Abstract
DNA geminiviruses are thought to be targets of RNA silencing. Here, we characterize small interfering (si) RNAs—the hallmarks of silencing—associated with Cabbage leaf curl begomovirus in Arabidopsis and African cassava mosaic begomovirus in Nicotiana benthamiana and cassava. We detected 21, 22 and 24 nt siRNAs of both polarities, derived from both the coding and the intergenic regions of these geminiviruses. Genetic evidence showed that all the 24 nt and a substantial fraction of the 22 nt viral siRNAs are generated by the dicer-like proteins DCL3 and DCL2, respectively. The viral siRNAs were 5′ end phosphorylated, as shown by phosphatase treatments, and methylated at the 3′-nucleotide, as shown by HEN1 miRNA methylase-dependent resistance to β-elimination. Similar modifications were found in all types of endogenous and transgene-derived siRNAs tested, but not in a major fraction of siRNAs from a cytoplasmic RNA tobamovirus. We conclude that several distinct silencing pathways are involved in DNA virus-plant interactions.
INTRODUCTION
Endogenous gene silencing can occur at the transcriptional and the post-transcriptional levels. In post-transcriptional gene silencing (PTGS), mRNA is degraded or repressed translationally; in transcriptional gene silencing (TGS), DNA and/or histones are modified leading to heterochromatization and transcriptional repression. PTGS and TGS are often correlated with the appearance of small interfering (si) RNAs of ∼21–24 nt in length, derived from silenced sequences. MicroRNA (miRNA)- and trans-acting siRNA (ta-siRNA) pathways, which play a crucial role in developmental gene regulation in plants (1,2), are PTGS-related processes, in which the respective small RNAs, miRNAs and ta-siRNAs are derived from separate genetic loci and act in trans to silence their target genes. In plants, epigenetic phenomena associated with all the aforementioned small RNAs (sRNAs) are collectively referred to as RNA silencing (2,3). Presumably, all types of silencing sRNAs are produced as duplexes from longer perfect or imperfect double-stranded (ds) RNA precursors by Dicer or Dicer-like (DCL) proteins (1–3). Then, one of the sRNA strands is channeled to the RNA-induced silencing complex (RISC) or an ‘RNA-induced initiation of TGS’ (RITS) complex to guide these effectors to their respective targets. Both RISC and RITS appear to contain a distinct Argonaute protein as an active component (4,5). In plants, nematodes and fungi, an RNA-dependent RNA polymerase (RDR) plays an important role in RNA silencing, most likely by converting single-stranded transcripts into dsRNA (2,6).
In plants, RNA silencing provides an adaptive immune system recognizing and inactivating pathogenic nucleic acids such as viruses and transposons (6). Indeed, virus-derived siRNAs have been detected in plants infected with various RNA viruses (7–11) and DNA geminiviruses (12–14). Furthermore, both RNA and DNA viruses encode distinct suppressors of RNA silencing that target different components of this system (6). In particular, tombusvirus p19 protein selectively sequesters 21 nt siRNA duplexes (15). Geminivirus suppressor protein AC4 appears to selectively bind single-stranded sRNAs including miRNAs (16). The latter observation is consistent with a hypothesis that not only the siRNA but also the miRNA pathway might restrict virus replication, as demonstrated for a mammalian retrovirus (17). Keeping in line with this idea, most viral silencing suppressors, when overexpressed in transgenic plants, interfere with production and/or action of miRNAs, thus leading to various abnormalities of plant development, often resembling viral symptoms (6).
Very little is known about the biogenesis and possible modification of virus-derived siRNAs. Based on biochemical studies of RNAi in animal systems (18) and wheat germ extracts (19), it is assumed that viral siRNAs are duplexes with 2 nt 3′-overhangs produced from longer perfect dsRNA by Dicer activity. However, recent work (8,11) has shown that predominantly the positive strand of RNA virus-derived sRNAs accumulates, suggesting that at least some sRNAs are produced as miRNA-like duplexes from secondary structure elements of the single-stranded viral genomes (i.e. imperfect dsRNA), rather than from the replicative intermediates (i.e. perfect dsRNA).
Genetic evidence suggests that the four Arabidopsis DCL genes (20) have diversified (10) partially redundant (21) functions. DCL1 is involved in the production of predominantly 21 and 22 nt miRNAs from hairpin-like precursor transcripts (22). DCL3 produces 24 nt repeat-associated siRNAs (ra-siRNAs), involved in TGS of the respective repetitive DNA loci, presumably from dsRNA precursors generated in a Pol IV- and RDR2-dependent pathway (10,21,23). DCL4 produces 21 nt ta-siRNAs from perfect dsRNA substrates generated by RDR6 on the miRNA-cleaved transcripts of ta-siRNA genes (21,24–26). The function of DCL2 is still unclear, but it seems to be a redundant DCL in the production of endogenous sRNAs (21,26).
It is still unclear which DCLs are involved in producing virus-derived siRNAs. Neither the DCL3 null mutation dcl3-1 nor the DCL1 weak mutation dcl1-7 compromised accumulation of RNA virus-derived siRNAs (10). Although turnip crinkle virus (TCV) siRNA production was compromised in the Arabidopsis DCL2 mutant dcl2-1 early in infection, at the late stages TCV siRNAs did accumulate to wild-type levels. Moreover, two other RNA viruses [cucumber mosaic virus (CMV) and turnip mosaic virus (TuMV)] produced wild-type levels of siRNAs in dcl2-1 plants at both early and late stages of infection (10).
Two other Arabidopsis genes in addition to DCL1 have been implicated in the biogenesis of miRNAs. The HYL1 (27,28) product has a dsRNA-binding motif and can physically interact with the DCL1 protein; other members of HYL1 gene family have also been proposed to interact with distinct DCLs (29). HEN1 (30,31) encodes a methyl transferase that methylates the last nucleotide of miRNAs at 2′-O- or 3′-O-position (32), with the 2′-OH claimed to be the major target of the modification (33). Recent evidence suggests that all endogenous sRNAs in Arabidopsis are methylated by HEN1, which protects them from a 3′ end uridylation activity (34). So far HEN1 has not been reported to methylate virus-derived sRNAs, albeit the bulk signal of CMV-derived siRNAs in Nicotiana benthamiana was shown to be resistant to β-elimination (33), suggesting a 3′ terminal nucleotide modification.
Geminiviruses are single-stranded DNA viruses with mono- or bipartite circular genomes of ∼2.5–2.7 kb (35). They replicate through double-stranded DNA intermediates that establish themselves as multiple minichromosomes in the nucleus of infected plant cell (36), which might be targets of TGS. The circular viral minichromosomes serve as templates for Pol II transcription driven by a bi-directional promoter located in the intergenic region (37). Bipartite begomoviruses possess an additional mono-directional promoter for the leftward gene AC2 (38) coding for a transactivator protein that activates viral and host transcription and suppresses PTGS (39). Geminiviruses do not obligatorily produce long dsRNA during their life cycle and their processed leftward and rightward transcripts overlap only in a short region (38). However, aberrant readthrough transcription on a circular viral DNA could potentially lead to production of longer antisense transcripts that might trigger RNA silencing. In fact, we detected such aberrant transcripts derived from the ‘non-transcribed’ promoter region of the Mungbean yellow mosaic begomovirus DNA-A (38).
In the present study, we characterized sRNAs associated with the begomoviruses, African cassava mosaic virus (ACMV) (40) in N.benthamiana and cassava, and Cabbage leaf curl virus (CaLCuV) (41) in Arabidopsis. We detected 21, 22 and 24 nt begomoviral sRNAs of both polarities representing both coding and intergenic regions. These viral siRNAs, similar to siRNAs derived from a dsRNA transgene, and endogenous ta-siRNAs and miRNAs were found to be phosphorylated at the 5′ end and modified at the 3′-terminal nucleotide. Genetic evidence indicated that DCL3, DCL2, at least one additional DCL activity and HEN1 are involved in the biogenesis of begomoviral siRNAs. This suggests that both TGS- and PTGS-related silencing pathways are involved in plant–geminivirus interactions.
MATERIALS AND METHODS
Plants and viruses
N.benthamiana plants were grown from seeds in soil at 26°C with 16 h day and 8 h night. About 3 to 4 weeks post-germination, seedlings were inoculated with ACMV using a biolistic particle delivery system (PDS-1000/He, BioRad): 750 µg of 1 µm gold particles were coated with a plasmid mixture (0.5 µg each) of the infectious clones of ACMV-KE DNA-A (GenBank accession NC_001467) (40) and ACMV-CM DNA-B (AF112353) (42) and delivered at 1100 psi, following the manufacturer's recommendations. Four weeks post-inoculation, systemic leaves of ACMV-infected plants were harvested in pools and titers of the virus were determined by a semi-quantitative duplex PCR (data not shown).
Cassava (Manihot esculenta Crantz cv. TMS 60444) plants were multiplied in vitro through nodal cuttings on CBM [Murashige and Skoog (MS) medium including vitamins (Duchefa) supplemented with 2% sucrose, 2 µM CuSO4, pH 5.8 and solidified with 0.3% Gelrite]. They were kept 8 weeks at 26°C with a 16 h photoperiod. Rooted plantlets were transferred to pots and grown 7–9 weeks in a greenhouse before being inoculated with ACMV-[NOg] (AJ427910, AJ427911) (43). The ACMV-[NOg] infectious clones were designed to contain partial tandem repeats of DNA-A and DNA-B in pBluescript (SK-) (H. Vanderschuren, P. Zhang, R. Akbergenov, M. M. Pooggin, T. Hohn and W. Gruissem, manuscript in preparation). Each cassava plant was bombarded twice with 250 µg of 1.5–3 µm gold particles coated with a mixture of the two ACMV-[NOg] infectious clones (200 ng each). The biolistic delivery was directed into the plant meristem as described by Zhang et al. (44). After inoculation, cassava plants were kept at 28°C with a 16 h photoperiod, and symptom development and virus accumulation were monitored as described previously (14). After 8 weeks, young emerging leaves of infected plants were harvested for analysis of virus-derived siRNAs.
Arabidopsis wild-type and mutant plants were grown from seeds in soil in a phytochamber (Sanyo) at 20°C with 12 h day and 12 h night. Four to five weeks post-germination, seedlings were inoculated with CaLCuV by biolistic delivery of a plasmid mixture (0.5 µg each) of pMTCbLCVA.008 and pCPCbLCVB.002 (45) as described above for N.benthamiana. Four weeks post-inoculation, CaLCuV-infected plants were harvested in pools and titers of the virus were determined a semi-quantitative duplex PCR using the CaLCV AC2 gene-specific PCR primers (data not shown).
The mutant line hen1-1 (30) in the La-er ecotype was kindly provided by Dr X. Chen. The mutant line dcl3-1 in the Col-0 ecotype, identical to the previously described one (10), was independently obtained from the SALK collection and characterized by us. The mutant dcl2-5 in the Col-0 ecotype was identified in the SALK collection (SALK 123586) and contains a T-DNA insertion in the 18th exon of DCL2 (At3g03300). The homozygous lines were selected using allele-specific PCR primers.
Mechanical inoculation with ORMV (46) of N.benthamiana and Arabidopsis plants at a stage of about 5 weeks post-germination was performed using celite 545 (Merck) and purified virions (kindly provided by Dr M. Heinlein) or sap of ORMV-infected N.benthamiana. Five and ten days post-inoculation, respectively, severely infected plants were harvested and total RNA was isolated for sRNA blot hybridization (see below) using a mixture of ORMV-specific short DNA probes (Table 1).
Table 1.
Probes for RNA blot hybridization
| Name | Sequence |
|---|---|
| ACMV NONA s | 5′-AGG GGC CAA CCG TAT AAT ATT ACC GGT-3′ |
| ACMV NONA as | 5′-ACC GGT AAT ATT ATA CGG TTG GCC CCT-3′ |
| ACMV-KE AC2 s | 5′-TGG AGG TAA TAT GAA CAT CCA CAG ACA-3′ |
| ACMV-KE AC2 as | 5′-TGT CTG TGG ATG TTC ATA TTA CCT CCA-3′ |
| ACMV-NOg AC2 s | 5′-TGG AGG TAA TAT GAA CAG CCA CAG ACA-3′ |
| ACMV-NOg AC2 as | 5′-TGT CTG TGG CTG TTC ATA TTA CCT CCA-3′ |
| CaLCuV AC2 s | 5′-TGG AGG AAG ATA GAA CAC CCG CAG TTC-3′ |
| CaLCuV AC2 as | 5′-GAA CTG CGG GTG TTC TAT CTT CCT CCA-3′ |
| ormv2 s | 5′-TAA CTA AAA GTG AGA GGT TCG AAT CCT-3′ |
| ormv3 s | 5′-ATC ACC TGT TAA CGT ACG CGT GGC GTA-3′ |
| ormv4 s | 5′-TTA GAT GAG GCC GTT GCC GAG GTC CAT-3′ |
| miR173 as | 5′-GTG ATT TCT CTC TGC AAG CGA A-3′ |
| siR1003 as | 5′-ATG CCT ATG TTG GCC TCA CGG TCT-3′ |
| siR255 as | 5′-TAC GCT ATG TTG GAC TTA GAA-3′ |
| gfp1 s | 5′-CCG GGG TGG TGC CCA TCC TGG TCG AGC TGG-3′ |
| gfp2 s | 5′-CGG CAA GCT GAC CCT GAA GTT CAT CTG CAC-3′ |
| gfp3 as | 5′-AGG GTG TCG CCC TCG AAC TTC ACC TCG GCG-3′ |
| gfp4 s | 5′-CAA CTA CAA CAG CCA CAA CGT CTA TAT CAT-3′ |
| gfp5 as | 5′-GTA GTG GTC GGC GAG CTG CAC GCT GCC GTC-3′ |
| gfp6 s | 5′-CCT GAG CAC CCA GTC CGC CCT GAG CAA AGA-3′ |
| gfp7 as | 5′-CGG CGG TCA CGA ACT CCA GCA GGA CCA TGC-3′ |
| U6-I | 5′-GGC CAT GCT AAT CTT CTC TGT ATC GTT-3′ |
| U6-II | 5′-CCA ATT TTA TCG GAT GTC CCC GAA GGG AC-3′ |
| β Internal control | 5′-CTT GAA GTT CAC CTT GAT GCC-3′ |
Transgenic plants
The ACMV dsRNA expression cassette was constructed based on an RNAi vector described by Pooggin et al. (47), which contains an inverted repeat of the MYMV sequences separated by a synthetic plant intron (syn7, positions 57–169) (48) under the control of the CaMV 35S promoter (228 bp, between XbaI and KpnI) and the CaMV terminator (202 bp, between BamHI and EcoRI). The MYMV sequences were replaced one-by-one with the ACMV-KE DNA-A sequence from position 21 to 277 (GenBank accession NC_001467) in the reverse and the forward orientations between the 35S promoter and the intron and the intron and the terminator, respectively. The resulting 1075 bp expression cassette was introduced between XbaI and EcoRI sites of the binary vector pCAMBIA-1300 (AF234296). The resulting construct was mobilized on Agrobacterium tumefaciens LBA4404 for transformation of N.benthamiana leaf discs and cassava cv. TMS 60444 embryogenic suspension cells (49), respectively. N.benthamiana primary transformants were selected on agar-solidified MS medium supplied with 250 µg/ml hygromycin and selfed. The resulting seeds were germinated on the selection medium and T1 transgenic plants were tested for the presence and integrity of the transgene by PCR using two pairs of primers specific for the 35S promoter and the intron, or the intron and the 35S terminator (data not shown). Putative transgenic cassava embryos were recovered from transformed somatic suspensions via embryogenesis under the step-wise hygromycin selection from 7.5 to 25 µg/ml as described by Zhang et al. (49). After a process of somatic embryo maturation and germination on the Gelrite-solidified MS medium supplemented with 0.4 mg/l BAP, new shoots were regenerated and multiplied on CBM as shoot cultures (see above). Transgenic cassava lines were verified by Southern blot hybridization (H. Vanderschuren, P. Zhang, R. Akbergenov, M. M. Pooggin, T. Hohn and W. Gruissem, manuscript in preparation). Young leaves of the line 1–2 carrying a single copy of the transgene were harvested and used for siRNA analysis (Supplementary Figure S1). The 27 nt NONA sense and antisense probes (Table 1) used for detection of the dsRNA transgene and virus siRNAs are derived from a part of the ACMV 257 bp ‘non-transcribed’ region (an inverted repeat in the transgene), 112 and 119 bp away from the leftward and the rightward transcription start sites, respectively.
Seeds of the Arabidopsis green fluorescent protein (GFP) transgenic line 8z2 (in the Col-0 background) (50) were germinated in soil at 20°C with 12 h day and 12 h night. The individual seedlings that had undergone spontaneous PTGS of the transgene at the two leaf-stage (50) were selected under UV-light and grown for 4 more weeks. Then, total RNA from three plants was extracted and GFP siRNAs were detected by RNA blot hybridization as described below, using a mixture of seven GFP transgene-derived oligonucleotides of both sense and antisense polarity (Table 1) as a probe.
Small RNA isolation and detection
Total RNA was extracted from 1 g plant tissue, derived from the pools of three plants ground in liquid nitrogen, using a Trizol reagent (Invitrogen) according to the manufacturer's protocol. In most cases, total RNA was fractionated using Midi RNeasy kit (Qiagen) and the RNA cleanup protocol. An aliquot of 20 µg RNA was re-suspended in 10 µl loading buffer (95% formamide, 20 mM EDTA, pH 8.0, 0.05% bromophenol blue and 0.05% xylene cyanol), heated at 95°C for 2 min and loaded on 15% polyacrylamide gel (a 19:1 ratio of acrylamide to bis-acrylamide, 8 M urea). The gel was run using the SE 600 electrophoresis machine (Hoefer) at 300 V for 4 h and then the RNA was transferred to a Hybond N+ membrane by electroblotting in 1× TBE buffer at 10 V overnight. The blot hybridization was performed at 35°C for 14–24 h in an UltraHyb-oligo buffer (Ambion) using, as a probe, one or several short DNA oligos (Table 1) end-labeled with 32P by T4 polynucleotide kinase (Roche) and purified through MicroSpin™ G-25 columns (Amersham) according to the manufacturers' recommendations. The blot was washed two times with 2× SSC, 0.5% SDS for 30 min at 35°C. The signal was detected after 1–3 days exposure to a phosphor screen using a Molecular Imager (BioRad). For repeated hybridization the membrane was stripped with 0.5× SSC, 0.5% SDS for 30 min at 80°C and then with 0.1× SSC, 0.5% SDS for 30 min at 80°C.
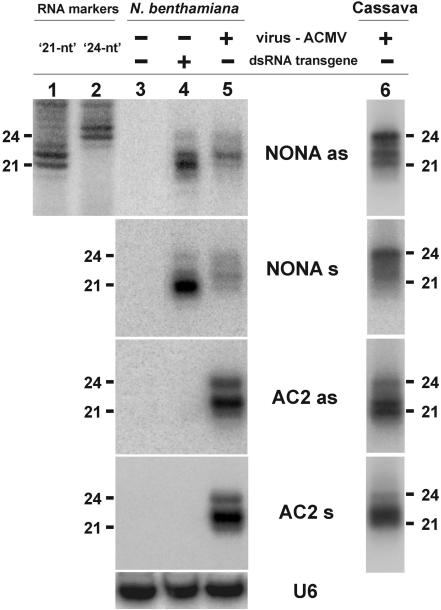
The RNA size-markers (‘21’ and ‘24 nt’, Figure 1) were prepared using T7 RNA polymerase (Promega) and a pair of DNA oligonucleotides as a template—the T7 promoter oligo 5′-TAATACGACTCACTATAG-3′ and for ‘21 nt’, 5′-ACGGTTGGCCCCTTGGTTTCCCTATAGTGAGTCGTATTA-3′, or for ‘24 nt’, 5′-ACGGTTGGCCCCTTGGTTGTCTCCCTATAGTGAGTCGTATTA-3′—according to the Promega protocol in the presence of [α-32P]rUTP (Hartman) and then purified through the G-25 columns (Amersham). The markers were verified by using synthetic RNA oligonucleotides (data not shown) as well as by comparing them to the Arabidopsis endogenous sRNAs, the sizes of which are known from the cloning data (miR173, siR255 and siR1003, Figure 4). Note that the 21 and the 24 nt bands of the respective markers become more intensive when the ‘NONA as’ probe, which is partially complementary to the marker RNA, is used for hybridization (Figure 1).
Figure 1.
Comparison of siRNAs derived from the geminivirus ACMV and the dsRNA transgene in N.benthamiana and cassava RNA gel blot analysis of 20 µg total RNA prepared from young leaves of wild-type (lane 3) and dsRNA-transgenic (lane 4) N.benthamiana, or from ACMV-infected wild-type N.benthamiana (lane 5) or cassava (lane 6) plants. The blots were successively probed with 32P-labeled 27 nt DNAs (Table 1) corresponding to the ACMV DNA-A complementary and virion strand sequences in the intergenic region (‘NONA as’ and ‘NONA s’, respectively) and the AC2 gene coding region (‘AC2 as’ and ‘AC2 s’). The N.benthamiana blot was stripped and re-probed with a mixture of two oligonucleotides complementary to U6 (Table 1) as a loading control. Positions of 21 and 24 nt RNAs from the in vitro synthesized, p32-labeled size-markers (lanes 1 and 2) are indicated. Note that the extra bands appear to be the result of untemplated extension by the T7 polymerase used for marker synthesis. The contrast change between marker lanes (1 and 2) and the adjacent part of the blot is due to a difference in exposure times. Marker lanes in panels below the top one are not shown, since the marker signal decayed during successive re-probing; sizes of viral siRNAs were determined by super-imposing the corresponding images with the topmost image.
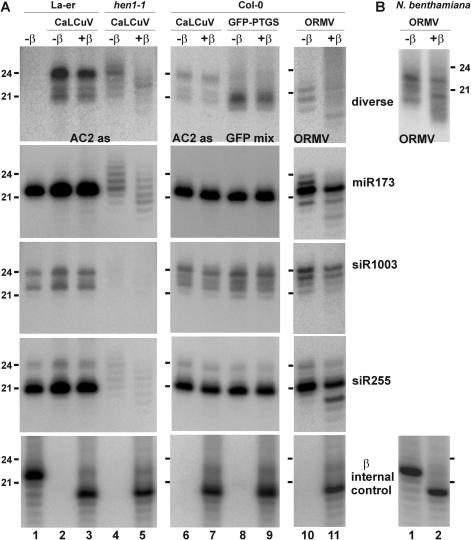
Figure 4.
DNA geminivirus-derived siRNAs are methylated in Arabidopsis, whereas a major fraction of RNA tobamovirus-derived siRNAs are not (A) RNA gel blot analysis of 20 µg total RNA prepared from the geminivirus CaLCuV-infected wild-type (La-er and Col-0) and HEN1 loss-of-function mutant (hen1-1) Arabidopsis plants, or from the tobamovirus ORMV-infected wild-type Arabidopsis, and from uninfected plants of the PTGS-silenced GFP transgenic line (GFP-PTGS), and treated (+β) or not (−β) with the oxidation and β-elimination reagents. The blots were successively probed with the virus- or the transgene-specific DNAs (‘AC2 as’, ‘ORMV’ or ‘GFP’, Table 1) and from the three endogenous loci (miR173, siR1003 and siR255). The blots were stripped and re-probed with an oligonucleotide complementary to a synthetic RNA (‘β internal control’) added to both the samples (lanes 3, 5, 7, 9 and 11) prior the β-elimination treatment and the non-treated control sample (lane 1). Positions of the 21 and 24 nt RNAs are indicated. (B) RNA gel blot analysis of 20 µg total RNA prepared from ORMV-infected N.benthamiana young leaves and treated (+β) or not (−β) with the oxidation and β-elimination reagents. The blot was probed with ORMV-specific DNAs (Table 1) and then stripped and re-probed with an oligonucleotide complementary to a synthetic RNA (‘β internal control’) added to the sample (lane 2) prior the β-elimination treatment and the non-treated control sample (lane 1). Positions of the 21 and 24 nt RNAs are indicated.
Phosphatase treatment and β-elimination
For dephosphorylation, 20 µg total RNA was treated with 1 U of calf intestine alkaline phosphatase (AP, Boehringer Mannheim) in 10 µl 1× AP buffer for 30 min at 37°C. For β-elimination, 20 µg total RNA was dissolved in 17.5 µl borax buffer, pH 8.6 [4.375 mM borax (Fluka), 50 mM boric acid (Fluka)] and 2.5 µl 0.2 M sodium periodate (Fluka) was added. The reaction mixture was incubated for 10 min at room temperature in the dark and, after addition of 2 µl glycerol, the incubation was repeated. The mixture was lyophilized using SpeedVac, dissolved in 50 µl borax buffer, pH 9.5 (33.75 mM borax, 50 mM boric acid, pH adjusted by NaOH) and incubated for 90 min at 45°C.
After the treatments, RNA samples were purified through the G-25 columns and used for RNA blot hybridization as described above.
RESULTS AND DISCUSSION
Three major size-classes of viral siRNAs are generated in begomovirus-infected N.benthamiana and cassava
Using high resolution RNA blot hybridization with reliable RNA size-markers and 32P-labeled 27 nt DNA oligonucleotide probes we detected three major size-classes of ACMV siRNAs, 21, 22 and 24 nt in length, in infected N.benthamiana and cassava plants. These siRNAs were of sense- and antisense polarities, derived from both coding (AC2) and non-coding (NONA) regions of the virus (Figure 1). Other 27 nt regions of the virus genome tested also gave rise to siRNAs of the three size-classes (data not shown). Together with earlier observations that siRNAs from begomovirus-infected plants could hybridize to most ∼500 bp segments of the virus genome (13), these results suggest that all begomovirus DNA sequences can be a source of the three size-classes of siRNAs.
To examine the possibility that perfect dsRNAs are intermediates in the formation of begomoviral siRNAs, we compared the properties of the ACMV siRNAs with those of siRNAs derived from a transgene expressing double-stranded ACMV sequences. The transgene encodes an inverted repeat of the 257 bp intergenic region of ACMV DNA-A interrupted with a synthetic intron and is regulated by a strong 35S promoter. Although this transgene gave rise to three major size-classes of siRNAs of both polarities in N.benthamiana, the relative abundance of the classes differed from that obtained with ACMV-infected plants. Comparison on the same gel using probes for the same viral sequences showed that the transgene generates predominantly 21 nt siRNAs, whereas the virus generates predominantly 22 and 24 nt siRNAs (Figure 1). Similarly, in cassava, ACMV generated relatively more large-sized siRNAs than did the transgene, especially from the non-coding region (Figures 1 and Supplementary Figure S1). Expression of the transgene in N.benthamiana produced small amounts of 20 nt siRNAs, which were barely detectable for the virus (Figures 1 and 2). In cassava, an additional 23 nt band was detected with some but not all the probes for both the virus and the transgene (Figures 1 and 3 and Supplementary Figure S1). These results are consistent with the hypothesis that perfect dsRNA is the precursor of most begomoviral siRNAs, but the difference in relative abundance of the different siRNA classes suggests that their modes of production (readthrough transcription versus RDRs) and processing (different DCLs, see below) differ somewhat. It could also be speculated that distinct phasing of cleavage by different DCL activities, as inferred from the ta-siRNA biogenesis by DCL4 (21,24–26), would account for differences in the relative signal intensities between the size-classes of siRNAs derived from transgenic versus viral dsRNA precusor(s).
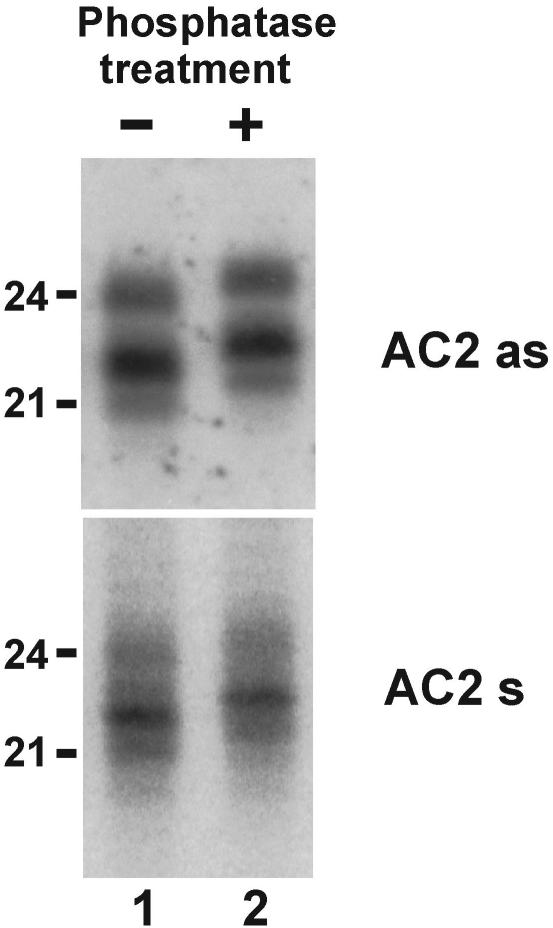
Figure 2.
ACMV-derived siRNAs are phosphorylated at the 5′ end RNA gel blot analysis of 20 µg total RNA prepared from ACMV-infected wild-type N.benthamiana and treated (+) or not (−) with alkaline phosphatase. The blot was successively probed with DNA oligonucleotides corresponding to the ACMV DNA-A complementary (AC2 as) and virion (AC2 s) strand sequences in the AC2 coding region. Positions of the 21 and 24 nt RNAs are indicated.
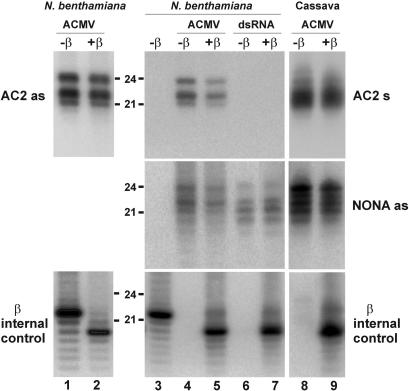
Figure 3.
ACMV- and dsRNA transgene-derived siRNAs are modified at the 3′ terminal nucleotide RNA gel blot analysis of 20 µg total RNA prepared from young leaves of wild-type (lane 3) and dsRNA-transgenic (lanes 6 and 7) N.benthamiana, or from ACMV-infected wild-type N.benthamiana (lanes 1, 2, 4 and 5) or cassava (lanes 8 and 9) and treated (+β) or not (−β) with the oxidation and β-elimination reagents. The blots were successively probed with 32P-labeled 27 nt DNAs (see Table 1) corresponding to the intergenic region (NONA as) and the AC2 gene coding region (‘AC2 as’ and ‘AC2 s’) of ACMV DNA-A. The blots were stripped and re-probed with an oligonucleotide complementary to a synthetic RNA (‘β internal control’) added to both the samples (lanes 2, 5, 7 and 9) prior the β-elimination treatment and some non-treated control samples (lanes 1 and 3). Positions of the 21 and 24 nt RNAs are indicated.
The 5′-nucleotide of siRNAs generated by Dicer activities is phosphorylated (18). Our results indicate that ACMV siRNAs of all the size-classes and polarities are phosphorylated at the 5′ end. After alkaline phosphatase treatment their apparent mobility on the gel was retarded by about a half-nucleotide (Figure 2), as expected for a decreased negative charge due to removal of a 5′-phosphate group. Using the same method, we found that all the CaLCuV siRNAs in Arabidopsis also possess a 5′-phosphate (see below), thus extending our finding to other begomoviruses.
The terminal 3′-nucleotide of Arabidopsis miRNAs, ta-siRNAs and transgene-derived siRNAs associated with PTGS, and ra-siRNAs involved in TGS of repetitive DNA loci has been shown to be modified by 2′- or 3′-O-methylation of ribose, which results in resistance to β-elimination (34). To test whether the 3′-nucleotide of begomoviral siRNAs has a similar modification, we treated total RNA samples from ACMV-infected or dsRNA-transgenic N.benthamiana and cassava plants with sodium periodate. Under these conditions, the 3′-nucleotide of RNAs with free 2′- and 3′-OH groups is lost leaving a 3′-phosphate. This results in products that migrate faster in gel electrophoresis by about 2 nt (32,34). As expected, a short synthetic RNA with free 2′-OH and 3′-OH, added to each sample as an internal control, was almost completely converted into a 2 nt-shorter-migrating form (Figure 3, β internal control). In contrast, we did not observe any change in the ACMV siRNA patterns derived from coding and non-coding regions (Figure 3, AC2 and NONA), indicating that all the types and size-classes of begomoviral siRNAs are modified at the 3′-nucleotide. Similarly, siRNAs derived from the dsRNA transgene in N.benthamiana (Figure 3) and cassava (Supplementary Figure S1) did not change their apparent mobility following the chemical treatment.
The 3′-nucleotide of begomoviral siRNAs in Arabidopsis undergoes HEN1-dependent methylation
The β-elimination test does not distinguish between the 3′-nucleotide methylation and other forms of modification at 3′- or 2′-OH group of the 3′-terminal nucleotide ribose. The activity of the miRNA methylase HEN1 is abolished in the Arabidopsis ecotype La-er mutant hen1-1 (32). To find out if begomoviral siRNAs are methylated at the 3′-nucleotide, we compared the sensitivity to β-elimination of CaLCuV siRNAs produced in wild-type La-er and hen1-1, which are susceptible to CaLCuV infection. Four weeks post-inoculation, CaLCuV-infected wild-type plants accumulated 21, 22 and 24 nt viral siRNAs of both polarities, with the 24 nt species being a predominant size-class (see La-er and Col-0 in Figures 4 and 5). In hen1-1 plants, accumulation of the three major classes of CaLCuV siRNAs was significantly reduced and, in addition, the appearance of new viral RNAs of 23 and 25 nt was observed (Figure 4). The CaLCuV siRNAs isolated from both the wild-type and hen1-1 plants exhibited the characteristic gel-shift following phosphatase treatment (data not shown), indicating that their 5′ ends are phosphorylated. Further comparison indicated that whereas CaLCuV siRNAs from wild-type plants are resistant to β-elimination, CaLCuV siRNAs from hen1-1 plants are sensitive to β-elimination and show the expected ∼2 nt increase in apparent mobility (Figure 4). Taken together, these results show that the 3′-terminal nucleotide of CaLCuV siRNAs is methylated at the 2′- or the 3′-OH group of ribose by HEN1 in Arabidopsis. Similar results were obtained for all types and size-classes of Arabidopsis endogenous sRNAs tested (Figure 4), including the 22 nt miRNA 173 (miR173), the 21 nt ta-siRNA 255 (siR255) and the 24 nt ra-siRNA 1003 (siR1003) (Figure 4). The ∼21 nt siRNAs associated with spontaneous PTGS of a GFP transgene (50) were also resistant to β-elimination in wild-type plants (Figure 4). Moreover, accumulation of both viral and non-viral sRNAs was reduced in hen1-1. Our results confirm earlier findings for miRNAs and show that methylation is likely to be a general feature of sRNA biogenesis in Arabidopsis. The reduced accumulation of sRNAs in hen1-1 is also consistent with the hypothesis that methylation helps stabilize sRNAs. Unmethylated sRNAs are susceptible to uridylation (34), which has been proposed to target the RNAs for degradation (51). We found faint bands of (apparent) 23 and 25 nt CaLCuV siRNAs in hen1-1 plants (Figure 4) that might arise by uridylation of the unmethylated viral siRNAs.
Figure 5.
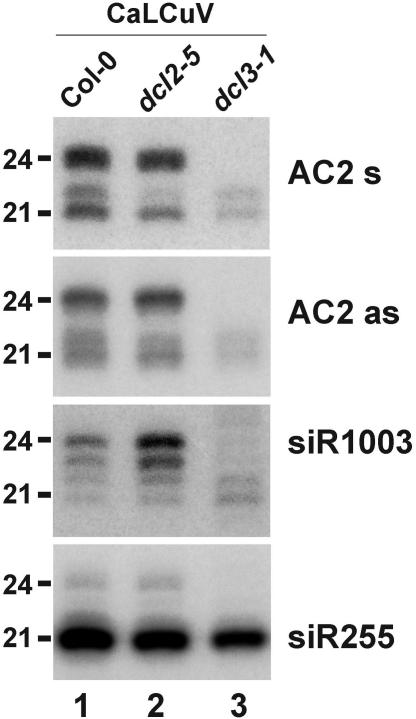
DCL2 and DCL3 produce distinct size-classes of CaLCuV siRNAs in Arabidopsis RNA gel blot analysis of 20 µg total RNA prepared from the geminivirus CaLCuV-infected wild-type (La-er and Col-0) and DCL2 and DCL3 loss-of-function mutant (dcl2-5 and dcl3-1) Arabidopsis plants. The blots were successively probed with 32P-labeled DNAs specific to CaLCuV (‘AC2 as’ and ‘AC2 s’, Table 1), or complementary to endogenous ra-siRNA (siR1003) and ta-siRNA (siR255). Positions of the 21 and 24 nt RNAs are indicated.
Begomoviral siRNAs differ from those derived from a cytoplasmic RNA virus
To find out if methylation of viral siRNAs is a special feature of DNA viruses with nuclear and cytoplasmic phases in their replication, we extended our study to Oilseed rape mosaic tobamovirus (ORMV) (46), which is an RNA virus that replicates in the cytoplasm. RNA blot hybridization using a mixture of 27 nt DNA probes derived from the ORMV 3′-region (see Table 1) detected ∼19, 20 and 21 nt ORMV siRNAs in Arabidopsis and ∼20, 21 and 22 nt siRNAs in N.benthamiana. Unexpectedly, ∼50% of each type of siRNA in Arabidopsis and N.benthamiana were resistant to β-elimination (Figure 4A and B). Although miR173, siR255, and siR1003 are fully resistant to β-elimination in wild-type plants, in ORMV-infected Arabidopsis a significant fraction of miR173 and siR255 but not siR1003 became sensitive to β-elimination. Moreover, in the case of miR173, extra bands are clearly visible, which resemble those accumulating in hen1-1, and are fully sensitive to β-elimination (Figure 4A). These results suggest that ORMV infection might suppress or, less likely, reverse HEN1-mediated modification of the viral siRNAs and a subclass of endogenous sRNAs. The fact that the 5S rDNA-derived ra-siRNA siR1003, which most likely functions in a RITS-like complex in the nucleus, is not affected suggests that the observed phenomenon takes place in the cytoplasm and would involve RISC-associated miRNAs and ta-siRNAs as well as ORMV siRNAs. The tobamoviral factor responsible for this effect remains to be identified. An attractive candidate would be a replication protein that has been shown to suppress PTGS in N.benthamiana (9).
Recently, it has been reported that the bulk of siRNAs derived from the cytoplasmic RNA virus, cucumber mosaic cucumovirus (CMV) and its Y-satellite RNA in N.benthamiana is resistant to β-elimination. But in the presence of the potyviral silencing suppressor HC-Pro, expressed from a transgene, this resistance was partially alleviated (33). These data support our hypothesis that a viral silencing suppressor can potentially interfere at the stage of sRNA modification. However, it remains to be seen whether the effect of HC-Pro is relevant in a context of the potyvirus infection. It is also obvious that not every viral silencing suppressor acts in a similar manner, because neither CMV [as could be deduced from ref. (33)], nor geminivirses (this work) are able to suppress or reverse the 3′-modification of sRNAs during the course of infection.
Begomoviral siRNAs are generated by more than two RNA silencing pathways
The accumulation of three distinct size-classes of begomoviral siRNAs in infected plants suggests that more than one DCL might be involved in the biogenesis of these RNAs. The candidates we considered were DCL3, which is required for production of 24 nt ra-siRNAs associated with TGS of endogenous repetitive DNA, and DCL2, which has been implicated in the biogenesis of siRNAs derived from the RNA virus TCV (12). We inoculated two presumed loss-of-function T-DNA insertion mutants of Arabidopsis, dcl2-5 and dcl3-1, with CaLCuV and 4 weeks later, small RNA fractions were analyzed by RNA blot hybridization. As controls, the blots were also hybridized with probes for 24 nt ra-siRNA siR1003 and for the 21 nt ta-siRNA siR255. The results show that accumulation of 24 nt CaLCuV siRNAs was blocked in dcl3-1, and accumulation of 22 nt CaLCuV siRNAs was reduced in dcl2-5 (Figure 5). These results lead us to conclude that DCL3 is required for production of the 24 nt begomoviral siRNAs and DCL2 is required for production of a substantial fraction of the 22 nt begomoviral siRNAs, whereas production of the 21 nt and the remaining 22 nt viral siRNAs requires other DCLs or combinations of DCLs. Production of 21 nt ta-siRNAs, which depends on DCL4 (21,26), was not affected in dcl2-5 or dcl3-1 (Figure 5), raising the possibility that 21 nt CaLCuV siRNAs also depend on DCL4. The DCL2-independent fraction of 22 nt viral siRNA might be produced by DCL1 that can generate both 21 and 22 nt (e.g. miR173) miRNAs.
Interestingly, we detected, in addition to the known 24 nt class, 23, 22 and 21 nt siR1003 sRNAs (Figure 5). Comparison of the accumulation of these ra-siRNAs in dcl2-5 and dcl3-1 suggest that production of both the 24 and 23 nt classes depends on DCL3, whereas production of the 21 and 22 nt classes is not dependent on either DCL2 or DCL3. Thus, siRNAs derived from repetitive DNA of both the multiple viral episomes and the plant chromosomes are produced by more than one DCL.
Taken together, we conclude that at least two RNA silencing pathways interact with begomoviruses. One pathway involves the production of the 24 nt siRNAs and depends on DCL3. Another pathway involves the production of the 22 nt siRNAs and depends on DCL2, as well as other as yet unidentified DCL activities. The fact that DCL2 is associated with the silencing of some RNA viruses, whereas DCL3 is associated with TGS at repetitive DNA loci suggests to us that targeting of begomoviruses involves both post-transcriptional and transcriptional silencing mechanisms. Our work in progress shows that mutations in individual DCL or HEN1 genes do not increase plant susceptibility to DNA viruses (T. Blevins, R. Rajeswaran, D. Beknazariants, P. V. Shivaprasad, A. Si-Ammour, H.-S. Park, D. Robertson, F. Meins, T. Hohn and M. M. Pooggin, manuscript in preparation). This suggests that the silencing pathways we propose are either redundant or can be effectively evaded or suppressed by the virus. The fact that begomoviruses encode at least two distinct types of silencing suppressors (52) that might function in the nucleus (39) and the cytoplasm (16,53) would support this hypothesis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
The authors thank Dominique Robertson and Sondra Lazarowitz for providing infectious clones of CaLCuV, John Stanley and Claude Fauquet for ACMV clones, and Xuemei Chen for hen1-1 seeds. The authors thank Johannes Fütterer for helpful advices and critical reading of the manuscript. The authors are grateful to Afzal Dogar for generating transgenic N.benthamiana and to Monika Fasler for excellent technical assistance. The authors thank Christian Körner and Thomas Boller for providing the laboratory space and infrastructure at the Institute of Botany. This work was supported by the European Union (EU) Framework V grant ‘Virus Induced Gene Silencing’ (G2-CT-2002-01637), a short-term EMBO fellowship to R.A., and in part by grants to the Gruissem group from the Swiss Centre for International Agriculture (ZIL) and the Eiselen-Foundation-Ulm, and to the Meins group from the Swiss Office for Education and Science as part of the EU Gene Silencing in Transgenic Plants Projects, and by the Novartis Research Foundation. Funding to pay the Open Access publication charges for this article was provided by Swiss National Science Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Meins F., Jr, Si-Ammour A., Blevins T. RNA silencing systems and their relevance to plant development. Annu. Rev. Cell Dev. Biol. 2005;21:297–318. doi: 10.1146/annurev.cellbio.21.122303.114706. [DOI] [PubMed] [Google Scholar]
- 3.Baulcombe D. RNA silencing. Trends Biochem. Sci. 2005;30:290–293. doi: 10.1016/j.tibs.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 4.Baumberger N., Baulcombe D.C. Arabidopsis ARGONAUTE1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. Proc. Natl Acad. Sci. USA. 2005;102:11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zilberman D., Cao X., Johansen L.K., Xie Z., Carrington J.C., Jacobsen S.E. Role of Arabidopsis ARGONAUTE4 in RNA-directed DNA methylation triggered by inverted repeats. Curr. Biol. 2004;14:1214–1220. doi: 10.1016/j.cub.2004.06.055. [DOI] [PubMed] [Google Scholar]
- 6.Voinnet O. Induction and suppression of RNA silencing: insights from viral infections. Nature Rev. Genet. 2005;6:206–220. doi: 10.1038/nrg1555. [DOI] [PubMed] [Google Scholar]
- 7.Hamilton A.J., Baulcombe D.C. A species of small antisense RNA in post-transcriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 8.Szittya G., Molnar A., Silhavy D., Hornyik C., Burgyan J. Short defective interfering RNAs of tombusviruses are not targeted but trigger post-transcriptional gene silencing against their helper virus. Plant Cell. 2002;14:359-372. doi: 10.1105/tpc.010366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kubota K., Tsuda S., Tamai A., Meshi T. Tomato mosaic virus replication protein suppresses virus-targeted posttranscriptional gene silencing. J. Virol. 2003;77:11016–11026. doi: 10.1128/JVI.77.20.11016-11026.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie Z., Johansen L.K., Gustafson A.M., Kasschau K.D., Lellis A.D., Zilberman D., Jacobsen S.E., Carrington J.C. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Molnar A., Csorba T., Lakatos L., Varallyay E., Lacomme C., Burgyan J. Plant virus-derived small interfering RNAs originate predominantly from highly structured single-stranded viral RNAs. J. Virol. 2005;79:7812–7818. doi: 10.1128/JVI.79.12.7812-7818.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lucioli A., Noris E., Brunetti A., Tavazza R., Ruzza V., Castillo A.G., Bejarano E.R., Accotto G.P., Tavazza M. Tomato yellow leaf curl Sardinia virus rep-derived resistance to homologous and heterologous geminiviruses occurs by different mechanisms and is overcome if virus-mediated transgene silencing is activated. J. Virol. 2003;77:6785–6798. doi: 10.1128/JVI.77.12.6785-6798.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chellappan P., Vanitharani R., Fauquet C.M. Short interfering RNA accumulation correlates with host recovery in DNA virus-infected hosts, and gene silencing targets specific viral sequences. J. Virol. 2004;78:7465–7477. doi: 10.1128/JVI.78.14.7465-7477.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang P., Vanderschuren H., Fütterer J., Gruissem W. Resistance to cassava mosaic disease in transgenic cassava expressing antisense RNAs targeting virus replication genes. Plant Biotech. J. 2005;3:385–397. doi: 10.1111/j.1467-7652.2005.00132.x. [DOI] [PubMed] [Google Scholar]
- 15.Lakatos L., Szittya G., Silhavy D., Burgyan J. Molecular mechanism of RNA silencing suppression mediated by p19 protein of tombusviruses. EMBO J. 2004;23:876–884. doi: 10.1038/sj.emboj.7600096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chellappan P., Vanitharani R., Fauquet C.M. MicroRNA-binding viral protein interferes with Arabidopsis development. Proc. Natl Acad. Sci. USA. 2005;102:10381–10386. doi: 10.1073/pnas.0504439102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lecellier C.H., Dunoyer P., Arar K., Lehmann-Che J., Eyquem S., Himber C., Saib A., Voinnet O. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- 18.Elbashir S.M., Lendeckel W., Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang G., Reinhart B.J., Bartel D.P., Zamore P.D. A biochemical framework for RNA silencing in plants. Genes Dev. 2003;17:49–63. doi: 10.1101/gad.1048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schauer S.E., Jacobsen S.E., Meinke D.W., Ray A. DICER-LIKE1: blind men and elephants in Arabidopsis development. Trends Plant Sci. 2002;7:487–491. doi: 10.1016/s1360-1385(02)02355-5. [DOI] [PubMed] [Google Scholar]
- 21.Gasciolli V., Mallory A.C., Bartel D.P., Vaucheret H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 2005;15:1494–1500. doi: 10.1016/j.cub.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 22.Kurihara Y., Watanabe Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl Acad. Sci. USA. 2004;101:12753–12758. doi: 10.1073/pnas.0403115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herr A.J., Jensen M.B., Dalmay T., Baulcombe D.C. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308:118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- 24.Vazquez F., Vaucheret H., Rajagopalan R., Lepers C., Gasciolli V., Mallory A.C., Hilbert J.L., Bartel D.P., Crete P. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell. 2004;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 25.Allen E., Xie Z., Gustafson A.M., Carrington J.C. MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 26.Xie Z., Allen E., Wilken A., Carrington J.C. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 2005;102:12984–1289. doi: 10.1073/pnas.0506426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han M.H., Goud S., Song L., Fedoroff N. The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc. Natl Acad. Sci. USA. 2004;101:1093–1098. doi: 10.1073/pnas.0307969100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vazquez F., Gasciolli V., Crete P., Vaucheret H. The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr. Biol. 2004;14:346–351. doi: 10.1016/j.cub.2004.01.035. [DOI] [PubMed] [Google Scholar]
- 29.Hiraguri A., Itoh R., Kondo N., Nomura Y., Aizawa D., Murai Y., Koiwa H., Seki M., Shinozaki K., Fukuhara T. Specific interactions between Dicer-like proteins and HYL1/DRB-family dsRNA-binding proteins in Arabidopsis thaliana. Plant Mol. Biol. 2005;57:173–188. doi: 10.1007/s11103-004-6853-5. [DOI] [PubMed] [Google Scholar]
- 30.Park W., Li J., Song R., Messing J., Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boutet S., Vazquez F., Liu J., Beclin C., Fagard M., Gratias A., Morel J.B., Crete P., Chen X., Vaucheret H. Arabidopsis HEN1: a genetic link between endogenous miRNA controlling development and siRNA controlling transgene silencing and virus resistance. Curr. Biol. 2003;13:843–848. doi: 10.1016/s0960-9822(03)00293-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu B., Yang Z., Li J., Minakhina S., Yang M., Padgett R.W., Steward R., Chen X. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ebhardt H.A., Thi E.P., Wang M.B., Unrau P.J. Extensive 3′ modification of plant small RNAs is modulated by helper component-proteinase expression. Proc. Natl Acad. Sci. USA. 2005;102:13398–13403. doi: 10.1073/pnas.0506597102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J., Yang Z., Yu B., Liu J., Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr. Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rybicki E.P., Briddon R.W., Brown J.K., Fauquet C.M., Maxwell D.P., Harrison B.D., Markham P.G., Bisaro D.M., Robinson D., Stanley J. Family Geminiviridae. In: van Regenmortel M.H.V., Fauquet C.M., Bishop D.H.L., et al., editors. Virus Taxonomy: Classification and Nomenclature of Viruses. NY: Academic Press; 2000. pp. 285–297. [Google Scholar]
- 36.Pilartz M., Jeske H. Mapping of abutilon mosaic geminivirus minichromosomes. J. Virol. 2003;77:10808–10818. doi: 10.1128/JVI.77.20.10808-10818.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanley-Bowdoin L., Settlage S.B., Orozco B.M., Nagar S., Robertson D. Geminiviruses: models for plant DNA replication, transcription, and cell cycle regulation. Crit. Rev. Plant Sci. 1999;18:71–106. [PubMed] [Google Scholar]
- 38.Shivaprasad P.V., Akbergenov R., Trinks D., Rajeswaran R., Veluthambi K., Hohn T., Pooggin M.M. Promoters, transcripts, and regulatory proteins of Mungbean yellow mosaic geminivirus. J. Virol. 2005;79:8149–8163. doi: 10.1128/JVI.79.13.8149-8163.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trinks D., Rajeswaran R., Shivaprasad P.V., Akbergenov R., Oakeley E.J., Veluthambi K., Hohn T., Pooggin M.M. Suppression of RNA silencing by a geminivirus nuclear protein, AC2, correlates with transactivation of host genes. J. Virol. 2005;79:2517–2527. doi: 10.1128/JVI.79.4.2517-2527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stanley J., Gay M.R. Nucleotide sequence of cassava latent virus DNA. Nature. 1983;301:260–262. [Google Scholar]
- 41.Hill J.E., Strandberg J.O., Hiebert E., Lazarowitz S.G. Asymmetric infectivity of pseudorecombinants of cabbage leaf curl virus and squash leaf curl virus: implications for bipartite geminivirus evolution and movement. Virology. 1998;250:283–292. doi: 10.1006/viro.1998.9366. [DOI] [PubMed] [Google Scholar]
- 42.Fondong V.N., Pita J.S., Rey M.E., de,Kochko A., Beachy R.N., Fauquet C.M. Evidence of synergism between African cassava mosaic virus and a new double-recombinant geminivirus infecting cassava in Cameroon. J. Gen. Virol. 2000;81:287–297. doi: 10.1099/0022-1317-81-1-287. [DOI] [PubMed] [Google Scholar]
- 43.Liu S.J., Bedford I.D., Briddon R.W., Markham P.G. Efficient whitefly transmission of African cassava mosaic geminivirus requires sequences from both genomic components. J. Gen. Virol. 1997;78:1791–1794. doi: 10.1099/0022-1317-78-7-1791. [DOI] [PubMed] [Google Scholar]
- 44.Zhang P., Legris G., Coulin P., Puonti-Kaerlas J. Production of stably transformed cassava plants via particle bombardment. Plant Cell Rep. 2000;19:939–945. doi: 10.1007/s002990000224. [DOI] [PubMed] [Google Scholar]
- 45.Turnage M.A., Muangsan N., Peele C.G., Robertson D. Geminivirus-based vectors for gene silencing in Arabidopsis. Plant J. 2002;30:107–114. doi: 10.1046/j.1365-313x.2002.01261.x. [DOI] [PubMed] [Google Scholar]
- 46.Aguilar I., Sanchez F., Martin,Martin A., Martinez-Herrera D., Ponz F. Nucleotide sequence of Chinese rape mosaic virus (oilseed rape mosaic virus), a crucifer tobamovirus infectious on Arabidopsis thaliana. Plant Mol. Biol. 1996;30:191–197. doi: 10.1007/BF00017814. [DOI] [PubMed] [Google Scholar]
- 47.Pooggin M., Shivaprasad P.V., Veluthambi K., Hohn T. RNAi targeting of DNA virus in plants. Nat. Biotechnol. 2003;21:131–132. doi: 10.1038/nbt0203-131b. [DOI] [PubMed] [Google Scholar]
- 48.Goodall G.J., Filipowicz W. The AU-rich sequences present in the introns of plant nuclear pre-mRNAs are required for splicing. Cell. 1989;58:473–483. doi: 10.1016/0092-8674(89)90428-5. [DOI] [PubMed] [Google Scholar]
- 49.Zhang P., Potrykus I., Puonti-Kaerlas J. Efficient production of transgenic cassava using negative and positive selection. Transgenic Res. 2000;9:405–415. doi: 10.1023/a:1026509017142. [DOI] [PubMed] [Google Scholar]
- 50.Glazov E., Phillips K., Budziszewski G.J., Schob H., Meins F., Jr, Levin J.Z. A gene encoding an RNase D exonuclease-like protein is required for post-transcriptional silencing in Arabidopsis. Plant J. 2003;35:342–349. doi: 10.1046/j.1365-313x.2003.01810.x. [DOI] [PubMed] [Google Scholar]
- 51.Shen B., Goodman H.M. Uridine addition after microRNA-directed cleavage. Science. 2004;306:997. doi: 10.1126/science.1103521. [DOI] [PubMed] [Google Scholar]
- 52.Vanitharani R., Chellappan P., Pita J.S., Fauquet C.M. Differential roles of AC2 and AC4 of cassava geminiviruses in mediating synergism and suppression of posttranscriptional gene silencing. J. Virol. 2004;78:9487–9498. doi: 10.1128/JVI.78.17.9487-9498.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang H., Buckley K.J., Yang X., Buchmann R.C., Bisaro D.M. Adenosine kinase inhibition and suppression of RNA silencing by geminivirus AL2 and L2 proteins. J. Virol. 2005;79:7410–7418. doi: 10.1128/JVI.79.12.7410-7418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.