Abstract
One of the primary sources of enzyme instability is protein oxidative modification triggering activity loss or denaturation. We show here that the side chain of Cys108 is the main site undergoing stress-induced oxidation in Trigonopsis variabilis d-amino acid oxidase, a flavoenzyme employed industrially for the conversion of cephalosporin C. High-resolution anion-exchange chromatography was used to separate the reduced and oxidized protein forms, which constitute, in a molar ratio of about 3:1, the active biocatalyst isolated from the yeast. Comparative analysis of their tryptic peptides by electrospray tandem mass spectrometry allowed unequivocal assignment of the modification as the oxidation of Cys108 into cysteine sulfinic acid. Cys108 is likely located on a surface-exposed protein region within the flavin adenine dinucleotide (FAD) binding domain, but remote from the active center. Its oxidized side chain was remarkably stable in solution, thus enabling the relative biochemical characterization of native and modified enzyme forms. The oxidation of Cys108 causes a global conformational response that affects the protein environment of the FAD cofactor. In comparison with the native enzyme, it results in a fourfold-decreased specific activity, reflecting a catalytic efficiency for reduction of dioxygen lowered by about the same factor, and a markedly decreased propensity to aggregate under conditions of thermal denaturation. These results open up unprecedented routes for stabilization of the oxidase and underscore the possible significance of protein chemical heterogeneity for biocatalyst function and stability.
Oxidases constitute a structurally diverse group of enzymes that catalyze a wide range of chemical reactions using molecular oxygen as an electron acceptor. Their ability to couple specific substrate dehydrogenation with controlled, stoichiometric reduction of dioxygen into H2O2 or water has been much exploited in biocatalysis (4) and the analytical sciences (1). The exactly reproducible performance of an oxidase-based bioreactor or analytical device over time strongly hinges on the operational stability of the biological component. The widely recognized fact that oxidases often fall short in their stabilities under conditions of use has stimulated research into factors causing the inactivation and methods of stabilizing the activity (9, 21). There is consensus that one of the primary sources of instability of oxidases is their susceptibility to protein oxidative modification, triggered by O2 or H2O2 and leading to activity loss or denaturation. However, to the best of our knowledge, there are presently only a few enzymes for which a defined chemical conversion of the critical side chain that is sensitive to oxidation has been correlated with consequences at the level of enzyme function. The transformation of Met222 to a sulfone in the protease subtilisin is a classical example of oxidative inactivation of an industrial biocatalyst (31). Bacterial tetrachlorohydrochinone dehalogenase is inactivated through the oxidation of its Cys13 to a sulfenic acid (36). A catalytic mutant of glucoamylase, Glu400→Cys, could be reactivated by oxidation of the cysteine to a sulfinic acid (18). In the physiological context, nitrile hydratase undergoes posttranslational oxidation at two active-site cysteines, thereby generating a unique Cys-SOH- and Cys-SO2H-containing catalytic center in the fully active enzyme (19). Likewise, the matrix metalloproteinase-7 is activated by oxidation of its Cys70 into Cys-SO2H (10).
Cytochrome c oxidase is oxidized by H2O2 at two Trp residues concomitant with loss of activity (20). In addition to its potential role in modulating the biological function of a protein through effects on its activity or stability, the site-specific oxidation of cysteines has recently attracted a lot of attention because of its proposed involvement in oxidant scavenging and redox signaling (3, 6, 8, 13, 26, 37). Furthermore, oxidation of sulfur-containing amino acids may cause protein abnormalities and is a likely factor in their lifespan in the cell (8, 24, 25, 30).
Detailed knowledge about possible oxidative modifications of the enzyme of interest is obviously vital to devise a targeted approach for improving the activity performance, especially stability. Disentanglement of the structural heterogeneity induced in an enzyme preparation by chemical oxidation is, however, a very challenging analytical task and requires that high-resolution tools of protein and peptide separation and structure elucidation be coupled in an appropriate manner.
Here, we have turned our attention to d-amino acid oxidase (DAO; EC 1.4.3.3) from the yeast Trigonopsis variabilis (TvDAO). The enzyme catalyzes with broad specificity the oxidative deamination of α-d-amino acids harboring natural and nonnatural side chains, producing the corresponding α-keto-acids, ammonia, and H2O2. Its activity depends on a flavin adenine dinucleotide (FAD) cofactor, which is bound noncovalently to each enzyme protomer. TvDAO is a functional oligomer composed of identical subunits (21, 29). The enzyme is a very interesting biocatalyst whose foremost importance is presently the conversion of cephalosporin C into glutaryl-7-aminocephalosporanic acid, the first intermediate en route to 7-amino-cephalosporanic acid. The 7-amino-cephalosporanic acid is produced commercially on a multiton-per-year scale, and use of immobilized TvDAO for this purpose is exemplary for the successful implementation of a flavo-oxidase in an industrial chemical process (27).
Other, also relevant applications of TvDAO include the analytical detection of α-d-amino acids in biological samples and foodstuffs, the resolution of racemic mixtures of amino acids, and the production of α-keto-acids from d-amino acids (for a review, see references 21 and 33).
A number of papers have reported the results of experiments designed to analyze and improve the stability of TvDAO (2, 14, 15). The persistence of soluble, carrier-bound, and physically entrapped forms of TvDAO against inactivation is not fully satisfactory and is seen as a major shortcoming of the enzyme (2, 27). Unfortunately, the available evidence is partly conflicting and cannot be consolidated to a clear focal point for the production of a more suitable TvDAO. The typically observed time courses of activity loss in the absence and presence of oxidative stress are complex (2, 23), suggesting that the denaturation mechanism involves more than a single species of active protein. We show here by combining high-resolution protein and peptide fractionation, structure determination by tandem mass spectrometry, and biochemical methods that site-specific oxidative conversion of Cys108 in TvDAO is a main source of structural and functional heterogeneity in the enzyme. The results reveal hitherto unprecedented routes for the stabilization of TvDAO and provide novel insights into the structure-function relationships. They strongly emphasize the potential value of identifying the site of chemical modification in biocatalysts that are sensitive to oxidation.
MATERIALS AND METHODS
Materials.
NADH was from Seppim S.A. (Sees, France), and l-lactate dehydrogenase was from Roche (Mannheim, Germany). All other chemicals, including proteomics grade modified trypsin, were obtained from Sigma-Aldrich (St.Louis, MO). Nanoflow liquid chromatography (NanoLC) eluents were prepared in deionized ultrapure water generated by the Nanopure Water system (Barnstead, Newton, WA). A biocatalyst preparation of TvDAO from strain Trigonopsis variabilis ATCC 10679 was kindly provided by Sandoz GmbH (Kundl, Austria). Its specific enzyme activity was 40 U/mg, determined by a reported coupled enzyme assay using d-alanine as the substrate (34). If not otherwise stated, all buffers contained 1 mM dithiothreitol (DTT).
Separation of native TvDAO and its modified protein forms.
The enzyme was stored in aliquots at −25°C for later processing. An aliquot was thawed, and precipitated protein was removed from it by centrifugation (Sorval centrifuge, rotor T865) at 80,000 × g (30 min; 4°C). The resulting supernatant was subjected to gel filtration (HiPrep 26/10 column [2.6 cm by 10 cm; Amersham Biosciences, Uppsala, Sweden], and BioLogic DuoFlow system [Bio-Rad, Hercules, CA]), and the buffer was exchanged for 5 mM Tris-HCl, pH 7.5, containing 1 mM DTT. Pooled eluate was concentrated to the original volume by ultrafiltration (Vivaspin ultrafiltration concentrator tubes; Vivascience AG, Hanover, Germany)at4,400 × g using molecular mass cutoff membranes of 10 kDa. The enzyme was loaded on an anion-exchange MonoQ HR 5/5 column (0.5 cm by 5 cm; Amersham Biosciences) using a BioLogic DuoFlow system (Bio-Rad). The column had been equilibrated with 1 mM DTT-10 mM Tris-HCl, pH 7.5 (buffer A). The bound protein was eluted with six steps of isocratic flow of 0.1 to 0.6 M KCl in buffer A (within 96 min; flow rate, 0.5 ml/min), followed by isocratic flow of 1 M KCl in buffer A (5 min; flow rate, 0.75 ml/min). UV detection at 280 nm was used. The collected fractions were pooled according to eluted protein peaks.
The fractions were checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), nondenaturing anionic PAGE, and isoelectric focusing using Coomassie blue for the staining of protein bands. Enzyme preparations which were not used immediately were aliquoted and stored at −25°C. Protein concentrations were determined by the Bradford method (protein microassay from Bio-Rad, Hercules, CA) using bovine serum albumin as a standard.
Size exclusion chromatography.
Size exclusion chromatography was performed at 10°C using a Superdex 200 HR 10/30 column and an Äkta FPLC system (both from Amersham Biosciences). Intact TvDAO or its modified form (500 μg; 100 μl) was loaded on the column equilibrated with 10 mM Tris-HCl (pH 7.5; 0.2 M KCl) containing 1 mM DTT. Elution was carried out isocratically with a flow rate of 0.5 ml/min and detection at 280 nm. Molecular mass determinations were referenced against appropriate protein standards (Bio-Rad).
Spectral measurements and enzyme assays.
Protein absorbance spectra were collected with a Beckmann DU800 UV-VIS spectrophotometer (Beckmann-Coulter, Inc., Fullerton, CA) at 20 ± 0.1°C using a 10-mm-path-length quartz cell. The enzyme concentration was 10 μM in 10 mM Tris-HCl, pH 7.5, containing 1 mM DTT.
Activity of TvDAO was measured with an assay based on the decolorization of 2,6-dichloroindophenol (DCIP) and a coupled enzymatic assay (TvDAO/l-lactate dehydrogenase) using conditions reported recently (34). d-Methionine (20 mM) or d-alanine (50 mM) was employed as the substrate. Enzymatic oxygen consumption rates were recorded with a fiber optic fluorescence microsensor, as reported elsewhere (34).
For stability measurements, purified preparations of TvDAO (20 μM) dissolved in 10 mM Tris-HCl, pH 7.5, were incubated at 50 ± 1°C on an orbital shaker operated at an agitation rate of 300 rpm. Samples (350 μl) were taken at certain incubation times, and part of the sample (50 μl) was used to determine the residual enzyme activity with the DCIP-based assay. The remainder volume of the sample was used for light-scattering measurements, which served as particular probes of protein aggregation. Light scattering was recorded with an F4500 spectrofluorometer (Hitachi, Tokyo, Japan) under the conditions described above, except that excitation and emission wavelengths were both set to 500 nm.
The number of free sulfhydryl groups in purified fractions of TvDAO were determined using colorimetric titration with Ellman's reagent, 5,5′-dithio-bis (2-nitrobenzoic acid) (DTNB) (12). Briefly, exchange of buffer (80 mM sodium phosphate, pH 8.0, containing 1.2 mM EDTA) and exhaustive removal of DTT were achieved by repeated steps of gel filtration with a HiPrep 26/10 column (Amersham Biosciences), followed by concentration through ultrafiltration. The protein solution (6 μM; 400 μl) was mixed with 50 μl DTNB solution (10 mM; 100 mM sodium phosphate buffer, pH 7.8), corresponding to a molar ratio of DTNB to protein subunit of 200, and incubated at 25 ± 1°C for up to 240 min. The time course of absorbance at 412 nm was monitored continuously, and a molar absorption coefficient of 13,600 M−1 cm−1 for the 2-nitro-5-thiobenzoate anion was assumed for the calculation of the concentration of protein sulfhydryl groups reacted. Appropriate controls were recorded in all cases, and the reported values are corrected for minor blank readings. The standard error of determination was ± 20% (n = 6), and the results obtained with different concentrations of DTNB were fully consistent. Thiol group titration in unfolded TvDAO was carried out as just described after denaturation of the protein in the presence of SDS (15 mg/ml) for 15 min.
MS.
Analysis by mass spectrometry (MS) was performed using an Agilent 1100 series ion trap (IT) mass spectrophotometer (Agilent Technologies, Palo Alto, CA). The instrument, equipped with an orthogonal electrospray or nanospray source, was coupled to a microinfusion pump (KD Scientific, St. Louis, MO) or on line to an Agilent 1100 series nanoLC system (Agilent Technologies), respectively. The mass spectrometer was tuned using a calibration mixture provided by the manufacturer. The spectral data were acquired and monitored using the Agilent ChemStation software package (Agilent Technologies).
(i) ESI-MS.
For mass analysis, the TvDAO samples were prepared in 60% (vol/vol) acetonitrile, 1% formic acid solution and then directly infused into an electrospray ionization (ESI) source using a microsyringe (100 μl) coupled to the microinfusion pump (KD Scientific) at a flow rate of 5 μl/min. The spectra were obtained in the automatic acquisition mode scanning from 200 to 2,200 m/z. The tip was held at 3.5 kV in a positive ion detection mode, and nebulization was assisted by nitrogen gas at a flow rate of 8 liters/min. Data processing was done using the deconvolution module of the ChemStation software.
(ii) In-solution tryptic digestion.
A suitable volume of protein solution was diluted in Tris buffer (100 mM, 6 M urea, pH 7.8) and reduced with 5 mM DTT for 1 h at room temperature. Iodoacetamide (IAM) was added to the mixture in a fourfold excess over DTT and allowed to react for 1 h at room temperature in the dark. Subsequently, unreacted IAM was consumed by addition of the equimolar amount of reducing reagent (DTT) during the next hour, and then the reaction mixture was diluted with purified water (Nanopure Water System) to yield a final concentration of urea of 0.6 M. Sequencing grade modified trypsin was prepared by adding 20 μl of 1 mM HCl to a vial containing 20 μg of trypsin, and this solution was added to the pretreated sample at a substrate-to-enzyme ratio of 50:1 (wt/wt). The mixture was incubated overnight at 37°C, and the reaction was stopped by addition of acetic acid. The digest was used without any additional processing steps before analysis.
(iii) NanoLC/ESI-MS/MS.
The peptide mixture (1 μl) was trapped on a precolumn (ZORBAX 300SB-C18 column; 0.3 mm; Agilent Technologies) with a total loading time of 5 min and a sample solution flow rate of 200 nl/min. The loaded mixture was then flushed to a 0.75-μm (inside diameter) nanoLC separation column (ZORBAX 300SB-C18; 50 mm; Agilent Technologies), where the mobile-phase flow rate was 250 nl/min. Ultrapure water was used as mobile phase A, with addition of 0.1% high-performance liquid chromatography grade trifluoroacetic acid (vol/vol), and high-performance liquid chromatography grade acetonitrile-trifluoroacetic acid (99.9:0.1 [vol/vol]) was used as mobile phase B. Peptides were eluted from the stationary phase with a 60-min linear gradient ranging from 2 to 60% mobile phase B at a constant flow (250 nl/min). The column effluent was directed to an etched fused-silica ESI tip (360-50-8-D-20; New Objective, Inc., Cambridge, MA), and ions formed by electrospray were focused on the entrance of the IT mass spectrometer. The voltage at the spray tip and the heated MS inlet capillary temperature were optimized at 1.3 kV and 235°C, respectively. The IT operated in an automated data-dependent acquisition mode under the control of the Agilent ChemStation software system. The system was run in positive ion mode with a maximal ion accumulation time of 100 ms; the collision gas was helium. In all MS/MS experiments, a full MS scan between 200 and 2,200 m/z was followed by two maximal-resolution scans and six full MS/MS scans for the two most abundant ions from each MS scan.
(iv) Data processing.
Protein sequences were obtained from the National Center for Biotechnology Information (NCBI) database. The proteolytic digest pattern derived from the purified TvDAO was compared with a virtual digest of the protein derived using Peptide Tools software (Agilent Technologies). The acquired spectra were analyzed with Mascot (Matrix Science, London, United Kingdom) and Spectrum Mill, provided by Agilent Technologies, employing peptide mass fingerprint, MS/MS search, and de novo sequencing. Peptides containing cysteine residues were manually interpreted for confirmation.
RESULTS AND DISCUSSION
Separation and identification of multiple forms of TvDAO.
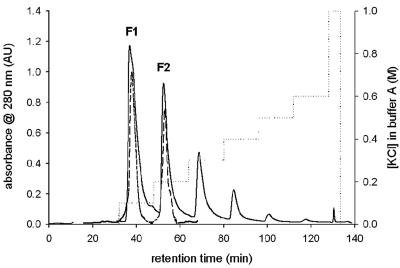
Figure 1 shows the protein elution profile during MonoQ anion-exchange chromatography of a representative TvDAO preparation that is used, after immobilization, in the conversion of cephalosporin C. Two of six distinct and baseline-separated protein peaks were found to contain significant yet different specific enzyme activities in the range of 40 to 220 U/mg (measured by the coupled enzyme assay using d-alanine as the substrate). The other peaks did not contain activity and were therefore not further analyzed. The pattern of multiple protein forms and their separation, as depicted in Fig. 1, was fully reproducible with different batches of the enzyme and did not change substantially regardless of the workup procedure used to obtain the technical-grade biocatalyst (data not shown). The recovery of the procedure, in respect to both enzyme activity and protein, was greater than 95%. Upon rechromatography on the MonoQ column, the isolated protein forms 1 (F1) and 2 (F2) eluted exactly as they did during fractionation of the protein mixture, as indicated by the superimposition of the respective UV traces in Fig. 1. It was proven that upon incubation at 4°C and 25°C in buffer (1 mM DTT-10 mM Tris-HCl, pH 7.5), isolated F1 and F2 were not interconverted into one another, clearly indicating that they represent stable protein forms of TvDAO.
FIG. 1.
Separation of different forms of TvDAO. Twenty milligrams of protein was applied on the MonoQ HR 5/5 column and separated (solid line) using seven steps of isocratic flow of 0.1 to 1 M KCl in buffer A (dotted line); TvDAO F1 and F2 were rechromatographed (dashed line) under the same conditions.
Molecular size determination of isolated forms of TvDAO.
The two separated, as well as rechromatographed, enzyme forms F1 and F2 migrated in SDS-PAGE as single protein bands with apparent molecular masses of about 39 kDa (data not shown). The molecular masses of F1 and F2 were calculated from ESI-MS spectra to be 39,214.1 ± 10 Da and 39,246.3 ± 10 Da, respectively. There are two different sequences of TvDAO reported in the NCBI database: entries Q99042 (or CAA90322) and AAR98816.
The primary structures of the TvDAOs described above differ at four positions: 87 (Ser in Q99042 and Asn in AAR98816), 102 (Val/Gly), 142 (Arg/Ala), and 344 (Val/Ile). The molecular masses of full-length TvDAOs, predicted from the genes including the initiator methionine without the bound cofactor, are 39,301.1 Da (Q99042) and 39,214.0 Da (AAR98816). According to full-mass scan spectra of both protein forms and molecular mass information, F1 is identified as the translated gene product of sequence AAR98816, and F2 appears to be a modified form of TvDAO. The mass difference of 32 Da (measured value, 32.2 Da) between F1 and F2 could occur if F2 had incorporated two additional oxygen atoms that are not present in F1 (Table 1). In nondenaturing anionic PAGE, F2 showed an unmistakably higher mobility (data not shown). Its pI value of ≈5.5 revealed in isoelectric focusing was slightly but significantly higher than that of F1 (ΔpI ≈ 0.12) (data not shown). This result, combined with the stronger retention of F2 in anion-exchange chromatography, suggests that the overall negative charge of F2 is probably greater than that of F1.
TABLE 1.
NanoLC/ESI-MS analysis of proteolytic digests of purified TvDAO protein formsa
| Peptide | Peptide sequenceb | Elemental composition | Predicted mass (Da) | Observed mass (Da) |
|---|---|---|---|---|
| 2 | 4IVVIGAGVAGLTTALQLLR22 | C85H153N23O23 | 1,865.2 | 1,865.2 |
| 4 | 24GHEVTIVSEFTPGDLSIGYTSPWAGANWLTFYDGGK59 | C179H254N42O55 | 3,873.8 | 3,874.0 |
| 12 (F1) | 100LEGAMSAICys108-CamQANPWFK115 | C79H120N20O22S2 (+C2H4NO) | 1,765.8 | 1,822.8 (+57) |
| 12 (F2) | 100LEGAMSAICys108-SO2HQANPWFK115 | C79H120N20O22S2 (+O2) | 1,765.8 | 1,798.0 (+32.2) |
| 15 | 130IVHDDEAYLVEFR142 | C73H108N18O23 | 1,604.8 | 1,604.8 |
| 16 | 143SVCIHTGVYLNWLMSQCLSLGATVVK168 | C126H204N32O35S3 | 2,822.4 | 2,822.5 |
| 20 | 176DANLLHSSGSRPDVIVNCSGLFAR199 | C107H174N34O35S | 2,528.3 | 2,528.4 |
| 25 | 221NSLPFMASFSSTPEK235 | C73H111N17O24S | 1,641.8 | 1,641.8 |
| 27 | 248FDGTSIIGGCFQPNNWSSEPDPSLTHR274 | C129H188N36O43S | 2,962.3 | 2,962.5 |
| 35 | 316IPGVGFVVHNYGAAGAGYQSSYGMADEAVSYVER349 | C157H229N41O50S | 3,522.6 | 3,522.8 |
Protein was digested with trypsin and subjected to nanoLC separation with electrospray ionization MS analysis, as described in Materials and Methods. The peptides listed here are all predicted and detected peptides that have masses higher than 1.5 kDa. Peptides 12 for TvDAO F1 and F2 are given separately, as indicated. The parameters for Spectrum Mill PMF search were as follows: maximum number of missed cleavages (1); cysteines modified by carboxamidomethylation; other possible modifications: peptide N-terminal Gln to pyroGlu, oxidation of Met, protein N-terminus acetylated; peptide mass tolerance of ±100 ppm; and monoisotopic peptide masses.
Boldface indicates the modifications of Cys108: carboxyamidomethylation (-cam), resulting from sample preparation for MS analysis; and oxidation to cysteine sulfinic acid (-SO2H).
Structural characterization of isolated forms of TvDAO.
The primary structure of TvDAO is predicted from the coding gene to contain six cysteines. Colorimetric titration with DTNB shows that all six thiol side chains are reduced in F1 and accessible for chemical derivatization in the folded state of the protein. In contrast, F2 has five reactive SH groups, suggesting that one cysteine may have been modified, most probably in an irreversible fashion. Tryptic peptide fingerprinting for F1 and F2 was carried out using ESI-MS, and the results are summarized in Table 1. The data reveal the identity of amino acids Asn87, Gly102, and Val344 and thus reinforce the above conclusion that the primary structure of F1 corresponds to the amino acid sequence predicted from gene AAR98816.
The peptides containing cysteine residues were identified unambiguously: Cys108, peptide 12; Cys145 and Cys159, peptide 16; Cys193, peptide 20; Cys257, peptide 27; and Cys298, peptide 32.
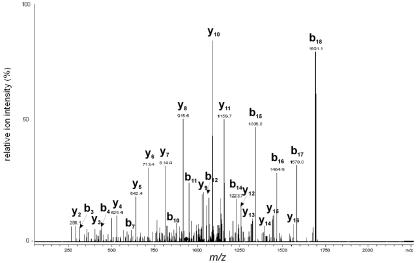
Tandem MS was used to clearly resolve conflicting published information regarding the N-terminal sequence of TvDAO (positions 4 to 22; peptide 2) (Fig. 2), which plays an important role in the function of the enzyme by providing contacts with the FAD cofactor. The N-terminal sequence of F1, I4VVIGAG10V11AGLT15T16A17L18Q19LLR22, corresponds to the sequences in the NCBI database but is different from another published sequence, I4VVIGAX10G11AGLA15G16X17G18L19LLR22 (22), as indicated by boldface letters. The overall sequence coverages obtained by MS sequencing were 95% and 90% for F1 and F2, respectively.
FIG. 2.
Determination of the N-terminal sequence of TvDAO by nanoLC/ESI-MS/MS. Parent ion at m/z 933.32+ (labeled with a diamond) was analyzed by collision-induced dissociation, and the sequence 4IVVIGAGVAGLTTALQLLR22 of the N-terminal peptide (peptide 2) was deduced.
Single-site oxidation of Cys108 distinguishes F2 from F1.
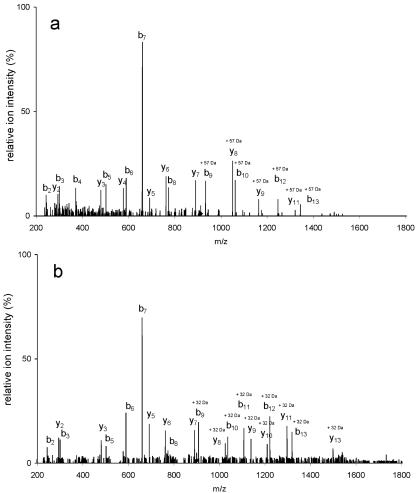
The MS analysis shown in Table 1 suggested that peptides 12 derived from F1 and F2 might be different, eventually causing the observed mass difference of 32.2 Da for the intact protein forms. Tandem MS was therefore used to characterize the molecular details of the peptides with the sequence L100EGAMSAIC108QANPWFK115, focusing in particular on the modification of Cys108 and using manual interpretation of the spectra. NanoLC-ESI-MS analysis of F1 detected a mass peak with an m/z value of 912.4. The calculated monoisotopic mass of the carboxamidomethyl adduct is 57.021, and the corresponding m/z value for the [M + 2H]2+ ion of peptide 12 with a carboxamidomethylated (Cam) Cys108 residue is 912.4. The mass of F1 peptide 12 was shifted upward by m/z 28.5 relative to the expected mass of the unmodified peptide.
To confirm the modification of the cysteine residue at position 108, we performed MS/MS sequencing of the precursor ion at m/z 912.4. The MS/MS spectrum of the ESI-produced [M + 57 + 2H]2+ ion consisted of typical fragment ions. A nearly complete series of b ions (b2 to b10, b12, and b13) and y ions (y2 to y9 and y11) demonstrated that Cys108 had gained 57 atomic mass units (Fig. 3a), fully consistent with the notion that the original thiol group of F1 had been alkylated with IAM during sample preparation for mass spectrometric analysis. In the mass spectrum of the tryptic digest of F1, we did not observe mass peaks at m/z 883.9 and 1766.8 that would correspond to, respectively, the doubly and singly charged ions of peptide 12 containing reduced Cys108. Also lacking were mass peaks of peptide 12 bearing oxidized Cys108 (i.e., Cys108-SOH, Cys108-SO2H, and Cys108-SO3H). We conclude, therefore, that the side chain of Cys108 is reduced in native F1 and fully carboxamidomethylated upon alkylation with IAM.
FIG. 3.
ESI tandem mass spectra of the modified peptide 12. (a) ESI-MS/MS spectrum of a precursor ion, [M + 57 + 2H]2+, at m/z 912.42+. The mass of Cys108 has increased by 57 mass units due to carboxamidomethylation. (b) ESI-MS/MS spectrum of a precursor ion, [M + 32 + 2H]2+, at m/z 900.02+. The mass of Cys108 has increased by 32 mass units (two oxygens) in F2.
In the mass spectrum of F2, a mass peak with m/z 900.0, which corresponds to the [M + 32 + 2H]2+ ion of peptide 12, was observed. The calculated m/z value for the doubly charged ion of peptide 12 plus two oxygens is 899.9. In contrast to the Cys108-containing peptide in F1, the series of b ions (b2 and b3 and b5 to b13) and y ions (y2 and y3 and y5 to y13) derived from the parent ion at m/z 900.0 demonstrated that the thiol residue at position 108 had gained 32 mass units, as shown in Fig. 3b. Therefore, this finding strongly suggests that Cys108 is oxidatively modified and present as cysteine sulfinic acid (Cys108-SO2H) in F2. Considering the currently available analytical tools (5, 7, 11), the results in Fig. 3b probably present the most direct determination of the site-specific modification of a protein cysteine, showing unambiguously the oxidation status of its thiol side chain.
An important conclusion that has general relevance for the analysis of oxidized protein cysteines with electrospray MS can be derived from Fig. 3b. The results reveal that the observed fragment ions of the Cys108-SO2H-containing peptide did not display loss upon collision-induced dissociation (32). As was already seen for alkylated Cys108 in Fig. 3a, the b7 ion was most abundant, while the b9 and y7 ions, which result from the cleavage of the peptide bond at the C-terminal side of Cys108-SO2H, had an abundance similar to that of the product ions emanating from cleavages at other sites within peptide 12. These findings are at variance with recently reported fragmentation patterns of synthetic peptides containing different oxidation levels of a cysteine side chain (17, 35), suggesting that gas phase collision-induced fragmentation of the F2-derived peptide bearing Cys108-SO2H may be affected by the particular sequence context.
Under the conditions of data acquisition used, we did not detect additional peptide fragments obtained from F2 in which Cys108 was reduced, alkylated, or oxidized to a different extent than that reported. The different modifications of sulfur-containing residues, especially Met104, and aromatic residues were analyzed manually by checking all MS/MS spectra that were acquired in data-dependent mode and not identified by Spectrum Mill. No detectable ions of modified peptides were found. Therefore, the results of the detailed mass spectrometric analysis of intact protein forms and their tryptic digests suggest that at a sequence coverage of >95%, microheterogeneity at only a single side chain, that of Cys108, distinguishes F2 from F1.
Structural and functional consequences of modification of Cys108.
A structural model of the TvDAO subunit was generated with the Swiss-PDBViewer using the X-ray structure of pig kidney DAO (PDB code 1kif) (16) as a template. From this model, Cys108 is proposed to be located within a flexible loop extending from the protein remote from the active center (>20 Å) (data not shown). F1 and F2 eluted from a Superdex 200 HR size exclusion column as single 80-kDa protein peaks that contained all the enzyme activity, suggesting that both forms of TvDAO are functional dimers.
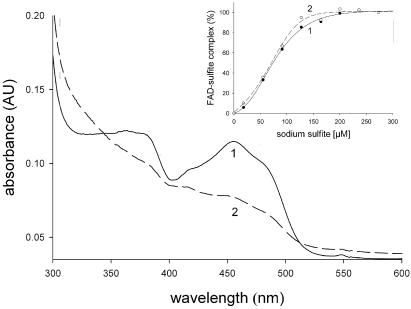
Absorption spectra of F1 and F2 are shown in Fig. 4. The spectrum of F1 exhibits two strong bands in the visible region, centered at 365 nm and 455 nm, which are characteristic of enzyme-bound FAD in the oxidized state. The value of 6.5 for the A280/A455 ratio is consistent with values reported in the literature (28). The spectrum of F2 is significantly altered compared with that of F1. It exhibits bands that are similar in position but far less pronounced than the corresponding bands in F2. Upon stepwise addition of sodium sulfite, the FAD bands in F1 and F2 were strongly quenched, reflecting the formation of a covalent N(5) adduct between the cofactor and sulfite, a feature characteristic of oxygen-reactive flavoenzymes (29). Figure 4 (inset) shows the dependence of the extent of band quenching in F1 and F2 on the molar excess of sulfite added. The results reveal that the reactivities of enzyme-bound FAD toward sulfite are identical in F1 and F2 and that at high levels of sulfite, the quenching is complete in both TvDAO forms. Therefore, this implies that within the limits of detection (±5%), FAD cofactors bound to F1 and F2 are present exclusively in their oxidized states. The observed changes in the absorption spectrum of F2 in comparison with that of F1 may thus be ascribed to long-range effects of the oxidative modification of Cys108 on the microenvironment of the FAD cofactor, which are manifested in particular functional properties of F2 (see below).
FIG. 4.
Absorption spectra of isolated TvDAO forms. The spectra (F1, solid line; F2, dashed line) were recorded at a protein concentration of 10 μM in 1 mM DTT-10 mM Tris-HCl, pH 7.5. The inset shows the formation of FAD-sulfite complexes of F1 (full circles) and F2 (open circles), expressed as percentages of the total protein concentration, upon the addition of sodium sulfite at 20°C (18, 55, 91, 127, 164, 200, 236, and 273 μM). The quenching of cofactor absorbance at 455 nm served as a reporter of complex formation. The lines (F1, solid line; F2, dashed line) represent sigmoidal fits to the data.
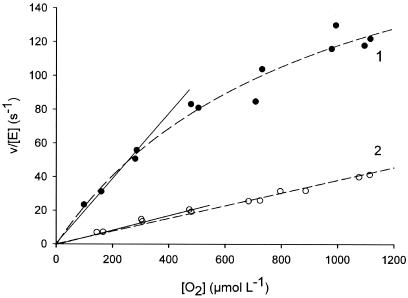
Using standard enzyme assays reported elsewhere (34), the activity of F2 was found to be approximately one-fourth that of F1, a significant difference. Initial rate measurements were carried out under conditions in which the natural cosubstrate dioxygen (220 μM) or the artificial electron acceptor DCIP (150 μM) was employed at a constant concentration and the substrate concentration was varied (d-alanine, 2 to 50 mM). Kinetic parameters were obtained from nonlinear least-squares fits of a rectangular hyperbola to the data. Within the limits of experimental error (±5%), the apparent Michaelis constants (Km) of F1 and F2 for d-alanine were 16 mM and showed no dependence on the cosubstrate used. The apparent turnover number (appkcat) of F1 for reaction with dioxygen (appkcat = 62 ± 2 s−1) was four times that of F2 (appkcat = 14 ± 2 s−1), in good agreement with the results of activity measurements using DCIP. Kinetic parameters of F1 and F2 for reduction of dioxygen were obtained from initial rate measurements in which the depletion of O2 was monitored directly (34). For both enzymes, the dependence of the O2 consumption rate showed a linear dependence on the O2 concentration in the range 50 to 500 μM (Fig. 5). The slopes of the linear plots were used to obtain second-order rate constants (kcat/Km) of 0.149 μM−1 s−1 and 0.040 μM−1 s−1 for F1 and F2, respectively. The results reveal that the catalytic efficiency of enzyme-bound FADH2 for reduction of O2 is decreased approximately fourfold as a result of the site-specific oxidation of Cys108, which fully explains the observed lower activity of F2. The data in Fig. 5 also show that the decrease in kcat/Km for F2, compared with F1, is partly due to an increase in the apparent Km for O2. The cofactor absorbance spectrum of F2, which is clearly different from that of F1, arguably reflects this significant decrease in reactivity.
FIG. 5.
Kinetic comparison of F1 and F2 in respect to the reduction of dioxygen by enzyme-bound FADH2. Initial rates were recorded by measuring the enzymatic O2 consumption under conditions in which d-methionine was saturating in the steady state (20 mM). The second-order rate constant (kcat/Km) of F1 (1, full circles) and F2 (2, open circles) was obtained from the part of the plot where the enzymatic rate is linearly dependent on the O2 concentration, as indicated in the figure.
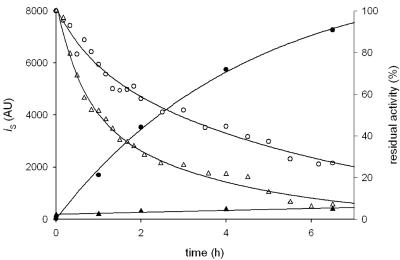
Oxidative modification of TvDAO alters the thermal denaturation pathway.
Under temperature conditions (50°C) that promote rapid inactivation of TvDAO, we compared the aggregation propensities of F1 and F2 using light scattering as a reporter of the aggregation event. Figure 6 shows time courses of aggregation of F1 and F2. The results reveal that the conversion of Cys108 into cysteine sulfinic acid causes a complete suppression of protein aggregation under the thermal-stress conditions. For F1, there is a clear kinetic correlation between loss of activity and aggregation. In the case of F2, inactivation and aggregation are partly uncoupled at 50°C so that a time-dependent decrease in activity, yet at a significant two- to threefold-higher rate than with F1, is seen even in the absence of detectable aggregation. Although the detailed comparison of inactivation pathways for F1 and F2 is beyond the scope of this paper, the results indicate clearly that Cys108 will be an important target for the stabilization of TvDAO under conditions in which the effect of protein aggregation is the main contributor to the inactivation rate. They provide a novel basis for the design of engineered enzyme variants which couple high activity (F1; Cys108 reduced) and suppressed aggregation (F2; Cys108 oxidized) to show improved performance in biocatalytic processes and analytical applications.
FIG. 6.
Time courses of aggregation and inactivation of native and oxidatively modified TvDAO under conditions of thermal stress. Activity of 20 μM F1 (open circles) and F2 (open triangles) was monitored at 50°C, and the intensity of scattered light (iS) at 500 nm of F1 (full circles) and F2 (full triangles) at the indicated times was measured. The lines show the trend of the data. AU, arbitrary units.
In summary, we have identified Cys108 as the prominent site of oxidative posttranslational modification of TvDAO and performed a detailed structure-function relationship analysis for the “chemical point mutation” Cys108→cysteine sulfinic acid. The evidence, aside from the obvious applied component associated with it, appears to be relevant in the context of TvDAO lifespan in the yeast. The enzyme is located in the peroxisomal compartment, and site-specific oxidation could mark it for turnover through one of the cellular protein degradation pathways (8, 30).
Finally, the reported analytical protocols could be useful for other researchers wishing to explore, on a defined structural basis, the stability and stabilization of oxidation-sensitive biocatalysts.
Acknowledgments
The expert technical assistance of Barbara Mautner is gratefully acknowledged.
REFERENCES
- 1.Alaejos, M. S., and F. J. G. Montelongo. 2004. Application of amperometric biosensors to the determination of vitamins and α-amino acids. Chem. Rev. 104:3239-3265. [DOI] [PubMed] [Google Scholar]
- 2.Betancor, L., A. Hidalgo, G. Fernandez-Lorente, C. Mateo, V. Rodriguez, M. Fuentes, F. Lopez-Gallego, R. Fernandez-Lafuente, and J. M. Guisan. 2003. Use of physicochemical tools to determine the choice of optimal enzyme: stabilization of d-amino acid oxidase. Biotechnol. Prog. 19:784-788. [DOI] [PubMed] [Google Scholar]
- 3.Biteau, B., J. Labarre, and M. B. Toledano. 2003. ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature 425:980-984. [DOI] [PubMed] [Google Scholar]
- 4.Burton, S. G. 2003. Oxidizing enzymes as biocatalysts. Trends Biotechnol. 21:543-549. [DOI] [PubMed] [Google Scholar]
- 5.Choi, M. H., I. K. Lee, G. W. Kim, B. U. Kim, Y.- H. Han, D.-Y. Yu, H. S. Park, K. Y. Kim, J. S. Lee, C. Choi, Y. S. Bae, B. I. Lee, S. G. Rhee, and S. W. Kang. 2005. Regulation of PDGF signalling and vascular remodeling by peroxiredoxin II. Nature 435:347-353. [DOI] [PubMed] [Google Scholar]
- 6.Claiborne, A., J. I. Yeh, C. Mallet, J. Luba, E. J. Crane III, V. Charrier, and D. Parsonage. 1999. Protein-sulfenic acids: diverse roles for an unlikely player in enzyme catalysis and redox regulation. Biochemistry 38:15407-15416. [DOI] [PubMed] [Google Scholar]
- 7.Clements, A., M. Johnston, B. S. Larsen, and C. N. McEwen. 2005. Fluorescence-based peptide labeling and fractionation strategies for analysis of cysteine-containing peptides. Anal. Chem. 77:4495-4502. [DOI] [PubMed] [Google Scholar]
- 8.Davies, M. J. 2005. The oxidative environment and protein damage. Biochim. Biophys. Acta 1703:93-109. [DOI] [PubMed] [Google Scholar]
- 9.Fagain, C. O. 2003. Enzyme stabilization—recent experimental progress. Enzyme Microb. Technol. 33:137-149. [Google Scholar]
- 10.Fu, X., S. Y. Kassim, W. C. Parks, and J. W. Heinecke. 2001. Hypochlorous acid oxygenates the cysteine switch domain of pro-matrilysin (MMP-7). A mechanism for matrix metalloproteinase activation and atherosclerotic plaque rupture by myeloperoxidase. J. Biol. Chem. 276:41279-41287. [DOI] [PubMed] [Google Scholar]
- 11.Griffiths, S. W., J. King, and C. L. Cooney. 2002. The reactivity and oxidation pathway of cysteine 232 in recombinant human α1-antitrypsin. J. Biol. Chem. 277:25486-25492. [DOI] [PubMed] [Google Scholar]
- 12.Habeeb, A. F. S. A. 1972. Reaction of protein sulfhydryl groups with Ellman's reagent. Methods Enzymol. 25:457-464. [DOI] [PubMed] [Google Scholar]
- 13.Jacob, C., J. R. Lancaster, and G. I. Giles. 2004. Reactive sulphur species in oxidative signal transduction. Biochem. Soc. Trans. 32:1015-1017. [DOI] [PubMed] [Google Scholar]
- 14.Ju, S.-S., L.-L. Lin, H. R. Chien, and W.-H. Hsu. 2000. Substitution of the critical methionine residues in Trigonopsis variabilis d-amino acid oxidase with leucine enhances its resistance to hydrogen peroxide. FEMS Microbiol. Lett. 186:215-219. [DOI] [PubMed] [Google Scholar]
- 15.Khang, Y.-H., I.-W. Kim, Y.-R. Hah, J.-H. Hwangbo, and K. K. Kang. 2003. Fusion protein of Vitreoscilla hemoglobin with d-amino acid oxidase enhances activity and stability of biocatalyst in the bioconversion process of cephalosporin C. Biotechnol. Bioeng. 82:480-488. [DOI] [PubMed] [Google Scholar]
- 16.Mattevi, A., M. A. Vanoni, F. Todone, M. Rizzi, A. Teplyakov, A. Coda, M. Bolognesi, and B. Curti. 1996. Crystal structure of d-amino acid oxidase: a case of active site mirror-image convergent evolution with flavocytochrome b2. Proc. Natl. Acad. Sci. USA 93:7496-7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Men, L., and Y. Wang. 2005. Further studies on the fragmentation of protonated ions of peptides containing aspartic acid, glutamic acid, cysteine sulfinic acid, and cysteine sulfonic acid. Rapid Commun. Mass Spectrom. 19:23-30. [DOI] [PubMed] [Google Scholar]
- 18.Mirgorodskaya, E., H.-P. Fierobe, B. Svensson, and P. Roepstorff. 1999. Mass spectrometric identification of a stable catalytic cysteinesulfinic acid residue in an enzymatically active chemically modified glucoamylase mutant. J. Mass Spectrom. 34:952-957. [DOI] [PubMed] [Google Scholar]
- 19.Murakami, T., M. Nojiri, H. Nakayama, M. Odaka, M. Yohda, N. Dohmae, K. Takio, T. Nagamune, and I. Endo. 2000. Post-translational modification is essential for catalytic activity or nitrile hydratase. Protein Sci. 9:1024-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Musatov, A., E. Hebert, C. A. Carroll, S. T. Weintraub, and N. C. Robinson. 2004. Specific modification of two tryptophans within the nuclear-encoded subunits of bovine cytochrome c oxidase by hydrogen peroxide. Biochemistry 43:1003-1009. [DOI] [PubMed] [Google Scholar]
- 21.Pilone, M. S., and L. Pollegioni. 2002. d-Amino acid oxidase as an industrial biocatalyst. Biocatal. Biotrans. 20:145-159. [Google Scholar]
- 22.Pollegioni, L., S. Buto, W. Tischer, S. Ghisla, and M. S. Pilone. 1993. Characterization of d-amino acid oxidase from Trigonopsis variabilis. Biochem. Mol. Biol. Int. 31:709-717. [PubMed] [Google Scholar]
- 23.Pollegioni, L., L. Caldinelli, G. Molla, S. Sacchi, and M. S. Pilone. 2004. Catalytic properties of d-amino acid oxidase in cephalosporin C bioconversion: a comparison between proteins from different sources. Biotechnol. Prog. 20:467-473. [DOI] [PubMed] [Google Scholar]
- 24.Poole, L. B. 2005. Bacterial defenses against oxidans: mechanistic features of cysteine-based peroxidases and their flavoprotein reductases. Arch. Biochem. Biophys. 433:240-254. [DOI] [PubMed] [Google Scholar]
- 25.Rhee, S. G., H. Z. Chae, and K. Kim. 2005. Peroxiredoxins: a historical overview and speculative preview of novel mechanisms and emerging concepts in cell signalling. Free Radic. Biol. Med. 38:1543-1552. [DOI] [PubMed] [Google Scholar]
- 26.Rhee, S. G., S. W. Kang, W. Jeong, T.-S. Chang, K.-S. Yang, and H. A. Woo. 2005. Intracellular messenger function of hydrogen peroxide and its regulation by peroxiredoxins. Curr. Opin. Cell Biol. 17:183-189. [DOI] [PubMed] [Google Scholar]
- 27.Riethorst, W., and A. Reichert. 1999. An industrial view on enzymes for the cleavage of cephalosporin C. Chimia 53:600-607. [Google Scholar]
- 28.Schräder, T., and J. R. Andreesen. 1993. Evidence for the functional importance of Cys298 in d-amino acid oxidase from Trigonopsis variabilis. Eur. J. Biochem. 218:735-744. [DOI] [PubMed] [Google Scholar]
- 29.Schräder, T., and J. R. Andreesen. 1996. Properties and chemical modification of d-amino acid oxidase from Trigonopsis variabilis. Arch. Microbiol. 165:41-47. [Google Scholar]
- 30.Stadtman, E. R., H. Van Remmen, A. Richardson, N. B. Wehr, and R. L. Levine. 2005. Methionine oxidation and aging. Biochim. Biophys. Acta 1703:135-140. [DOI] [PubMed] [Google Scholar]
- 31.Stauffer, C. E., and D. Etson. 1969. The effect on subtilisin activity of oxidizing a methionine residue. J. Biol. Chem. 224:5333-5338. [PubMed] [Google Scholar]
- 32.Stenn, H., and M. Mann. 2000. Similarity between condensed phase and gas phase chemistry: fragmentation of peptides containing oxidized cysteine residues and its implications for proteomics. J. Am. Soc. Mass. Spectrom. 12:228-232. [DOI] [PubMed] [Google Scholar]
- 33.Tishkov, V. I., and S. V. Khoronenkova. 2005. d-Amino acid oxidase: structure, catalytic mechanism, and practical application. Biochemistry (Moscow) 70:51-67. [PubMed] [Google Scholar]
- 34.Trampitsch, C., A. Slavica, W. Riethorst, and B. Nidetzky. 2005. Reaction of Trigonopsis variabilis d-amino acid oxidase with 2,6-dichloroindophenol: kinetic characterization and development of an oxygen-independent assay of the enzyme activity. J. Mol. Catal. B 32:271-278. [Google Scholar]
- 35.Wang, Y., S. Vivekananda, L. Men, and Q. Zhang. 2004. Fragmentation of protonated ions of peptides containing cysteine, cysteine sulfinic acid, and cysteine sulfonic acid. J. Am. Soc. Mass Spectrom. 15:697-702. [DOI] [PubMed] [Google Scholar]
- 36.Willett, W. S., and S. H. Copley. 1996. Identification and localization of a stable sulfenic acid in peroxide-treated tetrachlorohydroquinone dehalogenase using electrospray mass spectrometry. Chem. Biol. 3:851-857. [DOI] [PubMed] [Google Scholar]
- 37.Woo, H. A., H. Z. Chae, S. C. Hwang, K.-S. Yang, S. W. Kang, K. Kim, and S. G. Rhee. 2003. Reversing the inactivation of peroxiredoxins caused by cysteine sulfinic acid formation. Science 300:653-656. [DOI] [PubMed] [Google Scholar]