. J. Biol. Chem. 269:31701–31705. [PubMed] [Google Scholar]
- 36.Uptain, S. M., C. M. Kane, and M. J. Chamberlin. 1997. Basic mechanisms of transcript elongation and its regulation. Annu. Rev. Biochem. 66:117–172. [DOI] [PubMed] [Google Scholar]
- 37.Yarnell, W. S., and J. W. Roberts. 1999. Mechanism of intrinsic transcription termination and antitermination. Science 284:611–615. [DOI] [PubMed] [Google Scholar]
- 38.Zawel, L., K. P. Kumar, and D. Reinberg. 1995. Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev. 9:1479–1490. [DOI] [PubMed] [Google Scholar]
- 39.Zhang, G. Y., E. A. Campbell, L. Minakhin, C. Richter, K. Severinov, and S. A. Darst. 1999. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 Å resolution. Cell 98:811–824. [DOI] [PubMed] [Google Scholar]