Abstract
Polycomb group (PcG) proteins are epigenetic chromatin modifiers involved in heritable gene repression. Two main PcG complexes have been characterized. Polycomb repressive complex 2 (PRC2) is thought to be involved in the initiation of gene silencing, whereas Polycomb repressive complex 1 (PRC1) is implicated in the stable maintenance of gene repression. Here, we investigate the kinetic properties of the binding of one of the PRC1 core components, BMI1, with PcG bodies. PcG bodies are unique nuclear structures located on regions of pericentric heterochromatin, found to be the site of accumulation of PcG complexes in different cell lines. We report the presence of at least two kinetically different pools of BMI1, a highly dynamic and a less dynamic fraction, which may reflect BMI1 pools with different binding capacities to these stable heterochromatin domains. Interestingly, PRC2 members EED and EZH2 appear to be essential for BMI1 recruitment to the PcG bodies. Furthermore, we demonstrate that the maintenance DNA methyltransferase DNMT1 is necessary for proper PcG body assembly independent of DNMT-associated histone deacetylase activity. Together, these results provide new insights in the mechanism for regulation of chromatin silencing by PcG proteins and suggest a highly regulated recruitment of PRC1 to chromatin.
Polycomb group (PcG) proteins are part of a conserved cellular memory system that prevents changes in cell identity by repressing the transcriptional state of several loci in the genome. Biochemical and genetic studies indicate that PcG proteins exist in at least two separate protein complexes both in Drosophila and mammalian cells: Polycomb repressive complex 2 (PRC2) and Polycomb repressive complex 1 (PRC1) (30, 51). PRC2 consisting of EED, EZH2, YY1 and SU(Z)12, is thought to be required at the initiating stage of silencing, whereas PRC1, containing PcG proteins such as HPH, RING1, BMI1, and HPC, is continuously required for the stable maintenance of the initiated PcG repression on specific target loci. An additional EZH2/EED complex, Polycomb repressive complex 3 (PRC3), has recently been reported in human cells (22). PRC2 and PRC3 exhibit differential targeting of specific histones for lysine methylation that relies on the function of the distinct EED protein isoforms within each complex (22).
Although PRC1 and PRC2 complexes have different functions, EZH2/EED-mediated nucleosome methylation increases in vitro binding of the chromodomain protein Polycomb (Pc) to the chromatin (7, 10, 23), and it has been shown that both complexes can interact with one another at least transiently (46).
Polycomb response elements (PREs) have been identified in the regulatory regions of several genes in Drosophila (8, 61), and recent advances have clarified the sequence requirements for Drosophila PREs (51). However, the sequences that define the mammalian counterparts have been elusive, and the mechanisms involved in recruitment of PcG proteins and assembly of the repressive complexes remain unclear.
It has been shown that the maintenance of gene silencing by PcG proteins requires modifications in chromatin compaction (14) and locking of the inactive genes in a heterochromatin-like environment (42). A functional link between PcG-dependent gene silencing and the organization of inactive chromatin domains is shown in Drosophila and Saccharomyces cerevisiae (25). In mammals, members of the PRC1 not only localize to euchromatic regions but also accumulate at pericentric heterochromatin as discrete nuclear foci called PcG bodies (55, 67). In contrast to the broad HP1 association to constitutive heterochromatic regions, PcG bodies display preferential localization to pericentric chromosome territories, specifically, to the pUC1.77 probe-hybridizing domains of the human 1q12 region and to 1q12-related sequences on chromosomes 9, 15, and 16 (55, 67). This unique localization of PcG bodies suggests that the human complex might be specifically targeted to these repetitive DNA sequences. PcG bodies colocalize with these specific pericentric heterochromatin domains in a variety of transformed and primary human cell lines. The functional significance of these nuclear domains remains unknown, but pericentric heterochromatin integrity in general is essential for proper chromosome segregation, genomic stability, and transcriptional silencing (43, 64).
Heterochromatin domains generally are associated with a condensed appearance, late replication timing, low level of meiotic recombination, and highly methylated DNA. Such regions are gene poor (although not devoid of genes), being made up primarily of repetitive sequences (31). Methylation of lysine 9 on histone H3 (H3-K9) is required for the formation of heterochromatin from yeast to mammals (37, 49). Fundamental to histone H3-K9 methylation, providing a stable epigenetic mark is the ability to recruit the nuclear protein HP1 (3, 24). Similarly, the PRC2 component EZH2, containing histone methyltransferase (HMTase) activity, is able to deliver another specific histone methylation mark for inactive chromatin on lysine 27 of histone H3 (H3-K27), which can be recognized by chromodomain-containing Pc homologs in PRC1 (7, 10, 23).
Recent discoveries suggest that the PcG-mediated silencing is a dynamic process where activators and repressors coexist in the same region (41, 51). However, little is known about the mobility of the PcG proteins in the nucleus. BMI1 is one of the most studied PRC1 members and has been shown to be important not only for embryonic development but also for numerous other biological functions where flexible reprogramming of cellular transcription is crucial, such as stem cell fate decisions, differentiation, and tumorigenesis (5, 28, 65). The present study investigates the dynamic properties of BMI1 within PcG bodies and the factors involved in the recruitment of BMI1 to the pericentric heterochromatin-associated PcG bodies.
MATERIALS AND METHODS
Plasmid constructs.
A green fluorescent protein (GFP)-tagged BMI1 expression vector was generated using standard PCR methods and the pEGFP-N3 plasmid (Clontech) and verified by sequencing. RNA interference (RNAi) constructs carrying short hairpin RNA sequences expressed under the control of the U6 or the H1 promoter were essentially made as previously described (6, 63). The RNAi sequences for EZH2, EED, and MEL18 are available upon request. The expression construct for DNMT1 RNAi was obtained from Y. Shi (63). The EZH2-H694L mutant was made using a QuickChange site-directed mutagenesis kit (Stratagene).
Antibodies.
HPC2 polyclonal and EED monoclonal antibodies were obtained from A. P. Otte, EZH2 monoclonal antibody was from K. Helin, RING1B monoclonal antibody was from H. Koseki, and the di- and trimethyl-H3-K9 (mH3-K9) and trimethyl-H3-K27 polyclonal antibodies were from T. Jenuwein. The BMI1 monoclonal antibody was purchased from Upstate, and bromodeoxyuridine (BrdU) antibody (BU20a) was from DAKO. Myc antibody (A-14) was purchased from Santa Cruz. Secondary antibodies used for immunofluorescence were Alexa 568-conjugated anti-rabbit and Alexa 488-conjugated anti-mouse antibodies from Molecular Probes.
Cell culture, transfections, and retroviral transductions.
Parental U2OS and U2OS stably expressing BMI1-GFP cells were grown in Dulbecco's modified Eagle medium (GIBCO) supplemented with 10% fetal bovine serum (MP-Biomedicals) under standard conditions. Phoenix producer cells were used to generate retroviral stocks (calcium phosphate transfection method), and U2OS cells were transduced with viral supernatant in the presence of polybrene (4 μg/ml; Sigma). Puromycin selection (2 μg/ml) was started 24 h after transfection or transduction. Synchronization of the cells in G1 and G2 was attained by a double thymidine block or nocodazole and subsequently released, as previously described (67). When indicated, the cells were exposed to 5-aza-2′-deoxycytidine (5-Aza-dC; 4 days at 100 nM) or to trichostatin A (TSA; 4 days at 25 ng/ml).
FRAP and FLIP analysis.
Fluorescence recovery after photobleaching (FRAP) and fluorescence loss in photobleaching (FLIP) analysis were performed using a Leica TCS SP confocal laser scanning microscope equipped with an Ar/Kr laser. Green fluorescence was detected at 520 to 560 nm after excitation at 488 nm, as previously described (9, 12). For FRAP experiments, a prebleach image was made at 8% laser power, and subsequently photobleaching was acquired on a single spot for 1 s at 100% laser power, resulting in an 80 to 90% reduction of the fluorescent signal. Sequential images were then collected at 8% laser power at 1.6-s intervals for 40 frames and thereafter at 4-s intervals for 50 frames to minimize further photobleaching for a period up to 260 s. For FLIP experiments, prebleach and postbleach images were taken as described for FRAP experiments. Photobleaching was acquired on a region of interest, covering approximately 50% of the total nuclear surface area of the chosen focal plane, for 16 s at 100% laser power. Calculations and data analysis were done using MATLAB student version 6.0 software. Prebleach values were set at 1, and postbleach values were normalized to their corresponding prebleach values after correction for background and bleaching during measurements. FRAP data were fitted with an exponential curve. The best fits for the recovery curves were determined followed by calculations of half time of recovery (t1/2) values. The mobile fraction was calculated based on the predicted plateau from the best-fit analysis. For FLIP experiments we did not use the best-fit analysis because the loss of fluorescence with time was very slow and did not reach a plateau during the time measured. To quantify the FLIP data, the measured intensities at a fixed time point, 240 s postbleaching, were averaged relative to their corresponding prebleach values and plotted for G1 and G2, representing loss of fluorescence from PcG bodies with time.
Immunostainings.
U2OS cells were grown on glass slides and fixed in 4% paraformaldehyde for 10 min and permeabilized for 5 min in 0.2% Triton X-100 in phosphate-buffered saline (PBS). Subsequently, cells were incubated in blocking solution (5% goat serum, 0.2% fish skin gelatin, 0.2% Tween-20-PBS) for 30 min before incubating with primary antibody. Cells were then washed in 0.2% Tween 20-PBS and incubated with secondary antibody. Next the cells were washed with 0.2% Tween 20-PBS, stained with 4′-6-diamidino-2-phenylindole (DAPI), and mounted with Vectashield (Vector Laboratories).
To detect mH3-K27 throughout the cell cycle, unsynchronized U2OS cells were incubated for 1 h with medium containing 10 μM BrdU. After the immunostaining for mH3-K27, cells were refixed for 10 min in 4% paraformaldehyde at room temperature, washed with PBS, and incubated with blocking solution. DNA was denatured by a 30-min incubation with 2 N HCl-0.5% Triton X-100 at room temperature. Cells were subsequently washed once with 0.1 M Na2B4O7 (pH 8.5) for 5 min and twice with PBS and then incubated with BrdU antibody. After being washed with PBS and with blocking solution, cells were incubated with Alexa488-conjugated anti-mouse antibody. Cells were then washed with PBS, stained with DAPI, and mounted with Vectashield.
Fluorescence microscopy and image acquisition.
Stained cells and cells stably expressing BMI1-GFP were viewed by epifluorescence microscopy using a Zeiss microscope or by confocal laser microscopy using a Leica microscope. Epifluorescent images were captured with a cooled charge-coupled-device camera and processed using Image I software and Adobe Photoshop 3.0. Quantification of the percentage of cells was performed with a 40× objective. For mH3-K27 quantification throughout cell cycle, more than 200 cells positive for mH3-K27 were scored for BrdU profiles from at least eight random fields.
Western blot analysis.
Cells were lysed using lysis buffer (0.15 mM NaCl, 0.05 mM Tris-HCl [pH 7.2], 1% Triton X-100, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate) supplemented with 1 mM dithiothreitol and 50 μM phenylmethylsulfonyl fluoride (PMSF). Protein expression was analyzed using a conventional Western blotting protocol.
Histone methyltransferase assay (in vitro).
HEK293 cells were transfected using the calcium-phosphate precipitation method with Myc-EZH2-pBabe-puro or Myc-EZH2-H694L-pBabe-puro or were mock transfected and harvested 48 h posttransfection in mild lysis buffer (250 mM NaCl, 50 mM HEPES [pH 7.0], 5 mM EDTA, 0.1% NP-40) supplemented with 1 mM dithiothreitol and 50 μM PMSF, sonicated for 10 pulses of 50% at level 4, and centrifuged (20,000 × g) for 15 min at 4°C. Equal amounts of protein per condition were used for anti-Myc immunoprecipitation. Myc-tagged immunocomplexes were immobilized on a mixture of protein G- and A-agarose beads (Amersham Pharmacia) and washed four times with the lysis buffer and once with HMTase buffer (50 mM Tris [pH 8], 20 mM KCl, 10 mM MgCl2, 10 mM β-mercaptoethanol, 1 mM PMSF). The beads were subsequently incubated at 30°C for 1 h in the HMTase buffer supplemented to 10% glycerol and with 20 μg of core histones (Roche) as substrate and 2 μCi/reaction of S-adenosyl-l-[methyl-14C]methionine (Amersham Biosciences) as methyl donor, in a total volume of 25 μl. The reaction was stopped by the addition of sodium dodecyl sulfate sample buffer and then fractionated on 4 to 12% gradient NuPAGE Bis-Tris precast Western gel (Invitrogen). The upper part of the gel was separated and transferred onto a nitrocellulose membrane (Protran; Amersham Biosciences) used for further antibody probing. The histones on the lower part of the gel were visualized by Coomassie brilliant blue staining; the gel was treated with Amplify (Amersham Biosciences), dried, and used for an overnight exposure (Kodak Biomax MS).
RESULTS
Kinetics of BMI1 binding to the pericentric heterochromatin-associated PcG bodies.
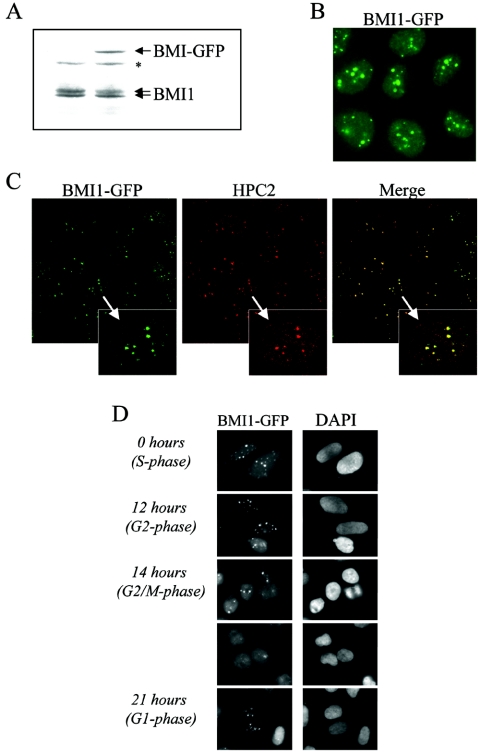
PcG bodies appear as stable domains that can be observed in many transformed cell lines and some primary cell types (55, 67). We questioned whether these sites of retention of PcG proteins form static or dynamic structures. To visualize BMI1 in living cells, we generated a U2OS cell line stably expressing BMI-GFP chimeric protein at approximately the same level of the endogenous BMI1 protein (Fig. 1A). As expected, the BMI1-GFP U2OS cell line showed nuclear GFP signal concentrated in several defined domains, the PcG bodies. A smaller amount of GFP was detected throughout the rest of the nucleus, likely representing euchromatin-bound and free BMI1 (Fig. 1B). The distribution of BMI1-GFP is identical to the previously described localization of endogenous BMI1 in U2OS and in other human tumor cell lines (55, 67). Furthermore the BMI1-GFP fusion protein colocalized with the endogenous HPC2 in PcG bodies (Fig. 1C) as described previously (54).
FIG. 1.
GFP-tagged BMI1 is correctly targeted to PcG bodies and closely resembles the endogenous BMI1 localization. (A) BMI1-GFP expression level is close to endogenous BMI1 expression level. Western blot analysis was conducted using extracts from parental and BMI1-GFP-expressing U2OS cells, hybridized with the BMI1 antibody. *, nonspecific band. (B) BMI1-GFP protein is targeted to PcG bodies, PcG-related pericentric heterochromatin foci, in U2OS cells as shown by GFP fluorescence. In addition, lower levels of green fluorescence in the regions between these foci are detected, indicating that BMI1-GFP is also present in euchromatic areas. (C) Confocal fluorescence microscopy demonstrating colocalization of BMI1-GFP with HPC2 (polyclonal anti-HPC2 immunostaining) in large nuclear domains (PcG bodies) of U2OS cells stably expressing BMI1-GFP. (D) Cell cycle-dependent chromatin association of BMI1-GFP in U2OS cells closely resembling endogenous BMI1 association (57). Cells were synchronized in early S phase by a double thymidine block and subsequently released. Representative fluorescence images were made at indicated time points after release.
To validate the use of the BMI1-GFP fusion protein for further analysis, we evaluated the behavior of this fusion protein during the cell cycle, as chromatin-association of BMI1 is cell cycle regulated (55, 67). BMI1-GFP-expressing U2OS cells were synchronized by a double thymidine block and subsequently released and analyzed at different time points. As Fig. 1D shows, BMI1-GFP bodies become reduced in size and number but remain associated to pericentric heterochromatic regions throughout mitosis and reassemble again as large intense foci in interphase, as previously described (55, 67). In addition, BMI1-GFP is able to complement growth defects of Bmi1-deficient mouse embryonic fibroblasts (data not shown), confirming that BMI1-GFP behaves like endogenous BMI1.
The changes in chromatin association of BMI1 during the cell cycle could reflect differences in the dynamic properties of PcG bodies. To examine this, we used the BMI1-GFP cell line to study the kinetics of BMI1 protein in living cells in two different stages of the cell cycle, G1 and G2, by FRAP and FLIP analysis. BMI1-GFP-expressing U2OS cells were either synchronized by a double thymidine block and released for 12 h in order to enrich for cells in G2 or blocked with nocodazole and released for 6 h in order to enrich for cells in G1.
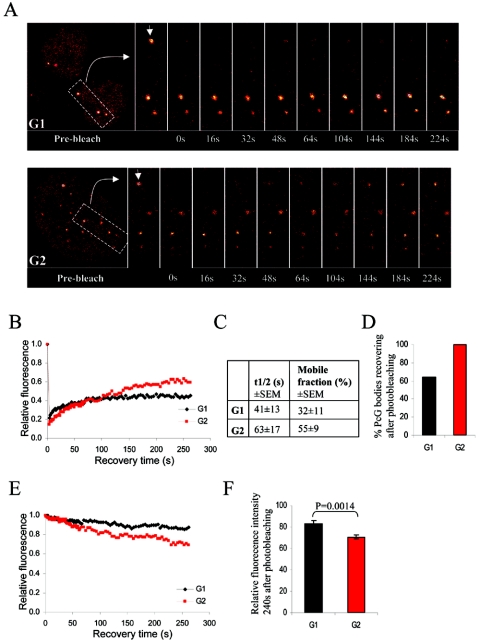
In FRAP experiments, we measured the time required for fluorescence to recover by time-lapse confocal microscopy after photobleaching individual PcG bodies. Photobleaching of BMI1-GFP destroys GFP and results in irreversible loss of fluorescence. Fluorescence recovery thus requires that mobile unbleached BMI1-GFP molecules from the surrounding environment move into the bleached region, replacing the bleached BMI1-GFP molecules (Fig. 2A). The recovery of fluorescence after photobleaching was plotted against time (Fig. 2B). Calculating the best-fit parameters allowed the determination of the half time of recovery (t1/2; the time in which the fluorescence intensity reached half of the final recovered intensity in the bleached spot) and the mobile fraction (the percentage of maximally recovered BMI1-GFP) (Fig. 2C) as described previously (50).
FIG. 2.
BMI1 is dynamically associated with PcG bodies. (A) Representative examples of a FRAP profile from BMI1-GFP-expressing U2OS cells. Cells were synchronized in G1 by a nocodazole treatment followed by release from the blockade or in G2 by a double thymidine block and subsequent release. Pseudo-colored images show the fluorescence signals and recovery with time. Arrowheads point to the bleached PcG bodies. (B) FRAP plots. Recovery of fluorescence in the bleached spots was plotted with time for the G1 and G2 cell cycle phases. The prebleach fluorescence was set at 100%, and the fluorescence intensity for each time point was averaged and normalized relative to the prebleach intensities after correcting for background and bleaching during the scanning procedure. (C) Quantifications of the recovery time (t1/2) and mobile fraction deduced from the best-fit analysis of the recovery plots. BMI1 has a slightly reduced t1/2 and mobile fraction in G1 versus G2 cell cycle phase. (D) Percentage of PcG bodies that showed any BMI1-GFP fluorescence recovery after photobleaching, indicating that BMI1-GFP is considerably less mobile in G1 than in G2. (E) FLIP plots. Loss of fluorescence with time from PcG bodies in a nonbleached region after photobleaching of a defined area of the nucleus was plotted for the G1 and G2 cell cycle phases. The prebleach fluorescence was set at 100%, and the fluorescence intensity for each time point was averaged and normalized relative to the prebleach intensities after correcting for background and bleaching during the scanning procedure. (F) Relative left fluorescence intensities at 240 s postbleaching are shown for G1 and G2 (±standard error of the means); prebleach fluorescence intensity was set at 100%. Loss of fluorescence in G2 is significantly faster than in G1, indicating the presence of a more mobile BMI1-GFP fraction on PcG bodies in G2.
No full recovery of the initial fluorescence is observed in any case, suggesting that a considerable fraction of the BMI1-GFP pool within photobleached PcG bodies shows relatively slow recovery kinetics and cannot be exchanged completely within the time measured. The BMI1-GFP fluorescence intensity in G2 recovered to 55% of the prebleach intensity, whereas the fluorescence recovery in G1 cells appeared to be less complete, reaching only 32% of the initial fluorescence intensity. Notably, the t1/2 of BMI1-GFP was reproducibly found to be relatively reduced in G1 compared to G2 (Fig. 2C). Although the differences between PcG bodies studied in G1 and G2 do not appear to be significant due to the high experimental variation, it is essential to mention that the measured intensities of approximately 35% of the studied PcG bodies in G1 could not be included in the t1/2 and recovery curve calculations, since no recovery after photobleaching was observed (Fig. 2D). This, however, was not the case for the studied PcG bodies in G2, which indicates that BMI1-GFP in the PcG bodies during G1 is remarkably less mobile.
The recovery plots deduced from the FRAP experiments (Fig. 2B) in addition suggest a biphasic recovery containing at least two mobile fractions behaving very differently: a fraction with fast recovery kinetics, which is responsible for the initial quick and accelerating recovery, and a fraction with slow recovery kinetics, which is responsible for the slow and gradual recovery of fluorescence seen at later time points.
The kinetics of BMI1-GFP was also tested in FLIP experiments. To this end, fluorescence loss from PcG bodies was determined in a nonbleached region after photobleaching of a defined area of the nucleus (corresponding to approximately 50% of the x-y plane). Using this technique, we were only able to study and confirm the kinetics of the slower mobile fraction revealed by FRAP experiments, since the recovery information of the fast mobile fraction is lost during the repeated region photobleaching. The loss of fluorescence with time was plotted (Fig. 2E). As expected, for cells arrested in both G1 and G2, a major part of the studied fraction (the fraction from which the slope of the fluorescence recovery curve in FRAP experiments was declined) of BMI1-GFP in PcG bodies seemed to be mobile but with very slow kinetics. To quantify the loss of fluorescence in FLIP experiments for G1 and G2, the fluorescence intensities measured at 240 s postbleaching were normalized to the prebleach fluorescence intensities which were set at 100%. As shown in Fig. 2F, in G1 PcG bodies retained 83% of their initial fluorescence intensity 240 s after bleaching, whereas in G2, PcG bodies contained only 71% of their initial fluorescence intensity (P = 0.0014).
Together, these results indicate the existence of at least two kinetically distinct populations of BMI1 in the observed PcG bodies. FRAP experiments show the exchange rate dynamics of both BMI1 pools in the observed biphasic recovery, the slow mobile fraction and the fast mobile fraction (regardless of the differences between G1 and G2). Concurrently, FLIP experiments show a significant difference in the off-rate of the slow mobile fraction between G1 and G2.
PRC2 is required for recruitment of BMI1 to PcG bodies.
Having assessed that PcG body formation is a dynamic and therefore possibly a regulated process, we next investigated which factors could be crucial for the recruitment of PRC1 members to PcG bodies.
The processes responsible for the transmission of epigenetic marks and higher-order structure of heterochromatin domains are not entirely clear, but protein deposition and enzymatic modifications coupled to semiconservative DNA replication are possible mechanisms that could faithfully maintain this higher-order organization after replication fork passage or mitosis (47). Previous studies have indicated that pericentric heterochromatin is relatively stable, requiring prolonged treatment of the cells with the histone deacetylase (HDAC) inhibitor TSA before HP1 is no longer localized to heterochromatic foci (29, 64), notwithstanding the recently proved high HP1 mobility (9, 12). Therefore, we used a retrovirus-based RNAi approach in order to stably silence endogenous gene expression of candidate proteins that could be involved in the recruitment of BMI1 to PcG bodies in U2OS cells.
H3-K9 methylation by Suv39h methyltransferases is a characteristic mark for heterochromatin and provides binding sites specifically for HP1, one of the major heterochromatin proteins (3, 13, 24). Likewise, Ezh2 has been shown to have HMTase activity, preferentially methylating H3-K27, which is specifically recognized by the chromodomain protein Pc (7, 10, 13, 23, 34, 36). This suggests a function for EZH2 within PRC2 to regulate the binding of the Pc/PRC1 to specific pericentric heterochromatic regions, even though EZH2 protein has not been described to specifically accumulate in the PcG bodies (57). To study this possibility, effective downregulation of EZH2 expression by RNAi in U2OS cells was verified (see Fig. S1A in the supplemental material). Subsequently, BMI1-GFP-expressing cells were transduced with EZH2 RNAi and were kept in culture under selection for 4, 7, or 10 days. As Fig. 3E shows, downregulation of the endogenous EZH2 protein led to a dispersion of the nuclear localization of BMI1-GFP, without any apparent decrease in total fluorescence levels. Notably, the dispersion of the BMI1-GFP signal was gradual from 4 to 10 days posttransduction and became most clear at later time points, with only occasional cells still showing some PcG bodies, possibly due to incomplete knockdown. The role of EZH2 and EED in cellular proliferation, acting as essential downstream mediators of E2F function, has recently been reported, and inhibition of EZH2 and EED expression by transfecting small interfering RNA oligonucleotides into U2OS cells results in reduced proliferation (4). In our hands, the number of cells that survived after selection was lower in the EZH2 RNAi than in the mock RNAi-treated cells (data not shown). This observation indicates that the BMI1-GFP-expressing cells can tolerate EZH2 downregulation to some extent. The reduced levels of EZH2 appear to be low enough to have an effect on BMI1 recruitment to PcG bodies without affecting cell viability.
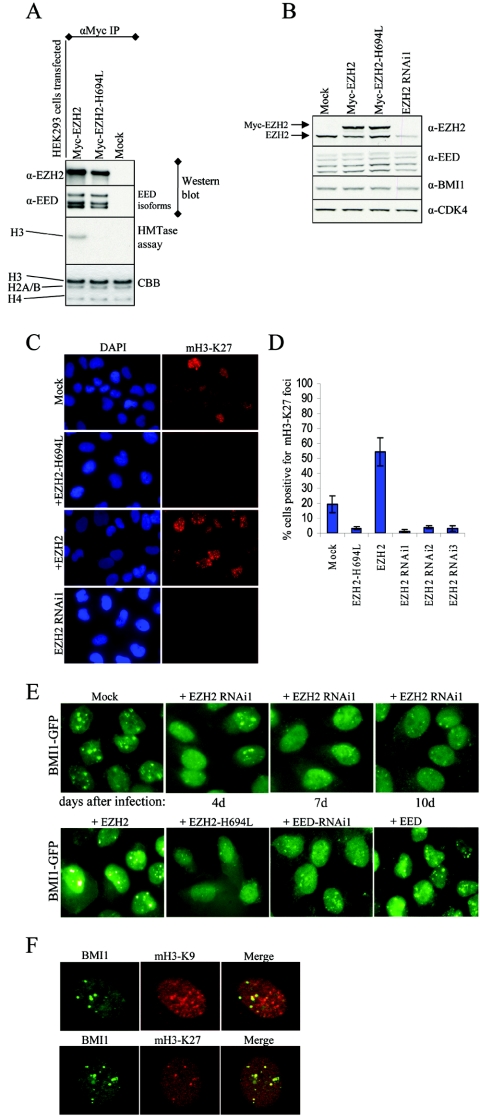
FIG. 3.
PRC2 member EZH2 is required for PcG body association of BMI1. (A) EZH2-H694L (SET domain mutant) is enzymatically inactive but still able to interact with EED. HEK293 cells were transiently transfected with EZH2 or EZH2-H694L or were mock transfected. Anti-Myc immunoprecipitated complexes were used for in vitro HMTase assays on core histone, shown in the third row from the top of the panel. The same immunoprecipitated complexes were probed with EZH2 and EED antibodies to confirm the pull-downs and functional PRC2 complex formation (the two upper rows). The bottom row shows Coomassie brilliant blue (CBB) staining for equal loading of the HMTase assay. (B) Western blot analysis showing expression levels of EZH2, EED, and BMI1 in U2OS cells used for immunostainings. Anti-CDK4 is used to show equal loading. Note that EZH2-H694L is moderately overexpressed compared to endogenous EZH2. (C) EZH2-H694L works in a dominant-interfering manner. Representative immunofluorescence images of U2OS cells transduced with EZH2, EZH2-H694L, or EZH2 RNAi1 or mock treated, using anti-trimethyl-H3-K27 (mH3-K27), indicating that EZH2-H694L is able to reduce the H3-K27 methylation levels in vivo by competing with endogenous wild-type EZH2. (D) Graph depicting quantifications of mH3-K27 immunofluorescence in U2OS cells transduced with EZH2, EZH2-H694L, or EZH2 RNAi1-3 or mock treated. Cells with mH3-K27foci were counted (n > 500 cells counted per condition from three independent experiments ± standard deviation). (E) Epifluorescence images of U2OS cells stably expressing BMI1-GFP, after transduction with EZH2 RNAi or mock treatment for the times indicated (upper row) or with EZH2, EZH2-H694L, EED, and EED RNAi1 10 days after transduction and selection with puromycin (lower row). (F) Immunofluorescence images of U2OS cells stained for di- and trimethylated H3-K9 or trimethylated H3-K27 and costained with anti-BMI1. Confocal single optical sections are shown. Note that PcG-associated heterochromatin is differentially recognized by antibodies against H3-K9 and H3-K27 methylation.
To rule out off-target effects of the EZH2 RNAi, the cells were transduced with an independent EZH2 RNAi, and the same dispersion in the BMI1-GFP nuclear signal was observed (see Fig. S1B in the supplemental material). Furthermore, overexpression of EZH2 induced a strong increase in the recruitment of BMI1 to PcG body-related pericentric regions that is reflected both in the number and in the fluorescence intensity of the PcG bodies (Fig. 3E, frame +EZH2). These results indicate that EZH2 is required for the spatial organization and maintenance of PcG bodies containing PRC1 proteins.
To test whether the PcG body-retaining ability of EZHZ relies on its catalytic activity, we made the EZH2-H694L mutant, which carries an inactivating mutation in the SET domain. The enzymatic activity of EZH2-H694L was tested in HMTase assays on core histones. HEK293 cells were transfected with Myc-tagged EZH2 or EZH2-H694L or were mock transfected, and Myc-immunoprecipitations were subsequently used for Western analysis and HMTase assays. As shown in Fig. 3A, both EZH2 and EZH2-H694L are able to form complexes with the regulating cofactor EED, which appears to be responsible for proper EZH2 function and the PRC2 HMTase activity (35, 38). EZH2 is able to form a functional complex with EED as demonstrated by histone H3 methylation (Fig. 3A). In contrast, EZH2-H694L, although perfectly able to bind to EED, is enzymatically inactive and unable to methylate H3-K27 (Fig. 3A). Hence, EZH2-H694L could potentially titrate away the endogenous EED from the endogenous PRC2 complex, thus acting in a dominant-interfering manner. This was further validated by quantifying the H3-K27 methylation levels using immunofluorescence experiments. U2OS cells transduced with EZH2, EZH2-H694L, or EZH2 RNAi or mock treated (expression levels shown in Fig. 3B) were stained with H3-K27 methyl-specific antibody (Fig. 3C) and quantified for cells positive for mH3-K27 nuclear foci. As shown in Fig. 3D, EZH2-H694L is able to decrease the mH3-K27 levels as efficiently as the different EZH2 RNAi's tested.
Once the EZH2-H694L mutant was characterized, we tested whether the catalytic activity of EZH2 is responsible for the PRC1 recruiting ability. BMI1 localization on PcG bodies was further tested using EZH2-H694L. BMI1-GFP-expressing U2OS cells that were transduced with EZH2-H694L and selected for 10 days displayed a fluorescence pattern that closely resembled the downregulation of EZH2 by RNAi, with very few and small PcG bodies and massive dispersion of the fluorescence throughout the nucleus (Fig. 3E). This indicates that the HMTase activity of EZH2 is essential for PRC1 recruitment to PcG bodies. Interestingly EZH2-H694L overexpression did not lead to any unusual phenotype or cell death, which we along with others (4) observed when EZH2 expression was substantially abrogated by RNAi.
It has recently been shown that Eed is required to direct the Ezh2 HMTase to the inactive X (Xi) chromosome. In addition, loss of methylation of H3-K9 or H3-K27 was observed on the Xi of embryos homozygous for an Eed mutation that disrupts the interaction between Eed and Ezh2 (60). We therefore investigated whether EED could act as a recruiting factor for BMI1 to PcG bodies. EED downregulation in the BMI1-GFP U2OS cell line did indeed reduce the BMI1 localization to the PcG bodies, leaving fewer and smaller PcG bodies. In contrast, cells overexpressing EED displayed bright PcG bodies similar to EZH2 overexpression (Fig. 3E). An independent RNAi for EED confirmed the specificity of the observed effect (see Fig. S1B in the supplemental material).
To confirm the BMI1-GFP results for endogenous BMI1, experiments similar to those mentioned above were carried out in the parental U2OS cell line, and the PcG bodies were visualized with antibodies specific for BMI1. The distribution of the endogenous BMI1 localization in the U2OS cells mirrored exactly the patterns found for BMI1-GFP fusion protein mentioned above for each condition (see Fig. S2 in the supplemental material). In addition, no changes could be observed in the immunofluorescence pattern of BMI1 in cells transduced with an RNAi targeted against MEL18, a PRC1 protein which is highly related to BMI1 (see Fig. S2 in the supplemental material), validating the specificity of the RNAi constructs used.
The reported changes in BMI1 fluorescence patterns do not result from a loss of BMI1 from nuclei, since total nuclear amounts were found to be unchanged (data not shown). Seemingly, they reflect a change in the pattern of histone modifications that do not allow BMI1 to be maintained at a high local concentration on pericentric heterochromatin. These data suggest that the EZH2/EED complex is actively required to place the mark that is necessary for stable maintenance of the PcG bodies.
BMI1 colocalizes with methylated H3-K27 in early S phase.
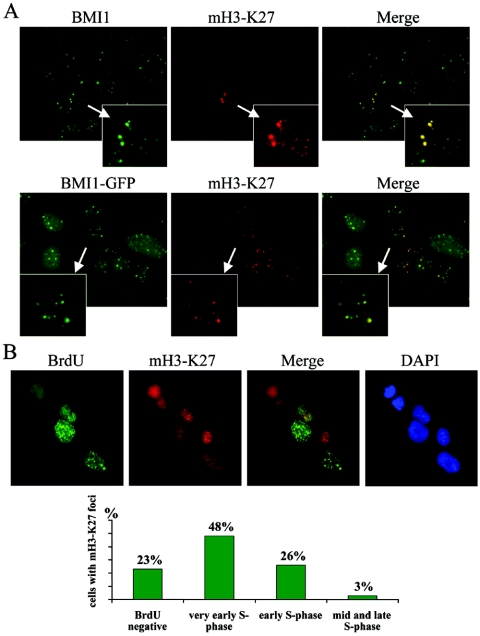
Next, we investigated the histone H3 methylation pattern underlying the PcG bodies. Whereas Suv39h-HP1 related pericentric heterochromatin is enriched in di- and trimethylated H3-K9, a robust hallmark of constitutive and facultative heterochromatin (17, 24, 32, 37, 43), EZH2/EED complexes catalyze preferentially the methylation of H3-K27. Moreover the H3-K27 methyl-specific antibody marks the Xi chromosome (7, 10, 23, 36, 45, 60). We therefore carried out immunofluorescence stainings using antibodies that detect di- and trimethyl-H3-K9 (mH3-K9) or trimethyl-H3-K27 (mH3-K27). U2OS cells revealed an overall nuclear mH3-K9 staining, which was granular and broad and with some nuclear dots in all the cells (Fig. 3F). In contrast, staining for mH3-K27 resulted in a diffuse nuclear pattern and only in a small subset of cells (12%; n > 400); prominent nuclear dots could be observed both in BMI1-GFP and the parental U2OS cells (Fig. 4A). Interestingly, all the mH3-K27-defined domains were underlying PcG bodies as detected by BMI1-GFP localization (Fig. 3F and 4A).
FIG. 4.
H3-K27 trimethylation colocalizes with Polycomb bodies in a cell cycle-dependent manner. (A) Asynchronously growing parental and BMI1-GFP-expressing U2OS cells were fixed and stained with mH3-K27 antibody and with the BMI1 antibody (upper row, parental U2OS cells). Merges of BMI1 and mH3-K27 signals show perfect overlap of the mH3-K27 with the PcG bodies. (B) Asynchronously growing U2OS cells were treated with 10 μM BrdU for 1 h. The cells were fixed and stained with antibodies against BrdU and mH3-K27 and with DAPI (see Materials and Methods). Based on the staining of the incorporated BrdU into the nascent DNA, different stages of S phase were recognized. In the figure representative images of different patterns of distribution of replication sites are shown. In early S phase, replication sites are distributed throughout the nucleus. As cells proceed from early to mid S phase, nuclei show no unlabeled areas and a fairly uniform staining of the nucleus. In the late S phase, replication of the bulk of heterochromatin follows, appearing as a pattern of large discrete foci. More than 200 cells positive for mH3-K27 foci were scored from at least eight random fields and classified into these different stages of the S phase.
The low percentage of cells with the mH3-K27 signal could indicate the cell cycle regulation of this mark. Since other proteins involved in maintenance of higher-order chromatin are recruited to the replication fork during S phase, we investigated this possibility by pulsing an asynchronous population of cells for 1 h with BrdU. Subsequently, cells were fixed and stained with BrdU and mH3-K27 antibodies. Cells with poor BrdU staining were profiled in very early S phase, those with dense BrdU labeling in early S phase, those with ring-shaped labeling in mid-late and late S phase, and those with few large dots mainly at the nuclear periphery in late S phase. Within the BrdU-positive cells with profiles typical of very early S phase, 48% of the cells displayed intense and bright mH3-K27 dots, whereas 26% of cells in early S phase had these nuclear domains. Only 3% of the cells in mid-late and late S phase displayed the mH3-K27 nuclear aggregates (Fig. 4B). Twenty-three percent of the cells with mH3-K27 failed to incorporate BrdU. These results reveal that H3-K27 methylation enrichment is not only limited to the Xi chromosome (45, 60) but also is present in a dynamic cell cycle-dependent manner in the pericentric heterochromatic regions associated with the PcG bodies.
DNMT1 is essential for BMI1 recruitment to PcG bodies.
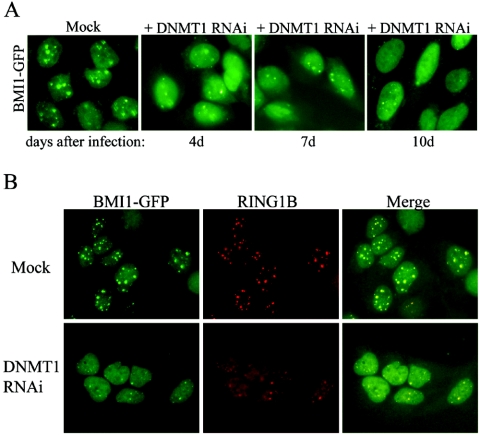
A hallmark of mammalian heterochromatin is CpG methylation within satellite DNA sequences, and the evolutionarily conserved link between the Suv39h-HP1 histone methylation system and DNA methylation has been demonstrated (26). Both layers of epigenetic control can stabilize repressed chromatin domains, thereby safeguarding the integrity and gene expression profiles of complex genomes. This DNA modification is mediated by DNA methyltransferases (DNMT) and three active DNMTs, namely DNMT1, DNMT3a, and DNMT3b, are required for the establishment and maintenance of genomic methylation patterns. DNMT3a and DNMT3b primarily methylate DNA de novo (40). In contrast DNMT1 principally maintains heritable methylation patterns during DNA replication, and Dnmt1-deficient embryonic stem cells have extensive loss of methylation at pericentric major satellite regions (27, 40). Therefore, we asked whether DNMT1 activity was crucial for the recruitment or maintenance of PcG bodies. To address this question we transduced BMI1-GFP-expressing and parental U2OS cells with DNMT1 RNAi, previously shown to downregulate DNMT1 protein in HeLa cells (63). Transduced cells were puromycin selected and kept in culture for 4, 7, or 10 days after the transduction. The achieved level of DNMT1 downregulation did not result in toxicity or cell death (data not shown). Interestingly, DNMT1 appeared to be required for BMI1 recruitment to the PcG body-associated pericentric regions, since downregulation resulted in diffuse distribution of the BMI1 fluorescence throughout the nucleus (Fig. 5A; see Fig. S2 in the supplemental material). Importantly, DNMT1 knockdown also led to mislocalization of endogenous RING1B, another PRC1 protein present in PcG bodies which is an essential binding partner of BMI1 (Fig. 5B), suggesting a general role for DNMT1 in PRC1 recruitment.
FIG. 5.
Reduced DNMT1 levels disrupt BMI1 and RING1B recruitment to PcG body-associated pericentric heterochromatin domains. (A) U2OS cells stably expressing BMI1-GFP were transduced with DNMT1 RNAi and selected with puromycin. Cells were fixed at different time points (4, 7, or 10 days posttransduction), and the fluorescence was analyzed. (B) U2OS cells stably expressing BMI1-GFP were transduced with DNMT1 RNAi, and selected with puromycin. Cells were fixed 10 days posttransduction and stained with anti-RING1B to detect endogenous RING1B localization.
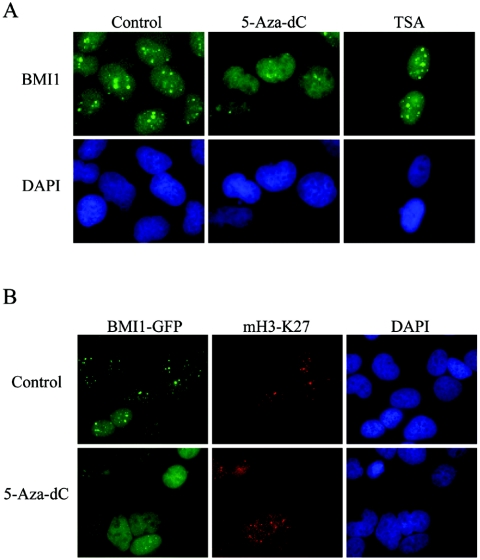
To further substantiate the role of the DNA methyltransferase, U2OS cells were cultured in the presence of low doses of the DNA methylation inhibitor 5-Aza-dC for 4 days. Cells treated with this inhibitor displayed an altered BMI1 pattern, with most of the signal dispersed in the nucleus, and loss of PcG body staining (Fig. 6A; see Fig. S2 in the supplemental material).
FIG. 6.
BMI1 localization to PcG bodies is disrupted by the DNA methylation inhibitor 5-Aza-dC, whereas H3-K27 methylation is not affected by DNA methylation inhibitor 5-Aza-dC. (A) Representative immunofluorescence images of parental U2OS cells treated with 5-Aza-dC or with TSA for 4 days, fixed, and stained with BMI1 antibody. BMI1 localization on PcG bodies is not affected by the HDAC inhibition. However, it is severely disrupted in cells treated with the DNA methylation inhibitor. (B) Untreated and 5-Aza-dC-treated BMI1-GFP-expressing U2OS cells were immunostained with mH3-K27 antibody. Representative immunofluorescence images show H3-K27 trimethylation persisting after 5-Aza-dC treatment. Whereas BMI1-GFP fluorescence is dispersed in the nucleus in the 5-Aza-dC-treated cells, the nuclear distribution of mH3-K27 is clearly not affected.
DNMTs also repress transcription independent of their methylation activity. This repression is partially dependent on HDAC activity, and prolonged treatment of mammalian cells with deacetylase inhibitors induces loss of recognition of pericentric heterochromatin by HP1 (16, 52, 53). To examine whether HDAC activity is involved in BMI1 recruitment, prolonged treatment of the U2OS cells with the HDAC inhibitor TSA was performed. However, even at high and sublethal doses, BMI1 staining revealed a normal localization to PcG bodies (Fig. 6A). These data suggest that DNMT1 activity is required for BMI1 recruitment to PcG bodies, independent from DNMT1-associated HDAC activity.
mH3-K27 signal is not affected by reduced levels of DNA methylation.
The interplay between DNA and histone methylation is supposedly a mechanism that insures a self-reinforcing repressive chromatin state, but the relative timing of these two events is currently under debate. H3-K9 methylation mediated by the histone methyltransferases Suv39h directs DNA methylation to major satellite repeats at pericentric heterochromatin (26). Consistently, the Suv39h double-null embryonic stem cells display an altered DNA methylation profile at pericentric satellite repeat, but not at other repeat sequences (26). In contrast, DNA methylation at centromeric repeats occurs independent of SUV39H methyltransferase function in human cancer cell lines, and treatment of these cell lines with the DNMT inhibitor 5-Aza-dC results in reversal of repressive histone methyl marks at silenced loci encoding tumor suppressor and cell cycle-related genes (11, 20, 39). We therefore analyzed whether H3-K27 trimethylation at pericentric heterochromatin is affected in the DNA-methylation inhibitor-treated cells. Comparative immunofluorescence analysis of interphase chromatin in control versus 5-Aza-dC-treated BMI1-GFP-expressing and parental U2OS cells confirmed reduced retention of BMI1 on PcG bodies but showed no changes in focal enrichment for H3-K27 trimethylation at these PcG-related pericentric heterochromatic loci (Fig. 6B; see Fig. S3 in the supplemental material).
Taken together, these results show that although DNMT1 activity is involved in stable maintenance of PcG bodies, it is dispensable for the H3-K27 trimethylation pool. Nevertheless, since a reduction in mH3-K27 also leads to reduced BMI1 localization to PcG bodies, BMI1 apparently requires both marks for proper localization to these heterochromatic loci.
DISCUSSION
Dynamics of BMI1 binding to PcG bodies associated with pericentric heterochromatin.
The packaging of silenced genes into repressive heterochromatin domains is an important mechanism for epigenetic gene regulation. Interestingly, recent reports demonstrated that the major heterochromatin protein HP1 is highly dynamic within stable pericentric heterochromatin domains (9, 12).
Several PcG proteins including BMI1, RING1, HPH1, and HPC2 are also associated with specific sequences in heterochromatic regions near the human chromosome 1 centromere and related sequences on several other chromosomes, forming the so-called PcG bodies (54, 55, 67). However, this PcG body-related pericentric heterochromatin is different from HP1-related constitutive pericentric heterochromatin. Pericentric heterochromatic regions that are decorated by PcG bodies are enriched for the H3-K27 methylation mark, whereas pericentric heterochromatin domains enriched for H3-K9 methylation coincide within Suv39h-HP1-silenced regions.
We have studied the kinetic properties of BMI1 association within PcG bodies in living cells by using FRAP and FLIP. Our data indicate that the BMI1 pool in the PcG bodies does not contain a static component, but, rather, it contains two kinetically different pools, one highly mobile and the other relatively less mobile which may reflect the chromatin-bound fraction of BMI1. FRAP experiments followed the exchange rate of BMI1 in both fractions, the very mobile and the relatively less mobile fractions. In addition, FLIP experiments followed the off-rate of BMI1, which was mostly determined by the less mobile fraction. Interestingly, the off-rate of the less mobile fraction appeared to be significantly faster in G2 than in G1 cells. This was reflected by the steeper slope of the loss of fluorescence curve with time in G2. Since the equilibrium rate is not different in both phases of the cell cycle, the on-rate should be faster in G2 as well, which is reflected by the relatively larger mobile fraction in FRAP experiments.
The dynamic nature of different BMI1 fractions in G1 and in G2 supports the notion that a high degree of plasticity in heterochromatin is required for appropriate reorganizations during cell cycle progression. In contrast to the extremely mobile character and continuous exchange of HP1 (9, 12), the recovery kinetics of the PcG body-associated BMI1 is much slower, again suggesting that PcG-silenced chromatin is distinct from HP1-guarded heterochromatin. Moreover, the two different dynamic fractions described here can reflect the flexible nature of PcG-induced silencing. The different components of the BMI1 pool could allow BMI1 to play diverse roles in PcG-related pericentric heterochromatin maintenance. This supports a model in which the relatively slow-moving BMI1 molecules are important for the stable maintenance of PRC1 on the PcG bodies. At the same time, the highly mobile BMI1 molecules dissociate faster from these silenced chromatin loci, allowing other possible binding partners to interact and form Polycomb complexes with different compositions and capacities. This implies that PcG bodies are more receptive for changes in their surrounding environment in G2, since the less mobile fraction is relatively more mobile in G2 than in G1, rendering PcG bodies more adaptable during this phase in the cell cycle.
Previous results from our group have shown that chromatin association of BMI1 correlates with its phosphorylation status in a cell cycle-dependent manner (67). Differential phosphorylation could account for the presence of two kinetically different BMI1 pools. Other posttranslational modifications such as ubiquitination and sumoylation could also be relevant for BMI1 kinetic properties within the PcG bodies. This possibility is further supported by reports showing the requirement of sumoylation of SOP-2 (the functional analogue of PcG protein polyhomeotic HPH) for in vivo Hox gene regulation and for proper localization of this PcG protein to distinct nuclear bodies in Caenorhabditis elegans (70). Furthermore, we have recently reported that BMI1 can be ubiquitinated (18). Together, these observations suggest that PcG recruitment to heterochromatic loci can be regulated during the cell cycle by posttranslational modifications of different members, thereby influencing the composition of the complexes.
The unique role of BMI1 in PcG complex composition and activity has been extensively illustrated by biochemical and genetic approaches. Mice deficient for Bmi1 display specific homeotic transformations of the axial skeleton, and a strictly controlled Bmi1 dose is required to regulate proper repression of Hox and other target genes (1, 19). BMI1 dosage affects the maintenance of cell identity and neoplastic transformation. This is essential for the proper function of stem cells or committed progenitors and in the pathogenesis of tumors originating from the neoplastic transformation of these cells (5, 28, 65).
In brief, our studies using FRAP and FLIP analysis on PcG bodies in U2OS cells are the first efforts to address the kinetics of PcG proteins in mammalian systems and shed new light on understanding the dynamic association of the critical core component BMI1 with other PRC1 members on PcG-related pericentric heterochromatin.
EZH2 and EED are essential for the recruitment of BMI1 to PcG bodies, and the EZH2-specific mH3-K27 signal colocalizes with PcG bodies in S phase.
Mammalian PcG proteins of the PRC1 maintenance complex harbor an intrinsic capacity to stabilize a repressive chromatin structure and counteract SWI/SNF chromatin remodeling complexes in vitro (15, 58). However, the in vivo mode of action is not well understood. Recent studies from our laboratory suggest a functional link between Ring1B-containing PRC1 protein complexes and Ezh2/PRC2-type complexes during early mammalian development, as is illustrated by highly similar loss-of-function phenotypes (66). Another link between PRC1 and PRC2 is the binding of Pc to the mH3-K27 mark set by the HMTase EZH2 (7, 10, 23). Currently, we report that downregulation of two PRC2 members, EED and EZH2, impairs the recruitment of BMI1 to the pericentric-associated PcG bodies. Remarkably, these two proteins have not been reported to accumulate specifically in PcG bodies but are diffusely localized through the nucleoplasm (57). Apparently, this diffuse subnuclear localization does not exclude more transient dynamic interactions of PRC2 components with PcG bodies. In agreement with a functional connection between PRC2 and PRC1, we also demonstrate that the catalytic core of EZH2, the SET domain, is crucial for the ability of EZH2 to recruit BMI1 and that EZH2 overexpression leads to the massive recruitment of BMI1 to pericentric PcG foci. The role of EZH2 in heterochromatin silencing is in agreement with previous data that suggested the direct structural involvement in the reorganization of repressive chromatin domains by E(z)-related proteins (25, 44, 48).
Using immunofluorescence, we have shown that PcG body distribution correlates with chromosomal regions enriched in H3-K27 methylation for a large subset of PcG bodies and that these defined nuclear domains enriched for this mark are present mainly in early S phase. Although we cannot exclude that EZH2/EED HMTase complexes act indirectly to maintain PcG bodies, the simplest explanation favors a direct role in PcG body maintenance as assayed by BMI1 localization on PcG bodies. In this model, during a very small window in early S phase, the PRC2 HMTase activity causes a transient wave of H3-K27 methylation on PcG-related pericentric heterochromatic regions. This facilitates PRC1 protein recruitment via chromodomains of Pc homologs in this complex and inheritance of this higher-order structure across many replication cycles. Although heterochromatic regions replicate late in S phase, our results indicate that the EZH2/EED-related histone methylation mark is deposited mainly early in S phase. We hypothesize that the addition of this posttranslational modification could be linked to replication-coupled histone deposition, as has been proposed for lysines 5 and 12 in histone H4 (33, 62). However, the exact timing of this event relative to DNA replication and histone deposition would need more thorough examination. Accordingly, we have observed that, in mammalian female cells, PRC1 recruitment to the Xi chromosome is also cell cycle regulated and takes place during S phase (18).
The transient activity of EZH2/EED complex during the early S phase versus the stable association of BMI1 with PcG bodies during the cell cycle could suggest the presence of other mechanisms involved in the maintenance of PcG bodies on PcG-related heterochromatic regions throughout the cell cycle. This would mean that PRC1 needs the H3-K27 methylation mark only temporarily to localize to regions of interest. Subsequent stabilization and maintenance of the PRC1 on PcG bodies throughout the cell cycle would then no longer depend on EZH2/EED HMTase activity but on other mechanisms, such as DNA methylation.
Furthermore, the transient nature of the H3-K27 methylation mark could formally be explained by epitope masking by PRC1, although the dynamic nature of PcG bodies shown inthis study argues against this concept. Since the H3-K27 methylation happens mainly in early S phase, histone exchange and/or demethylation of the methylated histone H3 could likely account for this short-lived posttranslational modification (2, 21, 59), although to this date no demethylating enzymes or mechanisms are known that could remove methyl groups from H3-K27.
These results also raise questions about the sequence context, conformation, or other recruiting marks or proteins that PRC2 might use to recognize these specific pericentric regions to subsequently allow their enrichment in PRC1 bodies. Alternatively, interactions between PcG-mediated and other gene silencing systems have been reported, which might also play a role in PRC1 recruitment (56, 68).
DNMT1 is essential for PRC1 maintenance on specific pericentric heterochromatin domains.
The data presented here indicate that downregulation of the maintenance DNA methyltransferase DNMT1 results in a substantial loss of PcG bodies, as detected by loss of localization of BMI1 and RING1B from these loci. In contrast, DNMT1 impairment did not account for a defective recruitment of the cell cycle-dependent H3-K27 methylation activity, ruling out major cell cycle alterations or toxic effects of DNMT1 RNAi that could affect chromatin conformation. Moreover, EZH2/EED activity is necessary but not sufficient to maintain all the observed PcG bodies, indicating that other mechanisms would additionally be required for PRC1 recruitment and maintenance on PcG bodies. Therefore, an attractive model is that the histone methyltransferase activity by PRC2/EZH2 would somehow recruit the DNA methyltransferase activity required for the replication and/or maintenance of the PcG-related heterochromatin domains. A clear precedent for such a model is the involvement of Suv39h for recruitment of DNMT activity to major satellite repeats at pericentric heterochromatin (26). Even though we could not visualize specific DNA methyltransferase enrichment at PcG bodies in U2OS cells (data not shown), similar to EZH2/EED localization, this does not discredit the model. Yet another explanation would be that the effect of DNMT1 depletion on PcG bodies reflects an indirect effect, such as by downregulation of the level of a critical PRC1 recruiting protein.
Finally, it is important to remark the absence of detectable DNA methylation in Drosophila except for a window during early fly development. This appears to rule out an essential role for DNA methylation in PRE function or recruitment of PRE-binding proteins that have been identified in flies but not in mammals (51). These observations, together with the redundancy of PcG proteins in mammals as a result of evolutionary gene amplification, indicate that the mechanisms responsible for PcG recruitment are more complex in mammals. Since no mammalian equivalents of Drosophila PREs have been identified so far, it remains possible that binding or recruitment of PRC1 complexes is fundamentally different between these species.
In summary, we describe two different pools of BMI1 in PcG bodies. One is highly mobile, allowing dynamic interactions, and the other is less mobile, presumably representing a relatively more strongly bound fraction within the PcG-related heterochromatin domains. We show that PRC2 members EED and EZH2 are required either directly or indirectly for the recruitment of PRC1 proteins to PcG-related pericentric heterochromatin and that the HMTase activity of EZH2 is essential for this recruiting ability. Immunostaining analysis confirmed the preferential localization of PcG bodies on trimethyl-H3-K27 foci, which is seen intriguingly mainly during early S phase. This highlights the dynamic nature of PcG complexes. In addition, we find that DNMT1 is necessary for proper PcG body assembly independent of the DNMT-associated HDAC activity. Interestingly, mH3-K27 is not influenced by DNA methylation. This could suggest a role for DNA methyl-ation as a link between histone methylation and PcG recruitment or maintenance, but it could also suggest an independent role for DNA methylation next to histone methylation in recruitment of PcG proteins to the target loci. On another note, a role for RNA intermediates in PcG-mediated silencing has been postulated before, where SOP-2 is shown to contain an RNA-binding domain which is essential for HOX gene repression (69). However, so far no direct evidence for the role of RNAs and RNAi in PcG recruitment or PcG-mediated silencing has been reported.
Together, these results suggest the existence of distinct types of heterochromatin, namely, the PcG-related silenced chromatin and the SUV39H-HP1-related silenced chromatin. Our results contribute to elucidate the PcG protein-mediated silencing mechanisms and open new, interesting areas for future research exploring the possible links between the DNA methyl-transferases and PRC1 recruitment.
Supplementary Material
Acknowledgments
I. H.-M. is supported by a 5th Framework EC grant. P.T. is supported by The Dutch Cancer Fund and The Netherlands Cancer Institute, through a grant to M.V.L.
We thank L. Oomen, L. Brocks and A. Griekspoor for their invaluable assistance with microscopy. We also thank T. Jenuwein for mH3-K9 and mH3-K27 antibodies, H. Koseki for RING1B antibody, A. Otte for HPC2 and EED antibodies, K. Helin for EZH2 antibody, Y. Shi for the DNMT1 RNAi, and B. van Steensel and P. van der Stoop for helpful discussions.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org./.
REFERENCES
- 1.Akasaka, T., M. van Lohuizen, N. van der Lugt, Y. Mizutani-Koseki, M. Kanno, M. Taniguchi, M. Vidal, M. Alkema, A. Berns, and H. Koseki. 2001. Mice doubly deficient for the Polycomb Group genes Mel18 and Bmi1 reveal synergy and requirement for maintenance but not initiation of Hox gene expression. Development 128:1587-1597. [DOI] [PubMed] [Google Scholar]
- 2.Bannister, A. J., R. Schneider, and T. Kouzarides. 2002. Histone methylation: dynamic or static? Cell 109:801-806. [DOI] [PubMed] [Google Scholar]
- 3.Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. [DOI] [PubMed] [Google Scholar]
- 4.Bracken, A. P., D. Pasini, M. Capra, E. Prosperini, E. Colli, and K. Helin. 2003. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 22:5323-5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruggeman, S. W., M. E. Valk-Lingbeek, P. P. van der Stoop, J. J. Jacobs, K. Kieboom, E. Tanger, D. Hulsman, C. Leung, Y. Arsenijevic, S. Marino, and M. van Lohuizen. 2005. Ink4a and Arf differentially affect cell proliferation and neural stem cell self-renewal in Bmi1-deficient mice. Genes Dev. 19:1438-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 7.Cao, R., L. Wang, H. Wang, L. Xia, H. Erdjument-Bromage, P. Tempst, R. S. Jones, and Y. Zhang. 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298:1039-1043. [DOI] [PubMed] [Google Scholar]
- 8.Chan, C. S., L. Rastelli, and V. Pirrotta. 1994. A Polycomb response element in the Ubx gene that determines an epigenetically inherited state of repression. EMBO J. 13:2553-2564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheutin, T., A. J. McNairn, T. Jenuwein, D. M. Gilbert, P. B. Singh, and T. Misteli. 2003. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science 299:721-725. [DOI] [PubMed] [Google Scholar]
- 10.Czermin, B., R. Melfi, D. McCabe, V. Seitz, A. Imhof, and V. Pirrotta. 2002. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111:185-196. [DOI] [PubMed] [Google Scholar]
- 11.Fahrner, J. A., S. Eguchi, J. G. Herman, and S. B. Baylin. 2002. Dependence of histone modifications and gene expression on DNA hypermethylation in cancer. Cancer Res. 62:7213-7218. [PubMed] [Google Scholar]
- 12.Festenstein, R., S. N. Pagakis, K. Hiragami, D. Lyon, A. Verreault, B. Sekkali, and D. Kioussis. 2003. Modulation of heterochromatin protein 1 dynamics in primary mammalian cells. Science 299:719-721. [DOI] [PubMed] [Google Scholar]
- 13.Fischle, W., Y. Wang, S. A. Jacobs, Y. Kim, C. D. Allis, and S. Khorasanizadeh. 2003. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 17:1870-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Francis, N. J., R. E. Kingston, and C. L. Woodcock. 2004. Chromatin compaction by a polycomb group protein complex. Science 306:1574-1577. [DOI] [PubMed] [Google Scholar]
- 15.Francis, N. J., A. J. Saurin, Z. Shao, and R. E. Kingston. 2001. Reconstitution of a functional core polycomb repressive complex. Mol. Cell 8:545-556. [DOI] [PubMed] [Google Scholar]
- 16.Fuks, F., W. A. Burgers, A. Brehm, L. Hughes-Davies, and T. Kouzarides. 2000. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat. Genet. 24:88-91. [DOI] [PubMed] [Google Scholar]
- 17.Guenatri, M., D. Bailly, C. Maison, and G. Almouzni. 2004. Mouse centric and pericentric satellite repeats form distinct functional heterochromatin. J. Cell Biol. 166:493-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hernandez-Munoz, I., A. H. Lund, S. P. van der, E. Boutsma, I. Muijrers, E. Verhoeven, D. A. Nusinow, B. Panning, Y. Marahrens, and M. van Lohuizen. 2005. Stable X chromosome inactivation involves the PRC1 Polycomb complex and requires histone MACROH2A1 and the CULLIN3/SPOP ubiquitin E3 ligase. Proc. Natl. Acad. Sci. USA. 102:7635-7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobs, J. J., K. Kieboom, S. Marino, R. A. DePinho, and M. van Lohuizen. 1999. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature 397:164-168. [DOI] [PubMed] [Google Scholar]
- 20.Kondo, Y., L. Shen, and J. P. Issa. 2003. Critical role of histone methylation in tumor suppressor gene silencing in colorectal cancer. Mol. Cell. Biol. 23:206-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kubicek, S., and T. Jenuwein. 2004. A crack in histone lysine methylation. Cell 119:903-906. [DOI] [PubMed] [Google Scholar]
- 22.Kuzmichev, A., T. Jenuwein, P. Tempst, and D. Reinberg. 2004. Different EZH2-containing complexes target methylation of histone H1 or nucleosomal histone H3. Mol. Cell 14:183-193. [DOI] [PubMed] [Google Scholar]
- 23.Kuzmichev, A., K. Nishioka, H. Erdjument-Bromage, P. Tempst, and D. Reinberg. 2002. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 16:2893-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lachner, M., D. O'Carroll, S. Rea, K. Mechtler, and T. Jenuwein. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116-120. [DOI] [PubMed] [Google Scholar]
- 25.Laible, G., A. Wolf, R. Dorn, G. Reuter, C. Nislow, A. Lebersorger, D. Popkin, L. Pillus, and T. Jenuwein. 1997. Mammalian homologues of the Polycomb-group gene Enhancer of zeste mediate gene silencing in Drosophila heterochromatin and at S. cerevisiae telomeres. EMBO J. 16:3219-3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lehnertz, B., Y. Ueda, A. A. Derijck, U. Braunschweig, L. Perez-Burgos, S. Kubicek, T. Chen, E. Li, T. Jenuwein, and A. H. Peters. 2003. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 13:1192-1200. [DOI] [PubMed] [Google Scholar]
- 27.Lei, H., S. P. Oh, M. Okano, R. Juttermann, K. A. Goss, R. Jaenisch, and E. Li. 1996. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development 122:3195-3205. [DOI] [PubMed] [Google Scholar]
- 28.Leung, C., M. Lingbeek, O. Shakhova, J. Liu, E. Tanger, P. Saremaslani, M. van Lohuizen, and S. Marino. 2004. Bmi1 is essential for cerebellar development and is overexpressed in human medulloblastomas. Nature 428:337-341. [DOI] [PubMed] [Google Scholar]
- 29.Liang, G., M. F. Chan, Y. Tomigahara, Y. C. Tsai, F. A. Gonzales, E. Li, P. W. Laird, and P. A. Jones. 2002. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol. Cell. Biol. 22:480-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lund, A. H., and M. van Lohuizen. 2004. Polycomb complexes and silencing mechanisms. Curr. Opin. Cell Biol. 16:239-246. [DOI] [PubMed] [Google Scholar]
- 31.Maison, C., and G. Almouzni. 2004. HP1 and the dynamics of heterochromatin maintenance. Nat. Rev. Mol. Cell Biol. 5:296-304. [DOI] [PubMed] [Google Scholar]
- 32.Maison, C., D. Bailly, A. H. Peters, J. P. Quivy, D. Roche, A. Taddei, M. Lachner, T. Jenuwein, and G. Almouzni. 2002. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat. Genet. 30:329-334. [DOI] [PubMed] [Google Scholar]
- 33.Mello, J. A., and G. Almouzni. 2001. The ins and outs of nucleosome assembly. Curr. Opin. Genet. Dev. 11:136-141. [DOI] [PubMed] [Google Scholar]
- 34.Min, J., Y. Zhang, and R. M. Xu. 2003. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 17:1823-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Montgomery, N. D., D. Yee, A. Chen, S. Kalantry, S. J. Chamberlain, A. P. Otte, and T. Magnuson. 2005. The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr. Biol. 15:942-947. [DOI] [PubMed] [Google Scholar]
- 36.Muller, J., C. M. Hart, N. J. Francis, M. L. Vargas, A. Sengupta, B. Wild, E. L. Miller, M. B. O'Connor, R. E. Kingston, and J. A. Simon. 2002. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111:197-208. [DOI] [PubMed] [Google Scholar]
- 37.Nakayama, J., J. C. Rice, B. D. Strahl, C. D. Allis, and S. I. Grewal. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292:110-113. [DOI] [PubMed] [Google Scholar]
- 38.Nekrasov, M., B. Wild, and J. Muller. 2005. Nucleosome binding and histone methyltransferase activity of Drosophila PRC2. EMBO Rep. 6:348-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen, C. T., D. J. Weisenberger, M. Velicescu, F. A. Gonzales, J. C. Lin, G. Liang, and P. A. Jones. 2002. Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2′-deoxycytidine. Cancer Res. 62:6456-6461. [PubMed] [Google Scholar]
- 40.Okano, M., D. W. Bell, D. A. Haber, and E. Li. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99:247-257. [DOI] [PubMed] [Google Scholar]
- 41.Orlando, V. 2003. Polycomb, epigenomes, and control of cell identity. Cell 112:599-606. [DOI] [PubMed] [Google Scholar]
- 42.Orlando, V., and R. Paro. 1995. Chromatin multiprotein complexes involved in the maintenance of transcription patterns. Curr. Opin. Genet. Dev. 5:174-179. [DOI] [PubMed] [Google Scholar]
- 43.Peters, A. H., D. O'Carroll, H. Scherthan, K. Mechtler, S. Sauer, C. Schofer, K. Weipoltshammer, M. Pagani, M. Lachner, A. Kohlmaier, S. Opravil, M. Doyle, M. Sibilia, and T. Jenuwein. 2001. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107:323-337. [DOI] [PubMed] [Google Scholar]
- 44.Platero, J. S., E. J. Sharp, P. N. Adler, and J. C. Eissenberg. 1996. In vivo assay for protein-protein interactions using Drosophila chromosomes. Chromosoma 104:393-404. [DOI] [PubMed] [Google Scholar]
- 45.Plath, K., J. Fang, S. K. Mlynarczyk-Evans, R. Cao, K. A. Worringer, H. Wang, C. C. de la Cruz, A. P. Otte, B. Panning, and Y. Zhang. 2003. Role of histone H3 lysine 27 methylation in X inactivation. Science 300:131-135. [DOI] [PubMed] [Google Scholar]
- 46.Poux, S., R. Melfi, and V. Pirrotta. 2001. Establishment of Polycomb silencing requires a transient interaction between PC and ESC. Genes Dev. 15:2509-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quivy, J. P., D. Roche, D. Kirschner, H. Tagami, Y. Nakatani, and G. Almouzni. 2004. A CAF-1 dependent pool of HP1 during heterochromatin duplication. EMBO J. 23:3516-3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rastelli, L., C. S. Chan, and V. Pirrotta. 1993. Related chromosome binding sites for zeste, suppressors of zeste and Polycomb group proteins in Drosophila and their dependence on Enhancer of zeste function. EMBO J. 12:1513-1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rea, S., F. Eisenhaber, D. O'Carroll, B. D. Strahl, Z. W. Sun, M. Schmid, S. Opravil, K. Mechtler, C. P. Ponting, C. D. Allis, and T. Jenuwein. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406:593-599. [DOI] [PubMed] [Google Scholar]
- 50.Reits, E. A., and J. J. Neefjes. 2001. From fixed to FRAP: measuring protein mobility and activity in living cells. Nat. Cell Biol. 3:E145-E147. [DOI] [PubMed] [Google Scholar]
- 51.Ringrose, L., and R. Paro. 2004. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu. Rev. Genet. 38:413-443. [DOI] [PubMed] [Google Scholar]
- 52.Robertson, K. D., S. Ait-Si-Ali, T. Yokochi, P. A. Wade, P. L. Jones, and A. P. Wolffe. 2000. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 25:338-342. [DOI] [PubMed] [Google Scholar]
- 53.Rountree, M. R., K. E. Bachman, and S. B. Baylin. 2000. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat. Genet. 25:269-277. [DOI] [PubMed] [Google Scholar]
- 54.Satijn, D. P., M. J. Gunster, J. van der Vlag, K. M. Hamer, W. Schul, M. J. Alkema, A. J. Saurin, P. S. Freemont, R. van Driel, and A. P. Otte. 1997. RING1 is associated with the polycomb group protein complex and acts as a transcriptional repressor. Mol. Cell. Biol. 17:4105-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saurin, A. J., C. Shiels, J. Williamson, D. P. Satijn, A. P. Otte, D. Sheer, and P. S. Freemont. 1998. The human polycomb group complex associates with pericentromeric heterochromatin to form a novel nuclear domain. J. Cell Biol. 142:887-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sewalt, R. G., M. Lachner, M. Vargas, K. M. Hamer, J. L. den Blaauwen, T. Hendrix, M. Melcher, D. Schweizer, T. Jenuwein, and A. P. Otte. 2002. Selective interactions between vertebrate Polycomb homologs and the SUV39H1 histone lysine methyltransferase suggest that histone H3-K9 methylation contributes to chromosomal targeting of Polycomb group proteins. Mol. Cell. Biol. 22:5539-5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sewalt, R. G., d. van, V., M. J. Gunster, K. M. Hamer, J. L. den Blaauwen, D. P. Satijn, T. Hendrix, R. van Driel, and A. P. Otte. 1998. Characterization of interactions between the mammalian Polycomb-group proteins Enx1/EZH2 and EED suggests the existence of different mammalian Polycomb-group protein complexes. Mol. Cell. Biol. 18:3586-3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shao, Z., F. Raible, R. Mollaaghababa, J. R. Guyon, C. T. Wu, W. Bender, and R. E. Kingston. 1999. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 98:37-46. [DOI] [PubMed] [Google Scholar]
- 59.Shi, Y., F. Lan, C. Matson, P. Mulligan, J. R. Whetstine, P. A. Cole, R. A. Casero, and Y. Shi. 2004. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119:941-953. [DOI] [PubMed] [Google Scholar]
- 60.Silva, J., W. Mak, I. Zvetkova, R. Appanah, T. B. Nesterova, Z. Webster, A. H. Peters, T. Jenuwein, A. P. Otte, and N. Brockdorff. 2003. Establishment of histone H3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev. Cell. 4:481-495. [DOI] [PubMed] [Google Scholar]
- 61.Simon, J., A. Chiang, W. Bender, M. J. Shimell, and M. O'Connor. 1993. Elements of the Drosophila bithorax complex that mediate repression by Polycomb group products. Dev. Biol. 158:131-144. [DOI] [PubMed] [Google Scholar]
- 62.Sobel, R. E., R. G. Cook, C. A. Perry, A. T. Annunziato, and C. D. Allis. 1995. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc. Natl. Acad. Sci. USA. 92:1237-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sui, G., C. Soohoo, e. B. Affar, F. Gay, Y. Shi, W. C. Forrester, and Y. Shi. 2002. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl. Acad. Sci. USA. 99:5515-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Taddei, A., C. Maison, D. Roche, and G. Almouzni. 2001. Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nat. Cell Biol. 3:114-120. [DOI] [PubMed] [Google Scholar]
- 65.Valk-Lingbeek, M. E., S. W. Bruggeman, and M. van Lohuizen. 2004. Stem cells and cancer; the polycomb connection. Cell 118:409-418. [DOI] [PubMed] [Google Scholar]
- 66.Voncken, J. W., B. A. Roelen, M. Roefs, S. de Vries, E. Verhoeven, S. Marino, J. Deschamps, and M. van Lohuizen. 2003. Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc. Natl. Acad. Sci. USA. 100:2468-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Voncken, J. W., D. Schweizer, L. Aagaard, L. Sattler, M. F. Jantsch, and M. van Lohuizen. 1999. Chromatin-association of the Polycomb group protein BMI1 is cell cycle-regulated and correlates with its phosphorylation status. J. Cell Sci. 112:4627-4639. [DOI] [PubMed] [Google Scholar]
- 68.Yamamoto, K., M. Sonoda, J. Inokuchi, S. Shirasawa, and T. Sasazuki. 2004. Polycomb group suppressor of zeste 12 links heterochromatin protein 1alpha and enhancer of zeste 2. J. Biol. Chem. 279:401-406. [DOI] [PubMed] [Google Scholar]
- 69.Zhang, H., A. Christoforou, L. Aravind, S. W. Emmons, H. S. van den, and D. A. Haber. 2004. The C. elegans Polycomb gene SOP-2 encodes an RNA binding protein. Mol. Cell 14:841-847. [DOI] [PubMed] [Google Scholar]
- 70.Zhang, H., G. A. Smolen, R. Palmer, A. Christoforou, S. van den Heuvel, and D. A. Haber. 2004. SUMO modification is required for in vivo Hox gene regulation by the Caenorhabditis elegans Polycomb group protein SOP-2. Nat. Genet. 36:507-511. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.