Abstract
The class II deacetylase histone deacetylase 4 (HDAC4) negatively regulates the transcription factor MEF2. HDAC4 is believed to repress MEF2 transcriptional activity by binding to MEF2 and catalyzing local histone deacetylation. Here we report that HDAC4 also controls MEF2 by a novel SUMO E3 ligase activity. We show that HDAC4 interacts with the SUMO E2 conjugating enzyme Ubc9 and is itself sumoylated. The overexpression of HDAC4 leads to prominent MEF2 sumoylation in vivo, whereas recombinant HDAC4 stimulates MEF2 sumoylation in a reconstituted system in vitro. Importantly, HDAC4 promotes sumoylation on a lysine residue that is also subject to acetylation by a MEF2 coactivator, the acetyltransferase CBP, suggesting a possible interplay between acetylation and sumoylation in regulating MEF2 activity. Indeed, MEF2 acetylation is correlated with MEF2 activation and dynamically induced upon muscle cell differentiation, while sumoylation inhibits MEF2 transcriptional activity. Unexpectedly, we found that HDAC4 does not function as a MEF2 deacetylase. Instead, the NAD+-dependent deacetylase SIRT1 can potently induce MEF2 deacetylation. Our studies reveal a novel regulation of MEF2 transcriptional activity by two distinct classes of deacetylases that affect MEF2 sumoylation and acetylation.
Precise temporal and spatial gene expression is critical for the execution of the differentiation program and other important biological processes. This tight regulation of gene expression is achieved by the interplay of transcriptional activation and repression controlled by transcription factors that recruit specific cofactor complexes capable of modifying histones and the local chromatin structure (reviewed in references 12 and 29). Among the cofactors that are involved in transcriptional repression, members of the histone deacetylase (HDAC) family are the most well characterized. Numerous studies have reported the recruitment of HDAC members by transcription factors to repress gene expression. Based on the long-established correlation between histone acetylation and gene transcription, it is generally thought that HDAC members repress gene transcription by catalyzing local histone deacetylation (reviewed in reference 21). However, whether this mechanism is universally applicable to HDAC-mediated transcriptional repression is not known.
Among HDAC family members involved in transcriptional regulation, HDAC4 and HDAC5 are closely related and belong to a subfamily that also includes HDAC7 and HDAC9 (reviewed in reference 37). This subfamily of HDACs share several unique properties. First, they all contain the N-terminal noncatalytic MITR (MEF2-interacting transcription repressor) homology domain (37). Second, they are all regulated by phosphorylation-dependent cytoplasmic-nuclear trafficking (18, 23, 45). Third, and most importantly, they are all critical regulators of MEF2, a family of transcription factors important in muscle cell differentiation and apoptosis (reviewed in reference 24). In skeletal muscle cells, MEF2 members collaborate with the basic helix-loop-helix (bHLH) protein MyoD to activate the genes critical for muscle cell differentiation (reviewed in reference 2). It was found that HDAC4 and related members bind to MEF2 via the MITR homology domain and potently repress MEF2-induced gene transcription (5, 22, 26, 39). Supporting the importance of this repression activity, the overexpression of HDAC4 and -5 can suppress skeletal muscle cell differentiation in a cell culture model (22), and the ablation of HDAC9 in mice leads to inappropriate and ectopic activation of MEF2-mediated gene transcription in cardiac muscle (43). These results identify members of the HDAC4 subfamily as critical negative regulators of MEF2. Because HDAC4 is a member of the histone deacetylase family, the transcriptional repression effect of HDAC4 on MEF2 has been naturally attributed to its ability to induce histone deacetylation (22, 26). However, deacetylase-independent transcription repression by HDAC4 has also been reported (3, 44; X. Zhao and T.-P. Yao, unpublished observation). This observation suggests that HDAC4 might control MEF2 activity by some other mechanisms that have yet to be characterized.
SUMO (small ubiquitin-related modifier) is a small polypeptide related to ubiquitin. Like ubiquitin, SUMO can be covalently attached to a target protein at specific lysine residues by the activities of an activating enzyme (E1), a conjugating enzyme (E2) (Ubc9), and a growing number of SUMO E3 ligases (reviewed in reference 35). Although the understanding of the function of protein sumoylation remains poor, recent studies have suggested that it might play a role in transcriptional regulation (reviewed in reference 9). Several reports indicate that lysine sumoylation negatively regulates transcription factors (reviewed in references 7 and 38), a function that is also dominantly associated with members of the HDAC family. However, it has not been determined previously whether HDAC members are functionally connected to transcription factor sumoylation.
In this report, we provide evidence that HDAC4 has a novel SUMO E3 ligase activity that regulates MEF2 sumoylation. We show that HDAC4 binds the SUMO-conjugating enzyme Ubc9 and potently stimulates MEF2 sumoylation in both cultured cells and an in vitro reconstituted system. Furthermore, we show that the MEF2 lysine acceptor for SUMO is also subject to dynamic acetylation mediated by a MEF2 coactivator, the acetyltransferase CBP (6, 33), and a NAD+-dependent class III deacetylase, SIRT1. Functionally, we found that a MEF2 sumoylation-deficient mutant is more potent than wild-type MEF2 in activating target gene transcription and promoting muscle cell differentiation, whereas a MEF2-SUMO fusion protein is transcriptionally inactive. Our results uncover the unexpected complexity of the regulation of MEF2 function by two different classes of deacetylases and reveal a novel mode of transcriptional regulation whereby the deacetylase HDAC4 controls MEF2 activity by promoting its sumoylation with SUMO E3 ligase activity.
MATERIALS AND METHODS
DNA constructs and antibodies.
HDAC4 and HDAC5 expression plasmids were generated by subcloning the coding sequences from PBJ-HDAC4 and -HDAC5 (kindly provided by S. Schreiber) into the pcDNA3 vector. The HDAC4 coiled-coil domain (amino acids 67 to 257) was cloned into the pGBKT7 vector (Clontech), and a yeast two-hybrid assay was performed according to the company-provided protocol. Ubc9 cDNA was amplified from a human brain cDNA library and cloned into the pcDNA3 vector. The SUMO expression construct was kindly provided by Michael Nevels (Princeton University). The MCK-luciferase and 3XMEF2-Luc reporters were kindly provided by Rhonda Bassel-Duby and Eric Olson (University of Texas, Southwestern Medical Center). To generate the SUMO-MEF2 fusion protein, SUMO-1 (amino acids 1 to 96) was amplified by PCR and ligated with the MEF2D N terminus into the pcDNA3 vector. Expression plasmids for HDAC4 K559R, MEF2D K424R, and MEF2D I423A mutants were created by a Quikchange site-directed mutagenesis kit (Stratagene). Anti-SUMO antibody and anti-MEF2D antibody were purchased from Zymed Laboratories and BD Biosciences, respectively. Anti-acetylated lysine antibody was kindly provided by Minoru Yoshida (RIKEN Institute, Japan). Anti-MHC (MF20) was used as described previously (6).
Tissue culture, transfections, and Western blots.
293T and 10T1/2 cells were maintained in Dulbecco's modified Eagle medium (DMEM) with 10% fetal bovine serum at 37°C in a humidified atmosphere with 5% CO2. Calcium phosphate-mediated transfections were performed as described previously to transfect 293T cells (41). Thirty-six hours after transfection, cells were harvested and lysed with RIPA buffer containing 100 mM N-ethylmaleimide. Five-hundred-microgram aliquots of protein were immunoprecipitated with anti-MEF2D antibody overnight, followed by immunoblotting using anti-SUMO or anti-acetylated lysine antibody. Endogenous MEF2 sumoylation was detected as described previously (17). Briefly, C2C12 cells were lysed in RIPA buffer containing 1% SDS. After a quick sonication, cell lysates were diluted 10-fold for immunoprecipitation with an anti-MEF2 antibody. For the endogenous MEF2 acetylation assay, C2C12 cells were induced to differentiate by serum withdrawal for different time periods, as indicated. The cells were treated with nicotinamide for 4 h before being harvested. The cells were lysed in 170 mM NETN (0.5% NP-40, 1 mM EDTA, 20 mM Tris-HCl, 170 mM NaCl), and MEF2 was immunoprecipitated and analyzed by Western blotting as stated above.
Coimmunoprecipitation assays.
For the interaction of HDAC4 and Ubc9, HDAC4 and Ubc9 were coexpressed in U2OS cells, which were then lysed in RIPA buffer. After a brief sonication, cell lysates were precleared with protein G beads for 1 h. The supernatants were then incubated with anti-Myc (9E10) antibody and protein G beads overnight. For the interaction of HDAC4 and SIRT1, 293T cells coexpressing SIRT1 and HDAC4 were lysed in 170 mM NETN and incubated with 9E10 antibody and protein G beads. In both cases, the immunoprecipitation products were analyzed by Western blotting using an anti-FLAG antibody to detect immunoprecipitated HDAC4.
Myogenic conversion assay.
Polyamine-based transfection (Genejammer; Stratagene) was carried out to transiently transfect 10T1/2 cells using company-provided instructions. Twenty-four hours after transfection, the growth medium (GM) was switched to differentiation medium (DM) (DMEM with 2% horse serum). Four days later, the cells were harvested and lysed in cracking buffer containing 4 M urea (27). The muscle cell differentiation marker myosin heavy chain (MHC) was detected in 30-μg aliquots of protein by a Western blot using anti-MHC antibody (MF20).
In vitro sumoylation assay.
Recombinant SUMO-1, SAE-1, and SAE-2 were expressed as glutathione S-transferase (GST) fusion proteins using the pGEX series of plasmids. H6-tagged Ubc9 was expressed from plasmid pET15b-Ubc9, which contains the Ubc9 cDNA as an XhoI/BamHI fragment. Recombinant HDAC4 was purified from a baculovirus expression system. In vitro sumoylation reactions were carried out in 20 μl of reaction buffer as described previously (36), supplemented with 2 mM ATP. One hundred nanograms of GST-SAE1, 150 ng of GST-SAE2, 50 ng of H6-Ubc9, 1 to 2 μg of SUMO-1, and 1 to 2 μl of in vitro-translated, 35S-labeled MEF2D were added to the reaction mix. Reactions were carried out for 45 min at 30°C and analyzed by SDS-polyacrylamide gel electrophoresis. Quantitation was performed using a PhosphorImager (Molecular Dynamics). After subtraction of the background, the sum of the signal for modified plus unmodified MEF2 in each lane was set to 100%, and the relative values are given accordingly.
Luciferase reporter assay.
10T1/2 cells were transfected by using the Genejammer transfection reagent (Stratagene) in 24-well plates with plasmids for the MCK-Luc reporter (0.4 μg), MEF2D or the MEF2D K424R (I423A) mutant (0.2 μg), MyoD (10 ng), and pCMX-βGal (0.1 μg). Twenty-four hours after transfection, cells were collected, and the luciferase activity was measured and normalized by the β-galactosidase activity as previously described (45).
RESULTS
HDAC4 interacts with Ubc9 and is subject to sumoylation.
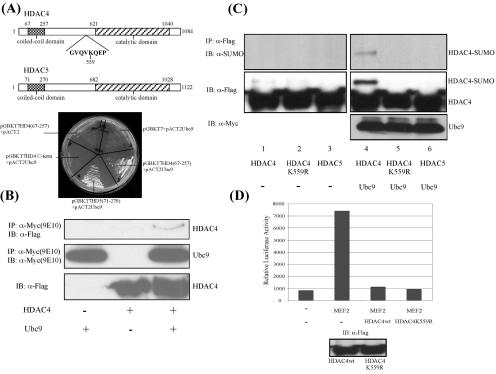
Sequence analysis identified a coiled-coil domain at the N termini of HDAC4 and the closely related HDAC5 (1) (Fig. 1A). This domain is located in a region previously shown to mediate HDAC4 binding to MEF2 and calmodulin (26, 42), suggesting that it might be important for protein-protein interactions. To identify novel HDAC4-interacting proteins, we performed a yeast two-hybrid screen, using this coiled-coil domain (amino acids 67 to 257) as bait. One of the positive clones was found to encode Ubc9, an E2 SUMO-conjugating enzyme (25). As shown in Fig. 1A, Ubc9 specifically interacts with the coiled-coil domains of HDAC4 and HDAC5, but not with the C terminus of HDAC4, in a yeast two-hybrid analysis. A coimmunoprecipitation assay also confirmed the association between Ubc9 and HDAC4 (Fig. 1B). Further supporting the observation that HDAC4 and Ubc9 functionally interact, the coexpression of HDAC4 and Ubc9 in 293T cells leads to specific sumoylation of HDAC4 at lysine 559 (Fig. 1C, lanes 4 and 5). However, the subsequent analysis failed to reveal obvious defects associated with an HDAC4 mutant that cannot be sumoylated (HDAC4 K559R). This sumoylation-deficient HDAC4 mutant appears to be indistinguishable from wild-type HDAC4 in its transcriptional repression activity (Fig. 1D) and deacetylase activity (data not shown). This observation indicates that sumoylation of HDAC4 might not be the key outcome of the HDAC4-Ubc9 interaction. Consistent with this idea, although HDAC5 also interacts with Ubc9, it is not subject to sumoylation (Fig. 1C, lane 6).
FIG. 1.
HDAC4 and HDAC5 interact with Ubc9. (A) (Upper panel) Schematic illustration of the structures of HDAC4 and HDAC5. The predicted N-terminal coiled coil (1) and the consensus sumoylation motif are shown. (Lower panel) Empty vectors or DNA constructs encoding different proteins, as indicated, were introduced into the AH109 yeast strain (Clontech). The protein interactions were revealed by growth of the transformed cells on −Trp −Leu −His −Ade dropout synthetic medium. Note the growth of yeast cells expressing HDAC4 (67-257) and HDAC5 (71-270) together with Ubc9. An HDAC4 C-terminal fragment was introduced as a negative control in this assay. (B) HDAC4 and Ubc9 were coexpressed in U2OS cells. The cell lysates were incubated with anti-Myc antibody to pull down the Ubc9 protein and probed with anti-Flag antibody to detect the interacting HDAC4. (C) Expression plasmids encoding HDAC4, the HDAC4 K559R mutant, and HDAC5 were transfected into 293T cells together with Ubc9. The sumoylation levels of HDAC4 and -5 were determined by immunoprecipitation followed by Western blotting using an anti-SUMO-1 antibody. (D) MEF2 was expressed with a luciferase reporter controlled by a tandem MEF2 binding site (3XMEF2-Luc). Wild-type (wt) HDAC4 or its K559R mutant was coexpressed to determine the repressive activity toward MEF2.
HDAC4 promotes MEF2 SUMO modification in vivo and in vitro.
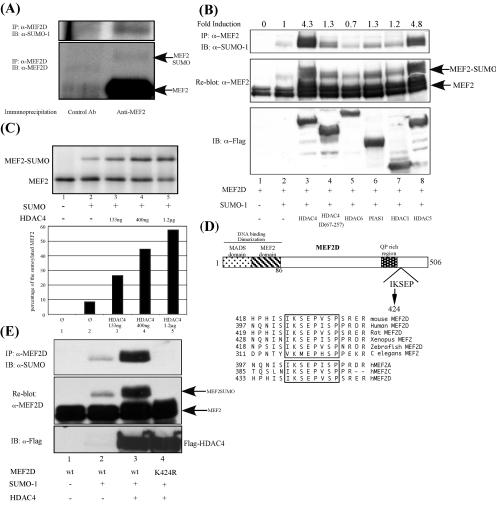
We therefore considered the alternative hypothesis that HDAC4 recruits the Ubc9 machinery to regulate the sumoylation of HDAC4-associated proteins such as MEF2. To this end, we first determined whether endogenous MEF2 is subject to sumoylation. As shown in Fig. 2A, the immunoprecipitation of endogenous MEF2D from C2C12 cells revealed a slower-migrating MEF2D species with the mobility predicted for SUMO-1-conjugated MEF2 (lower panel). Indeed, this MEF2D species can also be recognized by an anti-SUMO-1 antibody (upper panel), strongly indicating that endogenous MEF2 is subject to SUMO modification.
FIG. 2.
Characterization of MEF2 sumoylation. (A) Endogenous proteins were precipitated from C2C12 cell lysates with an anti-MEF2D antibody and a control antibody, as indicated, and analyzed by Western blotting using an anti-SUMO-1 antibody (upper panel) and a MEF2D antibody (lower panel). The position of sumoylated MEF2 is shown. (B) Expression plasmids encoding wt MEF2D were transfected into 293T cells together with SUMO-1 and different HDACs and PIAS1, as indicated. MEF2 sumoylation levels were determined by immunoprecipitation followed by Western blotting using an anti-SUMO-1 antibody (upper panel). The relative levels of MEF2 sumoylation were calculated by densitometry and normalized to the total amount of MEF2 in each lane. Sumoylation induction was determined by comparing the sumoylation level of each lane with that of lane 2, where only SUMO-1 was expressed with MEF2. Note the enhancement of MEF2 sumoylation in the presence of HDAC4 and HDAC5. The membrane was stripped and reprobed with anti-MEF2 antibody. A slower-migrating band corresponding to sumoylated MEF2 is shown (middle panel). The expression of different HDACs and the PIAS protein is also shown (lower panel). (C) (Upper panel) The in vitro sumoylation assay was performed as described in Materials and Methods. The proteins were separated by SDS-polyacrylamide gel electrophoresis and subjected to autoradiography. The unmodified and SUMO-1-modified MEF2 proteins are indicated. Note that the addition of an increased amount of purified HDAC4 enhanced the MEF2 sumoylation levels in a dose-dependent manner. (Lower panel) The relative level of MEF2 sumoylation was measured by a phosphorimager, and the percentage of the sumoylated population was calculated and plotted, with the total protein amount set to 100%. (D) Schematic diagram of MEF2D structure. The consensus sumoylation site is shown. An amino acid alignment reveals that this consensus site is highly conserved between different species and isoforms of MEF2. (E) An expression plasmid encoding wt MEF2D or the MEF2D K424R mutant was transfected into 293T cells together with SUMO-1 or HDAC4, as indicated. The MEF2 sumoylation level was determined as described above.
We then determined whether HDAC4 modulates MEF2 sumoylation. Similar to endogenous MEF2D, ectopically expressed MEF2D also becomes moderately sumoylated when it is coexpressed with SUMO-1 (Fig. 2B, lane 2, upper panel). Remarkably, the coexpression of HDAC4 dramatically increases the level of sumoylated MEF2D (Fig. 2B, lane 3, upper panel). Normalized quantification of the sumoylated MEF2 revealed that HDAC4 can stimulate MEF2D sumoylation more than fourfold. This effect is specific, as an HDAC4 mutant that does not have the coiled-coil domain is inactive in this assay (lane 4). Importantly, the closely related HDAC5, which also contains the coiled-coil domain and interacts with Ubc9, potently promotes MEF2D sumoylation as well (lane 8). In contrast, other HDAC members, such as HDAC1 and HDAC6, have little effect (lanes 5 and 7). Finally, PIAS1 and PIASy, which function as SUMO E3 ligases for p53 and LEF1 (32, 34), do not efficiently stimulate MEF2D sumoylation (Fig. 2B, lane 6, and data not shown). Together, these data demonstrate that MEF2D is a sumoylated protein and that HDAC4 can specifically stimulate MEF2 sumoylation in vivo.
To determine whether HDAC4 has the activity to promote MEF2 sumoylation directly, we examined the effect of recombinant HDAC4 on MEF2 sumoylation in a reconstituted system in vitro. To this end, in vitro-translated MEF2D was incubated with purified recombinant SUMO E1-activating enzymes (SAE1/SAE2), E2 Ubc9, and SUMO-1 in the presence or absence of recombinant HDAC4. As shown in Fig. 2C, a small population of MEF2D becomes sumoylated after incubation with the reconstituted SUMO machinery (upper panel, lanes 1 and 2). Significantly, the addition of purified recombinant HDAC4 to the reaction dramatically increases the production of sumoylated MEF2D, with a concomitant reduction in the unmodified species (Fig. 2C, lanes 3 to 5). Quantification of these experiments revealed that recombinant HDAC4 can stimulate MEF2D sumoylation more than fivefold in a dose-dependent manner (Fig. 2C, lower panel). These results strongly support a direct role for HDAC4 in promoting MEF2 sumoylation.
A sequence analysis of MEF2D revealed the presence of one consensus sumoylation motif surrounding lysine 424. Notably, this motif is highly conserved and can be found in most isoforms of MEF2 from different animal species (Fig. 2D). Supporting the hypothesis that lysine 424 is the key SUMO acceptor, the conversion of lysine 424 to arginine (K424R) completely abrogates HDAC4-dependent MEF2D sumoylation in cells (Fig. 2E) and in the in vitro assay (data not shown). Together, these results demonstrate that HDAC4 can stimulate MEF2 sumoylation on lysine 424 both in vivo and in vitro.
MEF2D lysine 424 is subject to reversible acetylation catalyzed by CBP and SIRT1.
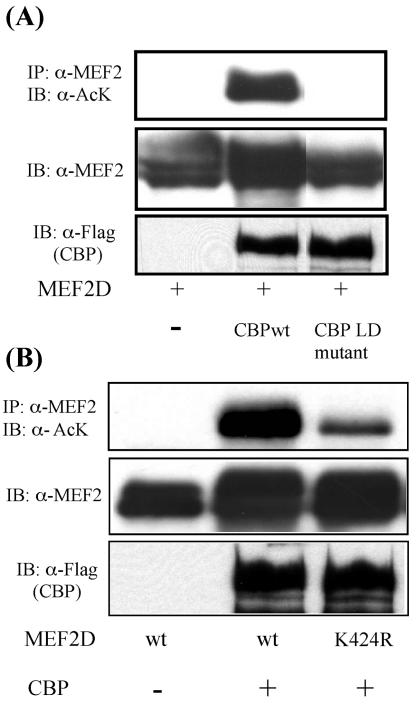
Recently, several lysine-targeting modifications, such as acetylation, sumoylation, and ubiquitination, have been reported as important modulators of transcription (7). Our observation that HDAC4, a protein deacetylase, can stimulate MEF2 sumoylation prompted us to ask whether MEF2 is also subject to lysine acetylation. To test this, we first determined whether MEF2 can be acetylated by the acetyltransferase CBP, which has been shown to promote MEF2 activity and is essential for C2C12 cell differentiation (6, 33). As shown in Fig. 3A, we found that wild-type CBP can potently induce MEF2D acetylation. An enzymatically deficient mutant of CBP fails to do so. Importantly, mutation of the SUMO acceptor lysine 424 dramatically reduces CBP-induced MEF2 acetylation (Fig. 3B), revealing lysine 424 as the major acceptor for both acetylation and sumoylation and suggesting a potential interplay of these two lysine-targeting modifications in regulating MEF2 function.
FIG. 3.
MEF2 is acetylated. (A) MEF2D was expressed with the acetyltransferase CBP and its enzyme-dead mutant (LD mutant) in 293T cells. MEF2 acetylation was detected by immunoprecipitation and Western blotting using anti-acetylated lysine (α-AcK) antibody (upper panel). The expression of MEF2D and CBP proteins in the whole-cell lysates is shown in the lower two panels. (B) The acetylation levels of wt MEF2D and the MEF2D K424R mutant were examined as described above. Note that the K424R mutant has reduced acetylation. The expression of MEF2D and CBP proteins is also shown below.
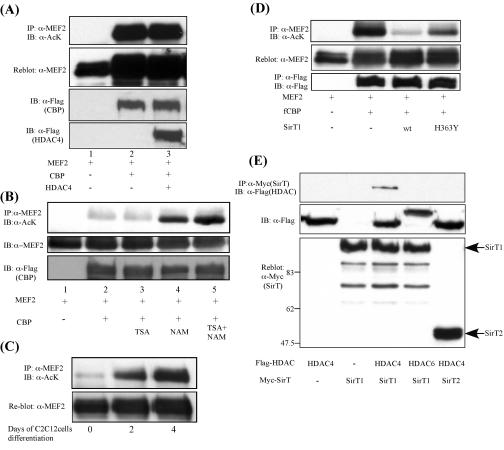
Since lysine 424 is targeted by both acetylation and sumoylation, we hypothesized that HDAC4 might promote lysine 424 sumoylation by deacetylating this lysine residue. Surprisingly, however, the ectopic expression of HDAC4 does not induce efficient MEF2 deacetylation (Fig. 4A, lane 3). Supporting this conclusion, treatment with a potent HDAC4 deacetylase inhibitor, trichostatin A (TSA), has little effect on the MEF2 acetylation level (Fig. 4B, lane 3), suggesting an HDAC4-independent deacetylation of MEF2. In contrast, a treatment with nicotinamide, an inhibitor of the class III NAD+-dependent Sir2 family deacetylases, dramatically increases MEF2 acetylation (Fig. 4B, lanes 4 and 5). Importantly, a differentiation-dependent acetylation of endogenous MEF2 can also be prominently induced by nicotinamide but not TSA treatment (Fig. 4C and data not shown). These results strongly suggest that MEF2 acetylation is dynamically regulated during muscle cell differentiation and that one or more of the Sir2 family proteins, but not HDAC4, function as the MEF2 deacetylase.
FIG. 4.
Class III deacetylase SIRT1 has MEF2 deacetylase activity. (A) MEF2D was expressed with CBP and HDAC4 in 293T cells. The MEF2 acetylation level was detected as described in the legend to Fig. 3A. Note that HDAC4 does not change MEF2 acetylation (top panel, lane 3). The expression levels of MEF2, CBP, and HDAC4 are also shown in the lower panels. (B) MEF2D was expressed in 293T cells with CBP to induce acetylation. Thirty-six hours after transfection, the cells were treated with either TSA or nicotinamide (NAM) for 5 h. MEF2 acetylation was detected as described above. (C) C2C12 cells were induced to differentiate by culturing in differentiation medium (DMEM with 2% horse serum). Cells were treated with nicotinamide for 4 h before being harvested. MEF2 was immunoprecipitated, and its acetylation level was shown with anti-acetylated lysine (anti-AcK) antibody. (D) 293T cells were transfected with expression constructs for MEF2, CBP, and SIRT1 or the SIRT1 H363Y mutant. Thirty-six hours later, cells were harvested, and the MEF2 acetylation level was determined by immunoprecipitation and Western blotting. MEF2 and CBP expression levels are also shown. (E) Different Sir2 proteins and HDACs were expressed in 293T cells. The cell lysates were incubated with anti-Myc antibody to pull down the Sir2 protein and were probed with anti-Flag antibody to detect the interacting HDAC. Note that only HDAC4 interacts with SIRT1.
Among the Sir2 family members, SIRT1 was shown to repress the muscle cell differentiation program (8). We thus tested whether SIRT1 can function as a MEF2 deacetylase. As shown in Fig. 4D, the cotransfection of SIRT1 potently reduces CBP-mediated MEF2D acetylation, whereas a catalytically inactive SIRT1 mutant (H363Y) and another Sir2 member, SIRT2, do not cause efficient MEF2D deacetylation (Fig. 4D and data not shown). Thus, SIRT1 can function as a MEF2 deacetylase. Interestingly, coimmunoprecipitation assays revealed that SIRT1 interacts with HDAC4 but not with HDAC6 (Fig. 4E). These results reveal that the class III deacetylase SIRT1 can function as a MEF2 deacetylase and can form complexes with the class II deacetylase HDAC4.
SUMO modification negatively regulates MEF2 activity.
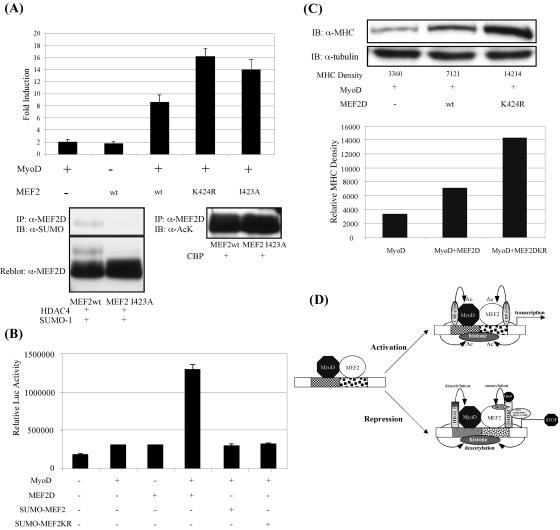
Since acetylation and sumoylation both target lysine 424 (Fig. 2E and 3B) and since acetylation positively correlates with MEF2 activity during the muscle cell differentiation process (Fig. 4C), we hypothesized that HDAC4-induced sumoylation negatively regulates MEF2 activity. To examine this, we coexpressed MyoD with wild-type or sumoylation-deficient mutant (MEF2D K424R) MEF2D and assessed their ability to activate a luciferase reporter controlled by the muscle creatine kinase (MCK) enhancer. Consistent with previous reports, wild-type MEF2D and MyoD activate the MCK reporter cooperatively (Fig. 5A, upper panel). Importantly, the sumoylation-deficient mutant MEF2D K424R is consistently more active than wild-type MEF2D in collaborating with MyoD to activate the MCK reporter (Fig. 5A, upper panel), suggesting that sumoylation negatively regulates MEF2 activity. A similar conclusion was made by assessing the transcriptional activity of another sumoylation-deficient mutant, MEF2D I423A (Fig. 5A, upper panel), in which the sumoylation consensus site (IKSE) was destroyed in order to eliminate only MEF2 sumoylation but still keep acetylation (Fig. 5A, lower panel). To further confirm that sumoylation negatively regulates MEF2D activity, we fused SUMO to MEF2D to mimic sumoylated MEF2. Both the SUMO-MEF2D and SUMO-MEF2DK424R fusion proteins fail to synergize with MyoD in activating the MCK-luciferase reporter (Fig. 5B) or a reporter driven by tandem MEF2 binding sites (data not shown). These data support the idea that sumoylated MEF2 is transcriptionally inactive. Lastly, we examined the activity of sumoylation-deficient MEF2 in converting 10T1/2 fibroblasts to myocytes. As shown in Fig. 5C, coexpression of the sumoylation-deficient MEF2D mutant with MyoD induces MHC in 10T1/2 fibroblasts more potently than that of wild-type MEF2. Together, our data indicate that the conjugation of SUMO down-regulates MEF2 activity.
FIG. 5.
Functional consequence of sumoylation on MEF2 activity. (A) (Upper panel) 10T1/2 fibroblasts were transfected with the MCK-luc reporter along with MyoD and MEF2 expression plasmids, as indicated. Luciferase activities relative to the basal level, which was set to 1, are shown. Note that the MEF2 K424R and I423A mutants have more potent, but comparable, transcription abilities. (Lower panel) wt MEF2 and the I423A mutant were either coexpressed with SUMO-1 and HDAC4 or coexpressed with CBP. The cell lysates were immunoprecipitated with anti-MEF2D antibody and analyzed with anti-SUMO-1 or anti-acetylated lysine (anti-AcK) antibody as indicated. Note that the I423A mutant cannot be sumoylated but can still be strongly acetylated by CBP. (B) Relative transcription activities of SUMO-MEF2 and SUMO-MEF2KR fusion proteins, as determined with the MCK-luc system as described in Materials and Methods. Note that the fusion of SUMO to MEF2 reduces MEF2's transcription ability. (C) 10T1/2 cells were transiently transfected with expression plasmids encoding MyoD and wt MEF2 or MEF2 K424R, as indicated. Myogenic conversion was induced as described in the text. MHC expression was determined by Western blot analysis of whole-cell lysates containing equal amounts of protein. Tubulin levels were also determined as a loading control. The relative density of each MHC band was calculated with NIH Image software and plotted on the lower panel. (D) Proposed models for the regulation of MEF2 and MyoD by sumoylation and acetylation. Acetyltransferases, including CBP and p300, can activate MEF2 and MyoD transactivation either by the induction of histone acetylation or by directly acetylating these two transcription factors. In the case of repression, HDAC4 functions as a repressor through several mechanisms, including histone deacetylation and the SUMO E3 ligase activity.
DISCUSSION
It is generally believed that HDACs repress gene transcription by catalyzing histone deacetylation. In this study, by identifying an interaction of HDAC4 with the SUMO E2 enzyme Ubc9, we uncovered a surprising SUMO E3 ligase activity associated with HDAC4 that catalyzes MEF2 sumoylation. We further showed that MEF2 is also subject to reversible acetylation controlled by the acetyltransferase CBP and a class III deacetylase, SIRT1, which target a common lysine acceptor modified by HDAC4-mediated sumoylation. Our studies reveal a novel regulation of MEF2 activity by two distinct classes of deacetylases through modulating MEF2 acetylation and sumoylation.
Supporting the idea that the interaction between HDAC4 and Ubc9 is functional, we found that HDAC4 can be sumoylated (Fig. 1C). This observation is consistent with two earlier reports (19, 36). However, unlike a previous report (19), our analysis of an HDAC4 sumoylation-deficient mutant did not uncover obvious defects, indicating that the sumoylation of HDAC4 itself might not be important (Fig. 1D and data not shown). Instead, we found that the HDAC4-Ubc9 interaction efficiently stimulates MEF2 sumoylation (Fig. 2B and C). Consistent with this notion, HDAC5, which also interacts with Ubc9 but is not itself sumoylated, can stimulate MEF2 sumoylation effectively (Fig. 2B). Importantly, HDAC members without the Ubc9-interactive coiled-coil domain, such as HDAC6 and HDAC1, cannot promote MEF2 sumoylation (Fig. 2B, lanes 5 and 7). Furthermore, well-characterized SUMO E3s, such as PIAS1 and PIASy, do not increase MEF2 sumoylation either (Fig. 2B and data not shown). Together, these data demonstrate that HDAC4 and its related family member HDAC5 can specifically stimulate MEF2 sumoylation.
Like protein ubiquitination, the efficient conjugation of SUMO to its protein substrates usually employs three enzymes. While the E1 activating enzyme and the E2 conjugating enzyme Ubc9 are sufficient to link SUMO to certain target proteins, an E3 ligase can significantly facilitate this process by simultaneously binding to both the E2 enzyme and the substrate protein (reviewed in reference 15). Thus, as with the RING finger E3 ubiquitination ligases, SUMO E3 enzymes function as adaptors to physically bring the E2 and substrate proteins together. In this report, we show that HDAC4 interacts with both SUMO E2 Ubc9 and MEF2. HDAC4 can significantly stimulate MEF2 SUMO modification both in vivo and in vitro (Fig. 2B and C). Like other SUMO E3 ligases, such as PIAS and RanBP2, HDAC4 itself is also sumoylated, probably as a by-product of its association with Ubc9 (19, 20, 30). These data reveal that HDAC4, a histone deacetylase, has all the properties expected for a MEF2 SUMO E3 ligase. These surprising findings suggest that HDAC4 and related members, such as HDAC5, possess SUMO E3 ligase activity and could regulate gene transcription via a novel mechanism by promoting protein sumoylation.
Recently, Gregoire and Yang described that HDAC4 can promote the modification of MEF2 by two other SUMO family members, SUMO-2 and -3 (10). However, the exact role of HDAC4 in this process is not known. Our study supports the conclusion that HDAC4 functions as a SUMO-1 E3 ligase, which can also be true for SUMO-2/3 conjugation. However, we have looked for a SUMO-2/3 modification of endogenous MEF2 but have not observed it in either undifferentiated or differentiated C2C12 cells. This discrepancy may be due to tissue type-specific modifications of MEF2 by different SUMO family members. Different signals could regulate the activity of HDAC4 to preferentially conjugate either SUMO-1 or SUMO-2/3 on MEF2 in different tissues. Alternatively, the desumoylation machinery may also be tissue specific. Regardless of the details, our observations suggest a complex regulation of MEF2 by sumoylation.
We have observed an inhibitory effect of SUMO modification on MEF2 activity. This is consistent with the role of HDAC4 as a transcription repressor. Similar results have also been reported by Gregoire and Yang for a Gal-4-MEF2D fusion protein (10). The exact mechanism(s) by which sumoylation regulates protein function remains unclear. Sumoylation has been shown to affect protein stability and to direct target proteins to promyelocytic leukemia nuclear bodies (reviewed in references 28 and 35). Our analysis, however, did not detect a significant effect of sumoylation on either the stability or subcellular localization of MEF2 (data not shown). SUMO has also been shown to recruit HDAC2 (40), and a similar mechanism could be involved in regulating MEF2 activity. For this report, we found that MEF2 is subject to both sumoylation and acetylation. MEF2 acetylation is positively correlated with its activation upon muscle cell differentiation and is catalyzed by its coactivator, CBP (Fig. 3A and 4C). In contrast to acetylation, sumoylation appears to inhibit MEF2 activity (Fig. 5). Importantly, we have found that both sumoylation and acetylation target lysine 424 of MEF2 (Fig. 2E and 3B). Together, these observations raise the possibility of a competition between these two modifications for this lysine residue, and this interplay might result in activation or repression of MEF2 activity. However, we have also found that both acetylation and sumoylation are very transient and dynamic processes. For example, endogenous MEF2 acetylation could only be detected when we treated C2C12 cells with the SIRT1 inhibitor nicotinamide (Fig. 4C). To detect the endogenous SUMO-1 modification of MEF2 in C2C12 cells, large amounts of cell lysates were needed (Fig. 2A). These observations argue that a sustained modification may not be required for the biological functions of acetylation or sumoylation. A similar concept has been proposed in several reports (9, 13, 35). Recently, it has also been suggested that SUMO can regulate the long-term fate of its target proteins despite being rapidly deconjugated from them (14). We believe that this potentially transient nature makes it technically very difficult to detect an inverse relationship between MEF2 acetylation and sumoylation. Nevertheless, on the biochemical level, since both acetylation and sumoylation are conjugated on the ɛ-amino group of the lysine side chain, once this lysine is occupied by one of these modifications, it cannot be available for others at any given time. Indeed, a recent report on Sp3 sumoylation reached a similar conclusion (31). In conjunction with the proposed competition between acetylation and ubiquitination on specific lysine residues in p53 and Smad7 (11, 16, 31), our study provides evidence further supporting the importance of the interplay between these lysine-targeting modifications in regulating gene transcription.
How acetylation changes MEF2 activity is not clear. It has been demonstrated that acetylation can modify transcription factor activity by increasing the DNA binding activity, stability, or interaction with other transcription components. While not excluding these possibilities, it is interesting that while the MEF2 K424R mutation affects both sumoylation and acetylation, another SUMO-deficient MEF2D mutant (I423A) can still be acetylated by CBP (Fig. 5A). However, the transcriptional activities of I423A and K424R mutant MEF2D are comparable (Fig. 5A), suggesting that acetylation on lysine 424 is not essential for full MEF2D transcriptional activity. We speculate that the main function of MEF2 lysine 424 acetylation might be to block sumoylation and prevent MEF2 from entering a repressed state.
Surprisingly, we found that HDAC4 cannot catalyze MEF2 deacetylation efficiently. Instead, we identified a class III deacetylase, SIRT1, as a potent MEF2 deacetylase that dynamically catalyzes MEF2 deacetylation during muscle cell differentiation (Fig. 4C). These observations could provide a potential mechanism by which SIRT1 negatively regulates myogenesis (8). Interestingly, we found that SIRT1 can form a complex with HDAC4, suggesting the possibility that MEF2-dependent myogenesis could be controlled by a dual-deacetylase complex. It is reasonable to speculate that HDAC4 might function to integrate sumoylation and deacetylation signals via its interaction with Ubc9 and SIRT1 (Fig. 5D).
Overall, our data uncover a new biological property of a deacetylase to act as a SUMO E3 ligase for MEF2. In addition, we have described CBP-induced acetylation as another novel modification of MEF2 and identified SIRT1 as the MEF2 deacetylase. Furthermore, our data showing that acetylation and sumoylation occupy the same lysine residue provide a potential mechanistic framework for understanding the interplay between sumoylation and acetylation in regulating MEF2 activity. Taken together, our results have provided a comprehensive understanding of the regulation of MEF2 function by two distinct classes of deacetylases. Given the potential roles of class II and class III HDACs in the pathogenesis of cardiac hypertrophy and muscle cell differentiation (4, 8, 43), it will be of great interest to explore the significance and therapeutic implications of these different regulatory mechanisms of MEF2 in the future.
Acknowledgments
We thank E. Olson and R. Bassel-Duby for providing MCK-luciferase and 3XMEF2-luciferase reporter constructs. We thank S. Schreiber and M. Nevels for HDAC4, HDAC5, and SUMO-1 expression plasmids. We thank M. Yoshida for the anti-acetylated lysine antibody. We thank A. VanDongen for pointing out the presence of a coiled-coil domain at the N terminus of HDAC4. We are grateful to J. Kovacs, A. Guardiola, and C. Hubbert for critically reading and editing the manuscript.
T.S. was supported by a grant from the Deutsche Forschungsgemeinschaft (Ste 1003/1) and the Howard Hughes Medical Institute (HHMI). X.Z. is supported by a Department of Defense Breast Cancer Research grant (DAMD 17-02-1-0380). T.A.B. is supported by a National Science Foundation graduate research fellowship. R.M.E. is an Investigator of the Howard Hughes Medical Institute at the Salk Institute and the March of Dimes Chair in Molecular and Developmental Biology. T.P.Y. is a Leukemia & Lymphoma Society Scholar.
REFERENCES
- 1.Berger, B., D. B. Wilson, E. Wolf, T. Tonchev, M. Milla, and P. S. Kim. 1995. Predicting coiled coils by use of pairwise residue correlations. Proc. Natl. Acad. Sci. USA 92:8259-8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black, B. L., and E. N. Olson. 1998. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell Dev. Biol. 14:167-196. [DOI] [PubMed] [Google Scholar]
- 3.Chan, J. K., L. Sun, X. J. Yang, G. Zhu, and Z. Wu. 2003. Functional characterization of an amino-terminal region of HDAC4 that possesses MEF2 binding and transcriptional repressive activity. J. Biol. Chem. 278:23515-23521. [DOI] [PubMed] [Google Scholar]
- 4.Chang, S., T. A. McKinsey, C. L. Zhang, J. A. Richardson, J. A. Hill, and E. N. Olson. 2004. Histone deacetylases 5 and 9 govern responsiveness of the heart to a subset of stress signals and play redundant roles in heart development. Mol. Cell. Biol. 24:8467-8476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dressel, U., P. J. Bailey, S. C. Wang, M. Downes, R. M. Evans, and G. E. Muscat. 2001. A dynamic role for HDAC7 in MEF2-mediated muscle differentiation. J. Biol. Chem. 276:17007-17013. [DOI] [PubMed] [Google Scholar]
- 6.Eckner, R., T. P. Yao, E. Oldread, and D. M. Livingston. 1996. Interaction and functional collaboration of p300/CBP and bHLH proteins in muscle and B-cell differentiation. Genes Dev. 10:2478-2490. [DOI] [PubMed] [Google Scholar]
- 7.Freiman, R. N., and R. Tjian. 2003. Regulating the regulators: lysine modifications make their mark. Cell 112:11-17. [DOI] [PubMed] [Google Scholar]
- 8.Fulco, M., R. L. Schiltz, S. Iezzi, M. T. King, P. Zhao, Y. Kashiwaya, E. Hoffman, R. L. Veech, and V. Sartorelli. 2003. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol. Cell 12:51-62. [DOI] [PubMed] [Google Scholar]
- 9.Gill, G. 2004. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 18:2046-2059. [DOI] [PubMed] [Google Scholar]
- 10.Gregoire, S., and X. J. Yang. 2005. Association with class IIa histone deacetylases upregulates the sumoylation of MEF2 transcription factors. Mol. Cell. Biol. 25:2273-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gronroos, E., U. Hellman, C. H. Heldin, and J. Ericsson. 2002. Control of Smad7 stability by competition between acetylation and ubiquitination. Mol. Cell 10:483-493. [DOI] [PubMed] [Google Scholar]
- 12.Grunstein, M. 1997. Histone acetylation in chromatin structure and transcription. Nature 389:349-352. [DOI] [PubMed] [Google Scholar]
- 13.Hardeland, U., R. Steinacher, J. Jiricny, and P. Schar. 2002. Modification of the human thymine-DNA glycosylase by ubiquitin-like proteins facilitates enzymatic turnover. EMBO J. 21:1456-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hay, R. T. 2005. SUMO: a history of modification. Mol. Cell 18:1-12. [DOI] [PubMed] [Google Scholar]
- 15.Hochstrasser, M. 2001. SP-RING for SUMO: new functions bloom for a ubiquitin-like protein. Cell 107:5-8. [DOI] [PubMed] [Google Scholar]
- 16.Ito, A., Y. Kawaguchi, C. H. Lai, J. J. Kovacs, Y. Higashimoto, E. Appella, and T. P. Yao. 2002. MDM2-HDAC1-mediated deacetylation of p53 is required for its degradation. EMBO J. 21:6236-6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kagey, M. H., T. A. Melhuish, and D. Wotton. 2003. The polycomb protein Pc2 is a SUMO E3. Cell 113:127-137. [DOI] [PubMed] [Google Scholar]
- 18.Kao, H. Y., A. Verdel, C. C. Tsai, C. Simon, H. Juguilon, and S. Khochbin. 2001. Mechanism for nucleocytoplasmic shuttling of histone deacetylase 7. J. Biol. Chem. 276:47496-47507. [DOI] [PubMed] [Google Scholar]
- 19.Kirsh, O., J. S. Seeler, A. Pichler, A. Gast, S. Muller, E. Miska, M. Mathieu, A. Harel-Bellan, T. Kouzarides, F. Melchior, and A. Dejean. 2002. The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. EMBO J. 21:2682-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kotaja, N., U. Karvonen, O. A. Janne, and J. J. Palvimo. 2002. PIAS proteins modulate transcription factors by functioning as SUMO-1 ligases. Mol. Cell. Biol. 22:5222-5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuo, M. H., and C. D. Allis. 1998. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays 20:615-626. [DOI] [PubMed] [Google Scholar]
- 22.Lu, J., T. A. McKinsey, C. L. Zhang, and E. N. Olson. 2000. Regulation of skeletal myogenesis by association of the MEF2 transcription factor with class II histone deacetylases. Mol. Cell 6:233-244. [DOI] [PubMed] [Google Scholar]
- 23.McKinsey, T. A., C. L. Zhang, J. Lu, and E. N. Olson. 2000. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 408:106-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McKinsey, T. A., C. L. Zhang, and E. N. Olson. 2002. MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem. Sci. 27:40-47. [DOI] [PubMed] [Google Scholar]
- 25.Melchior, F. 2000. SUMO—nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol. 16:591-626. [DOI] [PubMed] [Google Scholar]
- 26.Miska, E. A., C. Karlsson, E. Langley, S. J. Nielsen, J. Pines, and T. Kouzarides. 1999. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J. 18:5099-5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molkentin, J. D., B. L. Black, J. F. Martin, and E. N. Olson. 1995. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell 83:1125-1136. [DOI] [PubMed] [Google Scholar]
- 28.Muller, S., C. Hoege, G. Pyrowolakis, and S. Jentsch. 2001. SUMO, ubiquitin's mysterious cousin. Nat. Rev. Mol. Cell Biol. 2:202-210. [DOI] [PubMed] [Google Scholar]
- 29.Orphanides, G., and D. Reinberg. 2002. A unified theory of gene expression. Cell 108:439-451. [DOI] [PubMed] [Google Scholar]
- 30.Pichler, A., A. Gast, J. S. Seeler, A. Dejean, and F. Melchior. 2002. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108:109-120. [DOI] [PubMed] [Google Scholar]
- 31.Ross, S., J. L. Best, L. I. Zon, and G. Gill. 2002. SUMO-1 modification represses Sp3 transcriptional activation and modulates its subnuclear localization. Mol. Cell 10:831-842. [DOI] [PubMed] [Google Scholar]
- 32.Sachdev, S., L. Bruhn, H. Sieber, A. Pichler, F. Melchior, and R. Grosschedl. 2001. PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 15:3088-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sartorelli, V., J. Huang, Y. Hamamori, and L. Kedes. 1997. Molecular mechanisms of myogenic coactivation by p300: direct interaction with the activation domain of MyoD and with the MADS box of MEF2C. Mol. Cell. Biol. 17:1010-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schmidt, D., and S. Muller. 2002. Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc. Natl. Acad. Sci. USA 99:2872-2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seeler, J. S., and A. Dejean. 2003. Nuclear and unclear functions of SUMO. Nat. Rev. Mol. Cell Biol. 4:690-699. [DOI] [PubMed] [Google Scholar]
- 36.Tatham, M. H., E. Jaffray, O. A. Vaughan, J. M. Desterro, C. H. Botting, J. H. Naismith, and R. T. Hay. 2001. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J. Biol. Chem. 276:35368-35374. [DOI] [PubMed] [Google Scholar]
- 37.Verdin, E., F. Dequiedt, and H. G. Kasler. 2003. Class II histone deacetylases: versatile regulators. Trends Genet. 19:286-293. [DOI] [PubMed] [Google Scholar]
- 38.Verger, A., J. Perdomo, and M. Crossley. 2003. Modification with SUMO. A role in transcriptional regulation. EMBO Rep. 4:137-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, A. H., N. R. Bertos, M. Vezmar, N. Pelletier, M. Crosato, H. H. Heng, J. Th'ng, J. Han, and X. J. Yang. 1999. HDAC4, a human histone deacetylase related to yeast HDA1, is a transcriptional corepressor. Mol. Cell. Biol. 19:7816-7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang, S. H., and A. D. Sharrocks. 2004. SUMO promotes HDAC-mediated transcriptional repression. Mol. Cell 13:611-617. [DOI] [PubMed] [Google Scholar]
- 41.Yao, T. P., W. A. Segraves, A. E. Oro, M. McKeown, and R. M. Evans. 1992. Drosophila ultraspiracle modulates ecdysone receptor function via heterodimer formation. Cell 71:63-72. [DOI] [PubMed] [Google Scholar]
- 42.Youn, H. D., C. M. Grozinger, and J. O. Liu. 2000. Calcium regulates transcriptional repression of myocyte enhancer factor 2 by histone deacetylase 4. J. Biol. Chem. 275:22563-22567. [DOI] [PubMed] [Google Scholar]
- 43.Zhang, C. L., T. A. McKinsey, S. Chang, C. L. Antos, J. A. Hill, and E. N. Olson. 2002. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell 110:479-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, C. L., T. A. McKinsey, and E. N. Olson. 2002. Association of class II histone deacetylases with heterochromatin protein 1: potential role for histone methylation in control of muscle differentiation. Mol. Cell. Biol. 22:7302-7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao, X., A. Ito, C. D. Kane, T. S. Liao, T. A. Bolger, S. M. Lemrow, A. R. Means, and T. P. Yao. 2001. The modular nature of histone deacetylase HDAC4 confers phosphorylation-dependent intracellular trafficking. J. Biol. Chem. 276:35042-35048. [DOI] [PubMed] [Google Scholar]