Abstract
Enveloped viruses deliver their virions into the host cell by fusion with the cellular plasma or endosomal membrane, thus creating topological continuity between the cytosol and the inside of the viral envelope. Nonenveloped viruses are, by their very nature, denied this strategy and must employ alternative methods to breach their host cell's delimiting membrane. We show here that the compact icosahedral parvoviral virion gains entry by deploying a lipolytic enzyme, phospholipase A2 (PLA2), that is expressed at the N terminus of VP1, the minor coat protein. This region of VP1 is normally sequestered within the viral shell but is extruded during the entry process as a capsid-tethered domain. A single amino acid substitution in the active site of the VP1 PLA2 inactivates enzymatic activity and abrogates infectivity. We have used transencapsidation of a vector expressing green fluorescent protein to show that infection by this PLA2-defective mutant can be complemented by coinfection with wild-type or mutant full virions, provided they can express a functional PLA2. Even though wild-type empty capsids contain an active form of the enzyme, it is not externalized under physiological conditions, and such capsids are not able to complement the PLA2 mutant. Significantly, highly efficient rescue can be achieved by polyethyleneimine-induced endosome rupture or by coinfection with adenovirus as long as uptake of the two viruses is simultaneous and the adenovirus is capable of deploying pVI, a capsid protein with endosomolytic activity. Together, these results demonstrate a previously unrecognized enzymatic mechanism for nonenveloped virus penetration.
Keywords: endosomolysis, nonenveloped, parvovirus, phospholipase A2, viral entry
To gain entry into mammalian cells, viruses must deploy a mechanism for penetrating the plasma membrane of their host. Enveloped viruses achieve this through fusion of the viral membrane with that of the cell (1), but nonenveloped viruses must employ alternative strategies, and their methods for cell penetration are less well understood. Here we use the parvovirus minute virus of mice (MVM) as a model system for exploring bilayer transit by a nonenveloped virus. The MVM capsid is simple, assembling from 60 copies of two size variants of the capsid protein, VP1 and VP2, which have overlapping sequences but differ by the presence of an N-terminal extension in the minor partner VP1 (2). VP1 is dispensable for both assembly and DNA packaging, but it is absolutely required for infectivity (3). Its N terminus is sequestered within mature virions but can be exposed, along with the 3′ end of the genome, by limited heating in vitro (4), and cytoplasmic microinjection of antibodies directed against the VP1 N terminus blocks infection by the related canine parvovirus, suggesting that this peptide becomes exposed by the time the virus transits the host cell cytoplasm (5). The VP1-specific region contains nuclear localization signals (5, 6) and an active phospholipase A2 (PLA2) enzymatic core (7–9), both of which are essential for particle infectivity. Mutating catalytic residues within the PLA2 core of porcine parvovirus and a helper virus-dependent parvovirus, adeno-associated virus type 2, had no effect on the trafficking of virions into perinuclear compartments, apparently in late endosomes/lysosomes, demonstrating that initial binding and viral uptake were not affected by this mutation (7, 9). Confocal immunofluorescence microscopy of canine parvovirus virions during entry demonstrated that VP1 may be exposed in these perinuclear compartments (10) as was also shown for MVM in the presence of the proteosome inhibitor MG132 (11). However, it is difficult to assess whether this exposure of VP1 resulted from an entry-related process or from partial degradation of the virion.
Together these results demonstrate that PLA2 activity is required for a step after perinuclear accumulation but before early gene expression, suggesting endosomal escape, virion trafficking after vesicle release, or eicosanoid-mediated intracellular signaling (12) as possible functions for parvoviral PLA2. As of yet, there is no direct evidence distinguishing among these various possibilities, but several reports have suggested parvoviral PLA2 functions in endosomal escape. Although low molecular weight dextrans were released from endosomes during canine parvovirus infection (10), coendocytosed α-sarcin was not significantly released into the cytoplasm (13), indicating that extensive endosomal lysis does not occur during infection. Here we examine the complementation of parvoviral PLA2 function and explore whether two very different endosomolytic processes can substitute for its essential enzymatic function.
Materials and Methods
Cell Lines. A9 cells are A9 ouabr11, a ouabain-resistant derivative of the original HPRT- mouse fibroblast line A9 (14). A9-CAR cells, provided by Paul Freimuth (Brookhaven National Laboratory, Upton, NY), are a stable derivative of the original A9 cell line expressing CAR, the coxsackievirus–adenovirus receptor (15).
Virion and Vector Production and Entry Assays. Viral stocks were generated by transfection of A9 monolayers by using ExGen 500 transfection reagent (Fermentas, Hanover, MD). Virus was harvested after 72 h, extracted, and purified on iodixanol step gradients, as described in ref. 16. GFP vectors were packaged as described in ref. 17 and purified as described above. Ad5wt-GFP and Ad5-ts1 stocks were provided by Chris Weithoff and Glen Nemerow (The Scripps Research Institute, La Jolla, CA). Ad5wt-GFP was grown on 293 cells and purified as described by Weithoff et al. (18).
Semiconfluent monolayers of A9 or A9-CAR cells seeded on spot slides were incubated at 37°C for 18–20 h before being transfected with plasmid DNA for expansion assays or being infected with predetermined genome equivalents of virions or vector. Infectivity and complementation assays were performed as described in ref. 16 and examined by epifluorescence microscopy. Complementation indices were calculated as the ratio of cells expressing the “helped” virus marker in the presence versus the absence of helper.
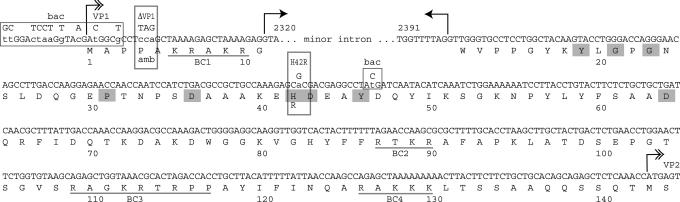
Baculovirus Production of Capsids for Phospholipase Analysis. Baculoviruses expressing VP1 and VP2 in the correct stoichiometric ratios were generated in the Bac-to-Bac baculovirus expression system (Invitrogen). To obtain optimum expression of VP2 relative to VP1, a leaky scanning strategy was exploited (19), as shown in Fig. 1, except that the VP1-specific cDNA version that eliminates the minor intron was used as a substrate. The first block of mutations (upstream of exon 1) mutates the MVM VP1 initiation sequence to the adeno-associated virus VP2 “Kozak” sequence, changing the start codon from ATG to ACG (19). The T → C mutation at nucleotide 2506 disrupts a potential translation initiation site downstream of the VP1 start codon. Baculovirus-expressed MVM capsids were produced in Sf9 insect cells, purified on iodixanol step gradients, and analyzed on discontinuous SDS/polyacrylamide gels (4). Secretory PLA2 assays were performed with a commercially available kit, according to the manufacturer's instructions (Cayman Chemical, Ann Arbor, MI). For high-pH disruption, 2–4 μg of capsids was exposed to 0.2 M NaOH for 5 min at room temperature and then neutralized with HCl, diluted into assay buffer, and monitored for neutralization before analysis.
Fig. 1.
The MVM genome sequence encoding the VP1-specific region. Double-hashed arrows depict translation starts for VP1 and VP2. Small arrows depict the minor splice donor and acceptor that give rise to the VP1 transcript, and most of the minor intron sequence is omitted. Mutations are shown in uppercase with the wild-type sequence in lowercase. Critical residues for PLA2 function are shaded in gray, and the basic clusters (BC1–BC4), with potential nuclear localization activity, are underlined. Nucleotides altered in the construction of the amb termination codon in ΔVP1 and the change in H42R are boxed. Boxes marked “bac” denote mutations introduced into the baculovirus construct described in Materials and Methods.
Results
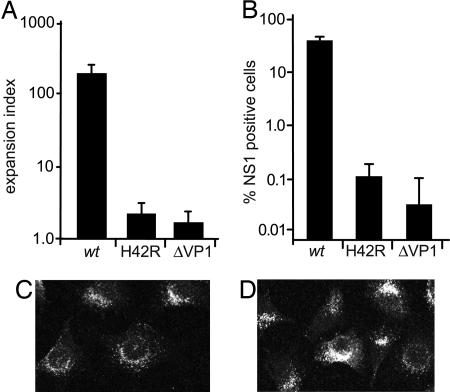
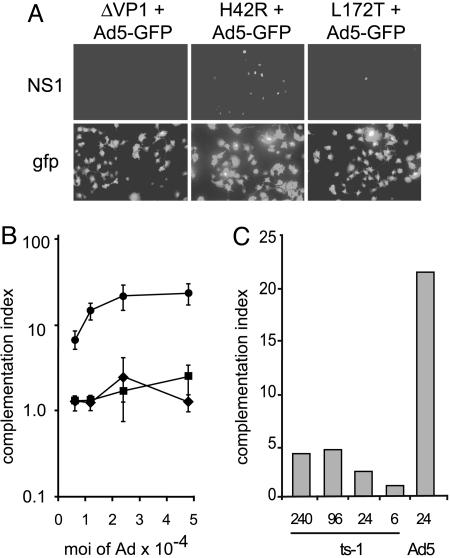
The H42R Substitution Abrogates MVM Infectivity and Disrupts PLA2 Activity. The substitution mutation within the PLA2 active site (H42R), and the premature termination mutation that abrogates VP1 expression (ΔVP1), shown in Fig. 1, were engineered into the MVMp infectious clone. As shown in Fig. 2A, neither mutant was capable of sustaining multiple rounds of infection after transfection, measured in a 50-h expansion assay (16). However, the VP1-specific region functions during the establishment of infection, allowing these mutants to be recovered as viruses after a single round of replication initiated by transfection. The infection efficiency in A9 cells for purified H42R or ΔVP1 virions was reduced from wild-type levels by 300- or 1,000-fold, respectively (Fig. 2B). Further analysis of mutant virion entry demonstrated that they enter cells and traffic to perinuclear compartments indistinguishably from wild type (Fig. 2 C and D), as has been reported for other parvoviral PLA2 mutants (7, 9). The virions detected were predominantly internalized because treatment of cells with neuraminidase 2 h after uptake did not affect staining, and uptake was receptor-dependent because neuraminidase pretreatment eliminated the signal (data not shown).
Fig. 2.
Mutation of the VP1 N terminus abrogates virus viability. (A) A9 cells transfected with wild-type or mutant infectious clones were stained for the nonstructural protein NS1 at 50 h, because this is the earliest detectable viral gene product, and expansion indices were calculated as described in ref. 16. A minimum of 1,000 cells were counted for each assay, which was performed in triplicate. (B) A9 cells were infected with purified wild-type or mutant virions at 3,000 genomes per cell, stained for NS1 at 24 h, and counted as described above. (C and D) A9 cells infected with 500,000 wild-type (C) or H42R (D) virions per cell, fixed 8 h after infection, and stained with a monoclonal antibody specific for intact capsids. Images (1-μm sections) were acquired on a Zeiss LSM 510 laser scanning confocal microscope.
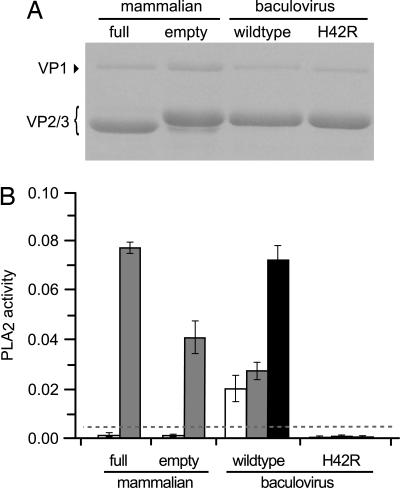
For analysis of their PLA2 activity, large-scale preparations of wild-type full virions and empty capsids were produced in mammalian cell culture and extensively purified. Obtaining sufficient yields of mutant capsids for this type of analysis, however, required development of a baculovirus expression system by using the differential VP1/VP2 translation initiation strategy described in Fig. 1. On iodixanol gradients, the profile of baculovirus-expressed capsids resembled that of authentic empty capsids, and analysis of purified capsids by SDS/PAGE showed that VP1 and VP2 were incorporated into particles with the correct stoichiometry (Fig. 3A). PLA2 activity associated with these particles was determined, as shown in Fig. 3B, in a colorimetric assay that measures hydrolysis of the sn-2-thioester bond of diheptanoyl 1,2-dithiophosphatidylcholine. No significant PLA2 activity could be detected for untreated full virions or empty capsids derived from mammalian cells, indicating that VP1 remained strictly internalized in these particles. When they were exposed briefly to 0.2 M NaOH, a dramatic increase in PLA2 activity was observed for both types of particle, indicating that the VP1 N terminus was exposed, but not denatured, by the high-pH treatment.
Fig. 3.
PLA2 activity associated with viral particles. (A) Purified authentic (mammalian) virions or capsids or baculovirus-expressed capsids were analyzed by SDS/PAGE and stained with Coomassie brilliant blue (4). (B) PLA2 activity, expressed as μmol of substrate hydrolyzed per min/ml of sample, of 2 μg of the capsids preparations in A above, before (open bars) or after (gray bars) exposure to 0.2 M NaOH. Black bars represent PLA2 activities determined for 4 μg of wild-type or H42R capsids treated with 0.2 M NaOH. Horizontal dashed line indicates the maximum standard error for the background determination.
Interestingly, when baculovirus-derived capsids containing wild-type VP1 were assayed for PLA2 activity, a significant level of activity was observed in the absence of NaOH treatment (Fig. 3B). After NaOH denaturation, PLA2 activity increased by 30% more than untreated levels. Together these results indicate that baculovirus-expressed capsids may not be perfectly assembled in insect cells, such that some VP1 is exposed on the particle surface. Analysis of the baculovirus-expressed H42R capsids showed that this mutation essentially abrogated PLA2 activity with or without NaOH treatment. No significant activity was found for up to 4 μg of the PLA2 active site mutant, whereas the activity of wild-type capsids increased proportionally to the amount of added capsid (Fig. 3B), indicating that the relatively conservative H42R mutation reduces PLA2 activity by more than 20-fold compared with that of the wild type.
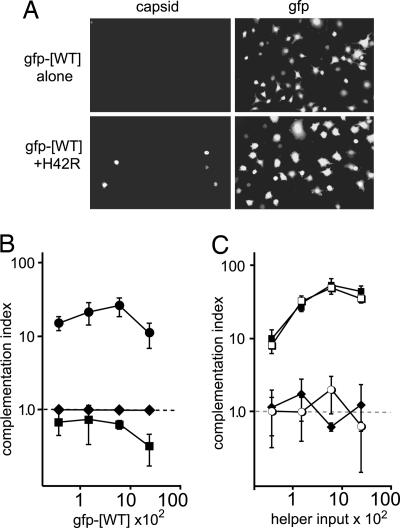
PLA2 Mutant Virions Can Be Complemented by Wild-Type Vector in Trans. To determine whether mutant virions, potentially trapped in perinuclear endosomes, could be helped by viruses carrying wild-type or mutant VP1 molecules, we developed a complementation assay that allows differential scoring of two separate viruses as they infect the same cell. For this assay we used an MVM vector in which the viral capsid genes had been replaced with GFP, and which can be packaged into a capsid of choice by cotransfection with the appropriate capsid-expressing helper plasmid (17). When titrated between 200 and 20,000 genomes per cell, GFP vectors packaged into wild-type capsids (gfp-[WT]) infect A9 cells as efficiently as wild-type MVMp when measured by both NS1 and GFP expression (Fig. 4A and data not shown). This vector does not contain a functional capsid gene; thus, no capsid expression can be detected in gfp-[WT]-infected cells (Fig. 4A), allowing coinfected cells to be monitored independently for vector or virion infection by GFP or capsid expression, respectively.
Fig. 4.
Wild-type vector rescues the mutant lacking PLA2 activity. (A) Coinfected cells were examined for GFP expression or by capsid staining at 30 h after infection. The micrographs show representative fields after infection with 6,000 genomes per cell of gfp-[WT] helper vector alone, or coinfection with 3,000 genomes per cell of mutant virions. (B) Complementation indices are shown as a function of helper vector multiplicity for coinfections with 3,000 genomes per cell of ΔVP1 (♦), L172T (▪), or H42R (•). In these assays, each mutant infected an average of 0.008%, 0.3%, or 0.05% of cells, respectively, in the absence of helper. Up to 12,000 cells were analyzed for each data point, and the experiments were performed in triplicate. (C) Complementation indices are shown for cells coinfected with 3,000 genomes per cell of gfp-[H42R] and increasing amounts of ΔVP1 (♦), L172T (▪), or wild-type (□) virions, or purified MVMp empty capsids (○), matched to virions by Coomassie blue-stained gel analysis. In the absence of helper virus, gfp-[H42R] infected 0.01% of cells.
In coinfections with mutant virions and gfp-[WT] vector, no complementation of ΔVP1 was detected at any vector input level (Fig. 4B), indicating that at least one function of VP1 is required in cis. To explore this result further, we asked whether gfp-[WT] could complement a virus carrying a mutation within the body of VP2, at residue 172. The L172T mutation has been shown to inactivate virions for infection in MVMi (16), and subsequent in vitro analysis of L172T in the MVMp background shows it to be temperature sensitive, due to premature extrusion of both its VP1 N terminus and genome after VP2 to VP3 cleavage (26). As expected, no complementation of L172T was observed, indicating that this mutant, like ΔVP1, is cis-defective. However, when cells were coinfected with H42R and gfp-[WT], 25-fold complementation of H42R was observed at an input multiplicity of 6,000 helper vector genomes per cell (Fig. 4B). Not surprisingly, at the highest dose tested there was an ≈2-fold competition for infection, as was also observed in coinfections with L172T and gfp-[WT] (Fig. 4B). In accord with this finding, increasing the effective dose of H42R and gfp-[WT] by 10-fold while maintaining the optimal ratio of virion to helper vector had no effect on complementation (data not shown).
Complementation of H42R Is VP1-Specific and Requires a Packaged Virion. To both confirm the above result and investigate PLA2 complementation further, we performed the converse experiment, the results of which are presented in Fig. 4C. Here, GFP vector genomes were packaged into capsids mutated in the PLA2 active site (gfp-[H42R]). As expected, this vector was unable to successfully establish infection on its own, and was, in fact, about 8-fold less infectious than H42R virions and >2,000-fold less infectious than wild type (Fig. 4C and data not shown). In accord with the previous experiments, wild-type virions readily rescued gfp-[H42R] infectivity, increasing the fraction of cells expressing GFP by up to 50-fold. As shown in Fig. 4C, complementation of PLA2 activity by coinfection was VP1-specific, because ΔVP1 virions had no effect of gfp-[H42R] infectivity, and required a packaged genome, because empty capsids also did not complement gfp-[H42R]. In contrast to its failure to be complemented by gfp-[WT], when L172T virus was used to complement gfp-[H42R] it proved to be as efficient as wild type at rescuing the PLA2-knockout vector. To analyze whether complementation of gfp-[H42R] coincided with the establishment of infection by the helper virus, we analyzed the results from Fig. 4C at the single-cell level. Interestingly, at a wild-type input of 6,000 genomes per cell, 95% (±2%) of GFP-expressing cells also stained positive for capsid, whereas a strikingly different pattern was observed in L172T and gfp-[H42R] coinfections, where this value dropped to 21% (±3%). Thus, L172T complementation of H42R reveals the complex phenotype of L172T, which can be inactivated by “untethering” of the VP1 N terminus or by loss of genome integrity (26), each of which could occur before or after the penetration step.
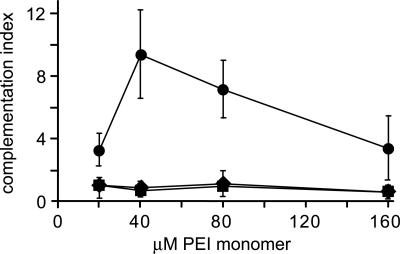
Endosome Lysis Is Sufficient to Rescue Entry of H42R. On the basis of the observation that PLA2 activity could be complemented in trans, we wished to determine whether PLA2 affected particle entry by directly affecting endosome integrity. Recently, it has become apparent that some synthetic polymer transfection reagents work by inducing the lysis of endocytic vesicles containing polymer/DNA complexes. For instance, polyethyleneimine (PEI) is believed to act as a “proton sponge,” inducing vesicle lysis by endosomal buffering that results in osmotic rupture (20). We found that PEI had little effect on the infectivity of ΔVP1 or L172T at any nontoxic concentration; however, coexposure to 40 μM PEI increased the infectivity of H42R virions more than 10-fold (Fig. 5), suggesting that these mutant virions are indeed trapped in endosomes.
Fig. 5.
Polymer-induced lysis of endosomes rescues PLA2 mutant virions. Various amounts of PEI (25 kDa; Sigma), prepared as a 10 mM monomer aqueous stock solution, were complexed with a GFP-expression plasmid to monitor PEI activity. These complexes were mixed with mutant virions and incubated with A9 cells at 24,000 genomes per cell for 4 h, after which the medium was changed. The cells were fixed 40 h later, stained for NS1 and capsid, and scored for both GFP and viral gene expression. Each set of infected/transfected cultures expressed GFP with equivalent efficiency. For example, 8–13% of cells were positive for GFP at 40 μM PEI. Complementation indices were determined for ΔVP1 (♦), L172T (▪), or H42R (•). The data represent two separate experiments in which up to 30,000 DAPI-positive cells were analyzed for each data point. In the absence of PEI, H42R, L172T, and ΔVP1 infection rates were 0.09%, 0.07%, and 0.003%, respectively.
We reasoned that the endosomolytic activity of adenovirus might also release incoming mutant PLA2 MVM virions if they were trapped in endosomes during entry. This approach required cells infectable by both MVM and adenovirus, and we found that A9 cell lines expressing CAR were efficiently infected with both wild-type MVM and Ad5-GFP, an E1-, E3-deleted adenovirus 5 vector expressing GFP (Fig. 6A and data not shown). Striking rescue of the H42R MVM mutant was observed in coinfections with the Ad5 vector, giving ≈25% MVM capsid-positive cells at the maximal Ad5-GFP input, whereas no significant rescue of ΔVP1 or L172T was observed over the same range of Ad5 vector input (Fig. 6B).
Fig. 6.
Adenovirus coinfection rescues PLA2 mutant virions. (A) A9 cells were coinfected with 24,000 genomes per cell of each MVM mutant and an equal amount of Ad5-GFP, fixed, and stained 24 h after infection. (Upper) Representative fields of cells stained for MVM NS1. (Lower) GFP expression from the adenovirus. (B) Complementation indices were determined for ΔVP1 (♦), L172T (▪), or H42R (•), at five different input multiplicities of helper. In these assays, each mutant infected an average of 0.007%, 1.2%, or 0.8% of cells, respectively, in the absence of adenovirus. Three separate experiments were performed and up to 14,000 cells were analyzed for each data point. (C) Complementation indices determined for MVM H42R upon coinfection at 24,000 genomes per cell with the indicated numbers (×10-3) of Ad5 ts1 or Ad5-GFP wild type particles.
No H42R complementation, or GFP expression, was observed in A9 cells lacking CAR (data not shown), indicating that it required binding of Ad5 to the cell surface. To ask whether just the interaction of adenovirus with CAR was sufficient to rescue H42R infection, we used the mutant adenovirus ts1, which efficiently binds to CAR and undergoes internalization but is defective for the disruption of endosomes (18). In contrast with the results with the entry-competent vector, no significant augmentation of H42R infectivity was observed for ts1 coinfection, even at a 10-fold higher multiplicity than the maximal input of Ad5-GFP (Fig. 6C).
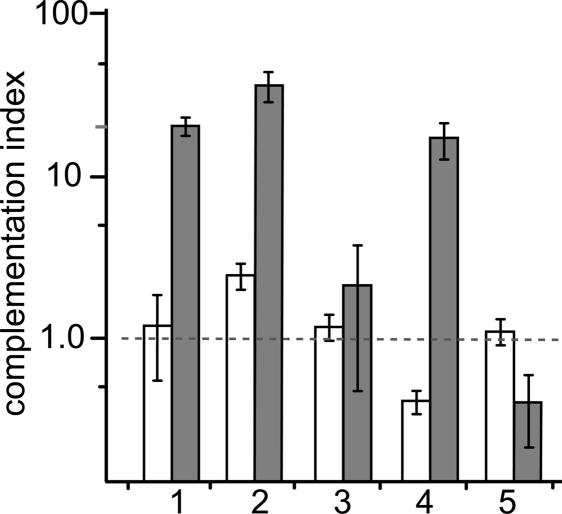
These experiments suggested that adenovirus-induced endosomal lysis affects the rescue of H42R, although it was possible that a downstream event, such as adenoviral early gene expression, might be responsible. Although we considered this possibility unlikely, because the Ad5-GFP vector is deleted for E1, which is necessary for adenovirus early gene expression in most cells, we asked whether complementation required the adenovirus to enter the cell in the same endosome as the H42R virion. As shown in bar 1 of Fig. 7, simultaneous coinfection resulted, as before, in a dramatic rescue of the H42R virus, whereas wild-type infection was unaffected. This result was not changed by the addition of anti-MVM antibody immediately after the infectious pulse (Fig. 7, bar 2), indicating that the MVM virions that will be complemented have entered the endosomal compartment during the pulse, as discussed below.
Fig. 7.
Ad5 complementation of H42R requires simultaneous coinfection. Complementation indices were determined for coinfections of MVM wild type (open bars) or H42R mutant (gray bars) with Ad5-GFP, each at 24,000 genomes per cell, and timed as follows: 1, simultaneous infection for 2 h; 2, as in 1, followed by medium containing 1% rabbit anti-MVM neutralizing antiserum for 2 h; 3, Ad5-GFP for 2 h, then H42R for 2 h; 4, H42R for 2 h, then Ad5-GFP for 2 h; and 5, as in 4, with anti-MVM antibody added with the adenovirus.
Most significantly, although the addition of Ad5-GFP 2 h before wild-type infection had no observable effect, when this infection order was repeated for H42R, no rescue was observed (Fig. 7, bar 3), strongly suggesting that both viruses must enter the cell at the same time for complementation to occur. Interestingly, if the timing of virus addition was reversed, with H42R preceding Ad5-GFP, rescue was similar to that observed for simultaneous infection (Fig. 7, bar 4). However, because some input MVM particles recycle to the surface still attached to their receptor (data not shown), we considered it possible that, during the second infection period, H42R particles might reappear at the cell surface and reenter simultaneously with the adenovirus. To test this possibility, after infection with H42R, we added anti-MVM antibody along with Ad5-GFP. In contrast to the previous result, this treatment completely abrogated any complementation by Ad5-GFP (Fig. 7, bar 5), suggesting that a significant fraction of the H42R virions had either remained at the surface, or recycled to it, during subsequent exposure to the adenovirus. Importantly, similar antibody treatment had no effect on the establishment of infection by wild type (Fig. 7, bar 5) or by the adenovirus (data not shown).
Discussion
The mechanisms by which nonenveloped viruses cross the delimiting membrane are less well understood than those used by enveloped viruses. Strategies that have been described to date involve deployment of hydrophobic proteins or capsid protein domains that destabilize the host membrane, either forming pores through which the genome accesses the cytosol (21) or leading to the disruption of the vesicle membrane, wholesale lysis, and release of the endosomal contents into the cytosol. Adenovirus capsid protein pVI and reovirus capsid protein μ1 have recently been identified as the latter type of endosome-disrupting molecules (18, 22). In this study, we establish that the parvoviral virion accomplishes this crucial step somewhat differently by using the lipolytic activity of a PLA2 enzyme embedded in an extension of its minor coat protein. Despite being tethered to the capsid shell, this enzyme can operate in trans to rescue infection by virions defective in PLA2 activity, presumably only if they occupy the same endosome.
As many as 19 distinct mammalian members of the PLA2 superfamily have been described (12), and a large subgroup of these, to which the parvoviral enzymes belong, is characterized by using a catalytic histidine residue. Although these enzymes can participate in membrane remodeling (23), they are predominantly involved in intracellular or intercellular signaling. PLA2-triggered signaling events are quite specific and feed into both the cyclooxygenase-1 and cyclooxygenase-2 pathways, regulating immune function, inflammation, and smooth muscle contraction through the generation of arachidonic acid and its metabolites (24). The low level of complementation observed in our assays and the lack of detectable rescue in previous experiments (7, 9) suggests that this type of signaling is not the primary function of the parvoviral PLA2s. The complete rescue of the MVM H42R mutant life cycle that results from PEI exposure or adenovirus coinfection indicates that a parvoviral PLA2-specific, arachidonic acid-mediated signaling event is not required for infection. However, it is possible that the incoming adenoviral capsid or early adenoviral gene expression compensates for the required signaling event. The former event is unlikely because Ad5 ts1 capsids did not significantly complement H42R, indicating that capsid interactions up to and including vesicle trafficking are not sufficient for this rescue. Adenoviral gene expression itself is also unlikely because we found that that cells actively infected by Ad5-GFP do not rescue PLA2-knockout virions unless the adenovirus is added simultaneously with MVM. Taken together, these results suggest that cooccupation of an individual endosome is required for complementation of MVM by adenovirus, that the PLA2 mutant is specifically defective in escaping from a membrane-bound compartment, and that, although mechanistically quite different, the parvoviral PLA2 and the adenovirus pVI protein perform an equivalent task during entry.
The parvovirus particle is a dynamic entity, undergoing multiple conformational changes during its life cycle, including, importantly, the externalization of the VP1 N terminus during the entry process. Empty capsids contain a functional VP1 PLA2 but are not primed to extrude the VP1 N terminus, as measured by in vitro assay (5). Significantly, we found that empty capsids did not rescue the mutant lacking PLA2 activity, suggesting that the VP1 N terminus must be deployed by extrusion for the PLA2 to exercise its critical function in entry. The coinfection experiments reported here demonstrate that the MVM PLA2 activity is capable of functioning in trans, whereas the ΔVP1 mutant was not complemented in any assay, indicating that at least one essential property of VP1 must function in cis. The VP1-specific N terminus contains four conserved basic clusters (BC1–BC4 in Fig. 1), two of which have been found to function as nuclear localization signals (6, 25). Because the VP1 N terminus is exposed in the cytosol during viral entry (5), it is very possible that it is required to transport the incoming virion, as an otherwise intact particle, into the nucleus (6). Because a nuclear localization signal must necessarily function in cis, it is reasonable to assume that the lack of complementation observed for ΔVP1 is due, at least in part, to the absence of these motifs.
Acknowledgments
We gratefully acknowledge the gifts of A9-CAR cell lines from Dr. Paul Freimuth (Brookhaven National Laboratories, Upton, NY) and of Ad5wt-GFP and ts1 from Drs. Chris Weithoff and Glen Nemerow (The Scripps Research Institute, La Jolla, CA), who generously shared their data on Ad pVI endosomolysis before publication. We also thank Dr. Susan Cotmore for critical reading of the manuscript, other members of the laboratory who contributed to these studies, and Dr. Walther Mothes for numerous helpful suggestions and discussions. DNA sequencing and synthesis of oligonucleotides were performed by the W. M. Keck Foundation Biotechnology Resource Laboratory at Yale University. G.A.F. was supported, in part, by National Institutes of Health Training Grant T 32 AI07640. This work was supported by Public Health Service Grant CA29303 from the National Cancer Institute.
Author contributions: G.A.F., L.-g.Z., and P.T. designed research; G.A.F. and L.-g.Z. performed research; G.A.F., L.-g.Z., and P.T. analyzed data; and G.A.F. and P.T. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: MVM, minute virus of mice; PLA2, phospholipase A2; PEI, polyethyleneimine; CAR, coxsackievirus–adenovirus receptor.
References
- 1.Smith, A. E. & Helenius, A. (2004) Science 304, 237-242. [DOI] [PubMed] [Google Scholar]
- 2.Tattersall, P., Cawte, P. J., Shatkin, A. J. & Ward, D. C. (1976) J. Virol. 20, 273-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tullis, G. E., Burger, L. R. & Pintel, D. J. (1993) J. Virol. 67, 131-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cotmore, S. F., D'Abramo, A. M., Jr., Ticknor, C. M. & Tattersall, P. (1999) Virology 254, 169-181. [DOI] [PubMed] [Google Scholar]
- 5.Vihinen-Ranta, M., Wang, D., Weichert, W. S. & Parrish, C. R. (2002) J. Virol. 76, 1884-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lombardo, E., Ramirez, J. C., Garcia, J. & Almendral, J. M. (2002) J. Virol. 76, 7049-7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zadori, Z., Szelei, J., Lacoste, M. C., Li, Y., Gariepy, S., Raymond, P., Allaire, M., Nabi, I. R. & Tijssen, P. (2001) Dev. Cell 1, 291-302. [DOI] [PubMed] [Google Scholar]
- 8.Dorsch, S., Liebisch, G., Kaufmann, B., von Landenberg, P., Hoffmann, J. H., Drobnik, W. & Modrow, S. (2002) J. Virol. 76, 2014-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Girod, A., Wobus, C. E., Zadori, Z., Ried, M., Leike, K., Tijssen, P., Kleinschmidt, J. A. & Hallek, M. (2002) J. Gen. Virol. 83, 973-978. [DOI] [PubMed] [Google Scholar]
- 10.Suikkanen, S., Antila, M., Jaatinen, A., Vihinen-Ranta, M. & Vuento, M. (2003) Virology 316, 267-280. [DOI] [PubMed] [Google Scholar]
- 11.Ros, C. & Kempf, C. (2004) Virology 324, 350-360. [DOI] [PubMed] [Google Scholar]
- 12.Balsinde, J., Winstead, M. V. & Dennis, E. A. (2002) FEBS Lett. 531, 2-6. [DOI] [PubMed] [Google Scholar]
- 13.Parker, J. S. & Parrish, C. R. (2000) J. Virol. 74, 1919-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tattersall, P. & Bratton, J. (1983) J. Virol. 46, 944-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howitt, J., Anderson, C. W. & Freimuth, P. (2003) Curr. Top. Microbiol. Immunol. 272, 331-364. [DOI] [PubMed] [Google Scholar]
- 16.Farr, G. A. & Tattersall, P. (2004) Virology 323, 243-256. [DOI] [PubMed] [Google Scholar]
- 17.Palmer, G. A., Brogdon, J. L., Constant, S. L. & Tattersall, P. (2004) J. Virol. 78, 1101-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiethoff, C. M., Wodrich, H., Gerace, L. & Nemerow, G. R. (2005) J. Virol. 79, 1992-2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Urabe, M., Ding, C. & Kotin, R. M. (2002) Hum. Gene Ther. 13, 1935-1943. [DOI] [PubMed] [Google Scholar]
- 20.Kichler, A., Leborgne, C., Coeytaux, E. & Danos, O. (2001) J. Gene Med. 3, 135-144. [DOI] [PubMed] [Google Scholar]
- 21.Schober, D., Kronenberger, P., Prchla, E., Blaas, D. & Fuchs, R. (1998) J. Virol. 72, 1354-1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chandran, K., Farsetta, D. L. & Nibert, M. L. (2002) J. Virol. 76, 9920-9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown, W. J., Chambers, K. & Doody, A. (2003) Traffic 4, 214-221. [DOI] [PubMed] [Google Scholar]
- 24.Kudo, I. & Murakami, M. (2002) Prostaglandins Other Lipid Mediat. 69, 3-58. [DOI] [PubMed] [Google Scholar]
- 25.Vihinen-Ranta, M., Kakkola, L., Kalela, A., Vilja, P. & Vuento, M. (1997) Eur. J. Biochem. 250, 389-394. [DOI] [PubMed] [Google Scholar]
- 26.Farr, G. A., Cotmore, S. F. & Tattersall, P. (2005) J. Virol., in press.