Abstract
PTF1 is a trimeric transcription factor essential to the development of the pancreas and to the maintenance of the differentiated state of the adult exocrine pancreas. It comprises a dimer of P48/PTF1a (a pancreas and neural restricted basic helix-loop-helix [bHLH] protein) and a class A bHLH protein, together with a third protein that we show can be either the mammalian Suppressor of Hairless (RBP-J) or its paralogue, RBP-L. In mature acinar cells, PTF1 exclusively contains the RBP-L isoform and is bound to the promoters of acinar specific genes. P48 interacts with the RBP subunit primarily through two short conserved tryptophan-containing motifs, similar to the motif of the Notch intracellular domain (NotchIC) that interacts with RBP-J. The transcriptional activities of the J and L forms of PTF1 are independent of Notch signaling, because P48 occupies the NotchIC docking site on RBP-J and RBP-L does not bind the NotchIC. Mutations that delete one or both of the RBP-interacting motifs of P48 eliminate RBP-binding and are associated with a human genetic disorder characterized by pancreatic and cerebellar agenesis, which indicates that the association of P48 and RBPs is required for proper embryonic development. The presence of related peptide motifs in other transcription factors indicates a broader Notch-independent function for RBPJ/SU(H).
One of the most intriguing properties of biological regulatory schemes is the certainty of evolutionary variations from an original theme. The definition of a canonical scheme nearly guarantees the discovery of an alternative in which a useful regulator is recruited for other purposes. In this regard, the canonical Notch-signaling pathway, which regulates cell fate decisions via a transcriptional off-on switch, is a useful example (2, 35). In the absence of signaling, a CSL-factor [CBF1/RBPJ/RBPSUH in mammals; Su(H) in Drosophila melanogaster; LAG-1 in nematodes] binds and represses target promoters by recruiting a corepressor complex. Binding of any of the family of DSL cell-surface ligands (Delta, Serrate/Jagged, and Lag-2) to the Notch receptor triggers cleavage of Notch and the release of its intracellular domain (NotchIC). The NotchIC enters the nucleus, binds CSL, displaces the corepressor, and recruits coactivators. Recently, a variation has been described in which the transcriptional effect is mediated independently of the CSL (29). Conversely, CSL appears to play a Notch-independent role in at least one developmental context in Drosophila (3). We describe here a novel Notch-independent function of the mammalian CSL (hereafter RBP-J) and its paralogue, RBP-L, by recruitment into PTF1, a basic helix-loop-helix (bHLH) transcription factor complex that controls pancreas-specific gene transcription.
The pancreas is a multifunctional gland composed of both endocrine and exocrine tissues. The exocrine tissue comprises more than 90% of the adult pancreas and is composed of acini, which secrete digestive enzymes, and ducts, which secrete fluid and transport the acinar enzymes to the duodenum. Massive synthesis of the digestive enzymes is reflected in the pancreatic mRNA population: nearly 90% of the mRNA from the entire gland encodes a small number (about 20) of acinar secretory enzymes, such as amylases, elastases, chymotrypsinogens, and carboxypeptidases (12). The selective transcription of the acinar specific genes at such a high level is controlled largely by the PTF1 complex (6, 32). However, the mechanism of target-gene activation by PTF1 is unknown.
Functional binding sites for the PTF1 complex are present in the 5′ promoter regions of all of the acinar digestive enzyme genes examined (6, 31). The binding site from the elastase 1 gene (Ela1) provides a model for the interaction of PTF1 with DNA and acinar cell-specific transcriptional activation. This site, known as the A element of the Ela1 enhancer, is located about 100 bp upstream of the 5′ end of Ela1, is necessary and sufficient to direct acinar specific expression in transgenic mice (30), and in situ cooperates with two nearby elements (B and C) to direct the high level of transcriptional activation characteristic of Ela1 (18).
PTF1 is an unusual heterotrimeric bHLH transcription factor composed of PTF1a/P48 (a pancreas and neural specific bHLH protein), one of the common class A bHLH proteins, and a previously unidentified subunit (32, 33). (For clarity, we retain the use of p48/P48 [gene/protein], rather than Ptf1a/PTF1A, to distinguish the P48 subunit from the PTF1 complex.) PTF1 binding sites are bipartite with an E-box (preferably CACCTG) and a TC-box (TTTCCCA) spaced one or two helical turns apart, center to center (6, 31). Targeted deletion of the p48 gene causes pancreatic and cerebellar agenesis (14, 17, 36), so understanding the mechanism of transcriptional activation by PTF1 in differentiated acini will likely give insights into PTF1 action during pancreas and brain development as well.
We show that the previously unidentified third subunit of PTF1 from adult pancreas is RBP-L, an organ-specific mammalian variant of the CSL proteins. RBP-L provides the high activation potential of the complex, which is dependent on contact of all three subunits of the complex with DNA. A similar transcriptionally active complex can be reconstituted with RBP-J, the mediator of Notch signaling. The interaction of P48 with the RBP subunits requires two peptide motifs conserved in P48s from insects to mammals. One or both of these peptides are deleted in families with heritable permanent neonatal diabetes mellitus, in which infants are born without a pancreas and cerebellum (36). The similar developmental consequences for neonatal mice without P48 and infants with mutant P48 unable to bind RBP-J or -L suggest that most or all of the developmental functions of P48 require its ability to recruit an RBP into a PTF1 complex. Motifs similar to the RBP-interacting sites of P48 are present in other transcription factors; therefore, PTF1 may be one of a new family of complexes that use RBP-J in a Notch-independent manner.
MATERIALS AND METHODS
Expression of PTF1 components.
The ds-cDNA for mouse RBP-L, human P48, and fruit fly FER1, DA, and SU(H) were derived by reverse transcription-PCR (RT-PCR) amplification. Myc-tagged human RBP-J cDNA and hemagglutinin (HA)-tagged mouse NotchIC cDNA were derived from plasmids SG5-myc-CBF1 and SG5-HA-mNOTCH (13), gifts from S. D. Hayward, Johns Hopkins Medical Center, Baltimore, Md. HEB, E47 (PAN1), and E12 (PAN2) cDNA plasmids have been described (32). All cDNAs were placed downstream of the 5′ untranslated region of the Xenopus laevis β-globin mRNA. The plasmid expressing the VP16-RBP-J fusion was created by inserting the VP16 activation domain at the N terminus of RBP-J. Transfection of 293 human embryonic kidney cells (ATCC CRL-1573) was performed as previously described (21). The minimal promoter construct (EIp.luc) has the Ela1 basal promoter linked to the 5′ end of the luciferase gene of PGL3-basic (Promega, Madison, WI). Each PTF1-binding site was tested by placing a tandem repeat of six sites upstream of EIp.luc (6A.EIp.luc). The RBP reporter 6R.EIp.luc contains six tandem copies of the consensus RBP-J site upstream of Elp.luc. The distribution of RNA transcripts for p48, Rbp-L, and Rbp-J was determined by RT-PCR analysis of RNA from 19 mouse organs.
Antibodies and immunofluorescence microscopy.
The rabbit anti-P48/PTF1a was described previously (32). The rabbit anti-RBP-L was prepared by AnaSpec, Inc. (San Jose, CA), against PNAQEPAPDADTLLE, a sequence near the C terminus of mouse RBP-L, and affinity purified using the synthetic peptide. For electrophoretic mobility shift assay (EMSA), anti-RBP-J was from the Institute of Immunology Co., Ltd. (Tokyo, Japan); for chromatin immunoprecipitation (ChIP), anti-RBP-J was from Santa Cruz Biotechnology. The anti-RBP-L did not cross-detect RBP-J, and the anti-RBP-J did not cross-detect RBP-L in EMSA and Western blotting experiments with in vitro-synthesized RBP proteins. Anti-HEB serum (34) was a gift from S. Sawada. Anti-E2.2, anti-E12/E2-2, and anti-E47 were from BD Biosciences Pharmingen (San Diego, CA), and anti-c-Myc was from Santa Cruz Biotechnology (Santa Cruz, CA). Immunofluorescent localization of P48 and RBP-L was performed with 5-μm tissue sections from paraformaldehyde-fixed, paraffin-embedded adult mouse pancreas.
EMSAs and antibody supershifts.
Nuclear extracts were prepared from rat pancreas, in vitro-translated (IVT) proteins were synthesized, and EMSAs including antibody supershifts were performed as previously described (30). Peptides for competition experiments were synthesized by the Protein Chemistry Technology Center (UT Southwestern, Dallas, TX). The double-stranded oligonucleotide for RBP-J binding had the sequence 5′-GTAGTAGTTGCTTTTCCCACG-3′.
RT-PCR analyses.
The organ distribution of RNA transcripts for P48, RBP-L, and RBP-J was determined by RT-PCR analysis of RNA from 19 mouse organs. Pancreatic RNA was isolated by the guanidine thiocyanate technique (22), whereas RNA from other organs was isolated with TRIzol (Invitrogen, Carlsbad, CA). cDNA from each RNA was synthesized by Superscript II reverse transcriptase (Invitrogen) with oligo(dT) primer. Aliquots of each cDNA derived from the equivalent of 25 ng of total RNA template were amplified in 40-μl reactions with the following gene-specific primers: p48, 5′-CGCGTCTTTGTGCATATTGT-3′ and 5′-CGGAGTTTCCTGGACAGAGT-3′; Rbp-L, 5′-GGAGCTGCACGGAGAAAA-3′ and 5′-GTGTGAACTCGTGGTGGATG-3′; Rbp-J, 5′-GAATTTCCACGCCAGTTCAC-3′ and 5′-ATACAGGGTCGTCTGCATCC-3′; and actin, 5′-AGCCATGTACGTAGCCATCC-3′ and 5′-ACATCTGCTGGAAGGTGGAC-3′. The amplification conditions were 30 cycles for p48, Rbp-L, and Rbp-J and 25 cycles for actin of 1 min at 94°C, 1 min at 60°C, and 1 min at 72°C. The amplification products were analyzed by agarose gel electrophoresis and staining with ethidium bromide.
Coimmunoprecipitation and ChIP.
Coimmunoprecipitation was performed with 75 μg of nuclear protein extract from rat pancreas, precleared and then incubated with 10 μg of anti-P48 or anti-cMyc immobilized on coupling beads.
Chromatin from rat pancreas and liver was prepared from formaldehyde cross-linked nuclei as described for rat liver chromatin (5) and sheared further by sonication. A total of 100 μl of purified chromatin in 900 μl of ChIP dilution buffer (Upstate, Lake Placid, NY) was incubated with 15 to 20 μg of antiserum. Blocked protein G-Sepharose beads (Upstate, Lake Placid, NY) were added to the chromatin. Bound chromatin was eluted from the beads, cross-linking was reversed, and the immunoprecipitated DNA was purified for PCR analysis. For sequential ChIP (9), half of the chromatin eluted from an initial immunoprecipitation was retained for the real-time PCR measurement of enrichment (below), and the remainder was used for a second immunoprecipitation. Ten percent of the immunoprecipitated DNA was amplified by 34 cycles of PCR for each PTF1 target gene. Quantification of ChIP enrichment of promoter regions was performed with SYBR Green Mastermix (Applied Biosystems, Foster City, CA) using the ABI prism 7700 (Applied Biosystems) and was calculated as the increase of Ela1 promoter DNA relative to that of the 28S rRNA gene.
RESULTS
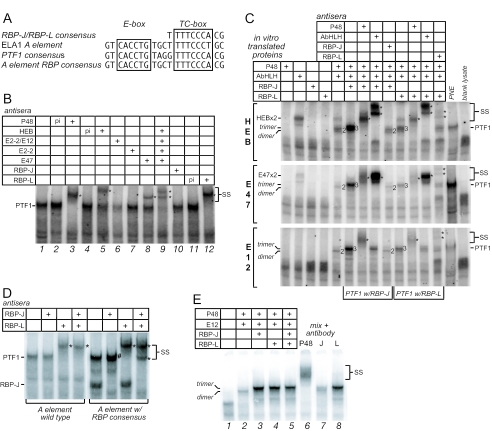
The well-characterized Ela1 PTF1-binding site, comprising an E-box and a TC-box separated by one turn (32), is closest to the consensus among known PTF1 sites; therefore, we use it for the binding site oligonucleotide in electrophoretic mobility shift experiments (Fig. 1A). Within the PTF1 complex, a heterodimer of P48 and a class A bHLH protein (P75; probably HEB/TCF12) binds the E-box and the unidentified protein P64 binds the TC-box (32, 33). P48 has been shown to interact also with RBP-J, the broadly expressed mediator of Notch signaling, in yeast two-hybrid assays (26). Moreover, the TC-box in PTF1 binding sites is similar to the consensus binding site for RBP-J (Fig. 1A) (41). The Notch-indifferent paralogue of vertebrate RBP-J, RBP-L (RBPSUH-L), shares 48% amino acid sequence identity and binds the same DNA consensus sequence (24). Thus, RBP-J and RBP-L were likely candidates for the P64 subunit.
FIG. 1.
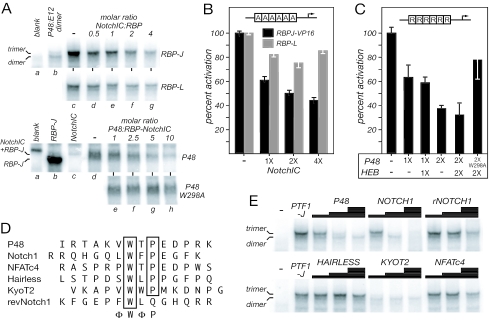
RBP-L is a subunit of the PTF1 complex. (A) The 21-bp Ela1 A element contains a TC-box similar to an RBP-J/L binding site one turn away from an E-box. Shown are the A element, the consensus RBP binding site, the PTF1-binding site consensus, and the Ela1 A element modified to contain the RBP consensus. (B) EMSA supershift analyses of complexes with nuclear extracts from rat pancreas with antisera to P48, HEB, E12, E47, E2.2, RBP-J, or RBP-L. p.i., preimmune; *, antibody-supershifted complexes (also indicated by SS). (C) IVT P48 forms heterodimeric complexes with HEB, E47, or E12, and trimeric complexes with the addition of RBP-L or RBP-J. Antibodies that recognize each component confirm their presence in the complex. (D) RBP-J is in pancreatic nuclear extract, but not as part of a PTF1 complex. Complexes from nuclear extracts, bound to either the wild-type Ela1 PTF1-binding site or a site in which the TC-box was changed to the consensus sequence for RBP-J, were incubated with antibody to either RBP-J or -L. RBP-L was detected only as part of the PTF1 complex. *, antibody-supershifted complexes; #, supershifted RBP-J monomer migrating with a slightly slower mobility than the authentic PTF1 complex. (E) RBP-J forms the PTF1 complex much more effectively than RBP-L does. PTF1 trimers were formed by mixing equimolar amounts of IVT P48, E12, RBP-J, and RBP-L. The relative amounts of trimer with P48 and either RBP-J or RBP-L were estimated from the amount of PTF1-band depletion with subunit-specific antibodies.
RBP-L is a stoichiometric component of the authentic PTF1 complex.
To determine whether RBP-J or RBP-L is part of the PTF1 complex, we tested whether antibodies against either could recognize the complex in EMSA experiments with nuclear extracts from adult rat pancreas and whether the complex could be reconstituted with IVT proteins. With the Ela1 binding site, the PTF1 complex from nuclear extracts migrates as a broad band, suggesting molecular heterogeneity (Fig. 1B, lane 1). Antibody to P48 eliminated the PTF1 band and generated a supershifted band (lane 3). Antibodies specific for individual AbHLH proteins each affected a fraction of the PTF1 complex (lanes 5 to 8), and a mix of all of the antibodies eliminated nearly all of the complex (lane 9). These results indicate that PTF1 is a population of similar complexes, each containing one AbHLH protein that can be HEB, E2-2, E12, or E47. Finally, antiserum specific for RBP-L supershifted all of the complex (lane 12), whereas several different antibodies specific for RBP-J were ineffective (e.g., lane 10). Thus, PTF1 from adult pancreas is a molecularly heterogeneous trimeric complex containing P48, RBP-L, and an AbHLH.
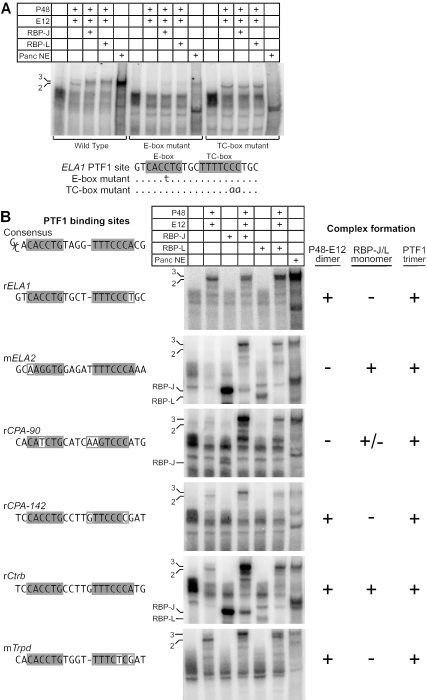
A complex with the same electrophoretic mobility as authentic PTF1 can be reconstituted with IVT P48, RBP-L, or RBP-J, and one of the common bHLH proteins (e.g., HEB, E12, or E47) (Fig. 1C). Whereas P48 alone cannot bind the PTF1 binding site, heterodimers of P48 and any of the three common bHLH proteins can. The heterodimers bind the E-box, because the binding can be disrupted by a mutation in the E-box and is insensitive to changes in the TC-box (Fig. 2A). All three of the possible P48 heterodimers have faster electrophoretic mobilities than does PTF1. A complex with the mobility of authentic PTF1 forms with the addition of either RBP-J or RBP-L (Fig. 1C). The ability to form a trimeric complex with an RBP may not be unique to P48, but it is not a common property of bHLH proteins. For example, although heterodimers of ASCL1 or NEUROD1 with E12 can bind the E-box of the PTF1 site, we were unable to detect the formation of trimeric complexes with RBP-J or RBP-L (data not shown).
FIG. 2.
PTF1 has unique binding requirements for both an E-box and a TC-box. (A) Binding of reconstituted and authentic PTF1 complexes to wild-type and mutant Ela1 PTF1 binding sites; (B) reconstituted and authentic PTF1 bind to PTF1 binding sites from the promoter regions of several digestive enzyme genes. The amount of complex formation depends on the particular sequence of the E-box or TC-box, but all bind the PTF1 trimer. 3, PTF1 trimer; 2, P48-E12 dimer.
Authentic and reconstituted PTF1 complexes have identical DNA binding requirements.
Unique DNA-binding properties distinguish PTF1 from other bHLH complexes. As for other bHLH complexes (4), P48-AbHLH heterodimers require an E-box with a bias for one or two flanking nucleotides. In contrast, the trimeric PTF1 complex requires, in addition, a TC-box positioned one or two DNA turns away (6, 32, 33). Either a single-base-pair change in the E-box or a two-base-pair mutation that disrupts the near RBP-consensus of the TC-box can prevent binding of the trimeric PTF1 (Fig. 2A). The unusual DNA-binding properties of the PTF1 complex are independent of the identities of the class A bHLH partner and the RBP isoform (Fig. 2 and data not shown). Changing the spacing between the boxes by a half turn also disrupts binding (data not shown). The reconstituted trimeric complexes have the same characteristic DNA-binding site requirements (Fig. 2A). Because the binding of a P48-AbHLH heterodimer alone is unaffected by changes in the TC-box, the association with RBP-L or -J must alter the binding of the heterodimer to an E-box in such a manner that binding now requires an RBP binding site as well. The identical idiosyncratic binding properties of the authentic and reconstituted complexes constitute further proof that the authentic complex also comprises P48, an AbHLH, and RBP-L or -J.
The binding of the three subunits to DNA is highly cooperative. For example, the PTF1 site of the rCPA-90 promoter does not bind a P48-E12 heterodimer, but the presence of a weak RBP site compensates and allows trimer binding via RBP-J/liter recognition of the TC-box (Fig. 2B). Conversely, the TC-boxes of the Ela1, Cpa-142, and Trpd promoter sites do not bind RBP-J/liter and yet are sufficient to recruit the PTF1 complex through collaborative binding to an E-box. Of the six PTF1-binding sites examined, only the Ctrb site could be bound independently by either a P48-AbHLH heterodimer or an RBP.
Cooperative binding may be facilitated by preformed trimeric PTF1 complexes. Anti-P48 coprecipitates RBP-L from nuclear extracts (data not shown), indicating that the trimeric PTF1 complex forms in vivo. Moreover, the addition of excess RBP-L or -J to an EMSA reaction prevents the binding of a P48-E12 heterodimer to an E-box in the absence of a paired TC-box (data not shown). However, RBP-J cannot inhibit the binding of a heterodimer containing a P48 with a mutation (W298A) that eliminates the interaction between P48 and RBP-J (see below, Fig. 4C). Thus, the ability of an RBP to inhibit the DNA binding of a P48-heterodimer requires its interaction with P48, and this interaction can occur in the absence of DNA binding.
FIG. 4.
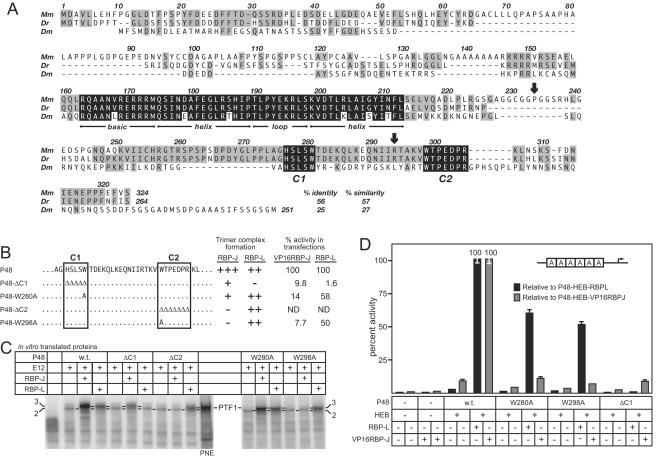
Two conserved peptide motifs near the C terminus of P48 mediate the interaction between P48 and the RBP-J/L. (A) Alignment of the sequences of the mouse (Mm) and zebra fish (Dr) P48s and FER1 from Drosophila (Dm) shows conservation of the bHLH domain (black shading) required for heterodimerization with class A bHLH proteins and DNA binding. The other significant conservation among all three is the two peptides (C1 and C2; black shading) near the C terminus. Gray shading highlights regions of lower sequence conservation, including the vertebrate-specific conservation between positions 246 and 275. The arrows indicate the relative positions of the human P48 mutations (see Fig. 8). (B) The sequences of the wild-type and mutant C1 and C2 regions of P48. The results from panels C and D are summarized at the right. (C) Ability of IVT wild-type and mutant P48 to form DNA-binding heterodimers with E12 or trimers with E12 plus RBP-L or RBP-J. All P48 mutants formed the heterodimer as effectively as wild-type P48. (D) Transcriptional activation of by PTF1 requires the interaction of P48 and RBP-L or RBP-J. The relative activity of the 6A-EIp.luc reporter construct in 293 cells was assayed in the presence or absence of cotransfected HEB, VP16RBP-J, or RBP-L, and wild-type or mutant P48, individually or in various combinations as indicated. All values are the means of at least three transfections ± the SEM.
RBP-J is excluded from the adult PTF1 complex.
Although a PTF1 complex can be reconstituted with either RBP, only RBP-L was detected in the PTF1 complex (PTF1-L) from adult pancreas (Fig. 1B). As a more sensitive assay for the RBP-J form of PTF1 (PTF1-J), we altered the TC-box and adjacent nucleotides of the Ela1 PTF1 site to create the consensus binding sequence shared by RBP-J and RBP-L. Only the PTF1-L complex was detected, even though RBP-J was present in the nuclear extract and bound to the consensus site much better than RBP-L did (Fig. 1D and data not shown). When the relative efficiencies of the formation of the two trimeric complexes were tested by mixing equimolar amounts of all four IVT subunits, the majority of the complex formed contained RBP-J rather than RBP-L (Fig. 1E). Because the reconstituted PTF1-J binds the Ela1 site more effectively than PTF1-L (Fig. 1C), the detection of only the PTF1-L form in nuclear extracts suggests that a cellular mechanism excludes RBP-J from the complex in favor of RBP-L.
RBP-L is essential for the high transcriptional activity of PTF1.
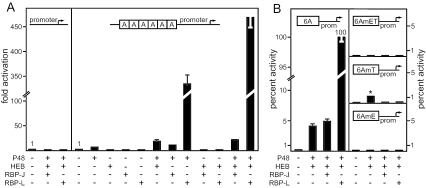
Forced expression of P48 and a common AbHLH (HEB, E47, or both) did not activate to high levels a cotransfected reporter driven by tandem repeats of a PTF1 binding site (26, 32). We tested whether RBP-L could supply the missing transcriptional activation. The human embryonic kidney cell line 293 has endogenous RBP-J and the common AbHLH proteins, but not P48 and RBP-L (data not shown). Coexpression of P48 and HEB in 293 cells activated a luciferase reporter driven by six copies of the Ela1 PTF1 binding site 18-fold higher than without the exogenous transcription factors (Fig. 3A). Overexpression of RBP-J did not change the extent of activation by P48 and HEB. Obata et al. showed previously that RBP-J could enhance activation by P48 on the PTF1-binding site of the Ctrb promoter threefold (26). In contrast, the addition of RBP-L boosted activation another 25-fold, to a total of 450-fold (Fig. 3A). P48 and RBP-L without the addition of exogenous HEB were nearly as effective (320-fold activation; Fig. 3A), due to the presence of ample endogenous AbHLH proteins. The supplemental activation provided by RBP-L depended on the incorporation of RBP-L into a PTF1 complex: a five-amino-acid deletion in P48 that prevents recruitment of RBP-L also eliminated the supplemental activation by RBP-L (see Fig. 4). Hence, RBP-L provides the high transcriptional activation of the PTF1 complex.
FIG. 3.
RBP-L is the critical subunit of PTF1 for high-level transcriptional activation of the Ela1 A element. (A) The relative activity of the 6A.EIp.luc reporter gene in 293 cells was assayed in the presence or absence of cotransfected P48, HEB, RBP-J, or RBP-L, individually or in various combinations as indicated. Transcription from the EIp.luc plasmid (containing only the Ela1 minimal promoter from −92 to +8 inserted upstream of the luciferase gene) was not affected by the addition of transcription factors via cotransfection. All values are the mean of at least four transfections ± the standard error of the mean (SEM). (B) Activation of the 6A reporter is dependent on both the E-box and the TC-box. Activation of wild-type A element (6A) was compared to activation of the E-box mutant (6AmE), the TC-box mutant (6AmT), or a mutant with both the E-box and TC-box mutated (6AmET). The asterisk in the 6AmT panel highlights the reduced ability of transfected P48 and HEB (with endogenous RBP-J) to activate the reporter in the absence of a TC-box. Values represent the percentage of wild-type activation and are the mean of least three transfections; error bars indicate the SEMs.
To verify that the trimeric PTF1 complex was responsible for the activation of PTF1-target genes, we tested whether the transcriptional activation had the same idiosyncratic DNA sequence requirements as the complex has for binding DNA. Indeed, the alterations of the PTF1 binding site that eliminated PTF1 binding in EMSA also eliminated the activation by transfected PTF1 components (Fig. 3B). Only the mutation of the TC-box that allows the binding of a P48-HEB heterodimer retained detectable, albeit very low, activation (Fig. 3B, asterisk 6AmT panel). This level of activation is fourfold less than of the heterodimer on a wild-type PTF1 binding site (Fig. 3A), suggesting that the endogenous RBP-J contributes to the binding or activity of P48-HEB when an effective TC-box is present. Consistent with this interpretation, the addition of RBP-J or RBP-L further reduced activation of the TC-box mutant (Fig. 3B, 6AmT panel), likely by forcing the formation of the trimeric complex, which then precludes binding to an E-box in the absence of a TC-box (Fig. 3B). Collectively, these results indicate that the association of either RBP enhances both DNA binding and transcriptional activation.
Two motifs in the C-terminal domain of P48 are essential for the interaction with RBPJ/L.
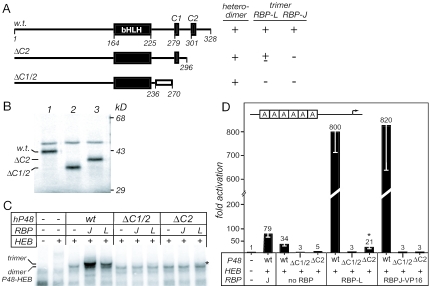
Comparison of the amino acid sequences for mouse, zebra fish, and fruit fly P48's revealed the conservation of two short peptide motifs, C1 (HSLSW) and C2 (WTPEDPR), in addition to the bHLH domain (Fig. 4A). To determine the function implied by this phylogenetic conservation, we tested the effects of mutations in C1 and C2 on the ability of P48 to form a trimeric DNA-binding complex (Fig. 4B). The deletion of either peptide or substitution of alanine for tryptophan in either peptide had no effect on the formation of DNA-binding heterodimers (Fig. 4C). In contrast, any of these changes did have pronounced and differential effects on the inclusion of RBP-J and RBP-L in a trimeric complex. The alterations of the C2 region prevented the association of RBP-J but had little or no effect with RBP-L. Conversely, deletion of the C1 peptide affected the recruitment of RBP-L more severely than that of RBP-J. Because tryptophan residues play key roles in the formation of other transcription factor complexes (15, 27), we tested whether the tryptophan residues in each of the peptides were necessary for P48 recruitment of the RBP-J or -L. Whereas substitution of alanine for either tryptophan 280 in C1 or 298 in C2 inhibited the recruitment of RBP-J, neither substitution detectably affected the recruitment of RBP-L. Deletion of amino acids 246 to 273, which are conserved among vertebrates but not to insects (Fig. 4A), severely affected the recruitment of RBP-L but much less so RBP-J (data not shown). These contrasting effects suggest that the binding requirements of the two RBPs emphasize different features in the C terminus of P48. The region for RBP-L binding is more extended than that for RBP-J and may correspond to the broad C-terminal region conserved among vertebrate P48 proteins.
PTF1 activity requires the interaction between P48 and RBP-L.
To verify that the transcriptional activity of P48 and RBP-L is dependent on their presence in the PTF1 complex, we tested whether the mutations of the C1 and C2 peptides also inhibited transcriptional activation in transfected cells. To derive an effective transfection assay for RBP-J with its weak activation potential, we created a strong, constitutively active form by fusing the VP16 activation domain to the N terminus of RBP-J, similar to that described by (19, 26). P48W298A, which could recruit RBP-L but not RBP-J (Fig. 4C), retained much of its transcriptional activity in combination with RBP-L but lost nearly all activity with VP16RBP-J (Fig. 4D). The effects of the W280A substitution were similar, though more modest. P48 lacking the C1 peptide region had little activity in combination with VP16RBP-J and was completely inactive with RBP-L (Fig. 4D), a finding consistent with its relative ability to interact with the RBPs (Fig. 4C). The congruence between the ability of the RBPs to interact with P48 and their ability to stimulate activation of a PTF1-reporter gene confirms that their activity on PTF1 sites derives from the trimeric complex.
The PTF1 complex is conserved from flies to mammals.
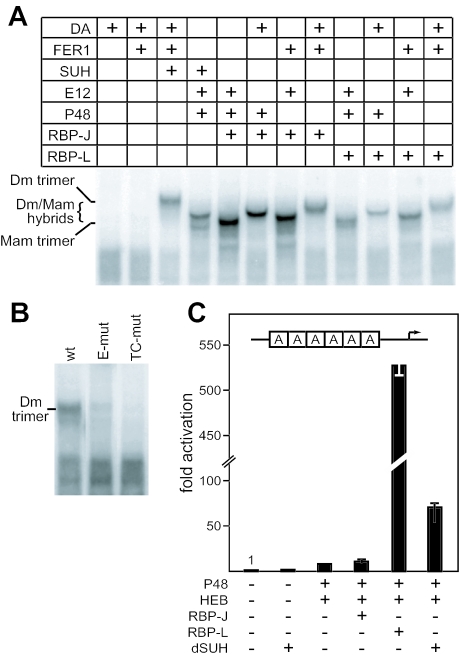
To determine whether a PTF1-like complex might be ancient, we tested whether the Drosophila orthologs of P48, E12, and RBP-J [FER1, DA and SU(H), respectively] could form a trimeric complex on the Ela1 PTF1-binding site. Indeed, the Drosophila proteins formed a DNA-binding complex that required all three proteins (Fig. 5A). Moreover, any combination containing the three types of subunits from Drosophila and mammals could form a trimeric DNA-binding complex. Like the mammalian PTF1, the Drosophila FER1-DA-SU(H) complex did not bind versions of the PTF1-site containing the diagnostic nucleotide substitutions in the E- and TC-boxes (Fig. 5B). Furthermore, coexpression of SU(H) with P48 and HEB in 293 cells activated the PTF1-responsive reporter to a level (70-fold over the basal) intermediate between PTF1-J and PTF1-L (Fig. 5C). The striking phylogenetic conservation of form and function indicates that the trimeric complex is an important regulator.
FIG. 5.
Structure and function of PTF1 is conserved between mammals and flies. (A) Formation of DNA-binding heterodimeric and heterotrimeric complexes using mammalian IVT P48, E12, and either RBP-J or RBP-L, Drosophila FER1, DA, and SU(H), or various combinations as indicated. The mobilities of the complexes depend on which mammalian or Drosophila proteins are used in the binding reactions. (B) Binding of the Drosophila protein complex is sensitive to same nucleotide changes in the E- and TC-boxes as the mammalian PTF1 complex. (C) The relative activity of the 6A.EIp.luc reporter gene in 293 cells was assayed in the presence or absence of cotransfected P48, HEB, RBP-J, RBP-L, or SU(H) in the combinations indicated. All values are the means of four transfections ± the SEM.
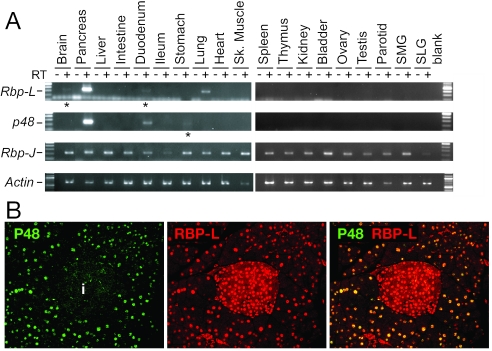
P48 and Rbp-L are selectively expressed at high levels in the pancreas.
p48 and Rbp-L have organ-restricted expression patterns (23, 24, 32). To determine whether the patterns of p48 and Rbp-L overlap, we used RT-PCR to survey the mRNAs for P48, RBP-L, and RBP-J in 19 adult mouse organs (Fig. 6A). p48 and Rbp-L are coexpressed at high levels in the pancreas and at low levels in the duodenum. Otherwise, p48 mRNA was detected in the stomach and Rbp-L mRNA in the brain and lung. Rbp-J is widely, if not universally, expressed. In the adult pancreas, P48 is restricted to acinar cell nuclei, whereas RBP-L is present in both acinar and islet nuclei (Fig. 6B). The presence of both proteins selectively in acinar nuclei is consistent with the acinar specific function of the PTF1 complex.
FIG. 6.
Sites of Rbp-L and p48 expression. (A) Rbp-L and p48 transcripts are present at high levels selectively in the pancreas. Amplified products from RT-PCR assays with RNA isolated from 19 adult mouse organs shows that Rbp-L transcripts, like p48 transcripts, are present at high levels in the pancreas. *, low level of Rbp-L mRNA in the duodenum and brain and p48 mRNA in the stomach. (B) Immunofluorescence with anti-P48 and anti-RBPL antibodies shows that P48 and RBP-L are specifically colocalized in the acini of the adult pancreas. In the left panel, green immunofluorescence indicates P48 is in acinar nuclei only. i, islet. In the center panel, red indicates RBP-L is in the nuclei of the acinar and islet cells. In the right panel, the merged image shows colocalization (yellow) only in acinar nuclei.
PTF1 components are bound to target promoters in vivo.
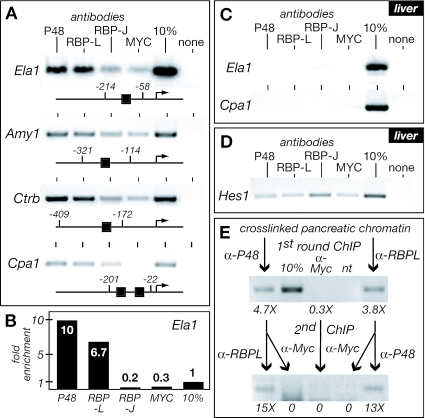
To determine whether the subunits of the PTF1 complex are bound to acinar genes, we performed ChIP with antibodies specific for P48, RBP-L, and RBP-J with cross-linked, sheared chromatin from adult rat pancreas and liver. The promoter region of Ela1 was enriched 32- and 22-fold when pancreatic chromatin was immunoprecipitated with anti-P48 and anti-RBP-L, respectively (Fig. 7A and B). The Ela1 promoter was not enriched from liver chromatin (Fig. 7C), in which acinar digestive enzymes genes are inactive (7, 37). Similar enrichment of the promoter regions from pancreatic chromatin for Amy1, Ctrb, and Cpa1 showed that the presence of P48 and RBP-L is common to acinar specific promoters with PTF1-binding sites. The lack of enrichment with anti-RBP-J (Fig. 7A) extends the exclusion of RBP-J to the active PTF1 complexes bound to promoters.
FIG. 7.
PTF1 components P48 and RBP-L are bound to acinar-specific promoters in vivo. (A) Fragments of pancreatic chromatin containing the PTF1 binding sites of the promoters for acinar-specific genes (Ela1; Amy2, amylase 2; Ctrb, chymotrypsinogen B; Cpa1, carboxypeptidase A1) were enriched with antibodies to P48 and RBP-L but not with antibodies to RBP-J or c-MYC. Schematic representations of the promoters show the locations of the PTF1 binding sites. cMYC antibody was used as a measure of nonspecific immunoprecipitation. (B) Fold enrichment (from panel A) of the Ela1 promoter upon immunoprecipitation of pancreas chromatin with P48, RBP-L, RBP-J, and cMYC antibodies and quantified by real-time PCR. Note that, relative to the anti-cMYC control, the enrichments with anti-P48 and anti-RBP-L were 32- and 22-fold, respectively. (C) ChIP of the Ela1 and Cpa1 promoters from liver chromatin with P48, RBP-L, RBP-J, and c-MYC antibodies. (D) The efficacy of the RBP-J antibody was verified by its ability to enrich the Hes1 promoter, a known Notch target, by ChIP of liver chromatin. The Hes1 promoter contains a pair of RBP-J consensus binding sites at −78 and −92. (E) Sequential ChIPs show that P48 and RBPL are present concurrently on the Ela1 promoter in chromatin from adult mouse pancreas. The numbers below the images of the PCR products from conventional PCR indicate the fold enrichment of the Ela1 promoter measured by real-time PCR after each of the two rounds of ChIP.
To verify that P48 and RBP-L reside concurrently on the same Ela1 promoter, we performed sequential ChIP with anti-P48 and anti-RBP-L (Fig. 7E). Indeed, enrichment with one antibody gave further enrichment with the next. The total enrichment approached the theoretical maximum if two proteins fully co-occupy a target DNA (9). Although these results do not show rigorously that P48 and RBP-L are present as part of a trimeric complex, the colocalization of both proteins to all four target promoters strongly suggests that the intact PTF1 complex is present.
Human p48 mutations disrupt the association of RBPs with the complex.
Two naturally occurring mutations in human p48 are associated with permanent neonatal diabetes mellitus, a genetic disorder characterized by the loss of pancreatic and cerebellar development (36). One of the mutations causes the deletion of the C terminus of P48, including peptide C2; the other deletes the region containing both C1 and C2 (Fig. 8A). We tested whether these deletions affect the ability of P48 to bind an RBP, as would be predicted. Normal human P48 readily formed PTF1 complexes with E12 and either RBP-J or RBP-L (Fig. 8C) and supported the characteristic high transcriptional activation in cell transfection assays (Fig. 8D). Although the mutant forms (which have more extensive alterations than the mouse mutants tested in Fig. 4) could form heterodimers with E12, they were unable to recruit RBP-J or -L into a trimeric complex (Fig. 8C). The small amount of the trimer formed with P48-ΔC2 (Fig. 8C, asterisk) provided less than 3% of the normal transcriptional activity (Fig. 8D). The developmental defects correlate with the inability of P48 to bind an RBP while retaining the ability to bind an AbHLH (Fig. 8C) and verify the importance of P48-RBP interactions in vivo.
FIG. 8.
Human disease truncations of the human P48 disrupt binding to RBP-J and RBP-L. (A) The truncated proteins resulting from the human R296X (ΔC2) and P236fsX270 (ΔC1/2) mutations. The open rectangle represents a 24-amino-acid frameshift extension. The ability of the wild-type and mutant human P48 proteins to form heterodimers with E12 and trimers with an RBP are summarized from the data in panel C. (B) Sodium dodecyl sulfate-polyacrylamide gel analysis of IVT human P48 proteins labeled by [35S]methionine incorporation. (C) Ability of the mutant human P48 proteins to form heterodimers with E12 and trimers with E12 and either RBP-J or RBP-L that bind the Ela1 PTF1-site. The asterisk indicates a small amount of PTF1-trimer formed with P48-ΔC2. (D) The human mutant P48 proteins cannot activate transcription of a PTF1 reporter gene in transfected 293 cells. The asterisk denotes a small amount of PTF1 activity with P48-ΔC2. Error bars indicate the SEM for three experiments.
The binding of P48 and the NotchIC to RBP-J is mutually exclusive.
The binding of NotchIC to RBP-J involves a conserved pentapeptide motif in the RAM domain of Notch that interacts with a hydrophobic pocket in RBP-J (16). The similarity of this motif (LWFPE in human Notch1) with the C2-peptide of P48 (VWTPE) suggested that the NotchIC and P48 bind to and compete for the same site on RBP-J. Indeed, NotchIC inhibits the formation of PTF1-J in a concentration-dependent manner (Fig. 9A) and decreases PTF1-J transcriptional activity in transfected cells (Fig. 9B). The lack of persistent effects of NotchIC on the formation or activity of PTF1-L (Fig. 9A and B) agrees with the evidence that the C2 peptide is neither necessary nor sufficient for the interaction of P48 with RBP-L (see Fig. 4B). Conversely, forced expression of P48 (with or without an AbHLH) inhibits the NotchIC enhancement of RBP-J activity on a reporter gene driven by a repeat of the consensus RBP-binding site (Fig. 9C). This interference by P48 is direct, because it is attenuated by the W298A mutation in the C2 peptide (Fig. 9C), which eliminates P48 binding to RBP-J (see Fig. 4B).
FIG. 9.
The NotchIC and peptides related to the Notch ΦWΦP-motif compete with P48 for binding to RBP-J. (A, top) NotchIC disrupts the trimeric PTF1 complex formed with RBP-J (upper set) much more than that with RBP-L (lower set). P48:E12:RBP complexes were formed with 1:1:1 molar ratios of the IVT proteins on the Ela1 PTF1 site alone (lane c) or in the presence of increasing IVT NotchIC (lanes d to g). Lane a, blank IVT reaction; lane b, dimer formation with 1:1 IVT P48 and E12. (A, bottom) P48 (upper set), but not the P48W298A mutant (lower set), disrupts the DNA-bound complex of RBP-J and NotchIC. RBP-J:NotchIC complexes were formed with 1:1 molar ratios of the IVT proteins on the consensus RBP-J binding-site (see Materials and Methods) alone (lane d) or in the presence of increasing P48 (lanes e to h). Lane a, blank IVT reaction; lane b, IVT RBP-J; lane c, IVT NotchIC. (B) The NotchIC inhibits the transcriptional activity of PTF1. Effects of increasing amounts of transfected NotchIC expression plasmid on the activation of the PTF1 reporter gene (6A.EIp.luc) in 293 cells. Error bars indicate the SEM for four transfections. (C) Conversely, P48 inhibits NotchIC-enhanced RBP-J activity. NotchIC-RBP-J activity was monitored with a luciferase reporter driven by six repeats of a consensus RBP-binding site (6R.EIp.Luc). (D) C2-like peptides from transcription factors containing the ΦWΦP-motif. RevNotch1 is a peptide with the Notch1 sequence reversed. (E) Inhibition of PTF1 complex formation with increasing concentrations (0.02, 0.2, and 2 mM) of various synthetic C2-like peptides. The relative inhibitory effects were as follows: KYOT2 ≫ NOTCH1 > P48 > revNOTCH1 > NFATc4 ≥ HAIRLESS. The P48:E12 dimer was not affected.
Synthetic C2-like peptides derived from P48 and NOTCH1 can interfere with the formation of PTF1-J in vitro (Fig. 9D and E). Both were more effective than a peptide with the NOTCH1 sequence reversed. The interfering activities of these similar peptides indicate that NotchIC and P48 bind the same site on RBP-J.
The presence of similar tryptophan-containing peptides in other transcription factors (Fig. 9D) suggests that Notch-independent functions of RBP-J/Su(H) may be more widespread. To determine whether these related motifs have the potential to mediate binding to RBP-J, we tested whether C2-like peptides we identified in HAIRLESS, KYOT2, and NFATc4 could disrupt the formation of the PTF1-J complex in vitro (Fig. 9E). Although the HAIRLESS and NFATc4 peptides were no more effective than the reversed-NOTCH1 peptide, the KYOT2 peptide was even more effective at low concentrations than those from P48 and NOTCH1.
DISCUSSION
The nature and the novelty of the PTF1 complex.
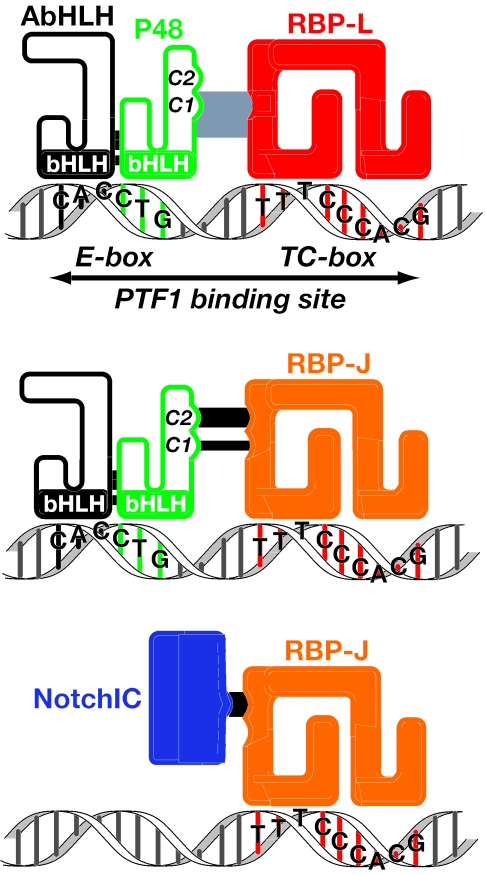
We showed here that PTF1 is an unusual compound transcription factor that incorporates RBP-J or RBP-L into an otherwise conventional bHLH factor complex (Fig. 10). The RBP subunit is tethered to the complex by direct contact with the C terminus of P48. RBP-L is the transcriptionally potent, Notch-unresponsive paralogue of RBP-J, the DNA-bound transducer of Notch signaling. RBP-L provides the strong transcriptional activity of the PTF1 complex that drives the high-level expression of the digestive enzyme genes of the adult pancreas. The DNA-binding properties of the complex are unusual as well. The trimeric complex forms in the absence of DNA, and the association of the RBP subunit and the P48-AbHLH heterodimer mutually alters the DNA binding of each. Whereas an E-box is sufficient to bind the bHLH heterodimer and a TC-box is sufficient for the RBP, the trimeric complex requires both. Moreover, the binding of the complex is highly cooperative; especially for binding sites with divergent E- and TC-boxes, the binding of the trimeric complex can be much greater than the sum of the binding of the individual parts. The formation of the complex creates a synergistic dependence on the presence of both DNA sites spaced appropriately. The nature of the complex explains the unusual binding properties of PTF1 and the ability to accommodate a wide variation in paired E- and TC-boxes in the PTF1 sites of acinar specific promoters (6, 32).
FIG. 10.
RBP-L and RBP-J in the PTF1 complex or associated with the NotchIC.
Whereas Obata et al. showed that RBP-J interacts with P48, they were unable to detect the formation of a trimeric complex (26). We demonstrated that RBP-J forms a specific, stable, and transcriptionally active trimeric complex with P48 that has the same DNA-binding characteristics as the RBP-L form (PTF1-L). The phylogenetic conservation of the RBP-J form from arthropods to mammals indicates that it plays an important, possibly developmental, role. The only previously known molecular function of RBP-J is its role as the DNA-bound transcriptional effector of Notch-signaling, although genetic evidence indicates that SU(H) has a Notch-independent activity during the development of the mechanosensory organs of the Drosophila peripheral nervous system (3). The incorporation of RBP-J into a PTF1 complex precludes its interaction with the activated form of the Notch receptor and thereby renders it insensitive to Notch-signaling. We propose that the participation of RBP-J in the PTF1 complex is but the first example of such a Notch-independent function.
The cooperative binding properties of the PTF1 complexes and their ability to form prior to binding DNA have important regulatory implications. The cooperativity allows the use of variant E- and TC-boxes that cannot be bound individually by bHLH dimers or RBPs and so remain unoccupied and inactive in the absence of the trimeric complex. In addition, the efficient formation of the trimeric complex may limit the amount of P48 heterodimers and free RBPs and thereby prevent their binding to isolated E-boxes and RBP-sites. For PTF1-sites with near consensus half-sites, transcriptional activity might vary with changes in the composition of the bound complexes. For example, with limiting amounts of RBPs, P48-AbHLH dimers might predominate; with limiting P48, free RBP-L (or RBP-J) would be available for binding RBP sites with or without a paired E-box. In the adult pancreas, a cellular process ensures the formation of the PTF1 complex with highest transcriptional activity by excluding RBP-J in preference for RBP-L and coincidentally ensures that most of the RBP-J is available for Notch signaling.
Neither PTF1 complex is dependent on Notch signaling.
The RBP subunits provide the transcriptional activity of the PTF1 complex. RBP-J and RBP-L are homologous proteins encoded by separate genetic loci; the amino acid sequences of the mouse proteins are 48% identical, despite highly divergent N- and C-terminal regions. High sequence conservation between the core regions of RBP-J and -L (67% for the mouse proteins), the retention of identical residues in RBP-L that in RBP-J contact DNA (36), and the same consensus DNA binding sequence indicate that the two isoforms have very similar structures and bind DNA in the same manner. The differences in the transactivation properties of RBP-J and -L likely lie in the divergent N- or C-terminal regions. The N and C regions of RBP-J augment the binding to NotchIC (39). The RBP-L C terminus is conserved across species and likely provides the potent transactivation that RBP-J lacks.
RBP-J mediates the transcriptional effects of Notch signaling by receiving the processed NotchIC fragment while bound to target genes (2, 35). The NotchIC binds RBP-J through a tryptophan-containing peptide motif that has been proposed to fit into a hydrophobic pocket on the beta-trefoil domain of the RBP-J (16, 38). This interacting motif has the core consensus ΦWΦP (Φ is a hydrophobic residue), is conserved in Notch from arthropods to mammals, and is present in two other known RBP-J binding proteins, EBNA2 (20) and KyoT2 (40). For RBP-J in the PTF1 complex to respond to Notch signaling, the hydrophobic pocket would have to remain accessible to the NotchIC. We showed that the C2-peptide of P48 is a variant of the core consensus and performs the same function. Because the P48 C2 peptide can displace NotchIC from its complex with RBP-J and the equivalent Notch peptide displaces P48, Notch and P48 must compete for the same site on RBP-J. Indeed, overexpression of P48 disrupts the transcriptional activity of the RBPJ-NotchIC complex. Consequently, the incorporation of RBP-J in PTF1 is incompatible with its activation by the NotchIC.
Unlike monomeric RBP-J, RBP-L is not bound and activated by the NotchIC (24). RBP-L is inherently indifferent to Notch signaling, within or without the PTF1 complex. Whereas the C2 peptide of P48 is most important for binding RBP-J, the C1 peptide is more important for binding RBP-L. The C1 peptide is near the end of an extended region conserved among vertebrate P48s but not in the fly FER1. The appearance of RBP-L in the vertebrate lineage suggests that the conservation may be driven by selection for maintaining the interaction with RBP-L, distinct from RBP-J. The restricted tissue distribution of RBP-L indicates that its function may be limited largely to the maintenance or development of pancreatic tissues, the tracheal glands of the lung, and regions of the forebrain.
Proof that RBP-L plays a nonredundant function awaits an effective inactivation of the gene. Mouse Rbp-L is a complex gene comprising 12 exons and two transcriptional start sites (23). The downstream start creates a translational initiation codon in exon 5 that makes a shorter protein missing the first 123 N-terminal amino acids of the full-length RBP-L. Insertion of an nlacZ neo cassette with the simian virus 40 transcriptional terminator into the first exon disrupted the production of the large transcript but not of the short transcript (23). Mice homozygous for the disruption showed no obvious defects, but pancreatic function was not investigated and the sufficiency of the short form of RBP-L remains untested.
Significance of the phylogenetic conservation of PTF1.
We showed that the Drosophila orthologues of P48 (FER1), E12/E47 (DA), and RBP-J [SU(H)] form a trimeric complex with identical DNA-binding characteristics and similar transcriptional activity as mammalian PTF1. Because Rbp-L appears in vertebrates, the original PTF1-like complex used the mediator of Notch signaling, although in a manner that excluded it from the Notch signaling pathway. Because arthropods do not have an organ homologous to the exocrine pancreas, the ancient function of the PTF1 complex cannot be pancreatic function or development. A Notch-independent function that may be analogous to the action of PTF1 has been ascribed for SU(H) during the development of the mechanosensory organs of the Drosophila peripheral nervous system (3). The requirement for a PTF1 complex for proper formation of the cerebellum (see below) suggests that the ancient role is nervous system development.
Role of RBP in cerebellar and pancreatic development.
Two mutations of human P48/PTF1A have been associated with the absence of pancreatic and cerebellar development in newborn infants (36). The functional defects caused by the mutations are now clear. One deletes a C-terminal region containing the C2 peptide; the other deletes the C1 peptide as well (see Fig. 4A). Sellick et al. showed that mouse P48 with the C2 deletion and a class A bHLH (E47) could not activate a reporter gene driven by a PTF1 site and concluded that the absence of activity was due to the inability of the mutant P48 to form the bHLH heterodimer. We showed instead that both truncated forms of human P48 can form DNA-binding heterodimers with class A bHLH proteins and that the transcriptional defect was due to the inability to recruit RBP-J or RBP-L into a PTF1 complex. Although the mutations delete more than just the peptides that mediate the interactions with RBP-J and -L, it seems apparent that the developmental defects in infants homozygous for the p48-truncations are largely, if not exclusively, due to the inability to form complexes containing RBP-J or RBP-L (or both). Because RBP-J is present in the developing cerebellum but RBP-L is not (10, 23; our unpublished results), the PTF1 complex required for cerebellar development must contain RBP-J, equivalent to the Drosophila complex of FER1:DA:SU(H). Because the human p48 mutations that eliminate the binding of P48 to RBPs appear to phenocopy the p48-null mouse mutations (36), the functions of P48 during cerebellar and pancreatic development may occur predominantly or solely within the context of the PTF1 complex.
As for most programs of organogenesis, pancreatic development requires proper Notch signaling (1, 8, 11, 25). Leach and coworkers showed that enforced activation of the Notch pathway by constitutive NotchIC expression can interfere with acinar development, possibly through the disruption of PTF1 binding activity (8). Our results now suggest that the interference may be due to competition between NotchIC and P48 for RBP-J. It is possible that Notch signaling affects acinar cell differentiation in part through this mechanism during normal pancreatic development as well.
Broad potential for RBP-J functions independently of Notch signaling.
RBP-J is ubiquitous and therefore available for recruitment by transcriptional regulatory schemes in a wide variety of developmental programs (see, for example, references 3 and 36). We have shown that other DNA-binding transcription factors, e.g., KYOT2 (FHL1/SLIM), contain peptides related to the RBP-J-interacting motifs of Notch and P48, suggesting that these may also bind RBP-J. Indeed, KYOT2 is known to bind RBP-J and, due to its ability (like P48) to interfere with NotchIC activation in cell transfection overexpression experiments, has been proposed to be a negative regulator of Notch signaling (40). Other LIM-only proteins similar to KYOT2 act as docking molecules that assemble transcription factor complexes via LIM domains (28). Rather than interfering with RBP-J function, we suggest that KYOT2 may be an LIM-only adapter that integrates RBP-J into select transcription factor complexes.
Acknowledgments
We thank Jane Johnson for helpful discussions; Susan Hayward for gifts of CBF1 and Notch1 plasmids and advice; Shinichiro Sawada for the anti-HEB serum; Jamie Zoloty for Drosophila RNA; Y. Megan Kong for advice and assistance; and Ken Zaret for advice on ChIP.
This study was supported by Public Health Service grants DK61220 and DK55266 from the National Institute of Diabetes and Digestive and Kidney Diseases.
REFERENCES
- 1.Apelqvist, A., H. Li, S. L., P. Beatus, D. J. Anderson, T. Honjo, M. Hrabe de Angelis, U. Lendahl, and H. Edlund. 1999. Notch signalling controls pancreatic cell differentiation. Nature 400:877-881. [DOI] [PubMed] [Google Scholar]
- 2.Artavanis-Tsakonas, S., M. D. Rand, and R. J. Lake. 1999. Notch signaling: cell fate control and signal integration in development. Science 284:770-776. [DOI] [PubMed] [Google Scholar]
- 3.Barolo, S., R. G. Walker, A. D. Polyanovsky, G. Freschi, T. Keil, and J. W. Posakony. 2000. A Notch-independent activity of Suppressor of Hairless is required for normal mechanoreceptor physiology. Cell 103:957-969. [DOI] [PubMed] [Google Scholar]
- 4.Blackwell, T. K., and H. Weintraub. 1990. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science 250:1104-1110. [DOI] [PubMed] [Google Scholar]
- 5.Chaya, D., and K. S. Zaret. 2003. Sequential chromatin immunoprecipitation from animal tissues. Methods Enzymol. 376:361-372. [DOI] [PubMed] [Google Scholar]
- 6.Cockell, M., B. J. Stevenson, M. Strubin, O. Hagenbuchle, and P. K. Wellauer. 1989. Identification of a cell-specific DNA-binding activity that interacts with a transcriptional activator of genes expressed in the acinar pancreas. Mol. Cell. Biol. 9:2464-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis, B. P., and R. J. MacDonald. 1988. Limited transcription of rat elastase I transgene repeats in transgenic mice. Genes Dev. 2:13-22. [DOI] [PubMed] [Google Scholar]
- 8.Esni, F., B. Ghosh, A. V. Biankin, J. W. Lin, M. A. Albert, X. Yu, R. J. MacDonald, C. I. Civin, F. X. Real, M. A. Pack, D. W. Ball, and S. D. Leach. 2004. Notch inhibits Ptf1 function and acinar cell differentiation in developiong mouse and zebra fish pancreas. Development 131:4213-4224. [DOI] [PubMed] [Google Scholar]
- 9.Geisberg, J. V., and K. Struhl. 2004. Quantitative sequential chromatin immunoprecipitation, a method for analyzing co-occupancy of proteins at genomic regions in vivo. Nucleic Acids Res. 32:e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gray, P. A., H. Fu, P. Luo, Q. Zhao, J. Yu, A. Ferrari, T. Tenzen, D. I. Yuk, E. F. Tsung, Z. Cai, J. A. Alberta, L. P. Cheng, Y. Liu, J. M. Stenman, M. T. Valerius, N. Billings, H. A. Kim, M. E. Greenberg, A. P. McMahon, D. H. Rowitch, C. D. Stiles, and Q. Ma. 2004. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science 306:2255-2257. [DOI] [PubMed] [Google Scholar]
- 11.Hald, J., P. Hjorth, M. S. German, O. D. Madsen, P. Serup, and J. Jensen. 2003. Activated Notch1 prevents differentiation of pancreatic acinar cells and attenuate endocrine development. Dev. Biol. 260:426-437. [DOI] [PubMed] [Google Scholar]
- 12.Harding, J. D., R. J. MacDonald, A. E. Przybyla, J. M. Chirgwin, R. L. Pictet, and W. J. Rutter. 1977. Changes in the frequency of specific transcripts during development of the pancreas. J. Biol. Chem. 252:7391-7397. [PubMed] [Google Scholar]
- 13.Hsieh, J. J., T. Henkel, P. Salmon, E. Robey, M. G. Peterson, and S. D. Hayward. 1996. Truncated Mammalian Notch1 activates CBF1/RBPJk-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol. Cell. Biol. 16:952-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawaguchi, Y., B. Cooper, M. Gannon, M. Ray, R. J. MacDonald, and C. V. E. Wright. 2002. The role of the transcriptional regulator PTF1a in converting intestinal to pancreatic progenitors. Nat. Genet. 32:128-134. [DOI] [PubMed] [Google Scholar]
- 15.Knoepfler, P. S., D. A. Bergstrom, T. Uetsuki, I. Dac-Korytko, Y. H. Sun, W. E. Wright, S. J. Tapscott, and M. P. Kamps. 1999. A conserved motif N-terminal to the DNA-binding domains of myogenic bHLH transcription factors mediates cooperative DNA binding with pbx- Meis1/Prep1. Nucleic Acids Res. 27:3752-3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kovall, R. A., and W. A. Hendrickson. 2004. Crystal structure of the nuclear effector of Notch signaling, CSL, bound to DNA. EMBO J. 23:3441-3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krapp, A., M. Knofler, B. Ledermann, K. Burki, C. Berney, N. Zoerkler, O. Hagenbuchle, and P. K. Wellauer. 1998. The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes Dev. 12:3752-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kruse, F., S. D. Rose, G. H. Swift, R. E. Hammer, and R. J. MacDonald. 1995. Cooperation between elements of an organ-specific transcriptional enhancer in animals. Mol. Cell. Biol. 15:4385-4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurooka, H., K. Kuroda, and T. Honjo. 1998. Roles of the ankyrin repeats and C-terminal region of the mouse Notch1 intracellular region. Nucleic Acids Res. 29:5448-5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ling, P. D., and S. D. Hayward. 1995. Contribution of conserved amino acids in mediating the interaction between EBNA2 and CBF1/RBPJk. J. Virol. 69:1944-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, Y., G. H. Swift, and R. J. MacDonald. 2001. The organization and function of the trimeric PDX1 homeodomain complex. J. Biol. Chem. 276:17985-17993. [DOI] [PubMed] [Google Scholar]
- 22.MacDonald, R. J., G. H. Swift, A. E. Przybyla, and J. M. Chirgwin. 1987. Isolation of RNA using guanidinium salts. Methods Enzymol. 152:219-227. [DOI] [PubMed] [Google Scholar]
- 23.Minoguchi, S., T. Ikeda, S. Itohara, T. Kaneko, H. Hokaichi, and T. Honjo. 1999. Studies on the cell-type specific expression of RBP-L, a RBP-J family member, by replacement insertion of beta-galactosidase. J. Biochem. 126:738-747. [DOI] [PubMed] [Google Scholar]
- 24.Minoguchi, S., Y. Taniguchi, H. Kato, T. Okazaki, L. J. Strobl, U. Zimber-Strobl, G. W. Bornkamm, and T. Honjo. 1997. RBP-L, a transcription factor related to RBP-Jk. Mol. Cell. Biol. 17:2679-2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murtaugh, L. C., B. Z. Stanger, D. M. Kwan, and D. A. Melton. 2003. Notch signaling controls multiple steps of pancreatic differentiation. Proc. Natl. Acad. Sci. USA 100:14920-14925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Obata, J., M. Yano, H. Mimura, T. Goto, R. Nakayama, Y. Mibu, C. Oka, and M. Kawaichi. 2001. p48 subunit of mouse PTF1 binds to RBP-Jk/CBF1, the intracellular mediator of Notch signalling, and is expressed in the neural tube of early stage embryos. Genes Cells 6:345-360. [DOI] [PubMed] [Google Scholar]
- 27.Piper, D. E., A. H. Batchelor, C. P. Chang, M. L. Cleary, and C. Wolberger. 1999. Structure of a HoxB1-Pbx1 heterodimer bound to DNA: role of the hexapeptide and a fourth homeodomain helix in complex formation. Cell 96:587-597. [DOI] [PubMed] [Google Scholar]
- 28.Rabbitts, T. H. 1998. LMO T-cell translocation oncogenes typify genes activated by chromosomal translocations that alter transcription and developmental processes. Genes Dev. 12:2651-2657. [DOI] [PubMed] [Google Scholar]
- 29.Ramain, P., K. Khechumian, L. Seugnet, N. Arbogast, C. Ackermann, and P. Heitzler. 2001. Novel Notch alleles reveal a Deltex-dependent pathway repressing neural cell fate. Curr. Biol. 11:1729-1738. [DOI] [PubMed] [Google Scholar]
- 30.Rose, S. D., F. Kruse, G. H. Swift, R. J. MacDonald, and R. E. Hammer. 1994. A single element of the elastase I enhancer is sufficient to direct transcription selectively to the pancreas and gut. Mol. Cell. Biol. 14:2048-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rose, S. D., and R. J. MacDonald. 1997. Evolutionary silencing of the human elastase I gene (ELA1). Hum. Mol. Gen. 6:897-903. [DOI] [PubMed] [Google Scholar]
- 32.Rose, S. D., G. H. Swift, M. J. Peyton, R. E. Hammer, and R. J. MacDonald. 2001. The role of PTF1-P48 in pancreatic acinar gene expression. J. Biol. Chem. 276:44018-44026. [DOI] [PubMed] [Google Scholar]
- 33.Roux, E., M. Strubin, O. Hagenbuchle, and P. K. Wellauer. 1989. The cell-specific transcription factor PTF1 contains two different subunits that interact with the DNA. Genes Dev. 3:1613-1624. [DOI] [PubMed] [Google Scholar]
- 34.Sawada, S., and D. R. Littman. 1993. A heterodimer of HEB and an E12-relataed protein interacts with the CD4 enhancer and regulates its activity in T cells. Mol. Cell. Biol. 13:5620-5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Scheisguth, F. 2004. Regulation of Notch signaling activity. Curr. Biol. 14:R129-R138. [PubMed] [Google Scholar]
- 36.Sellick, G. S., K. T. Barker, I. Stolte-Dijkstra, C. Fleischmann, R. J. Coleman, C. Garrett, A. L. Gloyn, E. L. Edghill, A. T. Hattersley, P. K. Wellauer, G. Goodwin, and R. S. Houlston. 2004. Mutations in PTF1A cause pancreatic and cerebellar agenesis. Nat. Genet. 36:1301-1305. [DOI] [PubMed] [Google Scholar]
- 37.Swift, G. H., R. E. Hammer, R. J. MacDonald, and R. L. Brinster. 1984. Tissue-specific expression of the rat pancreatic elastase I gene in transgenic mice. Cell 38:639-646. [DOI] [PubMed] [Google Scholar]
- 38.Tamura, K., Y. Taniguchi, S. Minoguchi, T. Sakai, T. Tun, T. Furukawa, and T. Honjo. 1995. Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-J kappa/Su(H). Curr. Biol. 5:1416-1423. [DOI] [PubMed] [Google Scholar]
- 39.Tani, S., H. Kurooka, T. Aoki, N. Hashimoto, and T. Honjo. 2001. The N- and C-terminal regions of RBP-J. interact with the ankyrin repeats of Notch1 RAMIC to activate transcription. Nucleic Acids Res. 29:1373-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taniguchi, Y., T. Furukawa, T. Tun, H. Han, and T. Honjo. 1998. LIM protein KyoT2 negatively regulates transcription by association with the RBP-J DNA-binding protein. Mol. Cell. Biol. 18:644-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tun, T., Y. Hamaguchi, N. Matsunami, T. Furukawa, T. Honjo, and M. Kawaichi. 1994. Recognition sequence of a highly conserved DNA binding protein RBP-Jk. Nucleic Acids Res. 22:965-971. [DOI] [PMC free article] [PubMed] [Google Scholar]