Abstract
Trigger factor (TF) is a ribosome-bound protein that combines catalysis of peptidyl-prolyl isomerization and chaperone-like activities in Escherichia coli. TF was shown to cooperate with the DnaK (Hsp70) chaperone machinery in the folding of newly synthesized proteins, and the double deletion of the corresponding genes (tig and dnaK) exhibited synthetic lethality. We used a detailed genetic approach to characterize various aspects of this functional cooperation in vivo. Surprisingly, we showed that under specific growth conditions, one can delete both dnaK and tig, indicating that bacterial survival can be maintained in the absence of these two major cytosolic chaperones. The strain lacking both DnaK and TF exhibits a very narrow temperature range of growth and a high level of aggregated proteins when compared to either of the single mutants. We found that, in the absence of DnaK, both the N-terminal ribosome-binding domain and the C-terminal domain of unknown function are essential for TF chaperone activity. In contrast, the central PPIase domain is dispensable. Taken together, our data indicate that under certain conditions, folding of newly synthesized proteins in E. coli is not totally dependent on an interaction with either TF and/or DnaK, and suggest that additional chaperones may be involved in this essential process.
Introduction
Folding of newly synthesized proteins in the cytosol of prokaryotes is assisted by three major molecular chaperones: GroEL, DnaK and trigger factor (TF) (Hartl & Hayer-Hartl, 2002). Recent studies showed that in Escherichia coli most de novosynthesized polypeptides may interact co-translationally with TF, nearly 15% co- and/or post-translationally with DnaK, and about 10% mainly post-translationally with GroEL (Ewalt et al, 1997; Deuerling et al, 1999; Houry et al, 1999; Teter et al, 1999).
TF (the tig gene product) is a 48 kDa protein, with both peptidyl-prolyl cis/trans isomerase activity (PPIase) and chaperone-like function (Crooke & Wickner, 1987; Hesterkamp et al, 1996). Approximately half the cellular TF in E. coli is bound to the ribosome, near the nascent peptide exit tunnel, while the other half is free in the cytosol (Hesterkamp et al, 1996; Kramer et al, 2002). TF associates co-translationally early with most nascent polypeptides of cytosolic and secreted proteins independently of proline residues (Valent et al, 1995; Hesterkamp et al, 1996; Patzelt et al, 2001). So far, there is no evidence for TF acting post-translationally. TF affinity for substrate is very low compared to most chaperones and is ATP independent, suggesting that rapid binding to and release from TF may be critical for elongating polypeptide chains (Maier et al, 2001).
TF comprises an N-terminal ribosome-binding domain, a central PPIase domain and a C-terminal domain of unknown function. Hesterkamp et al (1997) showed that the N-terminal 118 amino acids of TF are necessary for binding to the ribosome. Although the PPIase domain of TF is sufficient for the catalysis of peptidyl-prolyl isomerization, catalysis is greatly enhanced by the presence of the two other domains, suggesting that cooperation between the PPIase and the chaperone-like functions is critical (Scholz et al, 1997). Additional studies suggested that the N- and C-termini of TF may help to bind the substrate or modulate accessibility to the binding pocket (Scholz et al, 1997; Patzelt et al, 2001).
It has been shown that TF competes with DnaK in the chaperoning of newly synthesized proteins (Deuerling et al, 1999; Teter et al, 1999), because deletion of the tig gene increases the binding of DnaK to nascent polypeptides from 15% to about 40%. Deletion of the tig gene alone does not show any apparent growth defect, while the dnaK tig double mutant exhibits synthetic lethality, suggesting that at least one of these chaperones is required for bacterial survival (Deuerling et al, 1999; Teter et al, 1999).
Results and Discussion
Both DnaK and TF are dispensable for survival
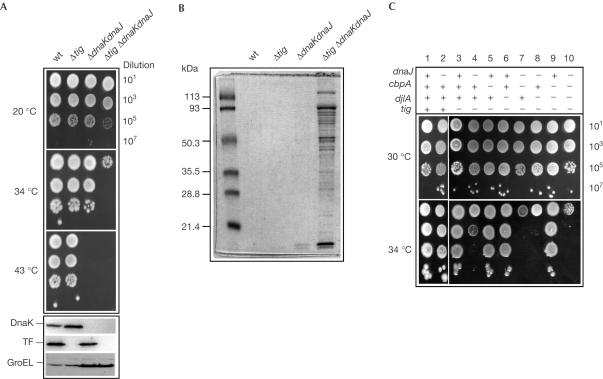
The TF and DnaK proteins cooperate in nascent chain chaperoning. In agreement with this, it has been reported that a Δtig ΔdnaK double mutant or a Δtig ΔdnaKdnaJ triple mutant exhibits synthetic lethality (Deuerling et al, 1999; Teter et al, 1999). We addressed the question of synthetic lethality further and found that both chaperone machines can be deleted, but at temperatures considerably lower than previously attempted. That is, the ΔdnaKdnaJ allele could be introduced at 20°C at the same frequency in either MC4100Δtig, MG1655Δtig or W3110Δtig bacteria, as in the corresponding isogenic wild-type derivatives. Furthermore, the observed dnaKdnaJ deletion frequencies were unchanged when the low-copy plasmid pDM38 carrying the wild-type dnaKdnaJ operon was present (data not shown). In addition, we also found a strain-backgroundspecific difference, since the triple Δtig ΔdnaKdnaJ mutant could also be obtained at 30°C in our MC4100 strain, but not in MG1655 or W3110. To confirm these results, the genetic experiments were repeated with two different tig deletion/replacement mutations made independently (data not shown). Immunoblot analysis confirmed the absence of both DnaK and TF. The growth defect was restored by low-copy expression of either DnaK and DnaJ or TF alone (Fig 1; data not shown).
Figure 1.
TF and DnaK are dispensable for E. coli survival. The genotype of the various bacterial strains is shown at the top. (A) Overnight cultures of wild-type (wt) MC4100 and its various mutant derivatives were serially diluted and spotted on LB agar plates at the indicated temperatures. Immunoblot analysis of endogenous steadystate expression levels of TF, DnaK and GroEL is shown at the bottom of the figure. (B) Aggregated proteins from either wild type or the indicated mutants grown for 4 h at 30°C were separated by 12% SDS–PAGE and stained with Coomassie blue. (C) Effect of various combinations of dnaJ, cbpA and djlA mutations on E. coli growth in the absence of TF, following overnight incubation at the indicated temperature. (+) indicates wild-type gene and (−) indicates null mutant.
The strains lacking TF, as well as DnaK and its co-chaperone partner DnaJ, were highly temperature sensitive (Ts) when compared to the corresponding isogenic strain that lacked only DnaK and DnaJ (Fig 1). Repeated passages of the Δtig ΔdnaKdnaJ mutant at higher temperature resulted in the rapid accumulation of unknown extragenic suppressors that improved growth considerably, but only up to 40°C (Fig 1A; data not shown).
To characterize further protein misfolding defects in the triple Δtig ΔdnaKdnaJ deletion strain, we analysed the level of total aggregated proteins using a method recently described by Tomoyasu et al (2001). We observed that the Δtig ΔdnaKdnaJ mutant exhibits a very high level of aggregated proteins, about ten times higher than that seen with the ΔdnaKdnaJ mutant alone (Fig 1B). These findings are consistent with the proposed cooperation between TF and DnaK in nascent polypeptide folding (Deuerling et al, 1999; Teter et al, 1999).
DnaJ specifically assists DnaK in the absence of TF
The Hsp70 family of molecular chaperones, such as DnaK, requires the action of co-chaperone members of the DnaJ protein family to both transfer specific substrates to DnaK and stimulate its ATPase activity. These co-chaperone functions are essential for the chaperone cycle of DnaK. DnaK can interact with three distinct, bona fide E. coli DnaJ co-chaperones, namely DnaJ, CbpA and DjlA (Genevaux et al, 2001). We asked which of the DnaJ co-chaperones is responsible for recruiting DnaK in the absence of TF. To answer this, we constructed multiple mutants lacking the tig gene and various combinations of dnaJ, cbpA and djlA, and tested them for growth at various temperatures (Fig 1C). We found that the single dnaJ mutation exhibited the most dramatic effect on E. coli growth (Ts above 30°C) in the absence of tig, whereas the single dnaJ mutation is only Ts at 43°C when tig is present. In addition, the H33Q substitution in the HPD signature motif of the DnaJ J-domain, known to abolish DnaJ/DnaK interaction without altering DnaJ chaperone properties, could not rescue bacterial growth. In a similar manner, DnaJ without co-expressed DnaK could not rescue the growth defect (data not shown). These results prove that DnaJ is the critical co-chaperone partner that efficiently recruits DnaK in the absence of TF. Whether DnaJ specifically recruits DnaK to nascent chains, as is the case with the yeast DnaJ homologue zuotin (Gautschi et al, 2002), or assists DnaK in preventing protein aggregation, or both, needs to be further examined.
TF domain requirements for chaperone function in vivo
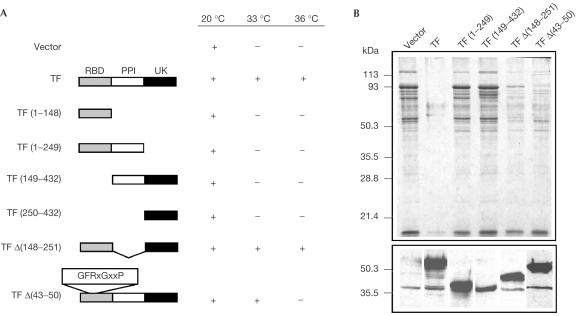
As stated earlier, the TF chaperone combines nascent chain chaperoning and peptidyl-prolyl isomerase activity. Since most of the functional analyses of TF have been performed in vitro, our tig dnaKdnaJ mutant represents a useful tool to study TF domain requirements in vivo. To do this, we constructed various tig mutants and tested their activity in our system. We found that neither the TF N-terminal ribosome-binding domain nor its C-terminal domain, alone or in combination with the PPIase domain, is sufficient to complement TF wild-type function in the absence of DnaK (Fig 2A). In addition, none of the tig deletion constructs prevents aggregation in the Δtig ΔdnaKdnaJ mutant (Fig 2B). Similar results were obtained with the various TF constructs expressed either from the tig native promoter or from a high-copy IPTG-inducible vector. Control immunoblot analysis showed comparable steadystate levels for TF expression between the low-copy plasmid-encoded TF under the control of its native promoter and the chromosomally encoded TF (data not shown).
Figure 2.
TF domain requirements. (A) Complementation of the Ts phenotype of MC4100Δtig ΔdnaKdnaJ on LB ampicillin agar plates by the various TF deletion constructs under the control of the tig native promoter. PPI, PPIase domain; RBD, ribosome-binding domain; UK, TF C-terminal domain of unknown function. (B) Complementation of protein aggregation in the same strain and by the same TF plasmid constructs. A western blot showing the steadystate expression levels of the various TF constructs used is shown at the bottom of the figure. The origin of the constructs is indicated at the top of the figure.
It is known that TF affinity for its substrates is dependent on its association with the ribosome (Patzelt et al, 2001). Since deletion of the TF N-terminal domain totally abolishes complementation in our assay, we asked whether mutations in the TF signature motif, shown to be critical for binding to the L23 protein located at the ribosome exit channel (Kramer et al, 2002), have a similar effect on TF chaperoning in vivo. Our results clearly show that the F44A and RK45–46AA substitutions, as well as the deletion of the TF GFRxGxxP signature motif, exert only a mild effect on TF function compared to the complete deletion of the N-terminal domain (Fig 2; data not shown). These results suggest that the N-terminal domain may provide an additional function(s) other than ribosome association, and may explain the interaction of the 144-amino-acid-long TF N-terminal domain with some nascent chains (Patzelt et al, 2001). It is also possible that deletion or substitutions in the TF signature motif may not completely abolish binding to the ribosome in vivo (Kramer et al, 2002).
TF binding to polypeptide substrates may occur in the PPIase domain (Patzelt et al, 2001). To test its importance in general chaperoning activity, we removed the entire central catalytic PPIase domain from TF, resulting in TF Δ(148–251), and tested it for complementation in our assay. Interestingly, TF without its PPIase domain could still efficiently complement the wild-type TF in the absence of DnaK, both for bacterial growth and protein folding, indicating that the TF PPIase domain is not essential for TF chaperone function in vivo (Fig 2). Nevertheless, slightly higher levels of aggregated proteins accumulated as compared to the wild-type TF (Fig 2B). A recent study showed that maturation of the Streptococcus pyogenes cysteine protease SpeB to its active form requires an intact TF PPIase domain (Lyon & Caparon, 2003). In this case, the need for an intact TF PPIase domain depended on the presence of one single proline residue in the protease prodomain. This result suggests that the PPIase domain of TF might be important for the biogenesis of some proteins.
Taken together, our data indicate that the bulk of the TF chaperone function is orchestrated by both the N- and C-terminal domains, but not by its PPIase domain.
TF overexpression leads to pre-OmpF aggregation
We observed that an approximately fourfold increase of TF is toxic, even in wild-type cells. This result is in agreement with the observation that high levels of TF affect the cellular division process (Guthrie & Wickner, 1990). In the absence of DnaK and DnaJ, this toxicity is further intensified (Fig 3A; data not shown).
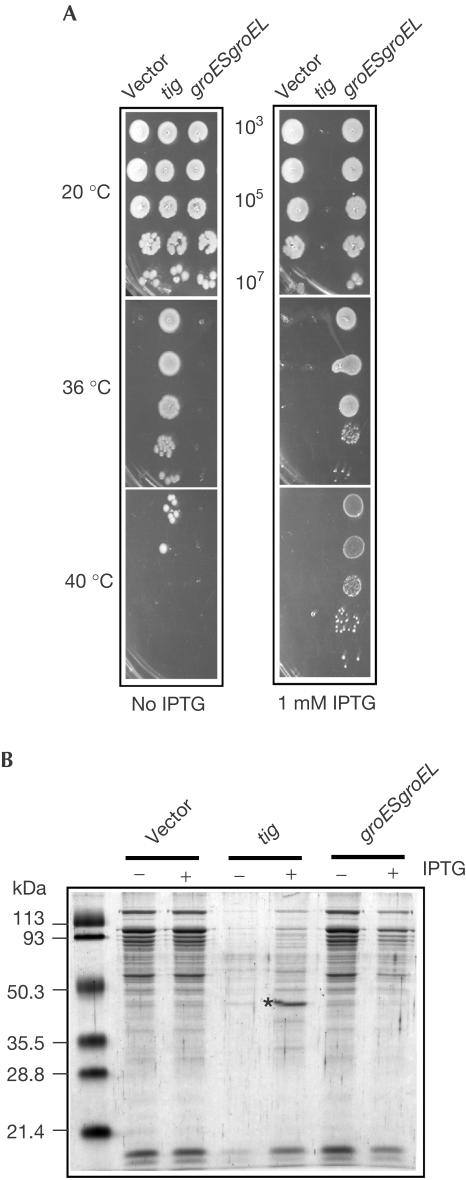
Figure 3.
Chaperonins GroES/GroEL partially compensate for the lack of TF and DnaK. Complementation of the Ts phenotype (A) and of protein aggregation (B) of the MC4100Δtig ΔdnaKdnaJ by p29SEN-based IPTG-inducible constructs carrying the genes indicated at the top of the figure. (−) indicates no IPTG, (+) indicates the presence of 1 mM IPTG and (*) indicates aggregated OmpF protein.
Interestingly, TF overexpression led to the rapid cytosolic accumulation of a major aggregated product of about 40 kDa (Fig 3B). Protein sequence analysis revealed that this specific aggregated protein is the unprocessed form of the major E. coli outer membrane porin OmpF. To see whether the cytosolic aggregation of OmpF has a role in TF toxicity, we overexpressed TF in an ompF-deleted strain and found that significantly higher levels of TF (eight- to tenfold) are required to detect bacterial toxicity (data not shown). Although toxicity is not completely abolished in the absence of OmpF, it is greatly reduced, suggesting that aggregation of OmpF is an important, contributory factor in TF-mediated toxicity.
Most secretory proteins of E. coli, such as OmpF, are exported through the SecA/SecB-dependent pathway, which is characterized by initial pre-protein binding to the SecB chaperone and subsequent transfer to the translocon-associated ATPase SecA (Muller et al, 2001). However, TF associates first with virtually all nascent chains of secretory and cytoplasmic proteins (Valent et al, 1995; Hesterkamp et al, 1996). Moreover, Lee & Bernstein (2002) recently showed that TF retards export of various outer membrane proteins, including OmpF. Consistent with our finding, in the presence of high levels of TF, newly synthesized OmpF precursor proteins may accumulate and aggregate in the cytosol. Presumably, TF prevents the timely interaction of pre-OmpF with the SecB/SecA machinery. In addition, the protective properties of DnaK and DnaJ, both in preventing protein aggregation and in directing protein secretion, are reflected in the enhanced TF toxicity in their absence. The fact that TF toxicity is not completely abolished in an ompF mutant suggests that additional polypeptide substrates may be deviated from their natural biogenesis pathways.
GroEL/GroES suppress the growth defect
Our finding that E. coli can survive in the absence of TF and DnaK suggested that other intracellular factors assist de novo protein folding. To address this possibility, we tested the effects of overexpressing the major heat shock chaperones GroEL/GroES (Hsp60/Hsp10), HtpG (Hsp90), IbpA/IbpB (small HSPs), ClpB (Hsp104), and proteases ClpP/ClpX, HslU/HslV, Lon, FtsH (Schlieker et al, 2002), as well as the other two Hsp70/Hsp40-like machines present in E. coli, HscA/HscB (Silberg et al, 2001) and HscC/DjlB/DjlC (Kluck et al, 2002). Among these, only the overproduction of GroEL/GroES could efficiently suppress the growth defect in the absence of TF and DnaK (Fig 3A; data not shown). Overexpression of GroEL/GroES (about six times above the cellular level at 30°C) could restore bacterial growth up to 40°C. However, although sufficient to allow bacterial growth at higher temperatures, the general protein folding defect was only partially restored by the overexpression of GroEL/GroES (Fig 3B). We also noted that the endogenous level of GroEL was not detectably changed in the Δtig ΔdnaKdnaJ mutant when compared to the ΔdnaKdnaJ mutant (Fig 1A).
In a separate study, we found that strains lacking both TF and DnaK could still tolerate the presence of a Ts allele of groEL (groEL44) at 20°C. Nevertheless, we observed some synergistic effects between the dnaKdnaJ deletion and the groEL44 and groES619 Ts alleles (data not shown).
Substrate sharing between DnaK and GroEL has previously been suggested (Gragerov et al, 1992; Tomoyasu et al, 2001), and a role for DnaK/DnaJ in substrate transfer to GroEL has been described (Langer et al, 1992; Teter et al, 1999). In addition, TF and DnaK are known to cooperate in the folding of newly synthesized proteins (Deuerling et al, 1999, 2003). Deuerling et al (1999) found that DnaK associates with two- to threefold more newly synthesized polypeptides in the absence of TF. In this case, a slightly higher level (about 1.3-fold) of newly synthesized proteins also associated with GroEL. The same laboratory recently showed that more than 300 protein species are common substrates for either TF or DnaK (Deuerling et al, 2003). In addition, a direct cooperation between TF and GroEL in protein degradation has been described for specific substrates (Kandror et al, 1995). The partially overlapping substrate specificity, the ability to interact with unfolded substrates, as well as the active role in protein degradation pathways shared between these E. coli chaperones reflect the synergy observed in our in vivo studies enumerated above.
In conclusion, our genetic studies have further detailed the functional cooperation in protein folding between three major cytosolic bacterial chaperones—GroEL, DnaK and TF—and showed that in spite of major protein folding defects, both DnaK and TF are dispensable for E. coli survival. Further studies are needed to identify additional cellular factors capable of assisting in the folding of newly synthesized proteins in the absence of TF and DnaK.
Materials and Methods
Bacterial strains and plasmids Genetic analyses were carried out in E. coli MG1655 (Blattner et al, 1997), W3110 (Bachmann, 1972) and MC4100 (Casadaban 1976). CG3212 is MC4100Δtig:Cmr and CG3214 is MC4100 ΔdnaKdnaJ:Kanr thr:Tn10 (Teter et al, 1999). The wild-type tig gene and its various deletion mutants were cloned on a low-copy pSC101ori vector under the control of either the tig native promoter or lac-inducible promoter, and on a high-copy CoREIori vector under the lac-inducible promoter (see supplementary information online).
Genetic analyses Construction of the Δtig ΔdnaKdnaJ triple mutants was performed as follows. First, the Δtig:Cmr allele was moved from strain CG3212 into either the MG1655, W3110 or MC4100 strain backgrounds by P1 transduction, selecting at 37°C on Luria–Bertani (LB) agar supplemented with chloramphenicol (15 μg/ml) and 5 × 10−3 M sodium citrate. Colonies were further purified on the same plates. P1 lysates were obtained after infection of the strain CG3214 carrying plasmid pDM38(dnaK+dnaJ+) and used to transduce subsequently the ΔdnaKdnaJ:Kanr allele into MG1655Δtig:Cmr, W3110Δtig:Cmr and MC4100Δtig:Cmr, as well as in the two isogenic parental strains. Selection was performed at 20°C on LB agar plates supplemented with 5 × 10−3 M sodium citrate and kanamycin (50 μg/ml). Candidate transductants were further purified under the same conditions. As a control, the same experiments were performed with MG1655Δtig:Cmr and MC4100Δtig:Cmr that also contained the plasmid pDM38(dnaK+dnaJ+).
Bacterial viability assays Bacteria were grown overnight at 20°C in LB broth (1% tryptone, 0.5% yeast extract, 0.5% NaCl, pH 7), electroporated with the appropriate plasmid constructs and plated at 20°C for 2 days on LB agar (1%) containing ampicillin (100 μg/ml). Fresh transformants were grown overnight at 20°C in LB supplemented with ampicillin, serially diluted, and spotted on LB agar plates containing ampicillin.
Detection of aggregated proteins Fresh colonies were grown overnight at 20°C in LB ampicillin, diluted 1:50 in the same medium and incubated for 4 h at 30°C. Aggregated proteins were further isolated as described by Tomoyasu et al (2001) and visualized by Coomassie blue staining after separation by 12% SDS–PAGE.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
ONLINE SUPPLEMENTARY DATA
Acknowledgments
We thank Carine Desplats for the ompF mutant, Joen Luirink for the anti-TF antibody and Severine Frutiger for protein sequencing. This work was supported by grant FN-31-065403 from the Swiss National Science Foundation and the Canton of Geneva.
References
- Bachmann BJ (1972) Pedigrees of some mutant strains of Escherichia coli K-12. Bacteriol Rev 36: 525–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner FR et al. (1997) The complete genome sequence of Escherichia coli K-12. Science 277: 1453–1474 [DOI] [PubMed] [Google Scholar]
- Casadaban MJ (1976) Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and mu. J Mol Biol 104: 541–555 [DOI] [PubMed] [Google Scholar]
- Crooke E, Wickner W (1987) Trigger factor: a soluble protein that folds pro-OmpA into a membrane-assembly-competent form. Proc Natl Acad Sci USA 84: 5216–5220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuerling E, Schulzespecking A, Tomoyasu T, Mogk A, Bukau B (1999) Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature 400: 693–696 [DOI] [PubMed] [Google Scholar]
- Deuerling E et al. (2003) Trigger factor and DnaK possess overlapping substrate pools and binding specificities. Mol Microbiol 47: 1317–1328 [DOI] [PubMed] [Google Scholar]
- Ewalt KL, Hendrick JP, Houry WA, Hartl FU (1997) In vivo observation of polypeptide flux through the bacterial chaperonin system. Cell 90: 491–500 [DOI] [PubMed] [Google Scholar]
- Gautschi M, Mun A, Ross S, Rospert S (2002) A functional chaperone triad on the yeast ribosome. Proc Natl Acad Sci USA 99: 4209–4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genevaux P, Schwager F, Georgopoulos C, Kelley WL (2001) The djlA gene acts synergistically with dnaJ in promoting Escherichia coli growth. J Bacteriol 183: 5747–5750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gragerov A, Nudler E, Komissarova N, Gaitanaris GA, Gottesman ME, Nikiforov V (1992) Cooperation of GroEL/GroES and DnaK/DnaJ heat shock proteins in preventing protein misfolding in Escherichia coli. Proc Natl Acad Sci USA 89: 10341–10344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie B, Wickner W (1990) Trigger factor depletion or overproduction causes defective cell division but does not block protein export. J Bacteriol 172: 5555–5562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M (2002) Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295: 1852–1858 [DOI] [PubMed] [Google Scholar]
- Hesterkamp T, Hauser S, Lutcke H, Bukau B (1996) Escherichia coli trigger factor is a prolyl-isomerase that associates with nascent polypeptide chains. Proc Natl Acad Sci USA 93: 4437–4441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesterkamp T, Deuerling E, Bukau B (1997) The amino-terminal 118 amino acids of Escherichia coli trigger factor constitute a domain that is necessary and sufficient for binding to ribosomes. J Biol Chem 272: 21865–21871 [DOI] [PubMed] [Google Scholar]
- Houry WA, Frishman D, Eckerskorn C, Lottspeich F, Hartl FU (1999) Identification of in vivo substrates of the chaperonin GroEL. Nature 402: 147–154 [DOI] [PubMed] [Google Scholar]
- Kandror O, Sherman M, Rhode M, Goldberg AL (1995) Trigger factor is involved in GroEL-dependent protein degradation in Escherichia coli and promotes binding of GroEL to unfolded proteins. EMBO J 14: 6021–6027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluck CJ et al. (2002) Structure–function analysis of HscC, the Escherichia coli member of a novel subfamily of specialized Hsp70 chaperones. J Biol Chem 277: 41060–41069 [DOI] [PubMed] [Google Scholar]
- Kramer GT et al. (2002) L23 protein functions as a chaperone docking site on the ribosome. Nature 419: 171–174 [DOI] [PubMed] [Google Scholar]
- Langer T, Lu C, Echols H, Flanagan J, Hayer MK, Hartl FU (1992) Successive action of DnaK, DnaJ and GroEL along the pathway of chaperone-mediated protein folding. Nature 356: 683–689 [DOI] [PubMed] [Google Scholar]
- Lee HC, Bernstein HD (2002) Trigger factor retards protein export in Escherichia coli. J Biol Chem 277: 43527–43535 [DOI] [PubMed] [Google Scholar]
- Lyon WR, Caparon MG (2003) Trigger factor-mediated prolyl isomerization influences maturation of the Streptococcus pyogenes cysteine protease. J Bacteriol 185: 3661–3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier R, Scholz C, Schmid FX (2001) Dynamic association of trigger factor with protein substrates. J Mol Biol 314: 1181–1190 [DOI] [PubMed] [Google Scholar]
- Muller M, Koch HG, Beck K, Schafer U (2001) Protein traffic in bacteria: multiple routes from the ribosome to and across the membrane. Prog Nucleic Acid Res Mol Biol 66: 107–157 [DOI] [PubMed] [Google Scholar]
- Patzelt HS et al. (2001) Binding specificity of Escherichia coli trigger factor. Proc Natl Acad Sci USA 98: 14244–14249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlieker C, Bukau B, Mogk A (2002) Prevention and reversion of protein aggregation by molecular chaperones in the Escherichia coli cytosol: implications for their applicability in biotechnology. J Biotechnol 96: 13–21 [DOI] [PubMed] [Google Scholar]
- Scholz C, Stoller G, Zarnt T, Fischer G, Schmid FX (1997) Cooperation of enzymatic and chaperone functions of trigger factor in the catalysis of protein folding. EMBO J 16: 54–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberg JJ, Hoff KG, Tapley TL, Vickery LE (2001) The Fe/S assembly protein IscU behaves as a substrate for the molecular chaperone Hsc66 from Escherichia coli. J Biol Chem 276: 1696–1700 [DOI] [PubMed] [Google Scholar]
- Teter SA et al. (1999) Polypeptide flux through bacterial Hsp70: DnaK cooperates with trigger factor in chaperoning nascent chains. Cell 97: 755–765 [DOI] [PubMed] [Google Scholar]
- Tomoyasu T, Mogk A, Langen H, Goloubinoff P, Bukau B (2001) Genetic dissection of the roles of chaperones and proteases in protein folding and degradation in the Escherichia coli cytosol. Mol Microbiol 40: 397–413 [DOI] [PubMed] [Google Scholar]
- Valent QA, Kendall DA, High S, Kusters R, Oudega B, Luirink J (1995) Early events in preprotein recognition in Escherichia coli: interaction of SRP and trigger factor with nascent polypeptides. EMBO J 14: 5494–5505 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ONLINE SUPPLEMENTARY DATA