Abstract
The Saccharomyces cerevisiae Paf1-RNA polymerase II (Pol II) complex is biochemically and functionally distinct from the Srb-mediator form of Pol II holoenzyme and is required for full expression of a subset of genes. In this work we have used tandem affinity purification tags to isolate the Paf1 complex and mass spectrometry to identify additional components. We have established that Ctr9, Rtf1, and Leo1 are factors that associate with Paf1, Cdc73, and Pol II, but not with the Srb-mediator. Deletion of either PAF1 or CTR9 leads to similar severe pleiotropic phenotypes, which are unaltered when the two mutations are combined. In contrast, we found that deletion of LEO1 or RTF1 leads to few obvious phenotypes, although mutation of RTF1 suppresses mutations in TATA-binding protein, alters transcriptional start sites, and affects elongation. Remarkably, deletion of LEO1 or RTF1 suppresses many paf1Δ phenotypes. In particular, an rtf1Δ paf1Δ double mutant grew faster, was less temperature sensitive, and was more resistant to caffeine and hydroxyurea than a paf1Δ single mutant. In addition, expression of the G1 cyclin CLN1, reduced nearly threefold in paf1Δ, is restored to wild-type levels in the rtf1Δ paf1Δ double mutant. We suggest that lack of Paf1 results in a defective complex and a block in transcription, which is relieved by removal of Leo1 or Rtf1.
RNA polymerase II (Pol II) requires both general transcription factors (GTFs) to form a stable initiation complex at promoters (reviewed in references 6, 24, and 60) and mediators and coactivators to communicate signals from transcriptional activators and repressors (reviewed in reference 36). Large, apparently preassembled forms of Pol II including many of these accessory factors may be the actual initiating forms of the enzyme in vivo. However, recent studies have demonstrated that mediator complexes may also function in the absence of Pol II (7, 3a, 40). The best characterized of these holoenzyme forms of Pol II is the Srb-mediator complex from Saccharomyces cerevisiae (reviewed in references 34 and 35). In addition to core Pol II, this complex contains the mediator, a collection of about 20 proteins including the Srb proteins, and the GTFs transcription factor IIB (TFIIB), TFIIF, TFIIE, and TFIIH. Complexes similar to the yeast Srb-mediator have also been described in multicellular organisms (reviewed in reference 13).
Our laboratory has isolated and characterized a Pol II complex from yeast containing Paf1, Cdc73, Hpr1, and Ccr4 that is biochemically distinct from the Srb-mediator complex (4, 47, 57). The Paf1 complex contains TFIIF and TFIIB and the coactivators Gal11 and Sin4 that are also present in the Srb-mediator complex. However, the Srb and Med proteins are not found in the Paf1 complex, and Paf1, Cdc73, Hpr1, and Ccr4 are not found with the proteins of the Srb-mediator complex. Although genes encoding Paf1 complex components are not essential, mutations in these genes exhibit a variety of phenotypes indicative of defects in gene expression (4, 47; J. L. Betz, M. Chang, T. M. Washburn, S. E. Porter, and J. A. Jaehning, submitted for publication). Consistent with these phenotypes, mutation of Paf1 complex genes results in changes in abundance of a small but significant subset of yeast transcripts (4, 47; M. Chang, J. Fostel, and J. A. Jaehning, unpublished data). In contrast, many of the genes encoding Srb and Med proteins are essential (reviewed in reference 34), and loss of at least one of the essential Srb proteins (Srb4) results in diminished expression of most, if not all, yeast genes (18).
Transcripts differentially expressed in strains lacking Paf1 include cell wall biosynthetic genes (4) and many cell cycle-regulated genes (S. E. Porter, R. M. Washburn, M. Chang, and J. A. Jaehning, submitted for publication). Loss of Paf1, Cdc73, or Ccr4 results in cell wall integrity defects, and loss of Paf1, Cdc73, or Hpr1 is correlated with elevated rates of recombination between direct repeats (4). The fact that these two otherwise unrelated phenotypes are also observed for mutations in genes in the protein kinase C-mitogen-activated protein kinase signaling pathway led us to establish that the Paf1 complex plays an important role in the transmission of signals from Pkc1 to downstream target genes (4). Although Paf1 and Cdc73 have not been identified in other complexes, both Hpr1 and Ccr4 are present in other non-Pol II-containing complexes. Hpr1 is part of the Hpr1/THO complex that influences transcription and recombination (5), and Ccr4 is part of the Ccr4/Caf1/NOT complex that functions in both transcript synthesis and turnover (27, 28, 52).
Recently Koch and coworkers identified Paf1 and Cdc73 as factors associated with Ctr9, a protein required for full expression of the G1 cyclin CLN2 (22). Loss of Paf1 or Ctr9 leads to similar severe pleiotropic phenotypes, and loss of both genes does not result in an enhanced phenotype. These genetic and biochemical observations are consistent with the idea that Ctr9 and Paf1 function in the same pathways. To further characterize the Paf1 complex, and to determine whether Ctr9 is also one of the Pol II-associated proteins in this complex, we have used the tandem affinity purification (TAP) (43) system to isolate the Paf1 complex. We have confirmed the presence of Ctr9 in the Pol II complex, and we have identified new components of the Paf1 complex, including Rtf1 and Leo1. The absence of these newly identified factors from the Srb-mediator complex further establishes the biochemical differences between these two forms of Pol II. Our observation that loss of Rtf1 or Leo1 suppresses many of the pleiotropic phenotypes caused by loss of Paf1 or Ctr9 has led us to a new model of the role of the Paf1 complex in transcription of yeast genes.
MATERIALS AND METHODS
Yeast strains, growth conditions, and genetic techniques.
The S. cerevisiae strains used in this study were derived from strain YJJ662 (MATa leu2Δ1 his3Δ200 ura3-52) (47) and are all isogenic except for the specific deletion or modification indicated: YJJ577 (paf1Δ::HIS3), YJJ1303 (rtf1Δ::kanr), YJJ1326 (rtf1Δ::kanr paf1Δ::HIS3), YJJ1336 (leo1Δ::kanr), YJJ1339 (trp1Δ::kanr), YJJ1361 (leo1Δ::kanr paf1Δ::HIS3), and YJJ1364 (ctr9Δ::kanr paf1 Δ::HIS3). Details of TAP-tagged and hemagglutinin (HA)-tagged strain construction are described below. YJJ1339 was used to create HA-tagged Leo1, resulting in strain YJJ1330 [LEO1::LEO1HA6(KlTRP1)]. The TAP-tagged strains were derived from YJJ662 and include YJJ1307 [CDC73::CDC73CBP-TEV-ProtA(KlURA3)], YJJ1308 [SRB5::SRB5CBP-TEV-ProtA(KlURA3)], YJJ1329 [CTR9::CTR9CBP-TEV-ProtA(KlURA3)], and YJJ1334 [HPR1::HPR1CBP-TEV-ProtA(KlURA3)]. Strains containing both Leo1-HA and TAP tags are YJJ1309 [CDC73::CDC73CBP-TEV-ProtA(KlURA3) LEO1::LEO1HA6(KlTRP1)], YJJ1331 [SRB5::SRB5CBP-TEV-ProtA(KlURA3) LEO1::LEO1HA6(KlTRP1)], and YJJ1332 [CTR9::CTR9CBP-TEV-ProtA(KlURA3) LEO1::LEO1HA6(KlTRP1)]. Strains were grown in YPD with 2% glucose using standard methods (15).
Construction of TAP-tagged strains.
Yeast chromosomal genes CDC73, SRB5, HPR1, and CTR9 were epitope tagged with a C-terminal TAP tag (43). The tagging cassette consists of two immunoglobulin G (IgG) binding domains of Staphylococcus aureus protein A and a calmodulin binding peptide separated by a tobacco etch virus (TEV) protease site. The Kluyveromyces lactis URA3 gene located at the 3′ end of the tag served as a selectable marker. PCR fragments were generated using a 5′ oligonucleotide corresponding to the last 51 bp of the gene of interest, excluding the stop codon, plus 17 bp matching the TAP tag, and a 3′ oligonucleotide corresponding to the 51 bp downstream of the stop codon plus 17 bases of the tagging cassette. Oligonucleotide sequences are available upon request. The PCR fragments were transformed into strain YJJ662, generating strains YJJ1307 (Cdc73-TAP), YJJ1308 (Srb5-TAP), YJJ1329 (Ctr9-TAP), and YJJ1334 (Hpr1-TAP). TAP-tagged constructs were confirmed by Western blotting and PCR.
Construction of Leo1-HA strains.
Leo1 was C-terminally epitope tagged using the pYM3 plasmid (19) containing a six-HA peptide and a K. lactis TRP1 marker. To permit selection on synthetic complete medium lacking Trp, TRP1 was deleted in YJJ662, using the KANMX4 cassette (54), resulting in YJJ1339. TAP-tagged constructs were generated as described above. The Leo1-HA PCR fragment was transformed into YJJ1339. The resulting strains containing Leo1-HA were YJJ1330 (Leo1-HA), YJJ1309 (Cdc73-TAP, Leo1-HA), YJJ1331 (Srb5-TAP, Leo1-HA), and YJJ1332 (Ctr9-TAP, Leo1-HA). HA-tagged constructs were confirmed by Western blotting and PCR.
Extract preparation and purification of TAP-tagged complexes.
The TAP-tagged strains were grown in YPD medium to a density of 2 × 107 cells/ml, and transcriptionally active whole-cell extracts were prepared as described previously (56). Briefly, protein pellets were resuspended after NH4SO4 precipitation in buffer A (20 mM HEPES-KOH [pH 7.9], 10% glycerol, 10 mM EGTA, 10 mM MgSO4, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF], 2 mM benzamidine hydrochloride, leupeptin [0.5 μg/ml], bestatin [0.35 μg/ml], pepstatin [0.4 μg/ml]) to a final protein concentration of 20 to 30 mg/ml. The resuspended proteins were extensively dialyzed against buffer A containing 100 mM NH4SO4. Approximately 1 g of total protein was incubated with 500 μl of rabbit IgG agarose (Sigma) at 4°C for 2 h. Beads were washed three times with 50 ml of buffer A and then equilibrated in TEV protease cleavage buffer (20 mM HEPES-KOH [pH 8.0], 10% glycerol, 0.5 mM EDTA, 1 mM DTT, 1 mM PMSF, 2 mM benzamidine hydrochloride, leupeptin [0.5 μg/ml], bestatin [0.35 μg/ml], pepstatin [0.4 μg/ml]). Proteins were eluted from the beads by adding 200 U of TEV protease in 1 ml of TEV protease cleavage buffer and incubating the mixture at ambient temperature for 2 h. The eluted proteins were concentrated with a Centricon YM-30 filter (Millipore) and applied at 0.2 ml/min to a Superose 6 HR 10/30 (Pharmacia) gel filtration column equilibrated in buffer B [30 mM HEPES-KOH (pH 7.9), 8% glycerol, 2 mM EDTA, 2 mM EGTA, 0.1 M (NH4)2SO4, 0.05% NP-40, 5 mM β-glycerophosphate, 1 mM DTT, 1 mM PMSF, 2 mM benzamidine hydrochloride, leupeptin (0.5 μg/ml), bestatin (0.35 μg/ml), pepstatin (0.4 μg/ml)]. Fractions (200 μl) were collected. Fractions were trichloroacetic acid (TCA) precipitated (38) and analyzed by Western blotting. Transcriptional activity was measured by a nonselective transcription assay as described elsewhere (58). To estimate the size of protein complexes, the following markers were used to calibrate the Superose 6 column: aggregated MCM protein (>2,000 kDa; gift from X. Chen, University of Colorado Health Sciences Center), thyroglobulin (669 kDa), apoferritin (443 kDa), and β-amylase (200 kDa).
Isolation of Leo1-HA complexes.
The C-terminally Leo1 HA-tagged strains were grown in 100 ml of YPD to a density of 1.4 × 107 cells/ml. Cells were pelleted and washed in sterile water, freeze-thawed in liquid nitrogen, and then resuspended in 1.0 ml of whole-cell extract lysis buffer [0.2 M Tris-HCl (pH 7.9), 0.39 M (NH4)2SO4, 10 mM MgSO4, 20% (vol/vol) glycerol, 1 mM EDTA, 1 mM DTT, and 1 mM PMSF, plus protease inhibitors as above]. Cells were lysed with the addition of 1 ml of glass beads and vortexed four times for 1 min while cooling. Lysates were clarified at 13,000 rpm with a Baxter Biofuge 13 for 20 min at 4°C. Protein concentrations were determined by Bradford assay (GibcoBRL). Approximately 1.5 mg of total protein was used for TAP tag purification or immunoprecipitation. TAP-tagged extracts were incubated with 25 μl of rabbit IgG agarose at 4°C for 2 h. Beads, equilibrated in TEV protease cleavage buffer, were washed three times with 1 ml of buffer B. Proteins were eluted from the beads with the addition of 25 U of TEV protease in 50 μl of TEV protease cleavage buffer. Eluates were TCA precipitated and analyzed by Western blotting.
Strains YJJ662 and YJJ1330 were used for immunoprecipitation studies. To reduce the binding of nonspecific proteins to the protein A-coupled beads, extracts were preincubated with 25 μl of Sepharose 4B (Sigma) for 1 h at 4°C. The beads were removed, 1 μl of 12CA5 antibody (Roche) was added, and extracts were incubated at 4°C for 1 h. To isolate Leo1-HA complexes, protein A immobilized on Sepharose 4B (Sigma) was added to the extract. After binding for 1 h at 4°C, the beads were pelleted and washed four times with buffer B. Beads were suspended in 20 μl of NuPAGE LDS Sample buffer (Invitrogen) and heated to 70°C for 10 min, and aliquots were resolved on a 4-to-12% Bis-Tris NuPAGE gel (Invitrogen).
Western blotting.
Proteins were resolved on a 4 to 12% bis-Tris NuPAGE gel and then transferred to a polyvinylidene difluoride (Millipore) membrane. Proteins were detected with the following primary antibodies: anti-Rpb1 (8WG16; D. Bentley); anti-Cdc73, anti-Paf1, and anti-TFIIB (X. Shi); anti-Srb5 (R. Young); anti-HA (Roche); anti-Spt5 (G. Hartzog); anti-Rtf1 (K. Arndt); and anti-Rpt5 and anti-Rpt6 (Sug1) (R. Deshaies). Horseradish peroxidase-conjugated antibody was the secondary antibody, and proteins were detected using a Chemiluminescent Reagent Plus kit from NEN.
Mass spectrometry analysis.
Fractions from the Superose 6 gel filtration column were pooled, TCA precipitated, and resolved on a 4-to-12% Bis-Tris NuPAGE gel. Proteins were visualized by staining with NOVEX colloidal blue. Protein bands were excised and processed as described previously (25). Briefly, samples were reduced and alkylated by incubating with 2.8 mM DTT in 100 mM NH4CO3 at 60°C for 15 min, followed by the addition of 10 μl of 100 mM iodoacetamide. After a 15-min incubation at ambient temperature in the dark, the gel slices were washed with 100 μl of 50% acetonitrile in 50 mM NH4CO3 with shaking for 20 min. To dehydrate the gel slices, 100 μl of 100% acetonitrile was added and incubated for 10 min. The solvent was removed, and the gel pieces were completely dried in a vacuum centrifuge. The gel pieces were swollen with the addition of 0.2 μg of modified trypsin (Promega) in 10 μl of 25 mM NH4CO3. The gel pieces were kept wet during enzymatic cleavage (37°C, overnight) with 25 mM NH4CO3. The supernatant containing the digested peptides was collected, and the remaining peptides were extracted from the gel twice with 20 μl of 1% trifluoroacetic acid. All supernatants were pooled and dried in a vacuum centrifuge. The sample was reconstituted in 3 to 5 μl of 1% trifluoroacetic acid and desalted using C18 Ziptips (Millipore). Samples were processed on a PE-Biosystems Voyager DE-PRO matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) spectrometer. The masses of the tryptic peptides were used to search for protein candidates using the programs Protein Prospector and ProFound. Up to two undigested trypsin sites and a mass tolerance of ±1.0 Da were allowed.
RNA isolation and analysis.
Cells were grown to a density of 1.3 × 107 cells per ml, and RNA was isolated as described (4). Fifteen micrograms of total RNA per sample was fractionated by formaldehyde gel electrophoresis according to standard methods (33). RNA was transferred to ZetaProbe GT membranes (Bio-Rad). The CLN1 probe, a gel-purified 500-bp fragment from a plasmid which contains the 1.2-kb CLN1 coding region (R. Sclafani), was labeled using the random prime method (RadPrime kit; GibcoBRL). The 18S rRNA oligonucleotide probe (5′-GCTTATACTTAGACATGCAT-3′) was 5′ end labeled. Hybridizations and washes were performed at either 64°C for the CLN1 probe or at 41°C for the 18S rRNA probe. Blots were exposed to a phosphorimaging screen, and bands were quantitated using Quanity One imaging software. Signals were normalized to 18S rRNA.
RESULTS
Affinity purification of the Paf1 complex.
Previous studies of the Paf1 complex used affinity purification techniques to establish the connection to Pol II and to identify some of the components of the complex. Paf1 and Cdc73 were originally identified as RNA Pol II-associated proteins using a monoclonal antibody directed against the C-terminal domain (CTD) of Pol II to isolate the enzyme from yeast transcription extracts (57). The RNA Pol II-associated proteins also included GTFs TFIIF and TFIIB. Subsequently, GST-tagged forms of Paf1, Cdc73, and TFIIF were used to demonstrate that the complex is distinct from the Srb-mediator form of Pol II and that Hpr1 and Ccr4 are also associated with the complex (47). Although the plasmid-borne GST-tagged constructs functionally complemented the genes they replaced (47), they were overexpressed relative to the natural chromosomal copies of the genes. In addition, dimerization of the GST tag and associated proteins (32, 41) causes difficulties in determining the size of the complex. Therefore, to further characterize the composition and function of the Paf1 complex, and to compare it to the Srb-mediator complex, we epitope-tagged the chromosomal copies of CDC73 and SRB5 with a TAP tag (43), as described in Materials and Methods. The TAP tag includes two IgG binding domains and a calmodulin binding peptide separated by a TEV protease site. This C-terminal tag should not alter the normal expression of the tagged genes from their native promoter. Consistent with this supposition, we found that the TAP-tagged strains did not display any of the pleiotropic phenotypes (including slow growth and temperature sensitivity) found for deletion mutants of these genes (data not shown).
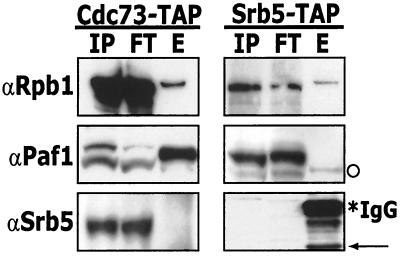
Yeast whole-cell transcription extracts were prepared from the tagged strains, the TAP-tagged constructs were isolated by binding to IgG agarose, and bound proteins were eluted by TEV protease digestion. We found that both the Cdc73-tagged Paf1 complex and the Srb5-tagged Srb-mediator complex could be isolated by this procedure (see below) but that neither complex was stable to subsequent purification on calmodulin beads. Apparently the interactions with Pol II are not stable to the second stringent affinity step, although they are maintained under the high-salt (100 mM NH4SO4) conditions used for binding to the IgG agarose. Figure 1 shows the results from the IgG agarose isolation of proteins associated with TAP-tagged Cdc73 and Srb5. The input, flowthrough, and eluate fractions from both preparations were probed with antibodies to the largest subunit of Pol II (αRpb1), Paf1 (αPaf1), and Srb5 (αSrb5). Pol II is clearly associated with both tagged proteins, Paf1 is only present in the TAP-tagged Cdc73 eluate, and Srb5 (in its tagged form) is uniquely found in the TAP-tagged Srb5 eluate (Fig. 1). These results establish that the TAP tag protocol specifically and efficiently isolates distinct Pol II complexes from tagged strains.
FIG. 1.
The Paf1 complex is distinct from the Srb-Med complex. Whole-cell extracts made from a TAP-tagged Cdc73 strain (YJJ1307) and a TAP-tagged Srb5 strain (YJJ1308) were incubated with rabbit IgG agarose. Unbound proteins were washed away, and proteins that associate with TAP-Cdc73 were eluted by the addition of TEV protease. Fractions (IP, input; FT, flowthrough; E, eluate) were loaded onto a 4-to-12% NuPAGE gel and analyzed by Western blotting with antibodies as indicated. The asterisk indicates cross-reacting IgG from the IgG agarose column, and the arrow indicates the TEV protease cleaved form of TAP-Srb5. We observe two cross-reacting bands with the α-Paf1 antibody (see also Fig. 5B and reference 47); however, the faint band indicated by the open circle in the TAP-tagged Srb5 eluate does not correspond to either form of Paf1, and its presence is not reproducible (see Fig. 5B).
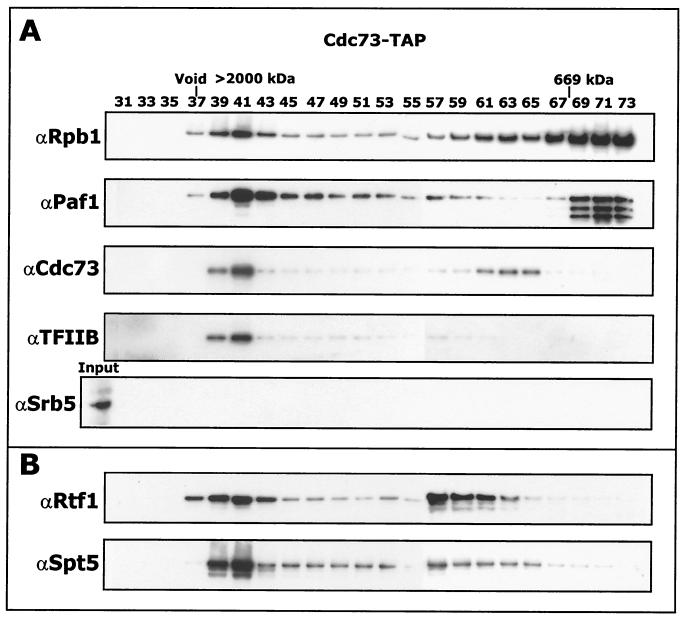
To further purify the Paf1 complex it was fractionated by chromatography on a Superose 6 gel filtration column (Fig. 2). The elution profile of Paf1 complex components was monitored by immunoblotting using the antibodies indicated in Fig. 2A. A high-molecular-mass complex containing Pol II, Paf1, Cdc73, and TFIIB, but not Srb5, was observed eluting from the column at approximately 1.7 MDa. Cdc73 is present in the high-molecular-mass complex and also in an intermediate position overlapping with a small amount of Pol II. Since Cdc73 is known to bind Pol II directly (47), this may represent a subcomplex of the larger Paf1 complex. We also observed significant amounts of free core Pol II in the lower-molecular-mass fractions (∼600 kDa), a position in the fractionation that also contained proteolyzed forms of Paf1 (Fig. 2, see also below). When the TAP-tagged Srb5 complex was fractionated on Superose 6 under identical conditions, we found that all of the Pol II was in the lower-molecular-mass position of free core, indicating that the Paf1 complex may be somewhat more stable to purification than the Srb-mediator complex.
FIG. 2.
The Paf1 complex is approximately 1.7 MDa. Proteins that associate with Cdc73-TAP after chromatography on IgG agarose and elution with TEV protease were subjected to gel filtration chromatography on a Superose 6 gel filtration column. Fractions were separated on a 4-to-12% NuPAGE gel and analyzed by Western blotting with the indicated antibodies. Sizes were determined by calibrating the gel filtration column with the indicated standards as described in Materials and Methods.
In addition to the antibody identification of Pol II, we also performed a nonspecific transcriptional assay for RNA polymerase activity on the Superose 6 fractions. Transcriptional activity was detected in fractions 39 to 47 corresponding to the high-molecular-mass Paf1 complex and also in fractions 69 to 73 corresponding to core Pol II (data not shown). Similar nonselective transcription assays performed on GST-tagged constructs (see reference 47) revealed that approximately 2% of the total Pol II was associated with the Paf1 complex (data not shown).
Identification of Paf1 complex components by mass spectrometry.
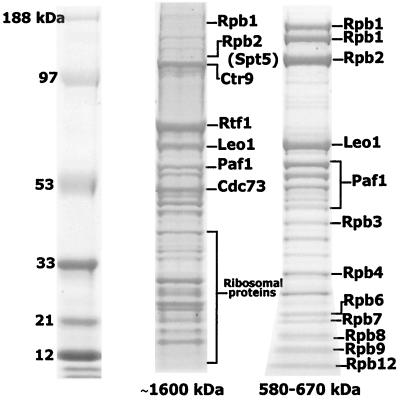
To identify proteins that associate with the 1.7-MDa TAP-tagged Cdc73 complex, we performed mass spectrometry on individual protein bands excised from a sodium dodecyl sulfate (SDS) gel (Fig. 3). Peptides from the trypsin-digested proteins were eluted and analyzed by MALDI-TOF mass spectrometry as described in Materials and Methods. The tryptic peptide masses were used to search for protein candidates in several Web-based data sets (ProFound, Protein Prospector, and MASCOT). Identification was considered positive if the top candidate had a probability score near 1.0 and the second-best candidate had a significantly lower probability. Proteins that were identified in this way are marked in Fig. 3. Newly identified proteins of the Paf1 complex include Ctr9, Rtf1, and Leo1. The ribosomal proteins identified in Fig. 3 are nonspecific contaminants in the high-molecular-mass fractions, as also observed by others isolating yeast complexes (28, 29, 45). A few peptides corresponding to Spt5 were also identified in the same gel band that contained Rpb2 and Ctr9, but the lower probability score precluded a confident match (indicated by the parentheses in Fig. 3). We also analyzed the proteins in the lower-molecular-mass fractions containing free Pol II (Fig. 3, 580 to 670 kDa). We were able to unambiguously identify most of the Pol II subunits, plus Leo1 and both intact and proteolyzed forms of Paf1.
FIG. 3.
Ctr9, Rtf1, Leo1, and Spt5 associate with the Paf1 complex. Fractions 37 to 41 and 69 to 73 from the Superose 6 gel filtration column were pooled, TCA precipitated, and separated on a 4-to-12% NuPAGE gel. Protein bands were visualized by colloidal blue staining and processed for MALDI-TOF analysis. Proteins that were accurately identified are marked.
Validation of newly identified Paf1 complex factors.
To confirm the identifications from mass spectrometry, we showed by Western blotting that Rtf1 and Spt5 were present in the same high-molecular-mass complex with Pol II and Paf1 (Fig. 2B). In addition, we observed Rtf1 and Spt5 in an intermediate fraction between the Pol II complex and free core Pol II, suggesting that they might exist in a separate subcomplex.
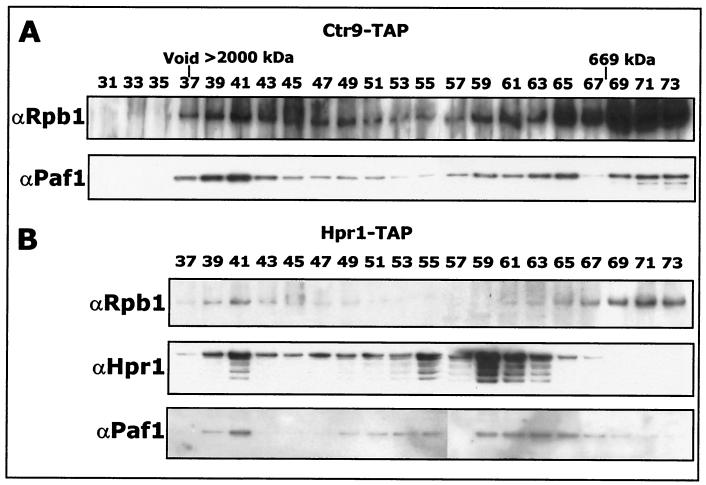
The Koch laboratory described an association between Ctr9 and Paf1 and Cdc73 but did not establish a connection to Pol II (22). To confirm that Ctr9 is part of the Paf1 complex, we constructed a strain bearing a TAP-tagged form of Ctr9 and confirmed that the tagged strain had a wild-type phenotype. We isolated TAP-tagged Ctr9-associated proteins as described above for TAP-tagged Cdc73. Fractions from the Superose 6 gel filtration column were immunoblotted with antibodies specific for Pol II and Paf1 (Fig. 4A). Both Paf1 and Pol II were found in these fractions, again in the high-molecular-mass complex and at the position of free core Pol II. Based on these observations, and the mass spectrometry identification, we conclude that Ctr9 is part of the Paf1 complex.
FIG. 4.
Both Ctr9 and Hpr1 associate with RNA Pol II and Paf1. (A) Ctr9-associated proteins were purified from a TAP-tagged Ctr9 strain (YJJ1329). Samples were fractionated on a Superose 6 gel filtration column and immunoblotted with the indicated antibodies. (B) Hpr1-associated proteins were purified from a TAP-tagged Hpr1 strain (YJJ1334). Samples were fractionated on a gel filtration column and immunoblotted with the indicated antibodies.
We have previously used a GST-tagged form of Hpr1 to establish that this protein is associated with Pol II and Paf1 (4). Because Hpr1 has also been found in another, non-Pol II-containing complex, the Hpr1/Tho complex (5), we confirmed the Pol II association by analyzing proteins associated with TAP-tagged Hpr1. The comigration of Pol II, Paf1, and Hpr1 in the high-molecular-mass fractions of the Superose 6 column (Fig. 4B) reaffirms that Hpr1 associates with the Paf1 complex.
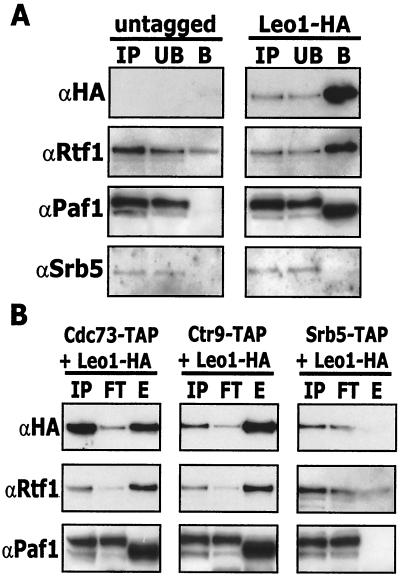
There is little information about Leo1 other than the fact that it is a very hydrophilic protein encoded by a nonessential gene (30). We therefore constructed strains containing chromosomally HA-tagged Leo1 in wild-type and in various TAP-tagged backgrounds (Materials and Methods). Using an anti-HA antibody to precipitate HA-tagged Leo1 we found both Paf1 and Rtf1, but not Srb5, in the bound fraction (Fig. 5A). However, although Paf1 was not present in the precipitate from the untagged strain, a small amount of Rtf1 was detected. We have observed low levels of nonspecific association of Rtf1 in many of the different affinity assays performed in these studies (see also below); however, the specific association of Rtf1 is clearly apparent in Fig. 5A over this background. To confirm that Leo1 associates specifically with the Paf1 complex, we used double-tagged strains (TAP tags plus Leo1-HA), first isolating the complexes with the TAP tag and then probing for the presence or absence of HA-tagged Leo1 (Fig. 5B). We found that Leo1, like Paf1 and Rtf1, associates with TAP-tagged Cdc73 and Ctr9, but not with TAP-tagged Srb5. These results confirm that Leo1 is part of the Paf1 complex and is not part of the Srb-mediator.
FIG. 5.
Leo1-HA interacts with the Paf1 complex but not with the Srb-Med complex. (A) Coimmunoprecipitation of Leo1-HA with Paf1 and Rtf1. Whole-cell extracts prepared from the wild type (YJJ662) or a Leo1-HA strain (YJJ1330) were subjected to immunoprecipitation with an anti-HA (12CA5) antibody as described in Materials and Methods. Immunoprecipitates were analyzed by Western blotting using the indicated antibodies. IP, input; UB, unbound fraction; B, bound fraction. (B) TAP tag purification of Leo1-HA. Whole-cell extracts prepared from strains containing both a TAP tag and a Leo1-HA tag (YJJ1309, YJJ1331, and YJJ1332) were incubated with IgG beads as described in Materials and Methods. After washing, proteins were eluted from the beads with the addition of 25 U of TEV protease. Eluates were TCA precipitated and analyzed by Western blotting with the indicated antibodies. IP, input; FT, flowthrough; E, eluate.
Proteasome components are not part of the Paf1 complex.
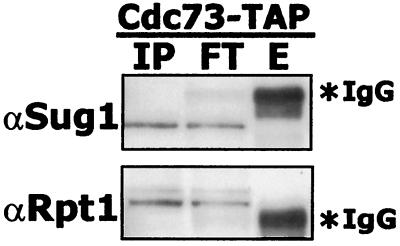
Paf1, Ctr9, Rtf1, and Leo1 were recently reported to associate with the 19S cap of the proteasome in the absence of ATP (53). However, we did not identify any proteasome components in the mass spectrometry analysis of the Paf1 complex. In addition, when we used antibodies specific to 19S proteasome components Sug1 and Rpt1 to probe TAP-tagged Cdc73 fractions, no proteasome components were found associated with the Paf1 complex (Fig. 6). The reported association of the Paf1 complex with the proteasome may therefore be an artifact of the affinity isolation used in these earlier studies. In particular, the nonspecific association of Rtf1 with many affinity matrices noted above (Fig. 5) may have led to these observations. Using antibodies directed against components of several other high-molecular-mass yeast complexes, we have found no evidence for the presence of Spt6, Bur2, Snf12, or Caf1 in the purified Paf1 complex (data not shown).
FIG. 6.
Cdc73-TAP does not associate with components of the proteasome. Extracts from a Cdc73-TAP strain were affinity purified using rabbit IgG agarose beads. Bound proteins were eluted from the resin by adding TEV protease. Eluates were resolved by SDS-PAGE and immunoblotted with antibodies specific to the proteasome. IP, input; FT, flowthrough; E, eluate.
Genetic interactions between PAF1, RTF1, and LEO1.
We have previously reported that although PAF1 is not an essential gene, its loss results in many phenotypic changes, including slow growth; increased sensitivity to low and high temperature, cell wall-damaging agents, and hydroxyurea; and abnormal morphology relative to wild-type cells (4, 48; Betz et al., submitted). Deletion of CTR9 results in an identical spectrum of phenotypes (22; Betz et al., submitted). Loss of CDC73 results in much more modest phenotypic changes that are in general a subset of those seen for paf1Δ (4, 47; Betz et al., submitted). Combining a PAF1 deletion with deletions of CDC73 or CTR9 does not result in any phenotypic enhancement (22, 47), consistent with the fact that they function in the same cellular processes. If Rtf1 and Leo1 are also involved in the same cellular process, then combining these mutations with mutations in other Paf1 complex components should not result in an enhanced phenotype (14). We therefore constructed leo1Δ and rtf1Δ mutants and used these to create double mutants by mating to an appropriate isogenic paf1Δ strain. The diploids were sporulated, and tetrads were dissected and tested for genotype and phenotype.
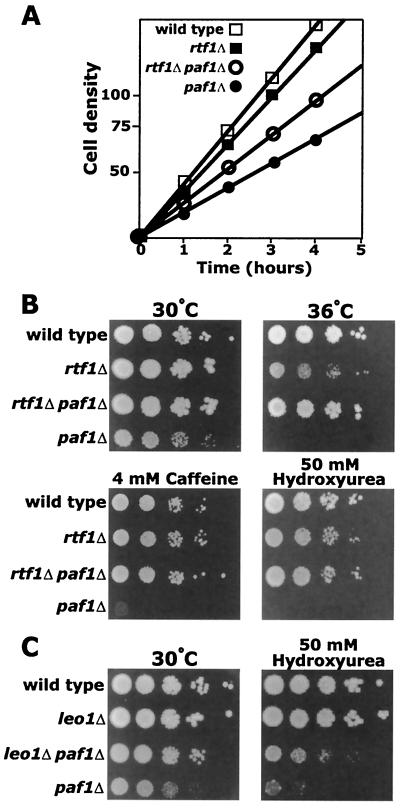
A point mutation in RTF1 was originally characterized as a suppressor of a mutation in TATA-binding protein (TBP) (48). Deletion of RTF1 also suppresses the same allele of TBP, and both the point mutation and the deletion result in alterations in transcriptional start sites. We found that rtf1Δ strains grew a little more slowly than wild type at 30°C in rich media (Fig. 7A) (doubling times, 97 versus 80 min) and were slightly temperature sensitive at 36°C (Fig. 7B). However, in contrast with a recent report (8), we observed that loss of Rtf1 in our yeast genetic background did not result in sensitivity to 6-azauracil (6AU) (data not shown). When we analyzed several rtf1Δ paf1Δ double mutants, we found that loss of Rtf1 dramatically suppressed many paf1Δ phenotypes. An rtf1Δ paf1Δ double mutant grew significantly faster than paf1Δ in rich media at 30°C (Fig. 7A) (doubling times of 145 versus 175 min), although more slowly than rtf1Δ. In addition, even though rtf1Δ alone is slightly temperature sensitive, the mutation suppressed the temperature-sensitive phenotype of paf1Δ (Fig. 7B). Loss of Rtf1 also restored growth of paf1Δ on caffeine and hydroxyurea (Fig. 7B). We also found that rtf1Δ suppressed many of these phenotypes when combined with ctr9Δ (data not shown). As previously reported (30), we found that leo1Δ strains did not demonstrate any obvious phenotype and grew essentially as well as wild type (Fig. 7C). However, we found that a leo1Δ paf1Δ double mutant was less temperature sensitive and slightly more resistant to hydroxyurea than a paf1Δ strain (Fig. 7C). Therefore, removal of either Rtf1 or Leo1 can suppress some of the pleiotropic defects caused by loss of Paf1.
FIG. 7.
A deletion of the RTF1 gene suppresses the defects caused by disruption of the PAF1 gene. (A) Growth curve of indicated deletion strains grown in YPD at 30°C. (B) Cultures of YJJ662 (wild type), YJJ1303 (rtf1Δ::kanr), YJJ1326 (rtf1Δ::kanr paf1Δ::HIS3), and YJJ577 (paf1Δ::HIS3) cells (approximately 107/ml) were spotted in 10-fold serial dilutions on YPD medium and grown at 30 and 36°C and on YPD containing 4 mM caffeine and 50 mM hydroxyurea and grown at 30°C. (C) Cultures of YJJ662 (wild type), YJJ1336 (leo1Δ), YJJ1361 (leo1Δ paf1Δ), and YJJ577 (paf1Δ) were grown on the indicated media as described in panel B. Photographs of plates were taken after 4 days of incubation.
Loss of Rtf1 restores CLN1 expression in paf1Δ.
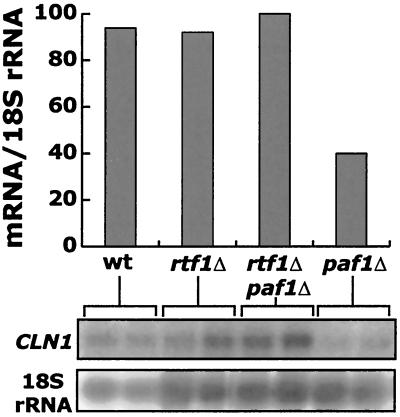
Loss of Paf1 results in changes in expression patterns of many yeast genes (4, 48; Betz et al., submitted). Since loss of Rtf1 does not completely suppress the paf1Δ phenotype, we would not expect that all transcription would be restored to normal levels. However, since the double mutant grows significantly faster than the paf1Δ strain, we analyzed the level of expression of the G1 cyclin CLN1 in these mutants. Koch and coworkers found that Paf1 and Ctr9 are required for full expression of CLN2, another G1 cyclin (29), and we have found that Paf1 is required for normal expression of many other cell cycle-regulated genes, including CLN1 (Betz et al., submitted). We found that in paf1Δ, CLN1 mRNA was reduced about threefold, whereas in an rtf1Δ paf1Δ strain CLN1 expression was restored to wild-type levels (Fig. 8). We have observed a similar restoration of mRNA levels for RNR1 and CYC1 in rtf1Δ paf1Δ relative to paf1Δ (data not shown; E. Amiott, personal communication). The restoration of RNR1 transcripts provides a potential molecular explanation for the suppression of hydroxyurea sensitivity seen in Fig. 7B. The fact that rtf1Δ, and to a lesser extent leo1Δ, can suppress the transcriptional defects caused by loss of Paf1 is consistent with the idea that a partially defective Paf1 complex may block transcription and that removal of additional complex components may relieve the block.
FIG. 8.
CLN1 message is restored to wild-type levels in an rtf1Δ paf1Δ double mutant strain. Duplicate cultures of the indicated deletion strains were grown to a density of 1.3 × 107 cells/ml, total RNA was harvested, and 15 μg of RNA per lane was fractionated. mRNA levels were detected using radiolabeled probes to CLN1 and normalized to 18S rRNA as described in Materials and Methods. The bars in the upper panel represent the average of the duplicate samples shown in the lower panel.
DISCUSSION
Yeast cells contain at least two complex forms of RNA Pol II. Ten to fifteen percent of Pol II is complexed with the Srb-mediator factors, and this form of Pol II is clearly important for initiation of transcription of most, if not all, yeast genes and for response to many activators and repressors (reviewed in reference 34). In this work we have established that Paf1, Cdc73, and Hpr1 are associated with only about 2% of the cellular Pol II in a high-molecular-mass (∼1.7-MDa) complex, distinct from the Srb-mediator. Although the Paf1 complex contains the general initiation factors TFIIB and TFIIF (Fig. 2) (47) and is found to be associated with the nonphosphorylated, initiating form of Pol II (57), it has not been established that it functions as an initiation complex. Loss of Paf1 complex genes causes alterations in the expression of only a small subset of yeast genes (4, 47; Chang et al., unpublished). Are these alterations due to changes in initiation or to changes in another transcriptional step? In these studies we have used affinity purification and mass spectrometry to identify three additional components of the Paf1 complex: Ctr9, Rtf1, and Leo1. These identifications suggest new and intriguing connections to both transcriptional initiation and elongation.
Ctr9, required for G1 cyclin expression, is part of the Paf1 complex.
CTR9, also known as CDP1, has been isolated in two genetic screens, one looking for genetic interactions with a centromere binding factor (10) and the second searching for genes required for G1 cyclin transcription (22). Loss of Ctr9 results in chromosome instability (10); reduction in the expression of G1 cyclins (21); slow growth; and sensitivity to extremes of temperature, hydroxyurea, and many cell wall-damaging agents, including caffeine and SDS (21; Betz et al., submitted). Except for the chromosome instability phenotype, which has not been tested, all other known phenotypes of ctr9Δ are shared by paf1Δ. Loss of Paf1 does, however, lead to an increase in recombination (4), which may be related to the loss of chromosome stability seen for ctr9Δ. Like ctr9Δ, paf1Δ strains have a severe defect in the expression of the G1 cyclins CLN1 and CLN2. In addition Paf1 is required for full expression of many other cell cycle-regulated genes, including HO and RNR1 (Porter et al., submitted). Paf1 is also critical for signaling from the protein kinase C-mitogen-activated protein kinase cascade to target genes required for maintenance of cell integrity (4). The requirement for Paf1 and Ctr9 in the expression of a significant subset of yeast genes is true both for the target genes in their chromosomal locations and for a variety of promoter-reporter constructs (4, 10, 22). In fact, CTR9 was originally identified as a factor required for G1 cyclin expression in a screen using cell cycle-regulated promoter elements fused to a reporter gene (22). Therefore, Paf1 and Ctr9 act through promoter elements to effect normal transcript levels and to respond to signals during the cell cycle and in response to cell damage.
Rtf1, important for both start site selection and elongation, is also part of the Paf1 complex.
RTF1 was originally identified in a genetic screen for mutations that would suppress a defective form of TBP (50). The spt15-122 allele of TBP has an altered specificity for TATA box recognition, which results in changes in transcriptional start sites (2). The rtf1-1 allele suppresses the phenotypes caused by the spt15-122 mutation and further alters start site selection but does not restore initiation sites to their wild-type positions (50). However, complete deletion of RTF1 also suppresses the spt15-122 defects and restores wild-type start site selection (50). It is interesting that mutations in Ccr4, also found in the Paf1 complex (4), also have extensive genetic interactions with TBP and the spt15-122 allele (3). Based on these studies it is clear that Rtf1, presumably as part of the Paf1 complex, plays an important role at an early stage of transcription.
In addition to its role in initiation, Rtf1 was recently reported to also have extensive genetic interactions with transcriptional elongation factors (8). In particular, rtf1Δ is synthetically lethal in combination with mutations in CTK1, a Pol II CTD kinase (8); FCP1, a CTD phosphatase (8); and POB3, a chromatin remodeling factor that plays roles in both initiation and elongation (11). In addition, RTF1 genetically interacts with other elongation factors, including TFIIS (PPR2) and SPT5 (8). The interaction with SPT5 is of particular interest since we detected peptides from Spt5 in our mass spectrometry analysis of TAP-tagged Cdc73, and an anti-Spt5 antibody detects a protein of the correct size for Spt5 in the high-molecular-mass Paf1 complex (Fig. 2B). In addition, Paf1, Cdc73, and Rtf1 have been identified as Spt5-interacting factors (G. Hartzog and K. Arndt, personal communication).
Like those in RTF1, mutations in SPT5 result in changes in transcription start sites and also have effects on elongation (16, 51), and Spt5 interacts with both the initiating and elongating forms of Pol II (26). Spt5 and Spt4, found together in a complex, are the yeast homologues of the human DSIF transcriptional elongation factor identified biochemically by Hartzog et al. (16). The DSIF elongation factor confers sensitivity to the drug DRB (5,6-dichloro-1-β-d-ribofuranosyl benzidazole) in vitro and is thought to play a negative role in transcription elongation in vivo (55). DRB inhibits the pTEFb kinase that phosphorylates the Pol II CTD and is required for an early stage of elongation in vitro (31). Some of the evidence that Spt5 and Rtf1 affect elongation comes from phenotypic analysis of mutants, specifically that mutation of either gene results in sensitivity to 6AU (8, 16). However, the connection between 6AU sensitivity and transcriptional elongation defects is complex. Mutation of the elongation factor TFIIS (PPR2) or the Rpb1 (1), Rpb2 (42) or Rpb9 (17) subunits of Pol II results in sensitivity to 6AU. In vitro, the 6AU-sensitive mutant of Rpb2 clearly has defects in elongation. However, in vivo the major effect of the various 6AU-sensitive mutants seems to be reduced expression of the PUR5 gene encoding IMP dehydrogenase (17, 46). Since the reduction in PUR5 expression can be transferred using a reporter construct fused to the promoter and 5′-untranslated region of the PUR5 gene (46), it is not yet clear what stage of transcription is affected by the 6AU-sensitive mutations in vivo.
Is the Paf1 complex involved in transcriptional elongation? Although paf1Δ strains are slow growing and sensitive to a variety of compounds, they are not sensitive to 6AU (Betz et al., submitted) although an isogenic ppr2 strain is strongly inhibited. In addition, rtf1Δ strains are not 6AU sensitive in our genetic background (C. Mueller, unpublished). Neither cdc73Δ nor hpr1Δ strains are 6AU sensitive, although ccr4Δ is very sensitive to the drug (9; Betz et al., submitted) and leo1Δ is slightly sensitive (C. Mueller, unpublished). As described above, sensitivity to 6AU neither proves nor disproves a protein's role in elongation. However, the many genetic connections between RTF1 and elongation factors (8) and the fact that the homologue of Spt5, DSIF, clearly is important for elongation in vitro are compelling evidence for a link.
The Paf1 complex is not the only factor that may straddle the line between initiation and elongation; many transcription factors seem to function at multiple stages of transcription. The GTF TFIIF is clearly required for initiation, promoter escape, and elongation (44, 59), and TFIIH's roles in both initiation and promoter escape can be traced to its action at different regions of downstream promoter DNA (49). Mutations in the Rpb9 subunit of Pol II reveal effects on both start site selection (12) and elongation (17). In addition, the Pob3-Spt16 chromatin remodeling complex, which has genetic links to RTF1 (8), also appears to be important for both initiation and elongation of transcription (11). Since initiation, promoter clearance, and entry into elongation are clearly a multistep process (39), perhaps it is not surprising that so many factors seem be involved in this critical transition. However, a central role in elongation for the Paf1 complex is not consistent with the fact that loss of Paf1 or other complex components affects the expression of only a small subset of genes. In addition, our observation that loss of both Paf1 and Rtf1 returns cells to a nearly wild-type phenotype does not support the concept that the Paf1 complex might be required for elongation of all yeast genes. Instead, our results fit a model wherein the Paf1 complex functions at a small subset of promoters to enhance expression in response to signaling pathways (i.e., protein kinase C, cell cycle). Partial defects in the complex, such as loss of Paf1 or Ctr9, could result in a defective complex that blocks transcription. Removal of a second component, such as Rtf1, may release the blocked complex and allow the return to nearly normal transcription patterns, perhaps through the action of the Srb-mediator form of Pol II. Testing this model will require the identification of the primary gene targets of the Paf1 complex.
Acknowledgments
We are grateful to Taylor Washburn and Connie Phernetton for technical assistance with many aspects of these studies. We also thank Beth Amiott, Joan Betz, Kristi Penheiter, Stephanie Porter, and other members of the laboratory for advice, sharing information, and comments on the manuscript. Natalie Ahn, Katheryn Resing, David Friedman, Kim Fong, Mark Duncan, and the UCHSC Mass Spectrometry Core facility are thanked for their help with protein identification. Karen Arndt and Grant Hartzog are thanked for reagents and for freely communicating information prior to publication. David Bentley provided much useful advice and comments on the manuscript.
These studies were supported by travel funds to C.L.M. from the UCHSC Molecular Biology Program Bolie Graduate Scholarship Fund and a grant from the NIH to J.A.J. (GM38101).
REFERENCES
- 1.Archambault, J., F. Lacroute, A. Ruet, and J. D. Friesen. 1992. Genetic interaction between transcription elongation factor TFIIS and RNA polymerase II. Mol. Cell. Biol. 12:4142-4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arndt, K. M., S. L. Ricupero, D. M. Eisenmann, and F. Winston. 1992. Biochemical and genetic characterization of a yeast TFIID mutant that alters transcription in vivo and DNA binding in vitro. Mol. Cell. Biol. 12:2372-2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Badarinarayana, V., Y. C. Chiang, and C. L. Denis. 2000. Functional interaction of CCR4-NOT proteins with TATAA-binding protein (TBP) and its associated factors in yeast. Genetics 155:1045-1054. [DOI] [PMC free article] [PubMed]
- 3a.Bhoite, L. T., Y. Yu, and D. J. Stillman. 2001. The Swi5 activator recruits the Mediator complex to the HO promoter without RNA polymerase II. Genes Dev. 15:2457-2469. [DOI] [PMC free article] [PubMed]
- 4.Chang, M., D. French-Cornay, H. Y. Fan, H. Klein, C. L. Denis, and J. A. Jaehning. 1999. A complex containing RNA polymerase II, Paf1p, Cdc73p, Hpr1p, and Ccr4p plays a role in protein kinase C signaling. Mol. Cell. Biol. 19:1056-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chavez, S., T. Beilharz, A. G. Rondon, H. Erdjument-Bromage, P. Tempst, J. Q. Svejstrup, T. Lithgow, and A. Aguilera. 2000. A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J. 19:5824-5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conaway, R. C., and J. W. Conaway. 1993. General initiation factors for RNA polymerase II. Annu. Rev. Biochem. 62:161-190. [DOI] [PubMed] [Google Scholar]
- 7.Cosma, M. P., S. Panizza, and K. Nasmyth. 2001. Cdk1 triggers association of RNA polymerase to cell cycle promoters only after recruitment of the mediator by SBF. Mol. Cell 7:1213-1220. [DOI] [PubMed] [Google Scholar]
- 8.Costa, P. J., and K. M. Arndt. 2000. Synthetic lethal interactions suggest a role for the Saccharomyces cerevisiae Rtf1 protein in transcription elongation. Genetics 156:535-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denis, C. L., Y. C. Chiang, Y. Cui, and J. Chen. 2001. Genetic evidence supports a role for the yeast ccr4-not complex in transcriptional elongation. Genetics 158:627-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foreman, P. K., and R. W. Davis. 1996. CDP1, a novel Saccharomyces cerevisiae gene required for proper nuclear division and chromosome segregation. Genetics 144:1387-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Formosa, T., P. Eriksson, J. Wittmeyer, J. Ginn, Y. Yu, and D. J. Stillman. 2001. Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J. 20:3506-3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furter-Graves, E. M., B. D. Hall, and R. Furter. 1994. Role of a small RNA pol II subunit in TATA to transcription start site spacing. Nucleic Acids Res. 22:4932-4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greenblatt, J. 1997. RNA polymerase II holoenzyme and transcriptional regulation. Curr. Opin. Cell Biol. 9:310-319. [DOI] [PubMed] [Google Scholar]
- 14.Guarente, L. 1993. Synthetic enhancement in gene interaction: a genetic tool come of age. Trends Genet. 9:362-366. [DOI] [PubMed] [Google Scholar]
- 15.Guthrie, C., and G. R. Fink. 1991. Guide to yeast genetics and molecular biology. Methods Enzymol. 194:1-931. [PubMed] [Google Scholar]
- 16.Hartzog, G. A., T. Wada, H. Handa, and F. Winston. 1998. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 12:357-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hemming, S. A., D. B. Jansma, P. F. Macgregor, A. Goryachev, J. D. Friesen, and A. M. Edwards. 2000. RNA polymerase II subunit Rpb9 regulates transcription elongation in vivo. J. Biol. Chem. 275:35506-35511. [DOI] [PubMed] [Google Scholar]
- 18.Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 19.Knop, M., K. Siegers, G. Pereira, W. Zachariae, B. Winsor, K. Nasmyth, and E. Schiebel. 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15:963-972. [DOI] [PubMed] [Google Scholar]
- 20.Kobor, M. S., J. Archambault, W. Lester, F. C. Holstege, O. Gileadi, D. B. Jansma, E. G. Jennings, F. Kouyoumdjian, A. R. Davidson, R. A. Young, and J. Greenblatt. 1999. An unusual eukaryotic protein phosphatase required for transcription by RNA polymerase II and CTD dephosphorylation in S. cerevisiae. Mol. Cell 4:55-62. [DOI] [PubMed] [Google Scholar]
- 21.Koch, C., A. Schleiffer, G. Ammerer, and K. Nasmyth. 1996. Switching transcription on and off during the yeast cell cycle: Cln/Cdc28 kinases activate bound transcription factor SBF (Swi4/Swi6) at start, whereas Clb/Cdc28 kinases displace it from the promoter in G2. Genes Dev. 10:129-141. [DOI] [PubMed] [Google Scholar]
- 22.Koch, C., P. Wollmann, M. Dahl, and F. Lottspeich. 1999. A role for Ctr9p and Paf1p in the regulation G1 cyclin expression in yeast. Nucleic Acids Res. 27:2126-2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, J. M., and A. L. Greenleaf. 1989. A protein kinase that phosphorylates the C-terminal repeat domain of the largest subunit of RNA polymerase II. Proc. Natl. Acad. Sci. USA 86:3624-3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, T. I., and R. A. Young. 2000. Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet. 34:77-137. [DOI] [PubMed] [Google Scholar]
- 25.Lewis, T. S., J. B. Hunt, L. D. Aveline, K. R. Jonscher, D. F. Louie, J. M. Yeh, T. S. Nahreini, K. A. Resing, and N. G. Ahn. 2000. Identification of novel MAP kinase pathway signaling targets by functional proteomics and mass spectrometry. Mol. Cell 6:1343-1354. [DOI] [PubMed] [Google Scholar]
- 26.Lindstrom, D. L., and G. A. Hartzog. 2001. Genetic interactions of Spt4-Spt5 and TFIIS with the RNA polymerase II CTD and CTD modifying enzymes in Saccharomyces cerevisiae. Genetics 159:487-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, H. Y., V. Badarinarayana, D. C. Audino, J. Rappsilber, M. Mann, and C. L. Denis. 1998. The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO J. 17:1096-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu, H. Y., Y. C. Chiang, J. Pan, J. Chen, C. Salvadore, D. C. Audino, V. Badarinarayana, V. Palaniswamy, B. Anderson, and C. L. Denis. 2001. Characterization of CAF4 and CAF16 reveals a functional connection between the CCR4-NOT complex and a subset of SRB proteins of the RNA polymerase II holoenzyme. J. Biol. Chem. 276:7541-7548. [DOI] [PubMed] [Google Scholar]
- 29.Liu, Y., J. A. Ranish, R. Aebersold, and S. Hahn. 2000. Yeast nuclear extract contains two major forms of RNA polymerase II mediator complexes. J. Biol. Chem. 276:7169-7175. [PubMed]
- 30.Magdolen, V., P. Lang, G. Mages, H. Hermann, and W. Bandlow. 1994. The gene LEO1 on yeast chromosome XV encodes a non-essential, extremely hydrophilic protein. Biochim. Biophys. Acta 1218:205-209. [DOI] [PubMed] [Google Scholar]
- 31.Marshall, N. F., and D. H. Price. 1995. Purification of P-TEFb, a transcription factor required for the transition into productive elongation. J. Biol. Chem. 270:12335-12338. [DOI] [PubMed] [Google Scholar]
- 32.Maru, Y., D. E. Afar, O. N. Witte, and M. Shibuya. 1996. The dimerization property of glutathione S-transferase partially reactivates Bcr-Abl lacking the oligomerization domain. J. Biol. Chem. 271:15353-15357. [DOI] [PubMed] [Google Scholar]
- 33.Murray, S., R. Udupa, S. Yao, G. Hartzog, and G. Prelich. 2001. Phosphorylation of the RNA polymerase II carboxy-terminal domain by the Bur1 cyclin-dependent kinase. Mol. Cell. Biol. 21:4089-4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Myer, V. E., and R. A. Young. 1998. RNA polymerase II holoenzymes and subcomplexes. J. Biol. Chem. 273:27757-27760. [DOI] [PubMed] [Google Scholar]
- 35.Myers, L. C., and R. D. Kornberg. 2000. Mediator of transcriptional regulation. Annu. Rev. Biochem. 69:729-749. [DOI] [PubMed] [Google Scholar]
- 36.Naar, A. M., B. D. Lemon, and R. Tjian. 2001. Transcriptional coactivator complexes. Annu. Rev. Biochem. 70:475-501. [DOI] [PubMed] [Google Scholar]
- 37.Nonet, M. L., and R. A. Young. 1989. Intragenic and extragenic suppressors of mutations in the heptapeptide repeat domain of Saccharomyces cerevisiae RNA polymerase II. Genetics 123:715-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ozols, J. 1990. Amino acid analysis. Methods Enzymol. 182:587-601. [DOI] [PubMed] [Google Scholar]
- 39.Pal, M., D. McKean, and D. S. Luse. 2001. Promoter clearance by RNA polymerase II is an extended, multistep process strongly affected by sequence. Mol. Cell. Biol. 21:5815-5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park, J. M., J. Werner, J. M. Kim, J. T. Lis, and Y. J. Kim. 2001. Mediator, not holoenzyme, is directly recruited to the heat shock promoter by HSF upon heat shock. Mol. Cell 8:9-19. [DOI] [PubMed] [Google Scholar]
- 41.Parker, M. W., M. Lo Bello, and G. Federici. 1990. Crystallization of glutathione S-transferase from human placenta. J. Mol. Biol. 213:221-222. [DOI] [PubMed] [Google Scholar]
- 42.Powell, W., and D. Reines. 1996. Mutations in the second largest subunit of RNA polymerase II cause 6-azauracil sensitivity in yeast and increased transcriptional arrest in vitro. J. Biol. Chem. 271:6866-6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Puig, O., F. Caspary, G. Rigaut, B. Rutz, E. Bouveret, E. Bragado-Nilsson, M. Wilm, and B. Seraphin. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24:218-229. [DOI] [PubMed] [Google Scholar]
- 44.Ren, D., L. Lei, and Z. F. Burton. 1999. A region within the RAP74 subunit of human transcription factor IIF is critical for initiation but dispensable for complex assembly. Mol. Cell. Biol. 19:7377-7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roguev, A., D. Schaft, A. Shevchenko, W. W. Pijnappel, M. Wilm, R. Aasland, and A. F. Stewart. 2001. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J. 20:7137-7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shaw, R. J., and D. Reines. 2000. Saccharomyces cerevisiae transcription elongation mutants are defective in PUR5 induction in response to nucleotide depletion. Mol. Cell. Biol. 20:7427-7437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi, X., M. Chang, A. J. Wolf, C. H. Chang, A. A. Frazer-Abel, P. A. Wade, Z. F. Burton, and J. A. Jaehning. 1997. Cdc73p and Paf1p are found in a novel RNA polymerase II-containing complex distinct from the Srbp-containing holoenzyme. Mol. Cell. Biol. 17:1160-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi, X., A. Finkelstein, A. J. Wolf, P. A. Wade, Z. F. Burton, and J. A. Jaehning. 1996. Paf1p, an RNA polymerase II-associated factor in Saccharomyces cerevisiae, may have both positive and negative roles in transcription. Mol. Cell. Biol. 16:669-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spangler, L., X. Wang, J. W. Conaway, R. C. Conaway, and A. Dvir. 2001. TFIIH action in transcription initiation and promoter escape requires distinct regions of downstream promoter DNA. Proc. Natl. Acad. Sci. USA 98:5544-5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stolinski, L. A., D. M. Eisenmann, and K. M. Arndt. 1997. Identification of RTF1, a novel gene important for TATA site selection by TATA box-binding protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:4490-4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swanson, M. S., E. A. Malone, and F. Winston. 1991. SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Mol. Cell. Biol. 11:3009-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tucker, M., M. A. Valencia-Sanchez, R. R. Staples, J. Chen, C. L. Denis, and R. Parker. 2001. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104:377-386. [DOI] [PubMed] [Google Scholar]
- 53.Verma, R., S. Chen, R. Feldman, D. Schieltz, J. Yates, J. Dohmen, and R. J. Deshaies. 2000. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell 11:3425-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wach, A., A. Brachat, R. Pohlmann, and P. Philippsen. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10:1793-1808. [DOI] [PubMed] [Google Scholar]
- 55.Wada, T., T. Takagi, Y. Yamaguchi, D. Watanabe, and H. Handa. 1998. Evidence that P-TEFb alleviates the negative effect of DSIF on RNA polymerase II-dependent transcription in vitro. EMBO J. 17:7395-7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wade, P. A., S. D. Shaffer, and J. A. Jaehning. 1993. Resolution of transcription factors from a transcriptionally active whole-cell extract from yeast: purification of TFIIB, TBP, and RNA polymerase IIa. Protein Expr. Purif. 4:290-297. [DOI] [PubMed] [Google Scholar]
- 57.Wade, P. A., W. Werel, R. C. Fentzke, N. E. Thompson, J. F. Leykam, R. R. Burgess, J. A. Jaehning, and Z. F. Burton. 1996. A novel collection of accessory factors associated with yeast RNA polymerase II. Protein Expr. Purif. 8:85-90. [DOI] [PubMed] [Google Scholar]
- 58.Wilcoxen, S. E., C. R. Peterson, C. S. Winkley, M. J. Keller, and J. A. Jaehning. 1988. Two forms of RPO41-dependent RNA polymerase. Regulation of the RNA polymerase by glucose repression may control yeast mitochondrial gene expression. J. Biol. Chem. 263:12346-12351. [PubMed] [Google Scholar]
- 59.Yan, Q., R. J. Moreland, J. Weliky Conaway, and R. C. Conaway. 1999. Dual roles for transcription factor IIF in promoter escape by RNA polymerase II. J. Biol. Chem. 274:35668-35675. [DOI] [PubMed] [Google Scholar]
- 60.Zawel, L., and D. Reinberg. 1992. Advances in RNA polymerase II transcription. Curr. Opin. Cell Biol. 4:488-495. [DOI] [PubMed] [Google Scholar]