Abstract
We report on the identification of Fes1p (yBR101cp) as a cytosolic homologue of Sls1p, an endoplasmic reticulum (ER) protein previously shown to act as a nucleotide exchange factor for yeast BiP (M. Kabani, J.-M. Beckerich, and C. Gaillardin, Mol. Cell. Biol. 20:6923-6934, 2000). We found that Fes1p associates preferentially to the ADP-bound form of the cytosolic Hsp70 molecular chaperone Ssa1p and promotes nucleotide release. Fes1p activity was shown to be compartment and species specific since Sls1p and Escherichia coli GrpE could not substitute for Fes1p. Surprisingly, whereas Sls1p stimulated the ATPase activity of BiP in cooperation with luminal J proteins, Fes1p was shown to inhibit the Ydj1p-mediated activation of Ssa1p ATPase activity in steady-state and single-turnover assays. Disruption of FES1 in several wild-type backgrounds conferred a strong thermosensitive phenotype but partially rescued ydj1-151 thermosensitivity. The Δfes1 strain was proficient for posttranslational protein translocation, as well as for the ER-associated degradation of two substrates. However, the Δfes1 mutant showed increased cycloheximide sensitivity and a general translational defect, suggesting that Fes1p acts during protein translation, a process in which Ssa1p and Ydj1p are known to be involved. In support of this hypothesis, Fes1p was found to be associated with ribosomes.
The 70-kDa heat shock proteins (Hsp70s) are a ubiquitous family of molecular chaperones found in all organisms and cellular compartments, and they are known to facilitate protein synthesis, folding, translocation, and degradation (2, 15, 22). This functional diversity relies on the intrinsic properties of these proteins as well as on their interaction with specific regulatory cofactors. Hsp70s are composed of a highly conserved N-terminal 44-kDa ATPase domain, an 18-kDa peptide-binding domain, and a C-terminal 10-kDa variable domain. In the ATP-bound state, Hsp70s display fast on and off rates of peptide binding, whereas in the ADP-bound state these constants are slowed (43, 57). The modulation of the intrinsic affinity for peptides is attributed to a cross talk between the ATPase domain and the C-terminal domain. Indeed, ATP hydrolysis triggers a conformational change in the C-terminal domain that is predicted to form a lid over the peptide-binding pocket and stabilizes the interaction with the substrate (74).
The weak ATPase activity of Hsp70s is stimulated by members of the DnaJ family (also known as Hsp40s), which share a common 70-amino-acid J domain required for binding and activation of Hsp70s (34). In Escherichia coli, the ATPase activity of the Hsp70 homologue DnaK is stimulated by the DnaJ chaperone which results in stable binding with polypeptide substrates (23, 61, 62). In addition, upon binding to the ATPase domain, the GrpE protein acts as a nucleotide exchange factor that promotes ADP release (9, 27, 42). Subsequently, ATP rebinding catalyzes the dissociation of the DnaK-substrate complex. Cycles of association and dissociation prevent aggregation of the polypeptide substrate and facilitate folding. Whereas members of the DnaJ family are found in all organisms and in all cellular compartments (34), homologues of GrpE are found only in prokaryotes and archea, or in organelles of prokaryotic origin such as mitochondria and chloroplasts. However, the antiapoptotic Bcl2-associated athanogene 1 (Bag-1) protein binds to Hsp70 and appears to be a nucleotide exchange factor for mammalian cytosolic Hsp70s and Hsc70s (9, 10, 29, 59, 64). Whereas GrpE exchanges both ATP and ADP from DnaK, Bag-1 promotes only ADP dissociation from Hsc70 (9), but the importance of the nucleotide exchange activity of Bag-1 in vivo is still subject to controversy (64). Moreover, Bag-1 was shown to either negatively or positively regulate Hsp70 and Hsc70 refolding activity depending on Bag-1:Hsp70 stoichiometry (24). Finally, there exist other cofactors that modulate the activity of Hsp70s chaperones in a manner different from that of J proteins or nucleotide exchange factors (4, 28, 30, 54). Overall, these data suggest that the regulation of eukaryotic Hsp70s is more complex than for bacterial DnaK homologues.
At least 10 members of the Hsp70 family are found in the yeast Saccharomyces cerevisiae (22). Ssa1p is the most extensively studied cytosolic Hsp70, as it plays essential functions in protein folding and translocation across the endoplasmic reticulum (ER) and mitochondrial membranes in combination with its J-domain partner Ydj1p and facilitates ER-associated degradation (ERAD) (21). Recently, a function for Ssa1p in translation initiation has been described (31). Ssa1p was found to be associated with translating ribosomes, and it associates with Pab1p, the poly(A) binding protein, and Sis1p, a DnaJ homologue. Depletion of Ssa1p results in a general translational defect as well as changes in the polysome profile (31). Moreover, efficient translation of heterologous proteins such as green fluorescent protein (GFP) or firefly luciferase depends upon functional Ydj1p (13), suggesting an involvement of the Ssa1p-Ydj1p pair in this vital cellular process. The Ssb1p and Ssb2p Hsp70s are also found to be associated with ribosomes, play a role in the elongation step of protein translation (49, 52), and interact with the Hsp40 homologues Sis1p and zuotin to facilitate translation (70, 73).
Another well-studied Hsp70 in yeast is Kar2p, the ER luminal homologue of mammalian BiP (26). Kar2p is also involved in protein translocation across the ER membrane, protein folding, and ERAD (21). Although three luminal J proteins mediate the recruitment and the regulation of Kar2p for specific functions (50), a GrpE or Bag-1 analogue was unknown until we showed that the ER localized protein Sls1p (yOL031cp; also called Sil1p [66] or Per100p [65]), identified as a homologue of a protein required for efficient cotranslational protein translocation in the yeast Yarrowia lipolytica (6), binds to the ATPase domain of the luminal Hsp70 BiP (Kar2p) and catalyzes nucleotide exchange (7, 32, 33). Sls1p is also involved in protein translocation and ERAD (32, 65, 66).
Because a nucleotide exchange factor for the yeast cytosolic Hsp70s had not been defined, we screened the yeast genome database for homologues of Sls1p and report here on Fes1p, the product of the yBR101c open reading frame (ORF), as a likely candidate. Fes1p binds to Ssa1p in an ATP-dependent manner and promotes nucleotide exchange. The FES1 gene is required for growth at high temperature, but its disruption suppresses the ydj1-151 thermosensitive phenotype, suggesting that Fes1p acts antagonistically to Ydj1p. Several assays excluded a function of Fes1p in protein translocation, ERAD, or folding, but instead suggest that this protein may be involved in protein translation.
MATERIALS AND METHODS
Strains and media.
E. coli strains used were DH5α (endA1 hsdR17 supE44 thi-1 recA1 gyrA relA1 Δ[lacZYA-argF] U169 deoR [φ80 dlacΔ lacZ M15]) and BL21 (F− ompT hsdS [rB− mB−] gal). Bacteria were grown in Luria-Bertani (LB) medium supplemented with ampicillin (50 μg/ml) for plasmid selection (3). Yeast strains used in this study are described in Table 1 and were grown on yeast extract-peptone-dextrose (YPD) medium or YNB-N0 lacking amino acids (Difco) but supplemented with the appropriate nutrients (1).
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| RSY801 | MATaade2-101 leu2-3,113 ura3-52 | 56 |
| W3031b | MATα ade2-101 his3 leu2-3,113 ura3-52 trp1 can1-100 | 16 |
| ydj1-151 (ACY17b) | W3031b ydj1-2::HIS3 LEU2::ydj1-151 | 16 |
| RSY801, Δfes1 | RSY801 fes1::KANMX4 | This study |
| W3031b, Δfes1 | W3031b fes1::KANMX4 | This study |
| ydj1-151, Δfes1 | ydj1-151 fes1::KANMX4 | This study |
| W3031b, GFP-FES1 | W3031b HIS3MX6-GAL1-GFP-FES1 | This study |
DNA manipulation techniques.
Standard techniques were used (3), and restriction enzymes were used according to the manufacturer's instructions. Ready-To-Go PCR beads (Pharmacia Biotech) or Taq DNA polymerase (Gibco BRL) and Crocodile III Thermocycler (Appligene Oncor) were used for PCRs.
To overexpress Fes1p for protein purification (see below), the yBR101c ORF was amplified by PCR of genomic DNA. The oligonucleotides used were MK1 (5′-CGCGGATCCGTGAAAAGCTATTACAGTGGT-3′) and MK2 (5′-CGCGGATCCTAATACATACTTTACGGC-3′), which allowed cloning into pBluescript SK− vector (Stratagene) via the underlined BamHI sites. The cloned PCR product was verified by DNA sequence analysis as described previously (33). The plasmid for glutathione S-transferase (GST)-Fes1p fusion protein expression was obtained by in-frame cloning of the yBR101c ORF into the BamHI site of pGEX-5X-1 (Amersham Pharmacia Biotech).
The GST-Fes1p coding sequence was amplified by PCR with pGEX-5X-1-yBR101c as a template. The oligonucleotides used were MK3 (5′-GCGCGCACTAGTATGTCCCCTATACTAGGTTATTGG-3′) and MK4 (5′-CCCATCGATTCATAATACATACTTTACGGCTAAATAATC-3′), which allowed cloning into pGPD425 (47) via the underlined SpeI and ClaI sites, respectively. This construct was transformed into the strains from which yBR101c had been deleted (see below) and fully complemented the thermosensitive phenotype of these mutants (see Results).
A yBR101c disruption cassette was obtained by amplifying pRS400 (8) with primers MK5 (5′-AACGAAAGAGTAAAATAGAAAAAAAAACACATACATAACTCTGTGCGGTATTTCACACCG-3′) and MK6 (5′-AAATGGTGAATGTAATATCATTTTATTTCTACGGACGTAAAGATTGTACTGAGAGTGCAC-3′), which contain a 40-bp sequence homologous to the promoter and terminator portions of yBR101c, respectively, along with a 20-bp region of homology with the KanMX4 cassette in pRS400. Disrupted mutants were then obtained by transforming the resulting PCR fragment into yeast and selecting for G418-resistant colonies at a final concentration of 200 μg/ml. Disruption of the yBR101c ORF was confirmed in resistant colonies by using primers MK7 (5′-CTCATCGTCAGTCAGAAAGC-3′) and MK8 (5′-ATAAATGTTCGAGATTCCGG-3′), which flank the yBR101c ORF. In order to exclude possible mutations arising from the transformation procedure, the phenotype (see Results) of the yBR101c disruption in each strain was analyzed in three independent clones.
Localization of a GFP-tagged Fes1p.
A GFP-tagged version of Fes1p was ob tained by using the method described by Longtine et al. (40). A HIS3MX6-GAL1-GFP cassette was obtained by PCR amplification using oligonucleotides JMB59 (5′- CCCTTGAATTCGCAATAGACCACTGTAATAGCTTTTCTTTGTATAGTTCATCCATGC-3′) and JMB68 (5′-GGTAAGCTTTTCAGCTACTGGTTGGTCAAGTTGGGCCCTATTAAGGCGAGCTCGTTTAAACTGGATGG-3′). The resulting cassette bears regions of homology to yBR101c (underlined) that allow integration into the yeast genome by double recombination. The W3031b yeast strain was transformed with this construct, and integrants expressing GFP-Fes1p under the control of the GAL1 promoter were selected on minimal medium lacking histidine and checked by PCR and Southern blot analysis. This strain was grown on raffinose-containing medium and then transferred into galactose-containing medium for 8 h, and the cells were observed with an Olympus BX 51 fluorescence microscope.
Cellular fractionation was performed using 20 A254 U of the lysate prepared for ribosome fractionation (see below). The lysate volume was brought up to 1 ml with polysome lysis buffer (PLB; see below) and centrifuged at 16,000 × g at 4°C for 15 min. The pellet was resuspended in 500 μl of PLB, and half of the supernatant was centrifuged at 150,000 × g at 4°C for 1 h. The pellet was resuspended in 500 μl of PLB. A 10-μl aliquot of each sample was resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by Western blotting using anti-GFP (a gift from S. Subramani, University of California, San Diego), anti-Sec61p (60), and anti-Sse1p antibodies (a gift from J. Goeckeler, University of Pittsburgh). Western blots were developed using chemiluminescence and ECL reagents (Pierce) according to the manufacturer's instructions.
Ribosome fractionation.
The yeast strains were grown in 200 to 400 ml of YPD (RSY801 and RSY801; Δfes1) or YP-galactose (W303; GFP-FES1) at 30°C to log phase (A600 = ∼1) and, where indicated, the cells were shifted for 30 min at 37°C. Cycloheximide was added at a final concentration of 50 μg/ml, and cells were pelleted and washed with ice-cold PLB (20 mM HEPES [pH 7.4], 100 mM NaCl, 20 mM MgCl2, 100 μg of cycloheximide/ml) containing 0.2 μl of diethylpyrocarbonate (Sigma) per ml. The cells were resuspended in 2 ml of the same buffer, and lysates were prepared by beating the cells with glass beads eight times for 15 s, with a 30-s period of cooling on ice between each vortexing step. The lysate was cleared by two centrifugation steps, each at 5,000 rpm for 5 min in a microcentrifuge at 4°C. A fraction of the supernatant corresponding to 50 to 100 A254 U was layered over a 30-ml linear sucrose gradient (7 to 47%, wt/vol) prepared in PLB. The sucrose gradient was centrifuged at 27,000 rpm for 4.5 h at 4°C in a Beckman SW28.1 rotor. Gradients were collected with continuous monitoring at 254 nm.
To determine the association of GFP-Fes1p and Ssa1p on ribosomes and polysomes, fractions representing the load, the cytosol (top of the gradient), fractions after the cytosol but before ribosome-containing fractions, the 80S ribosomes, and polysomes were collected and proteins were resolved by SDS-PAGE and analyzed by Western blotting using anti-GFP, anti-Ssa1p (a gift from C. Koehler, University of California, Los Angeles), and anti-L3 (a gift from J. Warner, Albert Einstein College of Medicine, New York, N.Y.) antisera.
Protein purification.
The GST-Fes1p protein was purified from E. coli strain BL21 transformed with the expression plasmid described above. A 50-ml culture in LB containing 50 μg of ampicillin per ml was grown overnight at 26°C and diluted into 2 liters of the same medium. After 2 h at 28°C, IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 0.5 mM, and cells were grown for an additional 4 h. Cells were harvested and washed once in water and once in phosphate-buffered saline (pH 7.4)-2 mM EDTA before the cell pellet was snap-frozen in liquid nitrogen and stored at −70°C. The cell pellet was thawed and resuspended in ∼20 ml of buffer A (phosphate-buffered saline [pH 7.4], 5 mM EDTA, 1 mM β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride, 1 μg of leupeptin per ml, 1 μg of pepstatin A per ml), and the cells were disrupted by sonication for 30 s six times at the highest setting with 1 min on ice between sonications. The broken cells were centrifuged at 13,000 rpm in a Sorvall SA600 rotor for 15 min, and the resulting supernatant was further centrifuged at 20,000 rpm in a Sorvall SA600 rotor for 20 min to obtain a cleared lysate. This lysate was loaded onto a 2-ml glutathione Sepharose 4B column (Pharmacia Biotech) equilibrated in buffer A containing 1% Triton X-100. The column was washed sequentially with 30 ml of the following: (i) buffer A containing 5% glycerol; (ii) buffer A containing 1 M NaCl and 5% glycerol; (iii) 50 mM Tris-Cl (pH 7.5), 10 mM magnesium acetate, 200 mM potassium acetate, 5 mM ATP, 5% glycerol; and (iv) buffer A containing 5% glycerol. GST-Fes1p was eluted with 10 ml of elution buffer (50 mM Tris-Cl [pH 8.0], 50 mM NaCl, 20 mM reduced glutathione, 5% glycerol), and 1-ml fractions were collected and analyzed by SDS-PAGE and Coomassie brilliant blue staining. Peak fractions were pooled, dialyzed against dialysis buffer (10 mM HEPES [pH 7.0], 50 mM NaCl, 10 mM dithiothreitol [DTT], 10% glycerol), and frozen in small aliquots in liquid nitrogen and stored at −70°C. GST-Fes1p was purified to near homogeneity as assessed by SDS-PAGE and Coomassie brilliant blue staining.
GST-Sls1p and Kar2p were purified as described before (32, 45), and purified Ssa1p and Ydj1p, prepared as published (63), were a kind gift from S. W. Fewell (University of Pittsburgh). Purified DnaK and GrpE proteins were obtained from StressGen.
Protein interaction assays.
GST pull-down assays were performed essentially as described in reference 32, with minor modifications. A total of 4 μg of GST-Fes1p or GST-Sls1p was incubated with 2 μg of either Ssa1p or Kar2p in the presence of 1 mM ATP or ADP in GST-binding buffer (20 mM HEPES [pH 6.8], 100 mM KCl, 5 mM MgCl2, 0.1% NP-40, 2% glycerol, 1 mM DTT, 1 mM EDTA, and 1 mM phenylmethylsulfonyl fluoride) for 1 h at 4°C. A 20-μl aliquot of glutathione-Sepharose 4B (50% slurry) (Amersham Pharmacia Biotech) was added, and reaction mixtures were rotated for 30 min at 4°C. Reaction mixtures were centrifuged for 2 min at 13,000 rpm in a microcentrifuge, the supernatant was removed, and the pellet was washed three times with 100 μl of GST-binding buffer. The pellet fraction was then analyzed by SDS-PAGE using 8% polyacrylamide gels followed by Coomassie brilliant blue staining.
Nucleotide exchange and ATPase assays.
An Hsp70-[α-32P]ATP complex (with Ssa1p or DnaK) was obtained as described previously (63). In brief, 25 μg of Hsp70 was incubated in complex buffer (25 mM HEPES-KOH [pH 7.5], 100 mM KCl, 11 mM magnesium acetate) containing 25 μM ATP and 100 μCi of [α-32P]ATP (3,000 Ci/mmol; NEN) for 30 min on ice. Complexes were purified from free nucleotide by gel filtration on G-50 Nick Columns (Amersham Pharmacia Biotech). Peak complex fractions were pooled, the glycerol concentration was adjusted to 10%, and the samples were frozen in small aliquots in liquid nitrogen. For nucleotide exchange assays, 25 μl of complex was quickly thawed and mixed with 25 μl of complex buffer with or without the indicated cofactors (at a threefold molar excess over Ssa1p) at 30°C. At the specified time points, a 15-μl aliquot was removed and purified from free nucleotide on G-50 microspin columns (Amersham Pharmacia Biotech), and the eluate was mixed with 5 μl of stop buffer (36 mM ATP, 2 M LiCl, 4 M formic acid). The amount of remaining nucleotide was measured by scintillation counting and analyzed by thin-layer chromatography as described previously (58).
Single-turnover ATPase assays were performed essentially as described elsewhere (63). In brief, 25 μl of Ssa1p-[α-32P]ATP complex (see above) was mixed with 25 μl of complex buffer containing or lacking the indicated cofactors (at a threefold molar excess over Ssa1p) at 30°C. At the specified time points, a 6-μl aliquot of the reaction mixture was removed and added to 2 μl of stop buffer on ice. Triplicate 2-μl aliquots of this mixture were spotted on a thin-layer chromatography plate and developed (63).
For steady-state ATPase assays, 4 μg of Ssa1p was mixed with a twofold molar excess of Ydj1p in ATPase buffer (50 mM HEPES [pH 7.4], 50 mM NaCl, 10 mM DTT, 2 mM MgCl2) containing 50 μM ATP and 0.2 μCi of [α-32P]ATP in a total volume of 20 μl. After 10 min at 30°C, a 4-μl aliquot was stopped with 4 μl of stop buffer, and 5-μl aliquots were added to 15 μl of buffer with or without the specified amount of cofactor (GST-Fes1p or GST-Sls1p). At the indicated time points, 4-μl aliquots were taken and added to 4 μl of stop buffer on ice. Triplicate 1-μl samples were analyzed by thin-layer chromatography as described above.
RESULTS
Identification of yBR101c as a homologue of SLS1 (yOL031c).
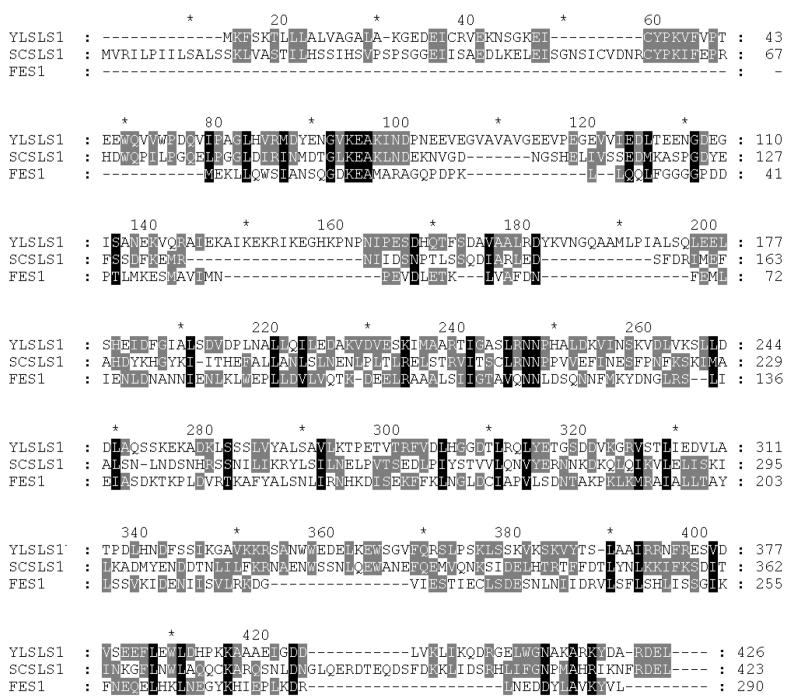
The identification of Sls1p as a nucleotide exchange factor for yeast BiP (6, 7, 32) raised the possibility for the existence of a similar factor for cytosolic Hsp70s. In order to address this question, we used the Psi-BLAST 2.0 software (http://www.ncbi.nih.nlm.gov) to identify Sls1p homologues in the yeast genome. The only gene uncovered in this search was yBR101c (Fig. 1), which encodes a predicted 290-amino-acid protein. The protein encoded by this gene, which we call Fes1p (mnemonic for factor exchange for Ssa1p), is significantly shorter than Sls1p and lacks a signal sequence, ER retention motif, nuclear localization sequence, and core glycosylation consensus sites, consistent with a cytosolic localization of Fes1p. Although the Sls1p homologues that we have found in the databases so far are poorly conserved, they share several clusters of amino acids that are present in Fes1p (Fig. 1).
FIG. 1.
Alignment of Fes1p with the amino acid sequences encoded by the Y. lipolytica and S. cerevisiae SLS1 genes. Black boxes represent conserved residues; shaded boxes represent similar residues. Amino acids 1 to 17 (6) and 1 to 19 correspond to the putative signal sequences of Y. lipolytica Sls1p and S. cerevisiae Sls1p, respectively. Neither signal sequences nor C-terminal ER retention motifs are found in Fes1p.
The Δfes1 mutant is thermosensitive.
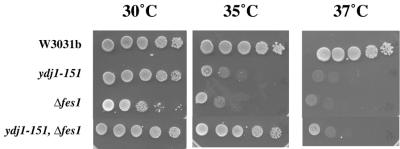
As a first step toward investigating the function of Fes1p, the encoding ORF was disrupted by using a kanamycin resistance cassette (8) in two unique wild-type genetic backgrounds (see Materials and Methods) (Table 1). In both cases, the disruption of FES1 did not affect viability but strongly impaired growth at high temperature (Fig. 2).
FIG. 2.
Genetic effects of the disruption of the FES1 gene. Serial dilutions of overnight cultures of the wild-type and indicated mutant strains were inoculated on YPD plates and incubated at the indicated temperatures for 3 days.
We also sought genetic interactions between Δfes1 and ydj1-151, a thermosensitive mutant of the Ydj1p J DnaJ homologue that is defective in its ability to activate Ssa1p (16). Genetic interactions were previously found between Δsls1 and a thermosensitive sec63 allele (32) which encodes a mutated form of the ER J protein Sec63p (55). Disruption of FES1 in the ydj1-151 strain partially suppressed the Δfes1 and ydj1-151 thermosensitive phenotypes (Fig. 2). Since several independent transformants were analyzed, we excluded the possibility that this result is due to a third mutation resulting from the transformation procedure. The observed genetic interactions suggest that Fes1p and Ydj1p may collaborate to facilitate a cellular pathway and/or coregulate another factor.
Fes1p is a soluble protein located in the cytosol.
In order to determine the subcellular localization of Fes1p, the protein was tagged at the N terminus with GFP. GFP-Fes1p is expressed under the control of the GAL1 promoter, and the corresponding strain grew as well as the isogenic wild type at 37°C (data not shown), suggesting that the GFP tag does not affect the function of Fes1p.
We used fluorescence microscopy to assess the localization of GFP-Fes1p after inducing its expression for 8 h in galactose-containing medium. As shown in Fig. 3A, the protein was found primarily in the cytosol, as expected from the primary sequence analysis (Fig. 1). The GFP-Fes1p protein appeared enriched at the nucleus (Fig. 3A), but whether this is physiologically relevant or whether this is an effect due to the overexpression of the fusion protein is not known. Indeed, the nuclear colocalization was more pronounced when the expression of the protein was induced for longer times (data not shown). In any event, GFP-Fes1p was excluded from the vacuole (Fig. 3A).
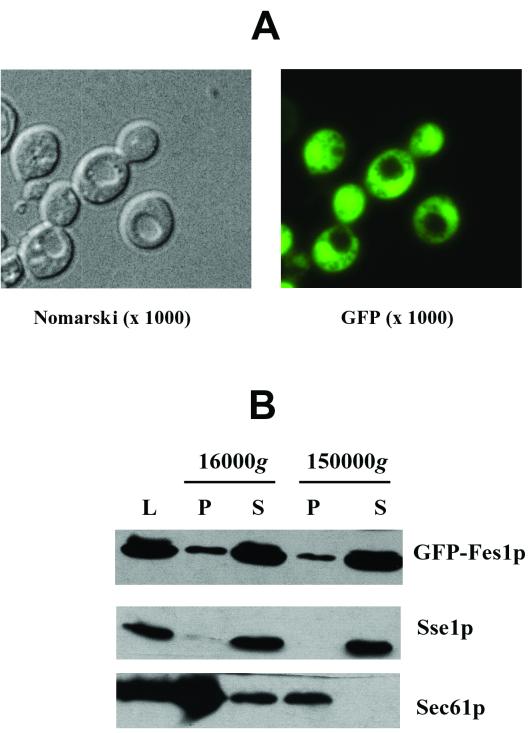
FIG. 3.
Subcellular localization of Fes1p. (A) A strain bearing a GFP-tagged version of Fes1p under the control of the GAL1 promoter was obtained as described in Materials and Methods. This strain was grown on raffinose-containing medium and then for 8 h in galactose-containing medium. The cells were then observed directly with a fluorescence microscope, and both Nomarski and GFP fluorescence images are shown (magnification, ×1,000). (B) Cell lysate (L) was subjected to 16,000 × g and 150,000 × g centrifugations. Equal amounts of proteins from pellet (P) and supernatant (S) fractions were resolved by SDS-PAGE and analyzed by Western blotting with anti-GFP (Fes1p), anti-Sse1p (cytosolic protein), and anti-Sec61p (integral membrane protein) antibodies.
To confirm the cytosolic localization of the protein, subcellular fractionation was performed on crude lysates as described in Materials and Methods. As shown in Fig. 3B, GFP-Fes1p was found in soluble fractions, as was Sse1p, a cytosolic Hsp110 (14) (Fig. 3B; supernatant fractions), and relatively minor amounts of GFP-Fes1p were found in the crude membrane fraction (Fig. 3B; 16,000 × g pellet), where Sec61p, the ER translocation pore protein, is detected. It should be noted that Ssa1p is also detected in both supernatant and pellet fractions (14). We conclude that Fes1p is a soluble cytosolic protein.
Fes1p interacts with Ssa1p in an ATP-dependent manner.
To determine if Fes1p, like Sls1p (32), binds to Hsp70, the protein was overexpressed as a GST fusion protein in E. coli and purified to near homogeneity by affinity chromatography (see Materials and Methods). The GST-Fes1p fusion protein fully complemented the thermosensitive phenotype of the Δfes1 strain when placed under the control of a strong constitutive promoter (see Materials and Methods), suggesting that the GST tag does not affect the function of Fes1p.
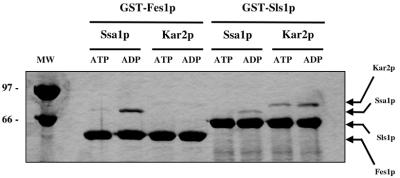
Although S. cerevisiae possesses six cytosolic Hsp70s (Ssa1 to Ssa4, Ssb1, and Ssb2; [22]), we first examined the interaction between Fes1p and the cytosolic Hsp70, Ssa1p, because of its defined roles in protein translation, translocation, folding, and ERAD (21). We also purified GST-Sls1p and Kar2p (32, 45) to provide negative and specificity controls for the binding reactions. Binding of GST-Fes1p and GST-Sls1p to the Hsp70s was assessed by GST pull-down assays in either 1 mM ATP or 1 mM ADP (see Materials and Methods). After 1 h at 4°C, the mixtures were centrifuged and washed, and the pellet fractions were analyzed by SDS-PAGE and Coomassie blue staining. As shown in Fig. 4, Fes1p binds to Ssa1p, but not to Kar2p, and conversely Sls1p binds preferentially to Kar2p, as shown previously (32), although some nonspecific binding was also observed between Sls1p and Ssa1p (Fig. 4); however, the Sls1p-Ssa1p binding is ∼10-fold lower than the interaction between Sls1p and Kar2p. Also previously described for the Sls1p-Kar2p interaction (32), Fes1p binds more efficiently to the ADP-bound form of Ssa1p, suggesting that the interaction is specific. It should be noted that the binding experiments shown in Fig. 4 were performed at pH 7.4 (with similar results at pH 8.0; data not shown) and under these conditions there is some interaction between Sls1p and the ATP-bound form of Kar2p, consistent with the ability of Sls1p to release ATP from Kar2p (32). The possibility that the Hsp70s recognize a population of misfolded GST fusion proteins is excluded by the specificity of interaction between each chaperone and either Fes1p or Sls1p and by control experiments using purified GST to which neither factor was fused (data not shown). Overall, the preferred interactions between Kar2p and Sls1p and between Fes1p and Ssa1p are consistent with the compartmentalization of these chaperone pairs in the ER lumen and cytoplasm, respectively.
FIG. 4.
Fes1p interacts with Ssa1p but not Kar2p. GST-Fes1p, GST-Sls1p, Ssa1p, and Kar2p were purified as described in Materials and Methods. GST-Fes1p or GST-Sls1p was incubated with Ssa1p or Kar2p in the presence of 1 mM nucleotide (ATP or ADP) and incubated at 4°C for 1 h. Glutathione-Sepharose 4B beads were added, and reactions were rotated for 30 min at 4°C. Proteins bound to the glutathione matrix were resolved by SDS-PAGE and visualized by Coomassie brilliant blue staining. Molecular weight standards (in thousands) are shown at the left.
Fes1p is a nucleotide exchange factor for Ssa1p.
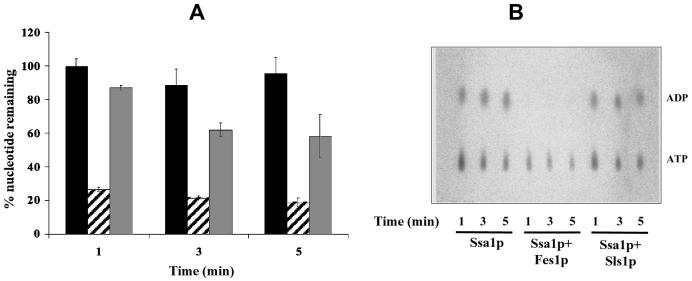
To determine whether Fes1p promotes nucleotide release upon binding to Ssa1p, we prepared an Ssa1p-[α-32P]ATP complex (63) and performed nucleotide exchange assays (see Materials and Methods). The Ssa1p-[α-32P]ATP complex was incubated at 30°C in the presence of buffer, GST-Fes1p, or GST-Sls1p, aliquots were removed over time, and Ssa1p-bound and free nucleotide were resolved by gel filtration chromatography. The amount of bound nucleotide was assessed by scintillation counting (Fig. 5A) and analyzed by thin-layer chromatography (Fig. 5B). In the presence of GST-Fes1p a rapid decrease (within 1 min) of bound nucleotide was observed compared to that of the buffer control. The thin-layer chromatography plate (Fig. 5B) shows that both ADP and, to a lesser extent, ATP were stripped by GST-Fes1p. Importantly, GST-Sls1p had very little effect on nucleotide release from Ssa1p (Fig. 5), a result which was consistent with the GST pull-down assays (Fig. 4). Also, the Fes1p-stimulated nucleotide release was most likely not due to its ability to act as a peptide substrate, which was previously shown to enhance nucleotide release from Ssa1p (75), because GST-Sls1p had less effect in this assay (Fig. 5A; compare striped bars to grey bars). These results show that Fes1p is a nucleotide exchange factor for Ssa1p.
FIG. 5.
Fes1p is a nucleotide exchange factor for Ssa1p. The [α-32P]ATP-Ssa1p complex was incubated with buffer (black bars), or a threefold molar excess of GST-Fes1p (striped bars) or GST-Sls1p (gray bars) at 30°C. At the indicated time points, an aliquot was taken and free nucleotide was removed on microspin G-50 columns. Further reaction in the eluate was stopped, and the amount of Ssa1p-bound nucleotide was determined by scintillation counting (A) or analyzed on thin-layer chromatography plates (B) as described previously (32). Error bars represent the standard deviations from three independent experiments.
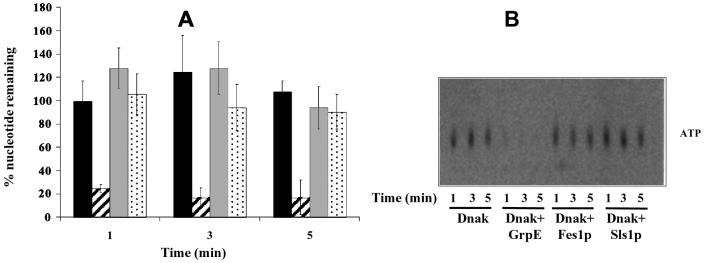
In order to validate our results and to gain more insight into the specificity of these cofactors, we performed similar assays with the E. coli DnaK protein and its nucleotide exchange factor GrpE, which lacks homology to Sls1p and Fes1p. We prepared a DnaK-[α-32P]ATP complex and performed nucleotide exchange assays as described for Ssa1p. As shown in Fig. 6A, GrpE induced the rapid release of nucleotide, consistent with previously published data (38, 42, 43), whereas Sls1p and Fes1p had no effect on the amount of nucleotide bound to DnaK (Fig. 6A; compare striped bars to grey and dotted bars). The thin-layer chromatography plate shows that GrpE quantitatively released ATP from DnaK (Fig. 6B). This result is consistent with previous findings that GrpE can release both ATP and ADP (9, 38, 42), although our preparation of the DnaK-[α-32P]ATP complex had insufficient specific activity that [α-32P]ADP was not detected by thin-layer chromatography. As described previously (37), we also found that GrpE was unable to trigger nucleotide release from Ssa1p (data not shown). These and previous results (32) suggest that the Sls1p and Fes1p class of exchange factors, like GrpE, bind preferentially to the ADP-bound form of Hsp70s, but can release both ADP and ATP from their respective chaperones. Further experiments will be required to determine if one nucleotide is more efficiently and/or rapidly released than the other by Fes1p and/or Sls1p.
FIG. 6.
Fes1p and Sls1p cannot substitute for GrpE as nucleotide exchange factors for DnaK. Nucleotide exchange reactions with E. coli DnaK were performed as described for Fig. 5. The [α-32P]ATP-DnaK complex was incubated with buffer (black bars) or a threefold molar excess of GrpE (dashed bars), GST-Fes1p (gray bars), or GST-Sls1p (dotted bars) at 30°C. At the indicated time points, an aliquot was taken and free nucleotide was removed on microspin G-50 columns. Reactions in the eluates were stopped, and radioactivity was analyzed by scintillation counting (A) and thin-layer chromatography (B). Error bars represent the standard deviations from three independent experiments.
Effects of Fes1p on the ATPase activity of Ssa1p.
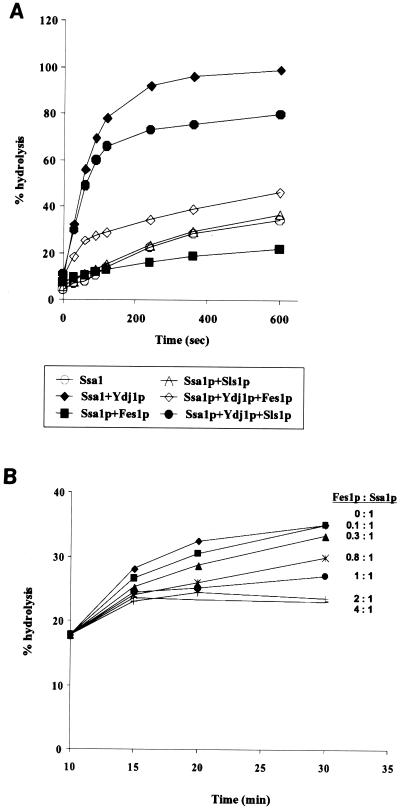
We then examined the effects of the Fes1p and Sls1p nucleotide exchange factors on Ssa1p single-turnover ATPase activity (63). The Ssa1p-[α-32P]ATP complex was incubated at 30°C in the presence or absence of GST-Fes1p, GST-Sls1p, and Ydj1p, which enhances ATP hydrolysis (16, 18, 19, 44, 45). The extent of ATP hydrolyzed over time was assessed by thin-layer chromatography and PhosphorImager analysis (63), and the results are shown in Fig. 7A. Ssa1p alone displayed a weak ATPase activity, which was efficiently stimulated by Ydj1p. Interestingly, Fes1p significantly inhibited the ATPase activity of Ssa1p both in the presence and in the absence of Ydj1p, whereas Sls1p had only a limited effect, probably due to its poor interaction with Ssa1p (Fig. 4). As discussed in another study (39), the observed inhibition of ATPase activity in single-turnover conditions is expected for a nucleotide exchange factor, since stripping the bound nucleotide will decrease the apparent rate of hydrolysis. Therefore, this result further supports the previous data showing that Fes1p is a nucleotide exchange factor for Ssa1p.
FIG. 7.
(A) Effects of Fes1p and Ydj1p on single-turnover ATPase activity. Single-turnover experiments were performed as described previously (63). The Ssa1p-[α32P]ATP complex (as in Fig. 5) was mixed with complex buffer with or without the indicated cofactors (at a threefold molar excess over Ssa1p) at 30°C. At the specified times, an aliquot was removed from the reaction mixture and added to stop buffer on ice. Triplicate aliquots of this mixture were analyzed on thin-layer chromatography plates. A representative experiment is shown. (B) Fes1p inhibits Ssa1p steady-state ATPase activity. Ssa1p was mixed with a twofold molar excess of Ydj1p in a reaction mixture containing 50 μM ATP and 0.2 μCi of [α-32P]ATP. After 10 min at 30°C, the indicated amount of Fes1p was added, and an aliquot of the reaction mixture was analyzed at the indicated times as described in Materials and Methods.
We next assessed the effect of Fes1p on the ATPase activity of Ssa1p under steady-state conditions, as it was previously shown that both GrpE and Sls1p stimulate the ATPase activity of their respective Hsp70s synergistically with J proteins (32, 38). Ssa1p was incubated for 10 min at 30°C in the presence of [α-32P]ATP and Ydj1p to allow ATPase activation. Then, following addition of either buffer or the indicated amounts of GST-Fes1p, ATP hydrolysis was monitored over time by thin-layer chromatography and PhosphorImager analysis (Fig. 7B). Surprisingly, Fes1p inhibited the steady-state ATPase activity of Ssa1p in a concentration-specific manner. Control experiments showed no effect of purified GST, to which no factor was fused, on the ATPase activity of Ssa1p (data not shown). It is important to state that although our nucleotide exchange experiments demonstrate that Fes1p is able to induce nucleotide dissociation from Ssa1p, they do not exclude the possibility that Fes1p also prevents nucleotide binding per se. In fact, inhibition of nucleotide binding by Fes1p could give rise to the observed effects on Ssa1p ATPase activity. For example, mammalian HspBP1 inhibits Hsp70 ATPase activity by preventing ATP binding (54). Regardless, we conclude that Fes1p has a negative effect on both steady-state and single-turnover Ssa1p ATPase activity.
Fes1p is dispensable for posttranslational protein translocation, ERAD, and luciferase refolding.
Since Ssa1p is required for posttranslational protein translocation (5, 17, 20, 44), ERAD of the cystic fibrosis transmembrane conductance regulator in yeast (72), and protein folding (14, 36, 37), we examined whether Fes1p is involved in the same pathways using well-established in vivo and in vitro assays.
Mutations in SSA1 affect the posttranslational translocation of pre-pro-α-factor (ppαF) (5, 17, 20, 44); therefore, we examined the Δfes1 strain for accumulation of ppαF. We also performed in vitro translocation assays (12) using microsomes purified from wild-type and Δfes1 strains. However, in neither case was a translocation defect apparent (data not shown). Next, cystic fibrosis transmembrane conductance regulator degradation was monitored in the Δfes1 strain by cycloheximide-chase analysis as described previously (71, 72), but no difference in the degradation rates of the Δfes1 mutant and of the wild type was observed (data not shown). We then performed in vitro ERAD assays using purified microsomes and cytosol from both wild-type and mutant strains (46, 68). In these experiments, ∼70% of the translocated ΔGpαF was degraded in a cytosol- and ATP-dependent manner, regardless of the origin of the microsomes or cytosol (data not shown). Finally, we failed to detect any defects in Ssa1p-dependent refolding activity of luciferase in vitro when using wild-type or Δfes1 mutant cytosol, or upon addition of excess purified Fes1p (reference 14 and data not shown).
From these data, we conclude that Fes1p is not a cofactor for every aspect of Ssa1p function but might be involved in another specific pathway. By analogy, function-specific cofactors for Hsp70s have been observed with DnaJ homologues (50, 67, 70, 73).
Evidence for a function of Fes1p in protein translation.
Many reports indicate the importance of molecular chaperones in protein translation. The Ssb1p and Ssb2p Hsp70s (49, 52) and Hsp40 family members (Sis1p, zuotin, Ydj1p [13, 70, 73]) associate with ribosomes and have been implicated in efficient protein synthesis or translation initiation (22). Similarly, Ssa1p was shown recently to be associated with ribosomes through its interaction with Sis1p, a cytosolic J protein, and Pab1p, the poly(A) binding protein (31). The thermosensitive ssa1-45 mutant is defective for protein translation at high temperature by affecting the interaction of Pab1p with the initiation factor eIF4G (31).
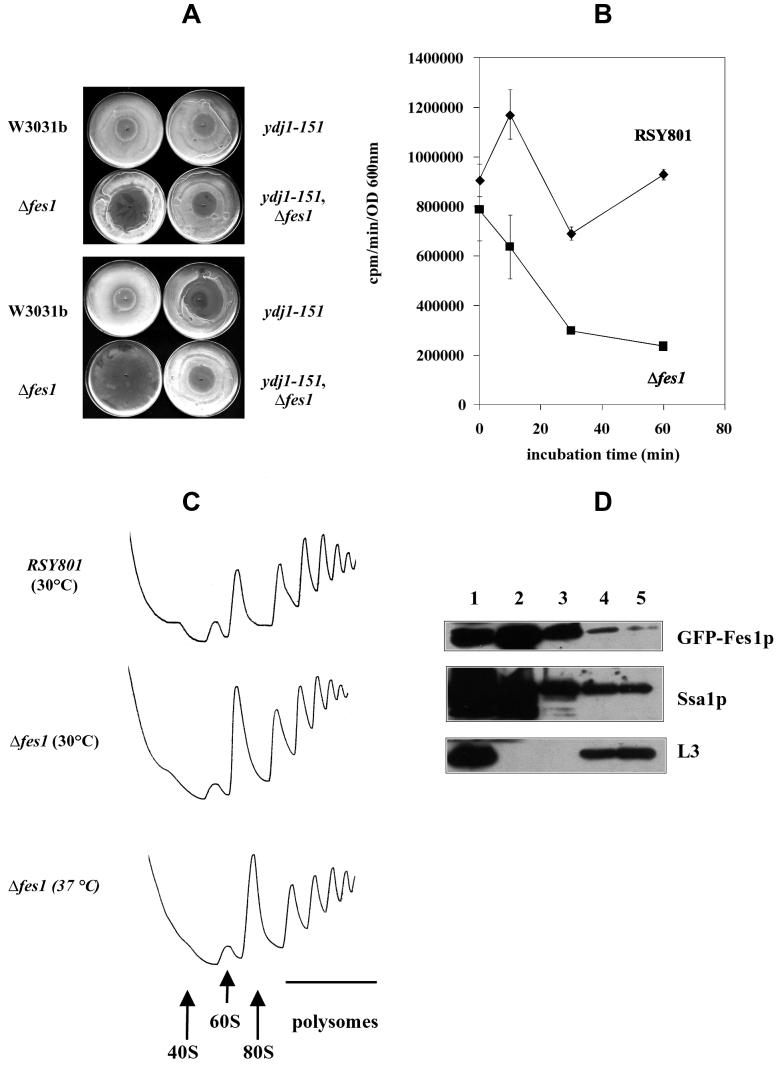
To determine if deletion of FES1 affected protein translation, we first assessed the sensitivity of wild-type and mutant strains to cycloheximide, an inhibitor of translation elongation, in a halo assay (for an example, see reference 51). As shown in Fig. 8A, the Δfes1 strain was more sensitive to cycloheximide than the wild-type control, as revealed by the formation of a larger halo of inhibition at 30°C. Interestingly, while the ydj1-151 strain showed increased cycloheximide sensitivity at 35°C, as expected from former studies (13), the ydj1-151 Δfes1 double mutant was almost indistinguishable from the wild-type strain at both temperatures, which might explain the suppressive effect of the FES1 disruption on the ydj1-151 thermosensitive growth (Fig. 2). It should be noted that the severe growth inhibition of the Δfes1 strain at 35°C probably results from both thermosensitivity and cycloheximide sensitivity. We were unable to observe significant differences in sensitivity between the wild-type and Δfes1 strains for anisomycin (data not shown), an inhibitor of peptidyltransferase activity, suggesting the specificity of Fes1p action in translation.
FIG. 8.
Fes1p plays a role in protein translation. (A) The indicated strains were grown overnight in YPD at 26°C and then plated on the same media and grown for 8 h at 26°C. Cycloheximide (20 μg) was inoculated in the middle of plates that were then incubated overnight at 30°C (top) or 35°C (bottom). (B) The indicated strains were grown at 26°C to mid-log phase and shifted to 37°C. At 0, 10, 30, and 60 min after incubation at 37°C, samples were taken and labeled with [35S]-Express Labeling mix for 2 min at 30°C. Proteins were trichloroacetic acid precipitated and assayed by scintillation counting. Error bars represent the standard deviations from three independent experiments. (C) The indicated yeast strains were grown at 30°C and, when indicated, shifted for 30 min at 37°C. Cycloheximide was added to a final concentration of 50 μg/ml, and cell lysates were prepared. Equal amounts of lysate (∼70 A254 U) were loaded onto 7 to 47% linear sucrose gradients and analyzed as described in Materials and Methods. The gradients were collected with continuous monitoring of A254. The positions of the 40S, 60S, 80S, and polysome peaks are shown. (D) Cells expressing GFP-Fes1p were grown in galactose-containing medium until log phase, and cell lysates were prepared as described for panel C. Approximately 100 A254 U were loaded onto 7 to 47% linear sucrose gradients and processed as described for panel C. Aliquots corresponding to the lysate (lane 1), the cytosol (top of the gradient; lane 2), ribosome-free fractions (lane 3), 80S ribosomes (lane 4), and the polysomes (lane 5) were resolved by SDS-PAGE and analyzed by Western blotting using anti-GFP, anti-Ssa1p, and anti-L3 antisera.
It was previously shown that the depletion of active Ssa1p affected protein translation, as the incubation of the ssa1-45 mutant at various times at the nonpermissive temperature of 37°C progressively reduced the ability of this strain to incorporate [35S]methionine into newly synthesized proteins (31). We recapitulated this experiment by measuring the incorporation of [35S]methionine-cysteine into newly translated proteins following a 2-min pulse at 0, 10, 30, and 60 min after incubation of the wild-type and Δfes1 strains at 37°C. Whereas the wild-type strain displayed little change in [35S]methionine-cysteine incorporation between 10 and 30 min, the translation rate decreased in the Δfes1 strain (Fig. 8B), similarly to what was observed for the ssa1-45 mutant (31). To verify that this effect was not a result of cell death of the Δfes1 mutant at 37°C, aliquots were taken at each time point, plated on complete media, and incubated at the permissive temperature of 26°C. Approximately equal numbers of colonies were obtained with wild-type and mutant strains at each time point; moreover, growth curves at 37°C showed that the Δfes1 mutant kept dividing after 2 h of incubation at 37°C, although at this time point a reduced rate compared to that of the wild-type strain was observed. However, until this time, similar growth rates were apparent (data not shown). Therefore, we conclude that the observed decrease in translation rate is due to a defect associated with the disruption of the FES1 gene.
Defects in protein translation may lead to changes in the ratio of free ribosomes to polysomes. The ssa1-45 strain showed an increase of 80S ribosomes concomitant with a decrease in polysomes when shifted to the restrictive temperature (31). To determine whether deletion of FES1 affected polysome profile, linear sucrose gradients (7 to 47%) were used to assess ribosome distribution in the wild-type and Δfes1 strains at 30°C and after a 30-min incubation at 37°C. As shown in Fig. 8C, the Δfes1 strain showed a slight increase in the 80S ribosomes relative to polysomes compared to the wild type at 30°C. At 37°C, the increase in 80S ribosomes was more pronounced (Fig. 8C; compare the 80S peak to the first polysome peak). The polysome profile for the wild-type strain after 30 min of incubation at 37°C was undistinguishable from that obtained at 30°C (data not shown).
To examine whether Fes1p is polysome associated, lysates prepared from the GFP-FES1 strain were fractionated through the sucrose gradient and proteins corresponding to the lysate (Fig. 8D, lane 1), the nonribosomal cytosolic fraction (top of the gradient, lane 2), the position between cytosol- and ribosome-containing fractions (lane 3), the 80S ribosomes (lane 4), and polysomes (lane 5) were separated by SDS-PAGE and analyzed by Western blotting. As expected, the L3 large subunit ribosomal protein was found in the lysate and ribosomal fractions and was excluded from soluble cytosolic and preribosomal fractions. Also, as published previously, Ssa1p was found in the 80S and polysome fractions (31). Finally, we identified Fes1p in these same fractions. These data are consistent with a role of Fes1p in protein translation and with its interaction with Ssa1p.
DISCUSSION
The work described here establishes that Fes1p, encoded by yBR101c, a previously uncharacterized yeast ORF, functions as a nucleotide exchange factor for the Hsp70 molecular chaperone Ssa1p. This protein was identified based on its homology to Sls1p, an ER protein previously shown to act as a nucleotide exchange factor for yeast BiP (32). Even though Fes1p is shorter than Sls1p, which is in part due to the absence of any targeting sequence or retention motif, it shares important features with the latter. The regions that are conserved between S. cerevisiae and Y. lipolytica Sls1p proteins are also found in Fes1p, whereas regions of weak homology are absent or are not conserved in Fes1p (Fig. 1). These proteins are also similar in the way they interact with their respective Hsp70 partner. Indeed, Sls1p was shown to preferentially interact with the ADP-bound form of Kar2p (32) and, using GST pull-down assays, we show here that Fes1p also preferentially interacts with the ADP-bound form of Ssa1p (Fig. 4). Both proteins release bound nucleotide from their respective Hsp70 chaperone partners specifically (Fig. 5) (32), as Sls1p could not substitute for Fes1p in the exchange assays. Moreover, whereas GrpE efficiently promoted nucleotide exchange on DnaK (38), thereby validating our experimental model, Fes1p and Sls1p were inactive (Fig. 6). Conversely, GrpE was unable to promote nucleotide release from Ssa1p (reference 37 and data not shown). Such specificity between chaperones and their cofactors has been observed for J protein-Hsp70 interactions (5, 11, 18, 35, 41, 45, 69). Because Hsp70 can be classified in several subclasses depending on the presence or absence of specific structural features within the highly conserved ATPase domain (9), it is reasonable to think that at least in some cases each chaperone and its cofactor(s) have evolved to fit one another. Structural analysis of several exchange factor-Hsp70 pairs would afford insights into this question and in defining the different mechanisms of nucleotide exchange, as GrpE, Bag-1, and Sls1p and Fes1p are unrelated.
The nucleotide exchange factors of the GrpE family can stimulate synergistically the ATPase activity of Hsp70s in cooperation with J proteins. For example, E. coli GrpE and DnaJ each stimulate the DnaK ATPase activity ∼2-fold and ∼3- to 13-fold, respectively (37, 38, 43), but up to ∼50-fold when added together in steady-state assays (38). Similarly, we showed that Sls1p synergistically stimulates the ATPase activity of Kar2p in the presence of Sec63p (32) and Jem1p (our unpublished results). Surprisingly, we found that Fes1p inhibited the steady-state ATPase activity of Ssa1p in a concentration-dependent manner (Fig. 7B). The GrpE protein was shown to act negatively on DnaK when added in excess (53), and recently, it was shown that Bag-1 positively or negatively modulates Hsp70 and Hsc70 refolding activity depending on its concentration (24). However, the inhibition of Ssa1p ATPase activity by Fes1p was observed even at substoichiometric concentrations of the cofactor, suggesting that Fes1p has evolved as a negative regulator of the Hsp70 chaperone. Similar negative regulation was observed with HspBP1, which inhibits ATP binding to Hsc70 (54). In addition, the Hsp70-interacting protein, HiP, regulates Hsp70 ATPase activity by stabilizing the ADP-Hsp70-peptide complex, which in turn promotes efficient folding of the substrate (30).
The Δfes1 mutant was viable at the permissive temperature but not at high temperature (Fig. 2), suggesting that this protein is important under conditions of elevated metabolism and stress. Using a functional GFP-tagged version of Fes1p, fluorescence microscopy, and subcellular fractionation, we were able to show that Fes1p is a soluble cytosolic protein (Fig. 3).
The Δfes1 mutant was proficient for protein translocation, ERAD, and protein folding, which are established Ssa1p-dependent processes (14, 17, 20, 36, 41, 44). Instead, Fes1p facilitates protein translation, a process in which both Ssa1p and Ydj1p have been shown to play a role (13, 31). Indeed, increased cycloheximide sensitivity is observed upon disruption of FES1 (Fig. 8A) and, as described for Ssa1p (31), a general translational defect is observed when the Δfes1 mutant is incubated at high temperature (Fig. 8B). Interestingly, the cycloheximide sensitivity is suppressed in the ydj1-151 Δfes1 double mutant, which is consistent with the suppression of the thermosensitive defect (Fig. 2). This suggests that Fes1p and Ydj1p play antagonist functions during translation, consistent with their respective negative and positive regulation of Ssa1p ATPase activity. Another important finding was that the absence of Fes1p leads to a slight decrease in polysomes and an increase in 80S ribosomes, a defect enhanced at high temperature (Fig. 8C). A similar phenotype was also observed with the ssa1-45 strain (31). Furthermore, Fes1p was found to be associated with 80S and polysome fractions, as was Ssa1p (Fig. 8D). Also, in support of a role of Fes1p in translation, the recent construction of a yeast proteome-wide interaction map indicated that Fes1p interacted with Tif1p, the yeast homologue of mammalian eIF4A, and with Hsp42, a small heat shock protein (25). Finally, Mutka and Walter found that the expression of FES1 was transiently activated during the cellular adaptation to the loss of the signal recognition particle (see Fig. 4 in reference 48). Since Fes1p does not seem to be involved in posttranslational protein translocation, its induction may alter the translation rate that is observed during the cellular adaptation to loss of cotranslational translocation (48). Although the deletion of Ydj1p did not significantly affect the adaptation (48), it will be interesting to examine if Fes1p and Ydj1p compete for binding to Ssa1p and/or if another protein (Sis1p or Pab1p, perhaps) or substrate regulates the temporal interaction of Ssa1p with its many partners. For example, Ssa1p is necessary for the association between Pab1p and initiation factor eIF4G, an association that is thought to be important for cap-dependent translation initiation (31). Although the sequential interaction of Ssa1p with Sis1p and Pab1p, as well as yet-unidentified substrates on translating ribosomes, is not well understood, an attractive hypothesis would be that Fes1p participates in the regulation and proper coordination of these different interactions.
Several homologues of Sls1p and Fes1p have been identified in the genome databases, with homologues in the human, mouse, Drosophila melanogaster, Schizosaccharomyces pombe, and Candida albicans genomes (M. Kabani, J.-M. Beckerich, and J. L. Brodsky, unpublished data). These proteins can be separated into two subclasses, one corresponding to ER Sls1p-related proteins, and the other to the cytosolic Fes1p-related proteins; each subclass displays the similarities and differences that are found between Sls1p and Fes1p (see above). While these proteins share features that might be important for binding with the same domain of their Hsp70 counterpart and promote nucleotide release, structural differences might be responsible for the different effects observed in steady-state ATPase assays. Clearly, much more work is warranted on this new class of chaperone modulators.
Acknowledgments
We thank Sheara W. Fewell for the generous gift of purified Ssa1p and Ydj1p proteins and for help and discussions and Jennifer Goeckeler for excellent technical assistance. We also thank John L. Woolford (Carnegie Mellon University, Pittsburgh, Pa.) for help with the ribosome fractionation procedure.
This work was supported by grant MCB-0110331 from the National Science Foundation to J.L.B.
J.-M.B. is a permanent investigator of the Institut National de la Recherche Agronomique (Centre de Versailles-Grignon, Thiverval-Grignon, France).
REFERENCES
- 1.Adams, A., D. E. Gottschling, C. A. Kaiser, and T. Stearns. 1997. Methods in yeast genetics. A Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 2.Agashe, V. R., and F. U. Hartl. 2000. Roles of molecular chaperones in cytoplasmic protein folding. Semin. Cell Dev. Biol. 11:15-25. [DOI] [PubMed] [Google Scholar]
- 3.Ausebel, F., R. Brent, R. Kingston, D. Moore, J. G. Seidman, et al. 1989. Current protocols in molecular biology. Wiley, New York, N.Y.
- 4.Ballinger, C. A., P. Connell, Y. Wu, Z. Hu, L. J. Thompson, L. Y. Yin, and C. Patterson. 1999. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell. Biol. 19:4535-4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker, J., W. Walter, W. Yan, and E. A. Craig. 1996. Functional interaction of cytosolic hsp70 and a DnaJ-related protein, Ydj1p, in protein translocation in vivo. Mol. Cell. Biol. 16:4378-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boisrame, A., J. M. Beckerich, and C. Gaillardin. 1996. Sls1p, an endoplasmic reticulum component, is involved in the protein translocation process in the yeast Yarrowia lipolytica. J. Biol. Chem. 271:11668-11675. [DOI] [PubMed] [Google Scholar]
- 7.Boisrame, A., M. Kabani, J. M. Beckerich, E. Hartmann, and C. Gaillardin. 1998. Interaction of Kar2p and Sls1p is required for efficient co-translational translocation of secreted proteins in the yeast Yarrowia lipolytica. J. Biol. Chem. 273:30903-30908. [DOI] [PubMed] [Google Scholar]
- 8.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 9.Brehmer, D., S. Rudiger, C. S. Gassler, D. Klostermeier, L. Packschies, J. Reinstein, M. P. Mayer, and B. Bukau. 2001. Tuning of chaperone activity of Hsp70 proteins by modulation of nucleotide exchange. Nat. Struct. Biol. 8:427-432. [DOI] [PubMed] [Google Scholar]
- 10.Briknarova, K., S. Takayama, L. Brive, M. L. Havert, D. A. Knee, J. Velasco, S. Homma, E. Cabezas, J. Stuart, D. W. Hoyt, A. C. Satterthwait, M. Llinas, J. C. Reed, and K. R. Ely. 2001. Structural analysis of BAG1 cochaperone and its interactions with Hsc70 heat shock protein. Nat. Struct. Biol. 8:349-352. [DOI] [PubMed] [Google Scholar]
- 11.Brodsky, J. L., M. Bauerle, M. Horst, and A. J. McClellan. 1998. Mitochondrial Hsp70 cannot replace BiP in driving protein translocation into the yeast endoplasmic reticulum. FEBS Lett. 435:183-186. [DOI] [PubMed] [Google Scholar]
- 12.Brodsky, J. L., J. Goeckeler, and R. Schekman. 1995. BiP and Sec63p are required for both co- and posttranslational protein translocation into the yeast endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 92:9643-9646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brodsky, J. L., J. G. Lawrence, and A. J. Caplan. 1998. Mutations in the cytosolic DnaJ homologue, YDJ1, delay and compromise the efficient translation of heterologous proteins in yeast. Biochemistry 37:18045-18055. [DOI] [PubMed] [Google Scholar]
- 14.Brodsky, J. L., E. D. Werner, M. E. Dubas, J. L. Goeckeler, K. B. Kruse, and A. A. McCracken. 1999. The requirement for molecular chaperones during endoplasmic reticulum-associated protein degradation demonstrates that protein export and import are mechanistically distinct. J. Biol. Chem. 274:3453-3460. [DOI] [PubMed] [Google Scholar]
- 15.Bukau, B., and A. L. Horwich. 1998. The Hsp70 and Hsp60 chaperone machines. Cell 92:351-366. [DOI] [PubMed] [Google Scholar]
- 16.Caplan, A. J., D. M. Cyr, and M. G. Douglas. 1992. YDJ1p facilitates polypeptide translocation across different intracellular membranes by a conserved mechanism. Cell 71:1143-1155. [DOI] [PubMed] [Google Scholar]
- 17.Chirico, W. J., M. G. Waters, and G. Blobel. 1988. 70K heat shock related proteins stimulate protein translocation into microsomes. Nature 332:805-810. [DOI] [PubMed] [Google Scholar]
- 18.Cyr, D. M., and M. G. Douglas. 1994. Differential regulation of Hsp70 subfamilies by the eukaryotic DnaJ homologue YDJ1. J. Biol. Chem. 269:9798-9804. [PubMed] [Google Scholar]
- 19.Cyr, D. M., X. Lu, and M. G. Douglas. 1992. Regulation of Hsp70 function by a eukaryotic DnaJ homolog. J. Biol. Chem. 267:20927-20931. [PubMed] [Google Scholar]
- 20.Deshaies, R. J., B. D. Koch, M. Werner-Washburne, E. A. Craig, and R. Schekman. 1988. A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature 332:800-805. [DOI] [PubMed] [Google Scholar]
- 21.Fewell, S. W., K. J. Travers, J. S. Weissman, and J. L. Brodsky. 2001. The action of molecular chaperones in the early secretory pathway. Annu. Rev. Genet. 35:149-191. [DOI] [PubMed] [Google Scholar]
- 22.Frydman, J. 2001. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu. Rev. Biochem. 70:603-647. [DOI] [PubMed] [Google Scholar]
- 23.Gässler, C. S., A. Buchberger, T. Laufen, M. P. Mayer, H. Schröder, A. Valencia, and B. Bukau. 1998. Mutations in the DnaK chaperone affecting interaction with the DnaJ cochaperone. Proc. Natl. Acad. Sci. USA 95:15229-15234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gässler, C. S., T. Wiederkehr, D. Brehmer, B. Bukau, and M. P. Mayer. 2001. Bag-1M accelerates nucleotide release for human Hsc70 and Hsp70 and can act concentration-dependent as positive and negative cofactor. J. Biol. Chem. 276:32538-32544. [DOI] [PubMed] [Google Scholar]
- 25.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141-147. [DOI] [PubMed] [Google Scholar]
- 26.Gething, M. J. 1999. Role and regulation of the ER chaperone BiP. Semin. Cell Dev. Biol. 10:465-472. [DOI] [PubMed] [Google Scholar]
- 27.Harrison, C. J., M. Hayer-Hartl, M. Di Liberto, F. Hartl, and J. Kuriyan. 1997. Crystal structure of the nucleotide exchange factor GrpE bound to the ATPase domain of the molecular chaperone DnaK. Science 276:431-435. [DOI] [PubMed] [Google Scholar]
- 28.Höhfeld, J. 1998. Regulation of the heat shock conjugate Hsc70 in the mammalian cell: the characterization of the anti-apoptotic protein BAG-1 provides novel insights. Biol. Chem. 379:269-274. [PubMed] [Google Scholar]
- 29.Höhfeld, J., and S. Jentsch. 1997. GrpE-like regulation of the hsc70 chaperone by the anti-apoptotic protein BAG-1. EMBO J. 16:6209-6216. (Erratum, 17:847, 1998.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Höhfeld, J., Y. Minami, and F. U. Hartl. 1995. Hip, a novel cochaperone involved in the eukaryotic Hsc70/Hsp40 reaction cycle. Cell 83:589-598. [DOI] [PubMed] [Google Scholar]
- 31.Horton, L. E., P. James, E. A. Craig, and J. O. Hensold. 2001. The yeast hsp70 homologue Ssa is required for translation and interacts with Sis1 and Pab1 on translating ribosomes. J. Biol. Chem. 276:14426-14433. [DOI] [PubMed] [Google Scholar]
- 32.Kabani, M., J. M. Beckerich, and C. Gaillardin. 2000. Sls1p stimulates Sec63p-mediated activation of Kar2p in a conformation-dependent manner in the yeast endoplasmic reticulum. Mol. Cell. Biol. 20:6923-6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kabani, M., A. Boisrame, J. M. Beckerich, and C. Gaillardin. 2000. A highly representative two-hybrid genomic library for the yeast Yarrowia lipolytica. Gene 241:309-315. [DOI] [PubMed] [Google Scholar]
- 34.Kelley, W. L. 1998. The J-domain family and the recruitment of chaperone power. Trends Biochem. Sci. 23:222-227. [DOI] [PubMed] [Google Scholar]
- 35.Kelley, W. L., D. M. Cyr, and M. G. Douglas. 1999. Molecular chaperones: how J domains turn on Hsp70s. Curr. Biol. 9:R305-R308. [DOI] [PubMed] [Google Scholar]
- 36.Kim, S., B. Schilke, E. A. Craig, and A. L. Horwich. 1998. Folding in vivo of a newly translated yeast cytosolic enzyme is mediated by the SSA class of cytosolic yeast Hsp70 proteins. Proc. Natl. Acad. Sci. USA 95:12860-12865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levy, E. J., J. McCarty, B. Bukau, and W. J. Chirico. 1995. Conserved ATPase and luciferase refolding activities between bacteria and yeast Hsp70 chaperones and modulators. FEBS Lett. 368:435-440. [DOI] [PubMed] [Google Scholar]
- 38.Liberek, K., J. Marszalek, D. Ang, C. Georgopoulos, and M. Zylicz. 1991. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc. Natl. Acad. Sci. USA 88:2874-2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu, Q., J. Krzewska, K. Liberek, and E. A. Craig. 2001. Mitochondrial Hsp70 Ssc1: role in protein folding. J. Biol. Chem. 276:6112-6118. [DOI] [PubMed] [Google Scholar]
- 40.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 41.Lu, Z., and D. M. Cyr. 1998. Protein folding activity of Hsp70 is modified differentially by the hsp40 co-chaperones Sis1 and Ydj1. J. Biol. Chem. 273:27824-27830. [DOI] [PubMed] [Google Scholar]
- 42.Mally, A., and S. N. Witt. 2001. GrpE accelerates peptide binding and release from the high affinity state of DnaK. Nat. Struct. Biol. 8:254-257. [DOI] [PubMed] [Google Scholar]
- 43.McCarty, J. S., A. Buchberger, J. Reinstein, and B. Bukau. 1995. The role of ATP in the functional cycle of the DnaK chaperone system. J. Mol. Biol. 249:126-137. [DOI] [PubMed] [Google Scholar]
- 44.McClellan, A. J., and J. L. Brodsky. 2000. Mutation of the ATP-binding pocket of SSA1 indicates that a functional interaction between Ssa1p and Ydj1p is required for post-translational translocation into the yeast endoplasmic reticulum. Genetics 156:501-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McClellan, A. J., J. B. Endres, J. P. Vogel, D. Palazzi, M. D. Rose, and J. L. Brodsky. 1998. Specific molecular chaperone interactions and an ATP-dependent conformational change are required during posttranslational protein translocation into the yeast ER. Mol. Biol. Cell 9:3533-3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCracken, A. A., and J. L. Brodsky. 1996. Assembly of ER-associated protein degradation in vitro: dependence on cytosol, calnexin, and ATP. J. Cell Biol. 132:291-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mumberg, D., R. Muller, and M. Funk. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156:119-122. [DOI] [PubMed] [Google Scholar]
- 48.Mutka, S. C., and P. Walter. 2001. Multifaceted physiological response allows yeast to adapt to the loss of the signal recognition particle-dependent protein-targeting pathway. Mol. Biol. Cell 12:577-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nelson, R. J., T. Ziegelhoffer, C. Nicolet, M. Werner-Washburne, and E. A. Craig. 1992. The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell 71:97-105. [DOI] [PubMed] [Google Scholar]
- 50.Nishikawa, S. I., S. W. Fewell, Y. Kato, J. L. Brodsky, and T. Endo. 2001. Molecular chaperones in the yeast endoplasmic reticulum maintain the solubility of proteins for retrotranslocation and degradation. J. Cell Biol. 153:1061-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ogg, S. C., and P. Walter. 1995. SRP samples nascent chains for the presence of signal sequences by interacting with ribosomes at a discrete step during translation elongation. Cell 81:1075-1084. [DOI] [PubMed] [Google Scholar]
- 52.Pfund, C., N. Lopez-Hoyo, T. Ziegelhoffer, B. A. Schilke, P. Lopez-Buesa, W. A. Walter, M. Wiedmann, and E. A. Craig. 1998. The molecular chaperone Ssb from Saccharomyces cerevisiae is a component of the ribosome-nascent chain complex. EMBO J. 17:3981-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pierpaoli, E. V., E. Sandmeier, H. J. Schonfeld, and P. Christen. 1998. Control of the DnaK chaperone cycle by substoichiometric concentrations of the co-chaperones DnaJ and GrpE. J. Biol. Chem. 273:6643-6649. [DOI] [PubMed] [Google Scholar]
- 54.Raynes, D. A., and V. Guerriero, Jr. 1998. Inhibition of Hsp70 ATPase activity and protein renaturation by a novel Hsp70-binding protein. J. Biol. Chem. 273:32883-32888. [DOI] [PubMed] [Google Scholar]
- 55.Rothblatt, J. A., R. J. Deshaies, S. L. Sanders, G. Daum, and R. Schekman. 1989. Multiple genes are required for proper insertion of secretory proteins into the endoplasmic reticulum in yeast. J. Cell Biol. 109:2641-2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sanders, S. L., K. M. Whitfield, J. P. Vogel, M. D. Rose, and R. W. Schekman. 1992. Sec61p and BiP directly facilitate polypeptide translocation into the ER. Cell 69:353-365. [DOI] [PubMed] [Google Scholar]
- 57.Schmid, D., A. Baici, H. Gehring, and P. Christen. 1994. Kinetics of molecular chaperone action. Science 263:971-973. [DOI] [PubMed] [Google Scholar]
- 58.Shlomai, J., and A. Kornberg. 1980. A prepriming DNA replication enzyme of Escherichia coli. II. Actions of protein n′: a sequence-specific, DNA-dependent ATPase. J. Biol. Chem. 255:6794-6798. [PubMed] [Google Scholar]
- 59.Sondermann, H., C. Scheufler, C. Schneider, J. Hohfeld, F. U. Hartl, and I. Moarefi. 2001. Structure of a Bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors. Science 291:1553-1557. [DOI] [PubMed] [Google Scholar]
- 60.Stirling, C. J., J. Rothblatt, M. Hosobuchi, R. Deshaies, and R. Schekman. 1992. Protein translocation mutants defective in the insertion of integral membrane proteins into the endoplasmic reticulum. Mol. Biol. Cell 3:129-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suh, W. C., W. F. Burkholder, C. Z. Lu, X. Zhao, M. E. Gottesman, and C. A. Gross. 1998. Interaction of the Hsp70 molecular chaperone, DnaK, with its cochaperone DnaJ. Proc. Natl. Acad. Sci. USA 95:15223-15228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suh, W. C., C. Z. Lu, and C. A. Gross. 1999. Structural features required for the interaction of the Hsp70 molecular chaperone DnaK with its cochaperone DnaJ. J. Biol. Chem. 274:30534-30539. [DOI] [PubMed] [Google Scholar]
- 63.Sullivan, C. S., J. D. Tremblay, S. W. Fewell, J. A. Lewis, J. L. Brodsky, and J. M. Pipas. 2000. Species-specific elements in the large T-antigen J domain are required for cellular transformation and DNA replication by simian virus 40. Mol. Cell. Biol. 20:5749-5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Takayama, S., J. C. Reed, V. R. Agashe, and F. U. Hartl. 2001. Molecular chaperone targeting and regulation by BAG family proteins. Nat. Cell Biol. 3:E237-E241. [DOI] [PubMed] [Google Scholar]
- 65.Travers, K. J., C. K. Patil, L. Wodicka, D. J. Lockhart, J. S. Weissman, and P. Walter. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101:249-258. [DOI] [PubMed] [Google Scholar]
- 66.Tyson, J. R., and C. J. Stirling. 2000. LHS1 and SIL1 provide a lumenal function that is essential for protein translocation into the endoplasmic reticulum. EMBO J. 19:6440-6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ungewickell, E., H. Ungewickell, S. E. Holstein, R. Lindner, K. Prasad, W. Barouch, B. Martin, L. E. Greene, and E. Eisenberg. 1995. Role of auxilin in uncoating clathrin-coated vesicles. Nature 378:632-635. [DOI] [PubMed] [Google Scholar]
- 68.Werner, E. D., J. L. Brodsky, and A. A. McCracken. 1996. Proteasome-dependent endoplasmic reticulum-associated protein degradation: an unconventional route to a familiar fate. Proc. Natl. Acad. Sci. USA 93:13797-13801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yan, W., and E. A. Craig. 1999. The glycine-phenylalanine-rich region determines the specificity of the yeast Hsp40 Sis1. Mol. Cell. Biol. 19:7751-7758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yan, W., B. Schilke, C. Pfund, W. Walter, S. Kim, and E. A. Craig. 1998. Zuotin, a ribosome-associated DnaJ molecular chaperone. EMBO J. 17:4809-4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang, Y., S. Michaelis, and J. L. Brodsky. 2002. CFTR expression and ER associated degradation in yeast. Cystic fibrosis methods and protocols, p. 257-265. In W. R. Skach (ed.). Methods in molecular medicine. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 72.Zhang, Y., G. Nijbroek, M. L. Sullivan, A. A. McCracken, S. C. Watkins, S. Michaelis, and J. L. Brodsky. 2001. Hsp70 molecular chaperone facilitates endoplasmic reticulum-associated protein degradation of cystic fibrosis transmembrane conductance regulator in yeast. Mol. Biol. Cell 12:1303-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhong, T., and K. T. Arndt. 1993. The yeast SIS1 protein, a DnaJ homolog, is required for the initiation of translation. Cell 73:1175-1186. [DOI] [PubMed] [Google Scholar]
- 74.Zhu, X., X. Zhao, W. F. Burkholder, A. Gragerov, C. M. Ogata, M. E. Gottesman, and W. A. Hendrickson. 1996. Structural analysis of substrate binding by the molecular chaperone DnaK. Science 272:1606-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ziegelhoffer, T., P. Lopez-Buesa, and E. A. Craig. 1995. The dissociation of ATP from hsp70 of Saccharomyces cerevisiae is stimulated by both Ydj1p and peptide substrates. J. Biol. Chem. 270:10412-10419. [DOI] [PubMed] [Google Scholar]