Abstract
Ubiquitin-conjugating enzymes (E2s) collaborate with the ubiquitin-activating enzyme (E1) and ubiquitin ligases (E3s) to attach ubiquitin to target proteins. RING-containing E3s simultaneously bind to E2s and substrates, bringing them into close proximity and thus facilitating ubiquitination. We show herein that, although the E3-binding site on the human E2 UbcH5b is distant from its active site, two RING-type minimal E3 modules lacking substrate-binding functions greatly stimulate the rate of ubiquitin release from the UbcH5b–ubiquitin thioester. Using statistical coupling analysis and mutagenesis, we identify and characterize clusters of coevolving and functionally linked residues within UbcH5b that span its E3-binding and active sites. Several UbcH5b mutants are defective in their stimulation by E3s despite their abilities to bind to these E3s, to form ubiquitin thioesters, and to release ubiquitin at a basal rate. One such mutation, I37A, is distant from both the active site and the E3-binding site of UbcH5b. Our studies reveal structural determinants for communication between distal functional sites of E2s and suggest that RING-type E3s activate E2s allosterically.
Keywords: mechanism, E2 enzymes, E3 enzymes
Ubiquitination is a major form of posttranslational modification of proteins in eukaryotes and generally leads to the degradation of target proteins by the proteasome (1, 2). Ubiquitination is catalyzed by a cascade of enzymes (1, 3, 4). The active-site cysteine of ubiquitin-activating enzyme (E1) forms a thioester with ubiquitin in an ATP-dependent reaction. Ubiquitin is then transferred to ubiquitin-conjugating enzymes (E2s) as a thioester. Ubiquitin ligases (E3s) facilitate the attachment of ubiquitin to ε-amino groups of lysines and, rarely, to the N-terminal amino group in substrates (1, 3–6). HECT (homology to E6AP C terminus) domain-containing E3s form intermediate thioesters with ubiquitin at their active-site cysteines before transferring the ubiquitin to substrates (3, 4, 7). In contrast, most E3s contain RING (really interesting new gene) finger domains and do not form thioesters with ubiquitin (3, 4, 8). They act as scaffolds that bind to E2s and substrates simultaneously, bringing them into close proximity and thus facilitating ubiquitination (9, 10). In this scenario, ubiquitin is transferred directly from E2s to substrates.
The mechanism of ubiquitin transfer from the thioesters of E2s to amino groups on target proteins is not completely elucidated (3, 4). However, it is generally believed that the ε-amino group of the acceptor lysine attacks the carbonyl of the E2–ubiquitin thioester bond, forming a tetrahedral intermediate. Elimination of the thiol group of the E2 from this intermediate results in the formation of an isopeptide bond between ubiquitin and the substrate (3, 4). A strictly conserved asparagine (N77 in UbcH5b) is located in the vicinity of the active-site cysteine and is required for ubiquitin transfer from E2s to substrates (3, 4, 11). This residue has been suggested to facilitate ubiquitin transfer by stabilizing the negatively charged oxyanion in the tetrahedral intermediate (3, 4, 11).
Intriguingly, RING-containing E3s stimulate the autoubiquitination of E2s and the synthesis of polyubiquitin chains in the absence of substrates (12–18). This finding suggests that, in addition to their scaffolding roles, RING-type E3s might activate the ability of E2s to transfer ubiquitin from their active-site cysteines to acceptor lysines. However, structural studies have revealed that the E3-binding site on an E2 is 15 Å away from its active site (19–21). It is thus unlikely that certain E3 residues contribute directly to the release of ubiquitin from the active sites of E2s. Furthermore, E2s do not undergo significant conformational changes upon binding to E3s (19, 20). These findings suggest that RING-type E3s activate E2s by perturbing the structures of E2s in a subtle, allosteric fashion. Allostery in this context is defined as the effect on the catalytic activity of an enzyme exerted by the binding of a cofactor at a distant site.
Using an algorithm that detects coevolving residues within a protein, called statistical coupling analysis (SCA) (22–25), we have identified clusters of coevolving and possibly energetically coupled residues within E2s that physically link their E3-binding and active sites. Mutations of a subset of these residues disrupt the allosteric communication between the E3-binding and active sites of E2s. We have chosen human UbcH5b, from the yeast Ubc4 family, as an E2 model for mutagenesis because of its strong activity with a variety of E3s and its simple domain structure without extensions and insertions. Our results suggest that E3 binding to UbcH5b triggers subtle long-range conformational changes on UbcH5b, which in turn enhances its ability to transfer ubiquitin from the thioester to substrates.
Materials and Methods
SCA. Nine E2s with known tertiary structures were used to search the National Center for Biotechnology Information nonredundant protein database by psi-blast to identify 345 E2s. Their sequences were aligned with clustalw and manually adjusted based on structural information. The SCA was performed as described in ref. 24 by using improved matlab scripts (R. Ranganathan, personal communication).
Protein Expression and Purification. His-tagged human E1 was expressed in Escherichia coli and purified as described in ref. 26. All UbcH5b mutants were generated by using the QuikChange mutagenesis kit (Stratagene). WT (wild-type) and mutant His-tagged UbcH5b were expressed in the BL21(DE3) strain and purified by a Ni2+-NTA (Qiagen) column. Apc2/11 contained human Apc11 (the RING-containing subunit) and the C-terminal fragment (residues 549–822) of human Apc2 (the cullin subunit) and was expressed and purified as described in ref. 15. A His-tagged, N-terminal fragment (residues 1–78) of CNOT4 containing its RING finger domain, referred to as CNOT4N, was expressed and purified as described in ref. 27.
UbcH5b Thioester Formation and Release Assays. The thioester formation reactions (10 μl) contained 1 μg of 0.9 μM E1, 1 μg of 5 μM UbcH5b, and 10 μg of 130 μM ubiquitin (Sigma) in a buffer containing 10 mM Hepes (pH 7.5), 100 mM NaCl, 40 μM ATP, and 2 mM MgCl2. Reaction mixture was incubated for 10 min at room temperature. In certain experiments, 1–5 mM NEM or 0.1 unit/μl apyrase (Sigma) was added to inactivate E1 or to deplete ATP, respectively. Apc2/11 (2.3 μM) or CNOT4 (44 μM) was then added to the reactions. Samples were taken at the indicated times, stopped by both reducing and nonreducing SDS sample buffers, separated by 4–20% gradient SDS/PAGE, and blotted with anti-UbcH5 (Boston Biochem, Cambridge, MA). Blots were quantified by a densitometer by using the software fluorchem. Reaction rates were modeled as single-exponential decays by using sigmaplot. Experiments were done in triplicates. Single-exponential decay curves fitted experimental data with R2 values >0.9. A cyclin B1 ubiquitination assay with Xenopus anaphase-promoting complex/cyclosome (APC/C) was performed as described in ref. 26.
NMR Spectroscopy. Unlabeled CNOT4N at 2–6 mM concentrations was titrated into samples of 15N-labeled UbcH5b, resulting in samples containing 150 μM UbcH5b and 0.7–1.2 mM CNOT4N at the end of each titration. For each titration point, a 15N/1H heteronuclear single quantum coherence spectrum was acquired. 1H and 15N backbone chemical-shift assignments of UbcH5b were kindly provided by Rolf Boelens (Utrecht University, The Netherlands) (21). Chemical-shift changes were plotted against the molar ratio of CNOT4N vs. UbcH5b and were fit to standard binding curves by using sigmaplot. For each UbcH5b protein, dissociation constants (Kd) with standard deviations were calculated by using chemical shifts for 10 UbcH5b residues that undergo changes upon binding to CNOT4N.
Results and Discussion
SCA Identifies Clusters of Coevolving Residues That Connect the E3-Binding and Active Sites of E2s. In this study, we sought to identify residues within E2s that mediate their allosteric activation by E3s using SCA (22–25). The key premise of SCA is that functionally or structurally coupled residues within a protein should coevolve and show covariation in a large, diverse multiple sequence alignment (MSA) of members of a given protein family (22–25, 28). If allostery is a conserved feature of E2s, residues of the E3-binding site and residues at or near the active site of E2s are expected to show covariation during evolution. We first obtained an MSA for 345 E2s from various organisms (Fig. 5A, which is published as supporting information on the PNAS web site) and then used SCA to analyze the covariation of pairs of residues in the MSA. As part of this analysis, the degree of covariation of a given residue pair is converted to a pseudoenergy term, allowing the use of hierarchical clustering to identify networks of residues that exhibit similar covariation patterns (24). These residues are considered coupled either structurally (e.g., pack against each other) or functionally (e.g., perform a common function).
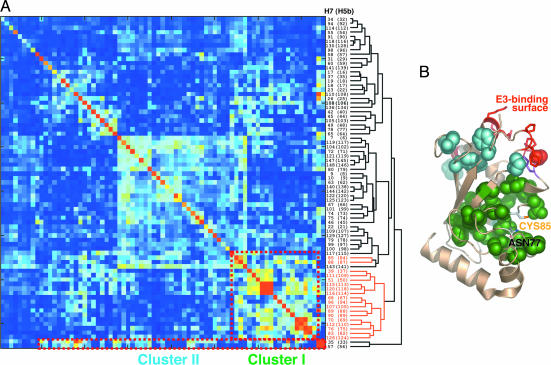
SCA reveals two clusters of coevolving residues within E2s (Fig. 1A; see also Fig. 6, which is published as supporting information on the PNAS web site). The residue numbers are based on UbcH5b. Mapping of these two clusters of residues on the structure of UbcH5b reveals that most cluster I residues are located in the hydrophobic core of E2s and form a contiguous patch surrounding the active site of E2s (Fig. 1B). Cluster II residues form more localized contacts surrounding the E3-binding site. The two clusters are connected through the common residue I88. The SCA results, revealing a connected set of residues stretching from the E3-interaction surface to an extensive active-site area, are consistent with the existence of long-range allosteric communication between the E3-binding and active sites of E2s.
Fig. 1.
SCA of E2s. (A) Hierarchical clustering of a submatrix of pairwise statistical coupling energies of E2 residues, which are plotted as a color gradient, with blue and red representing the lowest (0 kT*, with kT* being an arbitrary energy-like unit) and highest (2 kT*) energies, respectively. Both columns and rows of the matrix represent residue positions in UbcH7 and UbcH5b (in parentheses). The two clusters of residues that exhibit similar coupling patterns are boxed with dashed lines. (B) The two clusters of residues in A are shown as space-filling models and mapped onto the structure of UbcH5b. Cluster I and II residues are colored green and cyan, respectively. E3-binding residues are shown in red. The active-site cysteine and asparagine are labeled.
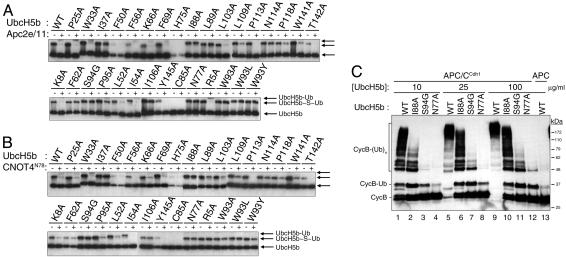
E3 Binding Enhances the Rate of Ubiquitin Release of E2–Ubiquitin Thioesters. We next mutated 11 coupled residues in cluster I, 4 coupled residues in cluster II, and 8 noncoupled neighboring residues as controls in UbcH5b (Fig. 5; see also Table 2, which is published as supporting information on the PNAS web site). We also mutated its active-site cysteine, the catalytic asparagine, and residues at or near its E3-binding site. We monitored the ability of UbcH5b to be charged with and to release ubiquitin from its active-site cysteine in the absence or presence of two RING-containing E3s: Apc2/11 (a subcomplex of APC/C) (15) or the N-terminal fragment of CNOT4 (CNOT4N) (21) (Fig. 2 A and B). This assay allowed us to evaluate the total rate of ubiquitin transfer from the UbcH5b–ubiquitin thioester to all possible ubiquitin acceptors, including H2O (i.e., hydrolysis). As expected, the WT UbcH5b, but not UbcH5b C85A, efficiently formed a thioester linkage (Fig. 2 A and B; see also Fig. 7A, which is published as supporting information on the PNAS web site). The ubiquitin thioester of UbcH5b WT was not observed in the presence of either Apc2/11 (15–18, 29) or CNOT4N (Fig. 2 A and B), because the rate of ubiquitin release exceeded the rate of ubiquitin charging under these conditions. These results confirmed that RING-type E3s activate the ubiquitin thioester of E2s in the absence of substrates. UbcH5b N77A efficiently formed a thioester with ubiquitin but failed to release ubiquitin in the absence or presence of E3s (Fig. 2), confirming the direct involvement of N77 in catalysis (11).
Fig. 2.
E3-stimulated release of ubiquitin from the UbcH5b–ubiquitin thioester. (A) WT and 31 mutants of UbcH5b were tested for their ability to release ubiquitin from their thioesters in the presence of buffer (–) or Apc2/11 (+) with continuous E1-catalyzed ubiquitin charging. The bands corresponding to free UbcH5b, UbcH5b–ubiquitin thioester, and monoubiquitinated UbcH5b (with isopeptide-linked ubiquitin) are indicated. (B) Same as in A except that CNOT4N was used as the E3. (C) UbcH5b I88A was defective in supporting APC/CCdh1-mediated ubiquitination of cyclin B1. Xenopus egg APC/C was incubated with buffer or Cdh1 and assayed for its ability to ubiquitinate an N-terminal fragment (residues 1–102) of human cyclin B1 in the presence of the varying concentrations of WT or mutant UbcH5b. Cyclin B–ubiquitin conjugates are indicated.
We also mutated several residues within the E3-binding site of UbcH5b. UbcH5b F62A and S94G retained their ubiquitin thioesters in the presence of E3s (Figs. 2 A and B and 7 B and C). Thus, the E3-stimulated ubiquitin release from the UbcH5b–ubiquitin thioester depended on UbcH5b's having an intact E3-binding site. Interestingly, autoubiquitination of UbcH5b R5A, a mutation shown to disrupt the UbcH5b–CNOT4N interaction (30), was not stimulated by CNOT4N (Figs. 2B and 7C) but was stimulated by Apc2/11 (Figs. 2 A and 7B). This finding suggests that different E3s might have slightly different binding surfaces on UbcH5b. UbcH5b K8A and P95A mutations presumably did not disrupt the binding of Apc2/11 or CNOT4N to UbcH5b. Our thioester ubiquitin release assay yielded predictable results with respect to the effects of various UbcH5b mutations, confirming its validity.
Several UbcH5b Mutants Are Refractory to E3-Stimulated Ubiquitin Release from the Thioester. Several mutations that target residues predicted by SCA to be energetically coupled, such as I37A, I88A, L89A, I106A, and N114A, formed ubiquitin thioesters efficiently and retained significant fractions of their ubiquitin thioesters in the presence of Apc2/11 or CNOT4N (Fig. 2). Because these mutations are not expected to disrupt the binding of UbcH5b to E3s, they most likely impair the ability of these E3s to stimulate UbcH5b by affecting the communication between the E3-binding and active sites of UbcH5b. Thus, these residues are probable structural determinants of allostery in E2s. Among other UbcH5b mutants, F50A, H75A, and Y145A expressed poorly in bacteria and did not efficiently form thioesters with ubiquitin, suggesting that these mutations affected the folding and/or stability of UbcH5b (Fig. 2 and data not shown). UbcH5b K66A and T142A expressed well in bacteria but did not appear to be efficiently charged with ubiquitin (Fig. 2).
E2s perform many functions, including (i) formation of the ubiquitin thioester, (ii) ubiquitin release from the thioester in the absence of E3s, (iii) binding to E3s, (iv) ubiquitin release from the thioester in the presence of E3s, and (v) E3-facilitated transfer of ubiquitin to target proteins. Mutations of allosteric residues of E2s should disrupt only the last two functions of E2s without affecting others. To test this, we repeated our assay in Fig. 2 A with 2-fold higher concentrations of Apc2/11 and with longer incubations. Among the putative allosteric mutants, only UbcH5b I88A retained a significant fraction of its ubiquitin thioester in the presence of Apc2/11 (Fig. 7D). This finding suggests that the I88A mutation is most detrimental to Apc2/11-stimulated ubiquitin release from the UbcH5b–ubiquitin thioester. We thus focused on UbcH5b I88A in subsequent experiments.
UbcH5b I88A Is Deficient in Supporting APC/C-Mediated Ubiquitination of Cyclin B1. We next tested whether UbcH5b mutants that were not stimulated by E3s to release ubiquitin from the thioester were also defective in supporting the ubiquitination of cyclin B1, a physiological substrate of APC/C (31). As anticipated, the S94G and N77A mutations that disrupted the E3-binding and catalytic activities of UbcH5b, respectively, largely abolished the ability of UbcH5b to support the ubiquitination of cyclin B1 by APC/CCdh1 (Fig. 2C). UbcH5b I88A was ≈10-fold less efficient in supporting cyclin B1 ubiquitination (Fig. 2C, compare lanes 1 and 10). These results indicate that, in the presence of E3s, the ability of UbcH5b to release ubiquitin from its thioester correlates well with its ability to support ubiquitination of substrates. This finding further validates the use of the ubiquitin-release assay to evaluate the allosteric activation of UbcH5b by E3s.
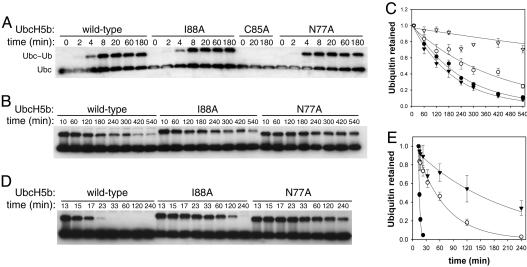
UbcH5b I88A Conjugates with and Releases Ubiquitin as a Thioester with Normal Kinetics. We next compared the rates of thioester formation of UbcH5b WT, N77A, and I88A. All three UbcH5b proteins formed ubiquitin thioesters with similar kinetics and efficiency (Fig. 3A). To rule out the possibility that I88 contributes directly to catalysis in a manner similar to N77, we measured the basal rate of ubiquitin release from the UbcH5b–ubiquitin thioester in the absence of E3s. As compared with UbcH5b WT, UbcH5b I88A released ubiquitin from its thioester with only slightly slower kinetics in the absence of an E3 (Fig. 3 B and C and Table 1). In contrast, UbcH5b N77A released ubiquitin at a much slower rate, consistent with its direct catalytic role in ubiquitin transfer.
Fig. 3.
Kinetics of ubiquitin charging and release from the ubiquitin thioester of UbcH5b WT, N77A, I88A, and S94G. (A) UbcH5b was incubated with E1, ubiquitin, and ATP for the indicated times, separated on nonreducing SDS/PAGE, and blotted with anti-UbcH5b. Bands for UbcH5b and UbcH5b–ubiquitin thioester are indicated. (B) After ubiquitin charging, NEM was added to the reactions to inactivate E1. Samples were taken at the indicated time points, separated on nonreducing SDS/PAGE, and blotted with anti-UbcH5b. (C) Quantitation of reactions in B. The average and standard errors are shown. Filled circles, WT; open circles, I88A; open triangles, N77A; filled triangles, S94G. (D) After ubiquitin charging, apyrase was added to deplete ATP and thus to inactivate E1-mediated ubiquitin charging of UbcH5b. CNOT4N was also added to the reactions after 3 min. Samples were taken at the indicated time points, separated on nonreducing SDS/PAGE, and blotted with anti-UbcH5b. (E) Quantitation of reactions in D. The average and standard errors are shown. Symbols are the same as in C.
Table 1. Kinetic and binding parameters of WT and mutant UbcH5b.
| UbcH5b | k, h-1* | kE3, h-1† | kE3/k‡ | Kd, mM§ |
|---|---|---|---|---|
| WT | 0.26 ± 0.01 | 22 ± 2 | 87 | 0.11 ± 0.05 |
| I37A | 0.26 ± 0.01 | 5.4 ± 0.4 | 20 | 0.11 ± 0.02 |
| N77A | 0.04 ± 0.01 | ND | ND | ND |
| I88A | 0.15 ± 0.02 | 0.97 ± 0.07 | 6.4 | 0.17 ± 0.03 |
| W93Y | 0.44 ± 0.03 | 8.9 ± 0.6 | 20 | 0.17 ± 0.06 |
| S94G | 0.31 ± 0.04 | 0.41 ± 0.16 | 1.3 | 1.2 ± 0.5 |
ND, not determined.
k denotes basal ubiquitin release rate of the UbcH5b—ubiquitin thioester in the absence of E3s. Errors indicate the SEM.
kE3 denotes CNOT4N-facilitated ubiquitin release rate of the UbcH5b—ubiquitin thioester. Errors indicate the SEM.
kE3/k is ratio of the two rates and represents the degree of UbcH5b activation by CNOT4N.
Kd is the dissociation constant for the binding between UbcH5b and CNOT4N. Errors indicate SD.
We next measured the rate of ubiquitin release from the UbcH5b thioester in the presence of CNOT4N (Fig. 3 D and E). This experiment allowed us to determine the degree of CNOT4N-dependent activation of UbcH5b, expressed as the ratio of E3-stimulated rate of ubiquitin release versus the basal ubiquitin-release rate (Table 1). CNOT4N increased the rate of ubiquitin release of UbcH5b WT by 87-fold. UbcH5b S94G, a mutant that did not bind to CNOT4N, was not activated by CNOT4N at all. UbcH5b I88A was activated only 6.4-fold by CNOT4N. Thus, as compared with UbcH5b WT, UbcH5b I88A was 13-fold less efficiently activated by CNOT4N, which correlated well with the fact that UbcH5b I88A was ≈10-fold less active in supporting APC/C-mediated ubiquitination of cyclin B1.
We then determined the crystal structures of UbcH5b WT, I88A, I37A, and S94G (Fig. 8 and Table 3, which are published as supporting information on the PNAS web site) and showed that the structures of UbcH5b mutants are nearly identical to that of UbcH5b WT. Thus, these mutations do not significantly perturb the structure of UbcH5b and are unlikely to alter the catalytic mechanisms and pathways of ubiquitin release of UbcH5b.
UbcH5b I88A Retains Its Binding to CNOT4N. To test whether UbcH5b I88A retained its binding to E3s, we measured the binding affinity between UbcH5b and CNOT4N using NMR. CNOT4N bound to UbcH5b WT and I88A with dissociation constants (Kd) of 110 μM and 170 μM, respectively, whereas CNOT4N bound to UbcH5b S94G extremely weakly (Kd > 1 mM) (Table 1; see also Fig. 9A, which is published as supporting information on the PNAS web site). Thus, UbcH5b I88A retained its ability to bind to CNOT4N. Binding of CNOT4N triggered chemical-shift perturbations of the same set of residues in UbcH5b WT and I88A, indicating that CNOT4N bound to UbcH5b WT and I88A at the same site (21).
Finally, we determined the rate of E3-facilitated ubiquitin release of UbcH5b–ubiquitin thioester as a function of the concentration of CNOT4N and estimated the binding affinity between CNOT4N and ubiquitin-charged UbcH5b (Fig. 9 B–E). CNOT4N bound to ubiquitin-charged UbcH5b WT or I88A with affinities between 100 and 200 μM, which was also similar to the affinity between CNOT4N and uncharged UbcH5b. This finding indicated that the I88A mutation did not significantly affect the binding affinity between CNOT4N and the ubiquitin-charged UbcH5b. Thus, the data presented so far indicate that the I88A mutation selectively disrupts the allosteric communication between the E3-binding and the active site of UbcH5b.
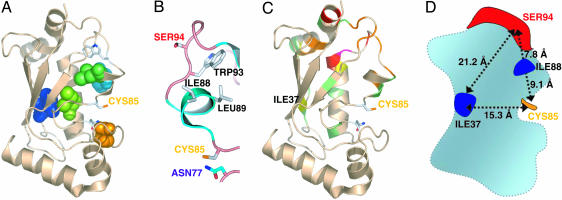
Other Allosteric Mutations That Disrupt Communication Between the E3-Binding and Active Sites of UbcH5b. Unlike other allosteric residues, I37 is distant from both the E3-binding site and the active site of UbcH5b (Fig. 4). We therefore also analyzed UbcH5b I37A in depth in a manner similar to UbcH5b I88A. Although UbcH5b I37A retained its binding to CNOT4N with a Kd of 107 μM, it was 4.4-fold less efficiently activated by CNOT4N to release ubiquitin from its thioester (Table 1). Thus, UbcH5b I37A was deficient in the allosteric communication between its E3-binding and active sites.
Fig. 4.
Allosteric determinants of UbcH5b. (A) UbcH5b residues involved in the long-range communication between the E3-binding and active sites of UbcH5b are shown in space-filling models. The E3-binding L1 and L2 loops, I37, and C85 are labeled. (B) A subset of allosteric UbcH5b residues is shown to illustrate the position of I88 relative to the E3-binding and active sites of UbcH5b. (C) The chemical-shift changes of UbcH5b upon binding to CNOT4N are mapped onto the structure of UbcH5b. The color schemes of the chemical-shift changes are as follows: red, >0.2 ppm; pink, 0.15–0.2 ppm; orange, 0.1–0.15 ppm; yellow, 0.075–0.1 ppm; green, 0.05–0.075 ppm. (D) Schematic drawing of UbcH5b to illustrate the relative positions of key residues. The inter-residue distances are between Cα atoms.
A shortcoming of SCA is its inability to examine pairwise coupling involving invariable residues in a given protein family (22). Because W93 is strictly conserved among E2s and because it is located near the E3-binding site, we mutated W93 to alanine, leucine, or tyrosine. UbcH5b W93A, W93L, and W93Y mutants were all defective in being activated by Apc2/11 or CNOT4N (Fig. 2 A and B). UbcH5b W93A did not bind to CNOT4N (Kd > 1 mM), whereas UbcH5b W93Y bound to CNOT4N almost as tightly as UbcH5b WT (Kd = 170 μM). Thus, UbcH5b W93Y reduces the allosteric activation of UbcH5b by 4.4-fold without significantly affecting the binding between UbcH5b and CNOT4N.
CNOT4N Binding Causes Chemical-Shift Perturbations at Sites Distal to the E3-Binding Site of UbcH5b. We also examined the possibility that E3 binding to UbcH5b might cause subtle conformational changes at I37 (Fig. 4). Even subtle conformational changes or changes in dynamics can lead to changes in the chemical shifts of the involved residues. CNOT4N binding indeed caused chemical-shift changes of residues that were distant from the E3-binding site of UbcH5b, including I37 and the 310 helix immediately C-terminal to the active-site cysteine (Fig. 4C). These results are consistent with the existence of long-range communication among the E3-binding site, the active site, and key allosteric residues of UbcH5b.
Mechanism of Allosteric Activation of UbcH5b by RING-Type E3s. Four of 11 cluster I residues tested in this study are allosteric residues, whereas none of the 3 cluster II residues and only 1 of 8 control residues (I106A) are allosteric. Therefore, a subset of cluster I residues mediates the allosteric communication between the E3-binding and active sites of UbcH5b. We do not know the exact mechanism by which the allosteric residues of UbcH5b propagate subtle structural changes at its E3-binding site to other parts of the protein, including the active site. On one hand, the spatial arrangement of some allosteric residues appears to suggest a linear pathway that consists of residues S94–W93–I88/L89–C85 for transducing the structural changes at the E3-binding site to the active site of UbcH5b (Fig. 4B). Furthermore, the W93A mutation disrupts E3 binding and presumably also allostery of UbcH5b whereas a more conservative W93Y mutation selectively impairs allostery, suggesting that the precise packing of certain side chains is required for allostery of UbcH5b. On the other hand, mutation of I37, a residue that is distant from both the E3-binding site and the active site of UbcH5b (Fig. 4), also significantly weakens the allosteric activation of UbcH5b by E3s. In addition, E3 binding to UbcH5b causes subtle long-range conformational changes at I37. These findings suggest that the allosteric communication between the E3-binding and active sites of UbcH5b relies on a complex structural unit formed by a large network of coevolving residues instead of a linear pathway consisting of a small set of residues.
An alternative explanation for our results is suggested by the non-RING RanBP2 system, a SUMO (small ubiquitin-like modifier) E3 ligase, which is believed to enhance sumoylation rates through its binding to SUMO (32). To examine a similar rationalization in our system, we tested and did not observe any binding between CNOT4N and free ubiquitin at mM concentrations (Fig. 10, which is published as supporting information on the PNAS web site). Although our results do not support a mechanism that employs strong affinity between CNOT4N and ubiquitin, we cannot rule out the possibility that CNOT4N, when bound to UbcH5b, directly contacts the ubiquitin molecule of the UbcH5b thioester, and thus facilitates ubiquitin release. Consistent with this notion, for several E2s, the 310 helix immediately C-terminal to their active site provides a major contacting surface for the thioester-bound ubiquitin (33, 34). Two allosteric residues identified in this study, I88 and L89, are located in the 310 helix. These residues might form direct contacts with ubiquitin in the E2–ubiquitin thioester. Thus, it is conceivable that the thioester-bound ubiquitin might also contribute to the allosteric communication between the E3-binding site and the active site of E2s.
Conclusion
RING-containing E3s do not themselves contain catalytic residues, but act as scaffolds to promote ubiquitin transfer from E2s to substrates. In addition to this role, they also stimulate the release of ubiquitin from their active-site cysteine in the absence of substrates. RING-type E3s bind to E2s at a site distal to the active site of E2s, and E3-binding does not significantly alter the structure of E2s. In the work reported here, we have identified mutations within UbcH5b that severely impair the allosteric communication between its E3-binding and active sites. These allosteric UbcH5b mutants retain their binding to E3s but are less efficiently stimulated by E3s to release ubiquitin from their active sites. Our findings provide mechanistic insight into the long-range communication between the E3-binding and active sites of E2s and further support the notion that RING-type E3s activate E2s in an allosteric fashion.
Supplementary Material
Acknowledgments
We thank R. Ranganathan and S. Lockless for assistance with SCA, J. Rizo and D. Araç for assistance with NMR experiments, B. Li for assistance with the APC/C ubiquitination assay, and M. Dorwart for assistance with densitometry. We are grateful to R. Boelens for providing the CNOT4N expression plasmid and the resonance assignment of UbcH5b. We also thank R. Ranganathan and J. Rizo for critical discussions. H.Y. is supported by the National Institutes of Health (Grant GM61542), the W. M. Keck Foundation, the March of Dimes Foundation, and the Leukemia and Lymphoma Society. J.D. is an Investigator at the Howard Hughes Medical Institute.
Author contributions: E.ö. and H.Y. designed research; E.ö. performed research; E.ö., H.Y., and J.D. analyzed data; and E.ö. and H.Y. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: SCA, statistical coupling analysis; APC/C, anaphase-promoting complex/cyclosome; E1, ubiquitin-activating enzyme; E2, ubiquitin-conjugating enzyme; E3, ubiquitin ligase.
Data deposition: Three-dimensional structure data have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2ESK, 2ESO, 2ESP, and 2ESQ).
References
- 1.Hershko, A. & Ciechanover, A. (1998) Annu. Rev. Biochem. 67, 425–479. [DOI] [PubMed] [Google Scholar]
- 2.Voges, D., Zwickl, P. & Baumeister, W. (1999) Annu. Rev. Biochem. 68, 1015–1068. [DOI] [PubMed] [Google Scholar]
- 3.Passmore, L. A. & Barford, D. (2004) Biochem. J. 379, 513–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pickart, C. M. & Eddins, M. J. (2004) Biochim. Biophys. Acta 1695, 55–72. [DOI] [PubMed] [Google Scholar]
- 5.Bloom, J., Amador, V., Bartolini, F., DeMartino, G. & Pagano, M. (2003) Cell 115, 71–82. [DOI] [PubMed] [Google Scholar]
- 6.Coulombe, P., Rodier, G., Bonneil, E., Thibault, P. & Meloche, S. (2004) Mol. Cell. Biol. 24, 6140–6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scheffner, M., Nuber, U. & Huibregtse, J. M. (1995) Nature 373, 81–83. [DOI] [PubMed] [Google Scholar]
- 8.Petroski, M. D. & Deshaies, R. J. (2005) Nat. Rev. Mol. Cell Biol. 6, 9–20. [DOI] [PubMed] [Google Scholar]
- 9.Zheng, N., Schulman, B. A., Song, L., Miller, J. J., Jeffrey, P. D., Wang, P., Chu, C., Koepp, D. M., Elledge, S. J., Pagano, M., et al. (2002) Nature 416, 703–709. [DOI] [PubMed] [Google Scholar]
- 10.Orlicky, S., Tang, X., Willems, A., Tyers, M. & Sicheri, F. (2003) Cell 112, 243–256. [DOI] [PubMed] [Google Scholar]
- 11.Wu, P. Y., Hanlon, M., Eddins, M., Tsui, C., Rogers, R. S., Jensen, J. P., Matunis, M. J., Weisman, A. M., Wolberger, C. P. & Pickart, C. M. (2003) EMBO J. 22, 5241–5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joazeiro, C. A., Wing, S. S., Huang, H., Leverson, J. D., Hunter, T. & Liu, Y. C. (1999) Science 286, 309–312. [DOI] [PubMed] [Google Scholar]
- 13.Seol, J. H., Feldman, R. M., Zachariae, W., Shevchenko, A., Correll, C. C., Lyapina, S., Chi, Y., Galova, M., Claypool, J., Sandmeyer, S., et al. (1999) Genes Dev. 13, 1614–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furukawa, M., Ohta, T. & Xiong, Y. (2002) J. Biol. Chem. 277, 15758–15765. [DOI] [PubMed] [Google Scholar]
- 15.Tang, Z., Li, B., Bharadwaj, R., Zhu, H., Ozkan, E., Hakala, K., Deisenhofer, J. & Yu, H. (2001) Mol. Biol. Cell 12, 3839–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leverson, J. D., Joazeiro, C. A., Page, A. M., Huang, H., Hieter, P. & Hunter, T. (2000) Mol. Biol. Cell 11, 2315–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gmachl, M., Gieffers, C., Podtelejnikov, A. V., Mann, M. & Peters, J. M. (2000) Proc. Natl. Acad. Sci. USA 97, 8973–8978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohta, T., Michel, J. J., Schottelius, A. J. & Xiong, Y. (1999) Mol. Cell 3, 535–541. [DOI] [PubMed] [Google Scholar]
- 19.Huang, L., Kinnucan, E., Wang, G., Beaudenon, S., Howley, P. M., Huibregtse, J. M. & Pavletich, N. P. (1999) Science 286, 1321–1326. [DOI] [PubMed] [Google Scholar]
- 20.Zheng, N., Wang, P., Jeffrey, P. D. & Pavletich, N. P. (2000) Cell 102, 533–539. [DOI] [PubMed] [Google Scholar]
- 21.Dominguez, C., Bonvin, A. M., Winkler, G. S., van Schaik, F. M., Timmers, H. T. & Boelens, R. (2004) Structure 12, 633–644. [DOI] [PubMed] [Google Scholar]
- 22.Lockless, S. W. & Ranganathan, R. (1999) Science 286, 295–299. [DOI] [PubMed] [Google Scholar]
- 23.Hatley, M. E., Lockless, S. W., Gibson, S. K., Gilman, A. G. & Ranganathan, R. (2003) Proc. Natl. Acad. Sci. USA 100, 14445–14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suel, G. M., Lockless, S. W., Wall, M. A. & Ranganathan, R. (2003) Nat. Struct. Biol. 10, 59–69. [DOI] [PubMed] [Google Scholar]
- 25.Shulman, A. I., Larson, C., Mangelsdorf, D. J. & Ranganathan, R. (2004) Cell 116, 417–429. [DOI] [PubMed] [Google Scholar]
- 26.Tang, Z. & Yu, H. (2004) Methods Mol. Biol. 281, 227–242. [DOI] [PubMed] [Google Scholar]
- 27.Albert, T. K., Hanzawa, H., Legtenberg, Y. I., de Ruwe, M. J., van den Heuvel, F. A., Collart, M. A., Boelens, R. & Timmers, H. T. (2002) EMBO J. 21, 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peterson, F. C., Penkert, R. R., Volkman, B. F. & Prehoda, K. E. (2004) Mol. Cell 13, 665–676. [DOI] [PubMed] [Google Scholar]
- 29.Yu, H., Peters, J. M., King, R. W., Page, A. M., Hieter, P. & Kirschner, M. W. (1998) Science 279, 1219–1222. [DOI] [PubMed] [Google Scholar]
- 30.Winkler, G. S., Albert, T. K., Dominguez, C., Legtenberg, Y. I., Boelens, R. & Timmers, H. T. (2004) J. Mol. Biol. 337, 157–165. [DOI] [PubMed] [Google Scholar]
- 31.Fang, G., Yu, H. & Kirschner, M. W. (1998) Mol. Cell 2, 163–171. [DOI] [PubMed] [Google Scholar]
- 32.Reverter, D. & Lima, C. D. (2005) Nature 435, 687–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamilton, K. S., Ellison, M. J., Barber, K. R., Williams, R. S., Huzil, J. T., McKenna, S., Ptak, C., Glover, M. & Shaw, G. S. (2001) Structure 9, 897–904. [DOI] [PubMed] [Google Scholar]
- 34.Hamilton, K. S., Ellison, M. J. & Shaw, G. S. (2000) J. Biomol. NMR 18, 319–327. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.